Abstract
