Abstract
The spice turmeric, with its active polyphenol curcumin, has been used as anti-inflammatory remedy in traditional Asian medicine for centuries. Many cellular targets of curcumin have been identified, but how such a wide range of targets can be affected by a single compound is unclear. Here, we identified curcumin as a pro-drug that requires oxidative activation into reactive metabolites to exert anti-inflammatory activities. Synthetic curcumin analogs that undergo oxidative transformation potently inhibited the pro-inflammatory transcription factor nuclear factor κB (NF-κB), whereas stable, non-oxidizable analogs were less active, with a correlation coefficient (R2) of IC50 versus log of autoxidation rate of 0.75. Inhibition of glutathione biosynthesis, which protects cells from reactive metabolites, increased the potency of curcumin and decreased the amount of curcumin-glutathione adducts in cells. Oxidative metabolites of curcumin adducted to and inhibited the inhibitor of NF-κB kinase subunit β (IKKβ), an activating kinase upstream of NF-κB. An unstable, alkynyl-tagged curcumin analog yielded abundant adducts with cellular protein that were decreased by pretreatment with curcumin or an unstable analog but not by a stable analog. Bioactivation of curcumin occurred readily in vitro, which may explain the wide range of cellular targets, but if bioactivation is insufficient in vivo, it may also help explain the inconclusive results in human studies with curcumin so far. We conclude that the paradigm of metabolic bioactivation uncovered here should be considered for the evaluation and design of clinical trials of curcumin and other polyphenols of medicinal interest.
Keywords: autoxidation, cysteine, electrophile, glutathione, protein adduction, polyphenol, flavonoid, quinone, NF-κB, redox regulation
Introduction
Curcumin is one of the most recognized and promising complementary alternative medicine agents (1, 2). It shares this status with other dietary phenolic compounds, for example, resveratrol (from grapes and red wine), quercetin (onions), and epigallocatechin gallate (green tea) (3–5). Preclinical evidence from cell culture and animal models has provided incentive for testing dietary polyphenols in randomized clinical trials (6, 7). In the form of an extract from the plant turmeric, curcumin has a rich history as an anti-inflammatory agent in traditional Asian medicine (8). Attempts to replicate these effects in clinical trials, however, have given mixed results, with therapeutic improvements limited to open-label studies (9, 10).
A key element to improve design and outcome of clinical trials is a molecular understanding of how curcumin and related polyphenols affect their targets (11, 12). Numerous cellular targets of curcumin have been identified but there is only limited insight into how such a wide range of targets can be affected by a single compound. The recognized molecular features of curcumin are its phenolic hydroxyl, mediating antioxidant activity (13), its α,β-unsaturated enone, mediating protein binding (14), and its central β-dicarbonyl, mediating metal ion chelation (15). These features are not unique to curcumin and appear insufficient to account for the differences in bioactivity compared with other dietary polyphenols.
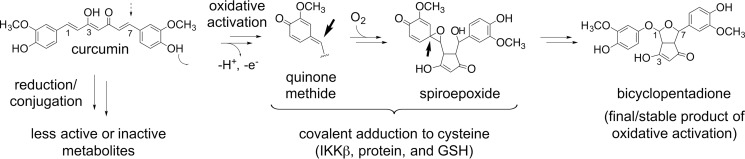
To explain the wide range of biological effects of curcumin we suggest a paradigm that takes into account the well recognized chemical instability of curcumin (16). Degradation of curcumin is initiated by H-abstraction from the phenolic hydroxyl, equivalent to its antioxidant reaction, and leads to rapid autoxidative transformation in buffer and cell culture medium (17). Oxidative degradation yields metabolites with reactive quinone methide, free radical, endoperoxide, and spiroepoxide moieties as intermediates in the transformation to the final bicyclopentadione product (18, 19). Here we provide evidence that the electrophilic metabolites, rather than parent curcumin or the final bicyclopentadione, adduct covalently to cellular protein, and specifically to IKKβ of the NF-κB pathway to exert anti-inflammatory activity. This provides a chemical–molecular paradigm relating the structure of curcumin and its autoxidative/antioxidant behavior to its anti-inflammatory activity.
Results
Inhibition of NF-κB correlates with oxidative transformation of curcumin
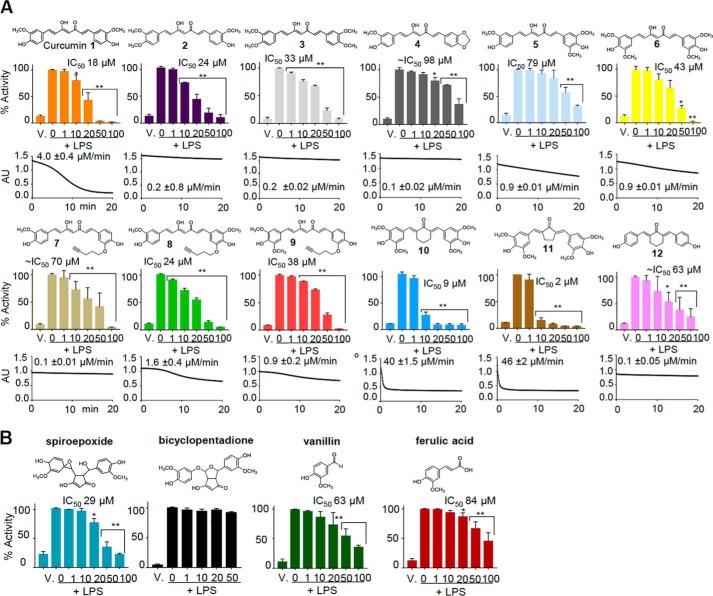
We synthesized analogs of curcumin that were designed to undergo or resist spontaneous autoxidation. Chemical stability was changed by blocking the phenolic hydroxyl through derivatization and modifying the methoxy groups at the aromatic rings. Based on the analysis of oxidative transformation of demethoxy- and bisdemethoxycurcumin (20) we had predicted that adding or removing methoxy groups from the aromatic rings would enhance or decrease, respectively, autoxidation. The rate of autoxidation of the compounds was quantified by monitoring the disappearance of the chromophore around 430 nm (Fig. 1A). Blocking one or both of the phenolic hydroxyl groups in compounds 2-4 increased the stability of the analogs. Adding a methoxy group in the meta-position of the aromatic ring increased autoxidation in 8 and 9 relative to 7 while decreasing autoxidation in 5 and 6 relative to curcumin. Autoxidation of analogs with a modified heptadienedione chain (10–12) was also increased by placing meta-methoxy groups in the aromatic rings. Six of the compounds (5, 6, and 8–11) were considered unstable, autoxidizing at rates slower or faster than curcumin. LC-MS analyses of autoxidation reactions of the unstable analogs showed the formation of products with an increase in molecular mass by 32 mass units, whereas the stable analogs did not form discernible dioxygenated products (Fig. S1). The increase by 32 mass units was consistent with the incorporation of two atoms of oxygen during the autoxidation, as is observed in the transformation of curcumin to bicyclopentadione (20, 21).
Figure 1.
Stability and inhibition of NF-κB activity of curcumin 1, analogs 2–12, metabolites, and related compounds. A, each panel shows the structure of the curcumin analog, UV-visible time-drive analysis and peak rate of autoxidative degradation at 430 nm, as well as inhibition and IC50 of LPS-induced activation of NF-κB in RAW264.7 cells. B, effect of isolated spiroepoxide and bicyclopentadione metabolites, vanillin, and ferulic acid on NF-κB activity.
The anti-inflammatory activity of curcumin and the synthetic analogs was tested in RAW264.7 cells stably expressing luciferase downstream of an NF-κB response element. The dose-dependent inhibition of LPS-induced luciferase activity by the analogs was expressed as IC50 values (Fig. 1A). Compounds 6, 8, and 9 had IC50 values and rates of autoxidation comparable with curcumin. Compounds 10 and 11 inhibited NF-κB at lower IC50 than curcumin, which correlated with their increased rate of autoxidation. Stable analogs 4, 7, and 12 were only weakly inhibitory such that their IC50 values were estimated. Compounds 2 and 3 had inhibitory activity similar to curcumin although they did not readily autoxidize. Because both compounds have methyl groups placed at the phenolic hydroxyl we reasoned that these may have been removed enzymatically during the cell incubations, reverting the compounds back to curcumin. In fact, LC-MS analyses showed that both 2 and 3 had lost one or both para-methoxy groups, respectively, during cell incubations (Fig. S2). Metabolically formed curcumin was present in the cell supernatants at about 20 and 75% abundance relative to 2 and 3, respectively, and likely contributed to the inhibition of NF-κB activity.
The HPLC-isolated spiroepoxide intermediate of curcumin autoxidation inhibited NF-κB activation with an IC50 of 29 μm (Fig. 1B), demonstrating activity of an unstable and reactive intermediate. The technically challenging aspects of isolating a reaction intermediate, including loss during sample preparation and quantification of the unstable spiroepoxide, likely contributed to the increased IC50 compared with curcumin. In addition, although at physiological pH the spiroepoxide has a half-life greater than curcumin it reacts much more readily with cellular thiols (19). Therefore, media and cellular environments likely contributed to scavenging of exogenously added spiroepoxide before it reached the inside of the cell and its protein targets. Nevertheless, the inhibitory potency of the spiroepoxide suggested that much of the anti-inflammatory activity of curcumin is mediated by this and other reactive intermediates of curcumin autoxidation. In contrast, the final bicyclopentadione oxidation product or the putative cleavage products, vanillin and ferulic acid (22–24), were unable or less able, respectively, to inhibit NF-κB (Fig. 1B).
Inhibition of IKKβ and p65
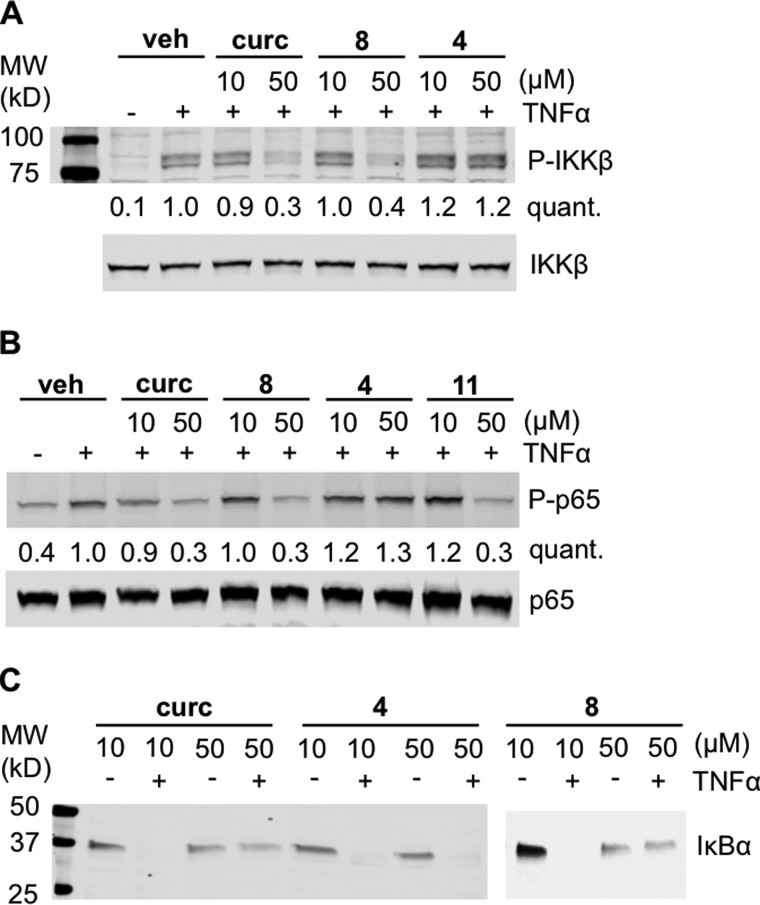
Both the upstream kinase IKKβ as well as the p65 subunit of NF-κB have been implicated as targets of the anti-inflammatory activity of curcumin (25, 26). We used TNFα-activated HeLa cells to determine targets of curcumin and its analogs within the NF-κB pathway. Curcumin reduced phosphorylation of IKKβ as well as p65 of the NF-κB complex and inhibited proteasomal degradation of IκBα (Fig. 2) (27). These results were consistent with inhibition of IKKβ (27). Oxidizable analog 8 had the same effect as curcumin, whereas non-oxidizable 4 did not inhibit phosphorylation of IKKβ and p65 and did not inhibit degradation of IκBα (Fig. 2), indicating that inhibition of NF-κB activity was dependent on oxidative activation. This was further supported by inhibition of p65 phosphorylation by highly active and oxidizable 11 (Fig. 2).
Figure 2.
Inhibition of NF-κB pathway proteins. HeLa cells were pretreated with curcumin or analogs 4, 8, or 11 for 1 h prior to stimulation with TNFα. Cells were harvested after 5 min in A or 20 min in B and C, and cellular protein was analyzed by SDS-PAGE/immunoblotting for total and phosphorylated IKKβ (A), p65 (B), and IκBα (C). The intensity of phosphorylated protein (p-IKKβ, p-p65) relative to total protein (IKKβ, p65) was quantified (quant.) and normalized to treatment with LPS. Veh, vehicle.
Oxidative status of the cell determines the response to curcumin
We hypothesized that cellular GSH scavenges and inactivates curcumin-derived electrophiles. A corresponding putative curcumin–GS adduct was detected in the supernatants of RAW264.7 cells in LC-MS analyses (Fig. 3A) (28). Therefore, depletion of GSH was predicted to enhance the potency of curcumin. Stably transfected RAW264.7 cells were depleted of GSH by preincubation with BSO (buthionine sulfoximine), an inhibitor of GSH biosynthesis (29), resulting in significantly reduced levels of curcumin–GS adducts in the cells (Fig. 3B). BSO treatment caused a leftward shift of the dose-response curve for curcumin, indicating increased potency in the absence of GSH (Fig. 3C), compatible with higher concentrations of electrophilic curcumin metabolites.
Figure 3.
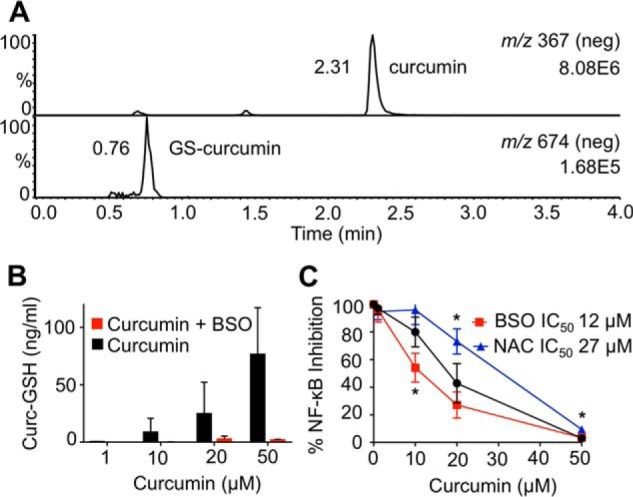
Modulation of glutathione changes the anti-inflammatory effect of curcumin. A, LC-MS detection of curcumin (top) and its glutathione adduct (bottom) in the supernatant of RAW264.7 cells. B, LC-MS quantification of curcumin-glutathione adducts with and without pretreatment with BSO. C, RAW264.7 cells were pretreated with BSO, NAC, or vehicle for 24 h prior to addition of curcumin, and the IC50 for inhibition of LPS-induced NF-κB activity was determined. *, p < 0.05 versus curcumin (n = 3).
We next pre-treated RAW264.7 cells with N-acetylcysteine (NAC),4 a GSH biosynthetic precursor (30). NAC shifted the dose-response curve for curcumin to the right, indicating a decrease in potency presumably due to increased protection of the cells from electrophilic oxidation products of curcumin (Fig. 3C).
Oxidative metabolites of curcumin adduct to IKKβ
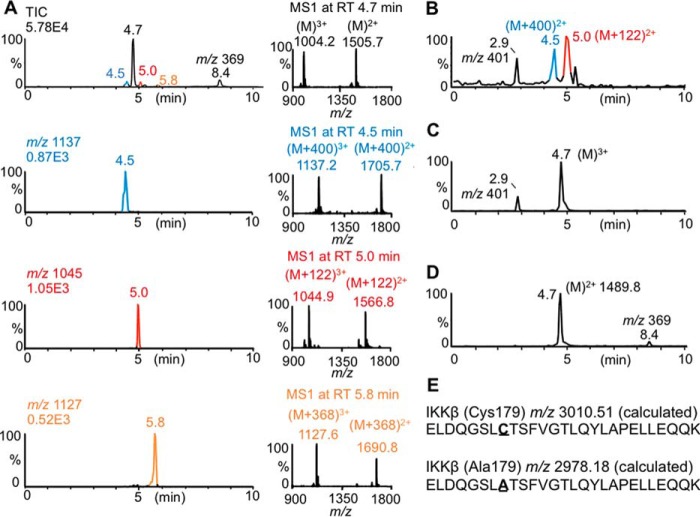
We aimed to obtain direct evidence for the covalent adduction of curcumin-derived electrophiles to IKKβ as a mechanism of inhibiting its kinase activity. We used a tryptic peptide of the activation loop of IKKβ as a model to analyze covalent adduction to the redox-sensitive residue, Cys179 (31). An equivalent but shorter peptide containing Cys179 has been shown to mimic redox regulation of IKKβ (32). Autoxidation of curcumin in the presence of the IKKβ-peptide gave adducted peptides with an increase of 400, 368, and 122 mass units, compatible with covalent adduction of oxidized curcumin (+400), curcumin or its quinone methide (+368), and a methoxyphenol cleavage product (+122), respectively (Fig. 4A). Using hexadeuterated curcumin (with the label placed in the methoxy groups) the adducts showed an additional shift of 6 (+406 and +374) or 3 (+125) mass units, respectively (Fig. S3). Incubation of HPLC-isolated spiroepoxide intermediate with the IKKβ-peptide gave abundant +400 as well as +122 adducts with no unadducted peptide remaining (Fig. 4B). In contrast, no adduct was detected when the IKKβ peptide was incubated with bicyclopentadione or stable dimethylcurcumin 3, or when a C179A mutant peptide was used in the autoxidation of curcumin (Fig. 4, C and D, and Fig. S3).
Figure 4.
Oxidative metabolites of curcumin covalently bind IKKβ. A, curcumin (50 μm) was autoxidized in the presence of a 27-amino acid peptide (10 μm) representing the activation domain of IKKβ for 1 h, and the mixture was analyzed by LC-MS. The total ion chromatogram, ion traces, and MS1 spectra of peptides increased by 400, 122, and 368 atomic mass units are shown. B and C, LC-MS analysis of the 27-amino acid peptide incubated with (B) HPLC-isolated spiroepoxide (Mr 400) or (C) bicyclopentadione (Mr 400). The peak with m/z 401 represents formed (B) or unreacted (C) bicyclopentadione, respectively. D, LC-MS analysis of a 27-amino acid C179A mutant peptide (10 μm) incubated with curcumin (50 μm). E, amino acids sequences of the 27-amino acid peptides Cys179 and Ala179.
Oxidative metabolites of curcumin adduct to cellular protein
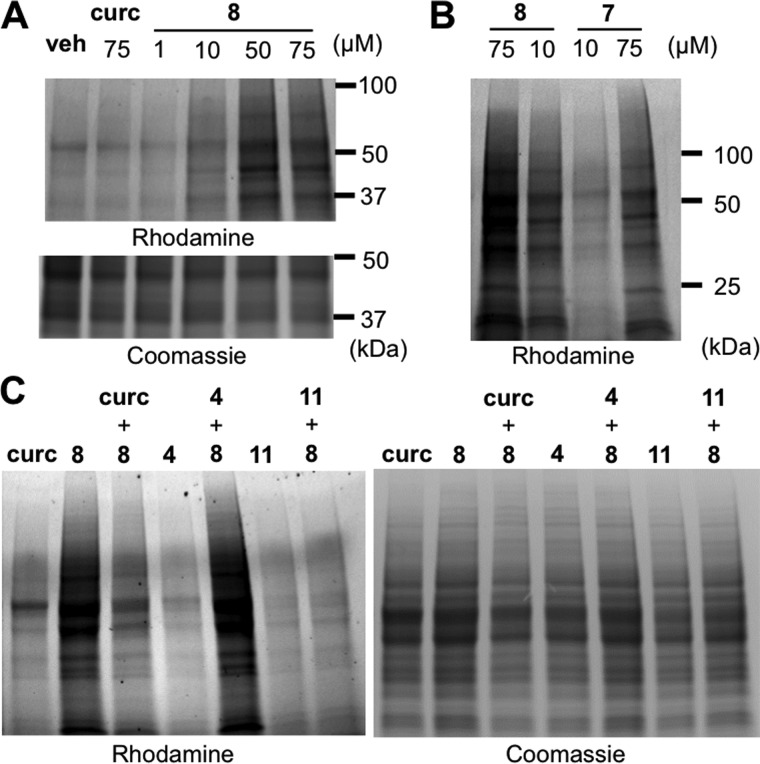
Alkynyl-tagged “click”-able derivatives have been used to analyze cellular protein targets of electrophilic small molecules (33, 34). To analyze adduction of curcumin and its oxidative metabolites to cellular protein we designed analogs in which the aromatic methoxy group was modified to contain an alkynyl tag. We prepared stable and oxidizable alkynyl-tagged analogs of curcumin to analyze the role of oxidative activation in protein adduction. 3′-O-Alkynyl-5′-O-methoxycurcumin 8 was most similar to curcumin in autoxidation rate (1.6 ± 0.4 μm/min versus 4.0 ± 0.4 μm/min) and IC50 for inhibition of NF-κB (24 versus 18 μm) (Fig. 1A). In contrast, alkynyl analog 7 was stable toward autoxidation and less active (IC50 ≈ 70 μm).
RAW264.7 cells were treated with test compounds, and adducted proteins were detected after click reaction with azide-linked rhodamine and resolution using SDS-PAGE. Untagged curcumin showed a weak signal, which may have been due to its intrinsic fluorescence activity. Oxidizable alkynyl analog 8 adducted to cellular proteins of a wide molecular weight range in a dose-dependent manner (Fig. 5A). In contrast, when cells were incubated with stable 3′-O-alkynylcurcumin 7 rhodamine staining was less intense, indicating that protein adduction was significantly less abundant by a probe that was resistant to oxidative activation (Fig. 5B).
Figure 5.
Adduction of alkynyl-analog 8 to cellular protein. A, RAW264.7 cells were treated with curcumin (75 μm) or alkynyl-analog 8 (1–75 μm). B, RAW264.7 cells were treated with alkynyl-analogs 8 (oxidizable) or 7 (non-oxidizable) at 10 and 75 μm, respectively. C, pretreatment of cells with curcumin, non-oxidizable 4, or oxidizable 11 (150 μm) followed by treatment with 8 (75 μm) or vehicle. Rhodamine-azide clicked protein samples (25 μg) were resolved by SDS-PAGE (4–20%) and analyzed by fluorescence or Coomassie stain.
To validate whether protein adduction by 8 was representative of curcumin we pretreated cells with curcumin and stable or unstable analogs. Pretreatment with curcumin prior to oxidizable 8 significantly reduced staining, and this effect was even more prominent by pretreatment with more readily oxidizable 11 (Fig. 5C). In contrast, preincubation with autoxidation-resistant 4 did not reduce adduct formation. These results imply that covalent adduction of curcumin to cellular protein mainly occurred by its reactive metabolites and less by the parent compound.
Discussion
The chemical instability of curcumin in aqueous buffer is well recognized (17) and, in addition to its low bioavailability, widely considered a key factor that limits successful therapeutic application of curcumin (35, 36). According to this hypothesis, loss of curcumin by degradation equals loss of the active compound (37). We propose the opposite. Our studies identify curcumin as a pro-drug that undergoes oxidative activation, i.e. degradation, into reactive metabolites as the mediators of its anti-inflammatory effects. The reactive metabolites, including quinone methide and spiroepoxide intermediates (19), covalently adduct to cellular protein thereby modulating function (Fig. 6).
Figure 6.
Oxidative bioactivation of curcumin. Bioactivation is initiated by hydrogen abstraction from a phenolic hydroxyl of curcumin. The resulting quinone methide radical forms a cyclopentadione ring and adds molecular oxygen to give a spiroepoxide intermediate that undergoes further transformation to the final bicyclopentadione as detailed in Ref. 19. The electrophilic carbons (indicated by arrows) are the proposed sites of reaction with nucleophilic Cys179 of IKKβ, cellular protein, and glutathione.
One of the targets of the anti-inflammatory activity of curcumin is IKKβ of the NF-κB pathway. Covalent modification of Cys179 of IKKβ prevents phosphorylation of neighboring serines 177 and 181 and inhibits kinase activity (38). Cys179 is the target of a number of lipid-derived small molecule electrophiles acting as endogenous anti-inflammatory mediators, including Δ12,14-15-deoxyprostaglandin J2, 4-hydroxy-nonenal, and nitro fatty acids (39–41). Michael addition to IKKβ as a mechanism for the anti-inflammatory activity has been attributed previously to the parent curcumin, and it was implied that the reacting electrophile is the α,β-unsaturated enone (42). The enone in curcumin, however, is only a weak electrophile due to a conjugated double bond system that spans the entire molecule, as is evident from the yellow–orange color. Using the IKKβ peptide probe we found that Cys179 was adducted by the spiroepoxide and possibly other oxidative curcumin metabolites (+400 atomic mass units) and a methoxyphenol cleavage product (+122 atomic mass units), whereas the reacting species for the +368 atomic mass unit adduct may be the parent curcumin or its quinone methide oxidative metabolite (Fig. 6). Whether the reaction occurs with curcumin or its quinone methide cannot readily be distinguished because the electrophilic carbon is identical in either molecule and thus predicted to result in the same adduct. The fact that two non-oxidizable analogs (3 and 4) containing the same α,β-unsaturated enone as curcumin were unable to adduct to the peptide (Fig. S3) provides indirect evidence favoring the quinone methide as the reacting species. The final product of oxidative transformation of curcumin, bicyclopentadione, was unable to bind to peptide and inhibit NF-κB (Figs. 1B and 4C, and Ref. 24), further supporting the importance of reactive intermediates.
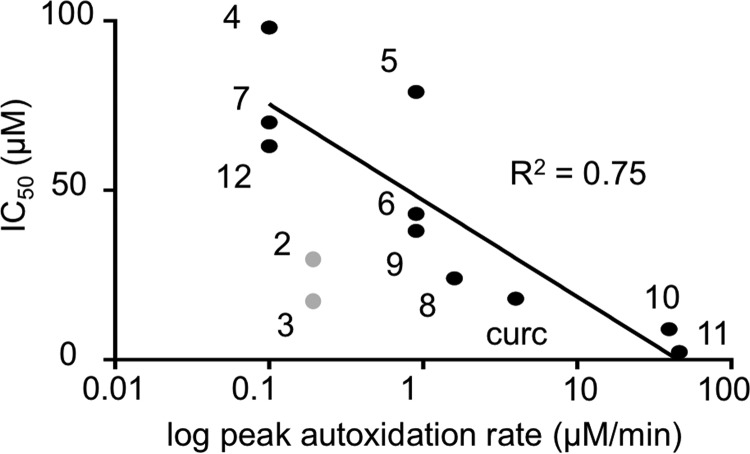
To illustrate how oxidative activation of curcumin and the different analogs contributed to their anti-inflammatory activity, we plotted the IC50 values for inhibition of NF-κB versus the rate of autoxidation and observed a positive correlation (R2 = 0.75) (Fig. 7). The more readily a compound autoxidized to form a reactive electrophile the greater was its ability to inhibit NF-κB. The relationship best fitted a hyperbolic curve, resulting in a straight line when the rate of autoxidation was plotted on a log scale. The non-linear relationship indicated that other factors contributed, including additional targets within the NF-κB pathway, possible enzymatic activation of the compounds by peroxidases (21, 43), as well as inhibitory effects independent of oxidative activation, conceivably due to the enone moiety present in 2-12. Last, the autoxidation rate is only a surrogate, albeit quantitative, measure for formation and reactivity of the electrophiles with protein thiols.
Figure 7.
Oxidizability of curcumin and its analogs correlates with inhibition of NF-κB. The IC50 values for inhibition of NF-κB are plotted versus the log of the peak autoxidation rate of curcumin, and the correlation coefficient (R2) for compounds 4-12 was determined. Compounds 2 and 3 (shown in gray) were not included in the quantitative analysis because they are resistant to autoxidation in vitro but undergo metabolic transformation to curcumin to gain inhibitory activity in cells, see Fig. S2.
The bioactivation hypothesis implies that the potency of curcumin to inhibit NF-κB is modulated by the redox status of the cell. The amount of GSH available to scavenge reactive electrophiles was inversely correlated to the activity of curcumin (Fig. 3). GSH protects cells from reactive metabolites of curcumin and limits their biologic effects that may also include oxidative damage to protein, membranes, and DNA. Inflammatory and cancer cells have an enhanced oxidative milieu (44), resulting in selective targeting by curcumin in a 2-fold manner. The oxidative milieu supports oxidative activation of curcumin, and the reduced levels of GSH weaken the ability of the cell to inactivate curcumin-derived electrophiles. Oxidative bioactivation is a novel paradigm to explain the cellular specificity of curcumin, which is known to target inflammatory as well as cancerous cells (45, 46).
Oxidative bioactivation of curcumin resulted in the covalent modification of a large number of cellular proteins, undoubtedly contributing to the polypharmacology of curcumin. Hundreds of cellular proteins are regulated, at least in part, through oxidative modification of crucial cysteine residues (47, 48), and these represent potential targets of bioactivated curcumin. An unstable, alkynyl-tagged probe mimicked protein binding by curcumin better than a stable probe (Fig. 5), although binding also by the parent curcumin could not be excluded. The alkynyl probes with differential stability developed here will be helpful in future studies to distinguish protein targets of oxidative metabolites and parent curcumin. Our results complement click chemistry approaches with the alkynyl tag placed at the phenolic hydroxyl of curcumin for the ease of chemical synthesis, and thus relying on the α,β-unsaturated enone as the sole electrophile (49–51). Based on our findings, these probes are predicted to have less anti-inflammatory and likely less overall biological activity. They are suited to detect protein targets of the parent curcumin but not of its reactive metabolites.
The biological and antioxidant activities of some of the compounds tested here (3, 6, and 10–12) have been evaluated previously in prototypical anti-inflammation assays (52, 53). In one study, 7 of 8 curcumin analogs that cannot oxidize due to blocking or absence of a phenolic hydroxyl were inactive in reducing rat paw edema, whereas 6 analogs that autoxidize or can be predicted to autoxidize showed activity comparable with curcumin (52, 54). There was a similar correlation for three oxidizable curcumin analogs (6, 10, and 11) in the transcriptional inhibition of cyclooxygenase-2 and inducible nitric-oxide synthase as well as inhibition of phorbol ester-induced ornithine decarboxylase activity in mouse skin (53, 54). Although metabolic events were not considered, these studies support the hypothesis of oxidative activation as a key mechanism mediating anti-inflammatory activities of curcumin.
There has been much interest in identifying metabolites as the active principle of dietary polyphenols and flavonoids in recent years (55, 56). The majority of investigations has focused on stable metabolites that can be detected in plasma and urine or degradation products generated via microbial transformation by intestinal bacteria (57, 58). In the case of curcumin, vanillin and ferulic acid have been invoked as potentially bioactive metabolites (22, 23). In contrast to these approaches, we have identified short-lived reactive metabolites as the main molecular species mediating anti-inflammatory effects and protein adduction of curcumin. This aligns with the concept of “biological reactive intermediates” generated from botanicals that mediate beneficial but also toxicological effects (59, 60). On a cautionary note, our studies have been conducted in cultured cells in vitro where oxidation of curcumin occurs readily. The extent of oxidative activation of curcumin in vivo is unknown. The known metabolism of curcumin in vivo entails reduction and/or conjugation with glucuronic acid and sulfate (61). Our attempts to provide evidence for oxidative transformation of curcumin in vivo, focusing on the detection of bicyclopentadione formation, have not been successful so far.5 In hindsight, this may not be surprising, considering the ready adduction of reactive intermediates to cellular thiols, resulting in metabolic redirection away from the final bicyclopentadione toward adduct formation. Nevertheless, our studies provide evidence that reactive metabolites from polyphenols are mediators of biological effects. A detailed understanding of the chemical reaction steps from the starting polyphenol to the detectable metabolites (19) will help in the identification of intermediate bioactive metabolites of other polyphenols and flavonoids of high medicinal interest.
Extracts of the plant turmeric, with its active polyphenol curcumin, have been used as an anti-inflammatory remedy in traditional Asian medicine for centuries. The widespread hopes for curcumin as a natural agent for the treatment of inflammatory and cancerous diseases have been fueled by positive results in numerous cell culture-based assays and animal models. Bioactivation occurs readily in vitro, which can explain the wide range of cellular targets of curcumin identified whereas their abundance bears the risk of overestimating its therapeutic potential. In turn, insufficient bioactivation in vivo may underlie the disappointing results in human studies with curcumin so far. Humans that are healthy and exhibit low levels of oxidative stress may be less likely to experience the benefits resulting from oxidative activation of curcumin but also be protected from potentially harmful effects. A judgment of the therapeutic potential of curcumin will be more adequate if the role of bioactivation is considered for the interpretation of in vitro data as well as for the selection of in vivo targets. The paradigm of metabolic bioactivation is likely to extent to other dietary polyphenols and flavonoids and should be considered for the evaluation and design of clinical trials.
Experimental procedures
Materials
Stock solutions of curcumin and analogs were prepared at 5 mm in methanol or at 10 mm in DMSO and stored at −20 °C. Lipopolysaccharide (LPS) from Escherichia coli serotype 0111:B4 was obtained from Sigma. HeLa and RAW264.7 cells were from ATCC.
Synthesis of curcumin and compounds 2–6
Curcumin and compounds 2-6 were synthesized following the general scheme of adding two aromatic aldehyde moieties to acetylacetone according to Pabon (62) using the appropriate aldehydes as described in Ref. 63.
Synthesis of compounds 7–9
Preparation followed the same procedure as for curcumin (62) but required synthesis of the appropriate starting aldehydes. We followed a procedure for the selective alkylation of the meta-position of 3,4-dihydroxybenzaldehyde in strongly basic solvent with some modifications (64, 65).
4-Hydroxy-3-(pent-4-yn-1-yloxy)benzaldehyde:3,4-dihydroxybenzaldehyde (501 mg, 3.86 mmol) was dissolved in a solution of 4 n NaOH in methanol (2.5 ml) and N,N-dimethylacetamide (10 ml). To this solution was added dropwise 5-iodo-pentyne (490 μl, 4.3 mmol) dissolved in toluene (5 ml) in about 45 min. The reaction was stirred at room temperature for 4 h. The solution was diluted with water and extracted with dichloromethane. 10 ml of 1 n HCl were added to the aqueous solution followed by extraction with dichloromethane (2 × 20 ml). The organic phase was washed with a saturated solution of K2CO3. 1H NMR (600 MHz, acetone-d6) δH 9.72 (s, 1H), 7.42 (m, 1H), 7.40 (d, J = 1.9 Hz, 1H), 7.06 (d, J = 8.4 Hz, 1H), 6.94 (d, J = 13.9 Hz, 1H), 4.20 (t, J = 6.2 Hz, 2H), 4.17 (t, J = 6.1 Hz, 1H), 2.42 (ddd, J = 14.1 Hz, 4.4 Hz, 2.6 Hz, 1H), 2.25 (t, 2.8 Hz, 2H), 1.99–2.06 (m, 4H).
4-Hydroxy-3-methoxy-5-(pent-4-yn-1-yloxy)benzaldehyde:3,4-dihydroxy-5-methoxybenzaldehyde (598 mg, 3.56 mmol) was dissolved in a solution of 4 n NaOH in methanol (2.5 ml) and N,N-dimethylacetamide (10 ml), and reacted with 5-iodopentyne as described above. The same extraction as with 4-hydroxy-3-(pent-4-yn-1-yloxy)benzaldehyde was performed. 1H NMR (600 MHz, acetone-d6) δH 9.79 (s, 1H), 8.28 (s, 1H), 7.22 (d, J = 12.7 Hz, 1.7 Hz, 2H), 4.20 (t, J = 6.1 Hz, 1H), 3.90 (s, 1H), 2.42 (ddd, J = 14.3 Hz, 7.1 Hz, 2.6 Hz, 1H), 2.35 (t, J = 2.6 Hz).
Compounds 7, 8, and 9 were prepared following the procedure for synthesis of curcumin and using an equimolar ratio of two aldehydes. The aldehydes were: for 7, vanillin and 4-hydroxy-3-(pent-4-yn-1-yloxy)benzaldehyde; for 8, vanillin and 4-hydroxy-3-methoxy-5-(pent-4-yn-1-yloxy)benzaldehyde; for 9, syringealdehyde and 4-hydroxy-3-(pent-4-yn-1-yloxy)benzaldehyde. The products were purified by RP-HPLC using a Waters SymmetryPrep 7-μm column (19 × 300 mm) eluted with a solvent of methanol/water/acetic acid, 70:30:0.01 (by volume), at a flow rate of 10 ml/min and UV-visible detection at 460 nm.
Compound 7 for LC-MS: 420.16; 1H NMR (600 MHz, acetone-d6) δH 8.15 (d, J = 16.8 Hz, 1H), 7.56 (d, J = 15.8, 2H), 7.32 (dd, J = 16.0 Hz, 1.9 Hz, 2H), 7.16 (m, 2H), 6.85 (dd, J = 5.28 Hz, 2.9 Hz, 2H), 6.68 (dd, J = 15.8 Hz, 4.9 Hz, 2H), 5.95 (s, 1H), 4.21 (t, J = 5.9 Hz, 2H), 3.90 (s, 3H), 3.01 (s, 1H), 2.35–2.45 (m, 3H), 1.26 (s, 1H).
Compound 8 for 1H NMR (600 MHz, acetone-d6): δH 8.19 (s, 1H), 7.97 (s, 1H), 7.62 (d, J = 15.8 Hz, 1H), 7.53 (d, J = 15.8 Hz, 1H), 7.35 (d, J = 1.9 Hz, 1H), 7.19 (dd, J = 8.2 Hz, 1.9 Hz, 1H), 6.91 (d, J = 1.9 Hz, 1H), 6.89 (d, J = 8.2 Hz), 6.86 (d, J = 1.9 Hz, 1H), 6.74 (d, J = 15.8 Hz, 1H), 6.72 (d, J = 15.8 Hz, 1H), 6.01 (s, 1H), 4.14 (t, J = 6.2 Hz, 2H), 3.92 (s, 3H), 3.91 (s, 3H), 2.42 (dt, J = 7.2 Hz, 2.7 Hz, 2H), 2.36 (t, J = 2.6 Hz), 1.93 (t, J = 6.3 Hz, 2H).
Compound 9 for 1H NMR (600 MHz, acetone-d6): δH 8.21 (s, 1H), 7.81 (s, 1H), 7.61 (d, J = 15.8 Hz, 1H), 7.59, (d, J = 15.8 Hz, 1H), 7.35 (d, J = 1.6 Hz, 1H), 7.19, (dd, J = 8.2 Hz, 1.6 Hz, 1H), 7.04 (s, 2H), 6.89 (d, J = 8.2 Hz, 1H), 6.72 (d, J = 15.8 Hz, 1H), 6.70 (d, J = 15.8 Hz, 1H), 5.98 (s, 1H), 4.23 (t, J = 6.0 Hz, 2H), 3.89 (s, 6H), 2.46 (dt, J = 7.1 Hz, 2.6 Hz, 2H), 2.40 (t, J = 2.7 Hz, 1H), 2.03 (t, J = 6.2 Hz, 2H).
Synthesis of compounds 10–12
Compounds 10-12 were synthesized according to Ref. 63 using the appropriate cyclohexanone or cyclopentanone and aldehyde starting materials.
Autoxidation of curcumin and analogs
Autoxidation rates of curcumin and its analogs were determined using a UV-visible spectrophotometer operated in the time-drive mode. Compounds were added at 50 μm to 500 μl of 20 mm ammonium acetate buffer, pH 7.5, and disappearance of the maximum absorbance for each compound (which was around 425 nm) was followed for 20 min. The rate of autoxidation was calculated using the part of the curve that showed the fastest decrease.
Cell culture and stable transfection
HeLa and RAW264.7 cells were cultured at 37 °C in 5% CO2 in DMEM containing 4.5 g/liter of d-glucose and 10% (v/v) fetal bovine serum (FBS). RAW264.7 cells at 70% confluence were stably transfected with pNFkB-TA-MetLuc with TB vector, 4.3 kb (Clontech), for expression of secreted luciferase using Lipofectamine 2000 (Invitrogen) following the manufacturer's guidelines. Stably transfected RAW 264.7 cells were supplemented with 1 mg/ml of geneticin.
NF-κB luciferase reporter assay
A Ready-to-glow Secreted Luciferase Reporter assay was used to quantify the effects of compounds on LPS-induced NF-κB activity. Stably transfected RAW267.4 cells were seeded in a 24-well plate at a concentration of 250,000 cells/well in 500 μl of DMEM. For some experiments, cells were incubated with N-acetyl-cysteine (3 mm) or buthionine sulfoximine (BSO; 0.5 mm) overnight. Cells were washed twice with cold PBS and pre-treated with curcumin or analogs (0–100 μm) in DMEM for 45 min. The cells were then stimulated with LPS (100 ng/ml). After 4 h, 50 μl of the medium was removed and luciferase activity was determined.
LC-MS analysis of autoxidation products
Curcumin and analogs (50 μm) were incubated in 20 mm ammonium acetate buffer, pH 7.5, for 20 min. Samples were extracted using Waters HLB cartridges and dissolved in 50 μl of acetonitrile/water (1:1) for LC-MS analysis. Samples were electrosprayed into a Thermo Vantage triple quadrupole mass spectrometer operated in the negative ion mode and coupled to a Waters Acquity UPLC system with a Waters symmetry Shield C18 3.5-μm column (2.1 × 100 mm) using a gradient of 5 to 95% acetonitrile in water, 0.1% formic acid within 3 min, and a flow rate of 0.4 μl/min.
LC-MS analysis of curcumin-glutathione adducts
Media samples of RAW264.7 cells treated with vehicle or BSO and curcumin were extracted using Waters HLB cartridges eluted with methanol. LC-MS analyses were performed in the negative ion mode. The same MS and UPLC system as for product analysis was used. The column was eluted with a gradient of acetonitrile in water, 0.1% formic acid changed from 30% acetonitrile to 60% in 2 min and further increased to 95% in 2 min at a flow rate of 0.4 ml/min. The selected reaction-monitoring transitions were for curcumin m/z 367 → 217, d6-curcumin m/z 373 → 220, and for curcumin-GSH adduct m/z 674 → 360.
LC-MS analysis of IKKβ peptide adducts
In silico tryptic digestion of IKKβ gave a peptide (172ELDQGSLCTSFVGTLQYLAPELLEQQK198) that contained Cys179 (underlined) (Mr 3012.43 g/mol). 10 μm Cys179 (or Ala179; Mr 2980.36 g/mol) IKKβ peptide was incubated with curcumin or analog (50 μm) at 37 °C in acetate buffer, pH 7.5, for 1 h. Aliquots were analyzed directly using a Waters Synapt G1 TOF instrument in positive ionization mode following separation of the sample using a Waters XSelect C18 column with a gradient of acetonitrile/water, 15:85 (by volume) to 65:35 (by volume), containing 0.1% formic acid within 10 min at a flow rate of 0.25 ml/min.
Immunoblot detection of NF-κB pathway proteins
HeLa cells were seeded in a 6-well plate in 2 ml of DMEM with 10% (v/v) FBS. Cells were pretreated with curcumin or analogs (0, 10, and 50 μm) for 1 h followed by stimulation with 20 ng/ml of human TNFα for 20 min. For analysis of IKKβ phosphorylation cells were stimulated for 5 min. Cells were washed with PBS and lysed using 100 μl of cell lysis buffer containing 1 mm PMSF, protease, and phosphatase inhibitor mixtures. Cell lysates (20 μg of protein) were resolved on a 10% SDS-PAGE gel and transferred to nitrocellulose. Primary phosphoprotein antibodies were used as 1:500 to 1:1,000 dilution and probed with IRDye secondary antibodies (1:20,000). Blots were stripped and re-probed for total protein. Band intensities were quantified using Image Studio Lite version 5.0 (LI-COR Biosciences). Primary antibodies used in this study were: NF-κB p65 (C22B4), phospho-NF-κB p65 (Ser536) (93H1), IκBα (44D4), phospho-IKKα/β (Ser176/180) (16A6) (Cell Signaling), and IKKβ (551920) (Pharmingen). Secondary antibodies used were IRDye 680LT goat anti-rabbit (926-68024) and IRDye 800CW donkey anti-goat (926-32214) (LI-COR Biosciences).
Detection of alkynyl-tagged curcumin-cellular protein adducts
RAW267.4 cells were seeded at 500,000 cells/ml in 6-well plates and allowed to adhere overnight. The cells were preincubated with DMSO, curcumin, or analogs (150 μm each) for 2 h, followed by incubation with 8 for 2 h. Cells were lysed using 100 mm sodium phosphate buffer, pH 7.5, with 0.1% Triton X-100 followed by centrifugation at 13,000 × g at 5 °C for 10 min. For click chemistry 50 μg of protein was incubated with rhodamine-B-azide (20 μm; Azide Fluor 545, Sigma), Tris(2-carboxyethyl)phosphine hydrochloride (1 mm), Tris((1-benzyl-1H-1,2,3-triazol-4-yl)methyl)amine (100 μm), and CuSO4 (1 mm) for 2 h. Excess reagents were removed by acetone precipitation at −20 °C overnight. Samples were centrifuged at 13,000 × g for 10 min, acetone was removed, and samples were left to air dry for 30 min. 25 μg of protein sample solubilized in loading buffer were resolved on a 4–20% SDS-PAGE gel. Protein adducts were visualized by scanning the gel using the Gel Doc EZ system (Bio-Rad) with settings for detection of ethidium bromide. The gel was then washed three times with water and stained with Coomassie Blue.
Statistical analysis
The statistical analysis in Figs. 1 and 3 was performed using GraphPad Prism 7.00 software using an unpaired non-parametric t test (Mann-Whitney) with significance of p < 0.05 (*, < 0.05; **, < 0.01).
Author contributions
R. L. E., P. B. L., P. V. V., A. I. J., and S. H. P. data curation; R. L. E., P. B. L., and C. S. formal analysis; R. L. E., P. B. L., P. V. V., R. C., and C. S. methodology; R. L. E. and C. S. writing-review and editing; A. I. J., S. H. P., and R. C. resources; C. S. conceptualization; C. S. supervision; C. S. funding acquisition; C. S. investigation; C. S. writing-original draft; C. S. project administration.
Supplementary Material
Acknowledgment
Mass spectrometric analyses were performed in part through Vanderbilt University Medical Center's Digestive Disease Research Center supported by the National Institutes of Health Grant P30DK058404 Core Scholarship.
This work was supported in part by National Institutes of Health Grant R01AT006896 from the National Center for Complementary and Integrative Health (NCCIH) and the Office of Dietary Supplements (ODS) and in part by pilot awards from the Vanderbilt Institute in Chemical Biology and National Cancer Institute SPORE in GI Cancer Grant 5P50CA095103 (to C. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
P. B. Luis and C. Schneider, unpublished data.
- NAC
- N-acetylcysteine
- BSO
- buthionine sulfoximine.
References
- 1. Esatbeyoglu T., Huebbe P., Ernst I. M., Chin D., Wagner A. E., and Rimbach G. (2012) Curcumin-from molecule to biological function. Angew. Chem. Int. Ed. Engl. 51, 5308–5332 [DOI] [PubMed] [Google Scholar]
- 2. Heger M., van Golen R. F., Broekgaarden M., and Michel M. C. (2014) The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol. Rev. 66, 222–307 [DOI] [PubMed] [Google Scholar]
- 3. Cottart C. H., Nivet-Antoine V., and Beaudeux J. L. (2014) Review of recent data on the metabolism, biological effects, and toxicity of resveratrol in humans. Mol. Nutr. Food Res. 58, 7–21 [DOI] [PubMed] [Google Scholar]
- 4. Russo M., Spagnuolo C., Tedesco I., Bilotto S., and Russo G. L. (2012) The flavonoid quercetin in disease prevention and therapy: facts and fancies. Biochem. Pharmacol. 83, 6–15 [DOI] [PubMed] [Google Scholar]
- 5. Singh B. N., Shankar S., and Srivastava R. K. (2011) Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 82, 1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williamson G., and Manach C. (2005) Bioavailability and bioefficacy of polyphenols in humans: II. review of 93 intervention studies. Am. J. Clin. Nutr. 81, 243S–255S [DOI] [PubMed] [Google Scholar]
- 7. Vauzour D., Rodriguez-Mateos A., Corona G., Oruna-Concha M. J., and Spencer J. P. (2010) Polyphenols and human health: prevention of disease and mechanisms of action. Nutrients 2, 1106–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta S. C., Sung B., Kim J. H., Prasad S., Li S., and Aggarwal B. B. (2013) Multitargeting by turmeric, the golden spice: from kitchen to clinic. Mol. Nutr. Food Res. 57, 1510–1528 [DOI] [PubMed] [Google Scholar]
- 9. Epstein J., Sanderson I. R., and Macdonald T. T. (2010) Curcumin as a therapeutic agent: the evidence from in vitro, animal and human studies. Br. J. Nutr. 103, 1545–1557 [DOI] [PubMed] [Google Scholar]
- 10. Daily J. W., Yang M., and Park S. (2016) Efficacy of turmeric extracts and curcumin for alleviating the symptoms of joint arthritis: a systematic review and meta-analysis of randomized clinical trials. J. Med. Food 19, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hatcher H., Planalp R., Cho J., Torti F. M., and Torti S. V. (2008) Curcumin: from ancient medicine to current clinical trials. Cell. Mol. Life Sci. 65, 1631–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Núuñez-Sanchez M. A., González-Sarrías A., Romo-Vaquero M., García-Villalba R., Selma M. V., Tomás-Barberán F. A., García-Conesa M. T., and Espín J. C. (2015) Dietary phenolics against colorectal cancer-From promising preclinical results to poor translation into clinical trials: Pitfalls and future needs. Mol. Nutr. Food Res. 59, 1274–1291 [DOI] [PubMed] [Google Scholar]
- 13. Masuda T., Hidaka K., Shinohara A., Maekawa T., Takeda Y., and Yamaguchi H. (1999) Chemical studies on antioxidant mechanism of curcuminoid: analysis of radical reaction products from curcumin. J. Agric. Food Chem. 47, 71–77 [DOI] [PubMed] [Google Scholar]
- 14. Dinkova-Kostova A. T., Abeygunawardana C., and Talalay P. (1998) Chemoprotective properties of phenylpropenoids, bis(benzylidene)cycloalkanones, and related Michael reaction acceptors: correlation of potencies as phase 2 enzyme inducers and radical scavengers. J. Med. Chem. 41, 5287–5296 [DOI] [PubMed] [Google Scholar]
- 15. Jiao Y., Wilkinson J. 4th, Christine Pietsch E., Buss J. L., Wang W., Planalp R., Torti F. M., and Torti S. V. (2006) Iron chelation in the biological activity of curcumin. Free Radic. Biol. Med. 40, 1152–1160 [DOI] [PubMed] [Google Scholar]
- 16. Wang Y. J., Pan M. H., Cheng A. L., Lin L. I., Ho Y. S., Hsieh C. Y., and Lin J. K. (1997) Stability of curcumin in buffer solutions and characterization of its degradation products. J. Pharm. Biomed. Anal. 15, 1867–1876 [DOI] [PubMed] [Google Scholar]
- 17. Schneider C., Gordon O. N., Edwards R. L., and Luis P. B. (2015) Degradation of curcumin: from mechanism to biological implications. J. Agric. Food Chem. 63, 7606–7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ketron A. C., Gordon O. N., Schneider C., and Osheroff N. (2013) Oxidative metabolites of curcumin poison human type II topoisomerases. Biochemistry 52, 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon O. N., Luis P. B., Sintim H. O., and Schneider C. (2015) Unraveling curcumin degradation: autoxidation proceeds through spiroepoxide and vinylether intermediates en route to the main bicyclopentadione. J. Biol. Chem. 290, 4817–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gordon O. N., Luis P. B., Ashley R. E., Osheroff N., and Schneider C. (2015) Oxidative transformation of demethoxy- and bisdemethoxycurcumin: products, mechanism of formation, and poisoning of human topoisomerase IIα. Chem. Res. Toxicol. 28, 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Griesser M., Pistis V., Suzuki T., Tejera N., Pratt D. A., and Schneider C. (2011) Autoxidative and cyclooxygenase-2 catalyzed transformation of the dietary chemopreventive agent curcumin. J. Biol. Chem. 286, 1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen L., and Ji H. F. (2012) The pharmacology of curcumin: is it the degradation products? Trends Mol. Med. 18, 138–144 [DOI] [PubMed] [Google Scholar]
- 23. Gordon O. N., and Schneider C. (2012) Vanillin and ferulic acid: not the major degradation products of curcumin. Trends Mol. Med. 18, 361–363; author reply 363–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanidad K. Z., Zhu J., Wang W., Du Z., and Zhang G. (2016) Effects of stable degradation products of curcumin on cancer cell proliferation and inflammation. J. Agric. Food Chem. 64, 9189–9195 [DOI] [PubMed] [Google Scholar]
- 25. Singh S., and Aggarwal B. B. (1995) Activation of transcription factor NF-κB is suppressed by curcumin (diferuloylmethane) [corrected]. J. Biol. Chem. 270, 24995–25000 [DOI] [PubMed] [Google Scholar]
- 26. Philip S., and Kundu G. C. (2003) Osteopontin induces nuclear factor κB-mediated promatrix metalloproteinase-2 activation through IκBα/IKK signaling pathways, and curcumin (diferulolylmethane) down-regulates these pathways. J. Biol. Chem. 278, 14487–14497 [DOI] [PubMed] [Google Scholar]
- 27. Sakurai H., Chiba H., Miyoshi H., Sugita T., and Toriumi W. (1999) IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274, 30353–30356 [DOI] [PubMed] [Google Scholar]
- 28. Awasthi S., Pandya U., Singhal S. S., Lin J. T., Thiviyanathan V., Seifert W. E. Jr., Awasthi Y. C., and Ansari G. A. (2000) Curcumin-glutathione interactions and the role of human glutathione S-transferase P1–1. Chem. Biol. Interact. 128, 19–38 [DOI] [PubMed] [Google Scholar]
- 29. Drew R., and Miners J. O. (1984) The effects of buthionine sulphoximine (BSO) on glutathione depletion and xenobiotic biotransformation. Biochem. Pharmacol. 33, 2989–2994 [DOI] [PubMed] [Google Scholar]
- 30. Commandeur J. N., Stijntjes G. J., and Vermeulen N. P. (1995) Enzymes and transport systems involved in the formation and disposition of glutathione S-conjugates: role in bioactivation and detoxication mechanisms of xenobiotics. Pharmacol. Rev. 47, 271–330 [PubMed] [Google Scholar]
- 31. Byun M. S., Choi J., and Jue D. M. (2006) Cysteine-179 of IκB kinase β plays a critical role in enzyme activation by promoting phosphorylation of activation loop serines. Exp. Mol. Med. 38, 546–552 [DOI] [PubMed] [Google Scholar]
- 32. Reynaert N. L., van der Vliet A., Guala A. S., McGovern T., Hristova M., Pantano C., Heintz N. H., Heim J., Ho Y. S., Matthews D. E., Wouters E. F., and Janssen-Heininger Y. M. (2006) Dynamic redox control of NF-κB through glutaredoxin-regulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl. Acad. Sci. U.S.A. 103, 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs A. T., and Marnett L. J. (2010) Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 43, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright M. H., and Sieber S. A. (2016) Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep. 33, 681–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metzler M., Pfeiffer E., Schulz S. I., and Dempe J. S. (2013) Curcumin uptake and metabolism. BioFactors 39, 14–20 [DOI] [PubMed] [Google Scholar]
- 36. Mahran R. I., Hagras M. M., Sun D., and Brenner D. E. (2017) Bringing curcumin to the clinic in cancer prevention: a review of strategies to enhance bioavailability and efficacy. AAPS J. 19, 54–81 [DOI] [PubMed] [Google Scholar]
- 37. Zhu J., Sanidad K. Z., Sukamtoh E., and Zhang G. (2017) Potential roles of chemical degradation in the biological activities of curcumin. Food Funct. 8, 907–914 [DOI] [PubMed] [Google Scholar]
- 38. Delhase M., Hayakawa M., Chen Y., and Karin M. (1999) Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science 284, 309–313 [DOI] [PubMed] [Google Scholar]
- 39. Straus D. S., Pascual G., Li M., Welch J. S., Ricote M., Hsiang C. H., Sengchanthalangsy L. L., Ghosh G., and Glass C. K. (2000) 15-Deoxy-Δ12,14-prostaglandin J2 inhibits multiple steps in the NF-κB signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 97, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji C., Kozak K. R., and Marnett L. J. (2001) IκB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J. Biol. Chem. 276, 18223–18228 [DOI] [PubMed] [Google Scholar]
- 41. Villacorta L., Chang L., Salvatore S. R., Ichikawa T., Zhang J., Petrovic-Djergovic D., Jia L., Carlsen H., Schopfer F. J., Freeman B. A., and Chen Y. E. (2013) Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc. Res. 98, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jobin C., Bradham C. A., Russo M. P., Juma B., Narula A. S., Brenner D. A., and Sartor R. B. (1999) Curcumin blocks cytokine-mediated NF-κB activation and proinflammatory gene expression by inhibiting inhibitory factor IκB kinase activity. J. Immunol. 163, 3474–3483 [PubMed] [Google Scholar]
- 43. Luis P. B., Gordon O. N., Nakashima F., Joseph A. I., Shibata T., Uchida K., and Schneider C. (2017) Oxidative metabolism of curcumin-glucuronide by peroxidases and isolated human leukocytes. Biochem. Pharmacol. 132, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grivennikov S. I., Greten F. R., and Karin M. (2010) Immunity, inflammation, and cancer. Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kunwar A., Barik A., Mishra B., Rathinasamy K., Pandey R., and Priyadarsini K. I. (2008) Quantitative cellular uptake, localization and cytotoxicity of curcumin in normal and tumor cells. Biochim. Biophys. Acta 1780, 673–679 [DOI] [PubMed] [Google Scholar]
- 46. Aggarwal B. B., and Sung B. (2009) Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol. Sci. 30, 85–94 [DOI] [PubMed] [Google Scholar]
- 47. Lo Conte M., and Carroll K. S. (2013) The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 288, 26480–26488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poole L. B. (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 80, 148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Angelo L. S., Maxwell D. S., Wu J. Y., Sun D., Hawke D. H., McCutcheon I. E., Slopis J. M., Peng Z., Bornmann W. G., and Kurzrock R. (2013) Binding partners for curcumin in human schwannoma cells: biologic implications. Bioorg. Med. Chem. 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 50. Wang J., Zhang J., Zhang C. J., Wong Y. K., Lim T. K., Hua Z. C., Liu B., Tannenbaum S. R., Shen H. M., and Lin Q. (2016) In situ proteomic profiling of curcumin targets in HCT116 colon cancer cell line. Sci. Rep. 6, 22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luo Z., Tikekar R. V., and Nitin N. (2014) Click chemistry approach for imaging intracellular and intratissue distribution of curcumin and its nanoscale carrier. Bioconj. Chem. 25, 32–42 [DOI] [PubMed] [Google Scholar]
- 52. Nurfina A. N., Reksohadiprodjo M. S., Timmerman H., Jenie U. A., Sugiyanto D., and vander Goot H. (1997) Synthesis of some symmetrical curcumin derivatives and their antiinflammatory activity. Eur. J. Med. Chem. 32, 321–328 [Google Scholar]
- 53. Gafner S., Lee S. K., Cuendet M., Barthélémy S., Vergnes L., Labidalle S., Mehta R. G., Boone C. W., and Pezzuto J. M. (2004) Biologic evaluation of curcumin and structural derivatives in cancer chemoprevention model systems. Phytochemistry 65, 2849–2859 [DOI] [PubMed] [Google Scholar]
- 54. Portes E., Gardrat C., and Castellan A. (2007) A comparative study on the antioxidant properties of tetrahydrocurcuminoids and curcuminoids. Tetrahedron 63, 9092–9099 [Google Scholar]
- 55. Edwards M., Czank C., Woodward G. M., Cassidy A., and Kay C. D. (2015) Phenolic metabolites of anthocyanins modulate mechanisms of endothelial function. J. Agric. Food Chem. 63, 2423–2431 [DOI] [PubMed] [Google Scholar]
- 56. Amin H. P., Czank C., Raheem S., Zhang Q., Botting N. P., Cassidy A., and Kay C. D. (2015) Anthocyanins and their physiologically relevant metabolites alter the expression of IL-6 and VCAM-1 in CD40L and oxidized LDL challenged vascular endothelial cells. Mol. Nutr. Food Res. 59, 1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Giménez-Bastida J. A., González-Sarrías A., Larrosa M., Tomás-Barberán F., Espín J. C., and García-Conesa M. T. (2012) Ellagitannin metabolites, urolithin A glucuronide and its aglycone urolithin A, ameliorate TNF-α-induced inflammation and associated molecular markers in human aortic endothelial cells. Mol. Nutr. Food Res. 56, 784–796 [DOI] [PubMed] [Google Scholar]
- 58. González-Sarrías A., Núñez-Sánchez M. Á., Tomé-Carneiro J., Tomás-Barberán F. A., García-Conesa M. T., and Espín J. C. (2016) Comprehensive characterization of the effects of ellagic acid and urolithins on colorectal cancer and key-associated molecular hallmarks: microRNA cell specific induction of CDKN1A (p21) as a common mechanism involved. Mol. Nutr. Food Res. 60, 701–716 [DOI] [PubMed] [Google Scholar]
- 59. Rietjens I. M., Al Huseiny W., and Boersma M. G. (2011) Flavonoids and alkenylbenzenes: new concepts in bioactivation studies. Chem. Biol. Interact. 192, 87–95 [DOI] [PubMed] [Google Scholar]
- 60. Dietz B. M., and Bolton J. L. (2011) Biological reactive intermediates (BRIs) formed from botanical dietary supplements. Chem. Biol. Interact. 192, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pan M. H., Huang T. M., and Lin J. K. (1999) Biotransformation of curcumin through reduction and glucuronidation in mice. Drug. Metab. Dispos. 27, 486–494 [PubMed] [Google Scholar]
- 62. Pabon H. J. J. (1964) A synthesis of curcumin and related compounds. Recl. Trav. Chim. Pays Bas 83, 379–386 [Google Scholar]
- 63. Ligeret H., Barthelemy S., Zini R., Tillement J. P., Labidalle S., and Morin D. (2004) Effects of curcumin and curcumin derivatives on mitochondrial permeability transition pore. Free Radic. Biol. Med. 36, 919–929 [DOI] [PubMed] [Google Scholar]
- 64. Schneider S., and Rolando C. (1992) One step synthesis of vanillin d3 (4-hydroxy-3-(methoxy-d3)-benzaldehyde). J. Labelled Comp. Radiopharm. 31, 489–492 [Google Scholar]
- 65. Gordon O. N., Graham L. A., and Schneider C. (2013) Facile synthesis of deuterated and [14C]labeled analogs of vanillin and curcumin for use as mechanistic and analytical tools. J. Labelled Comp. Radiopharm. 56, 696–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.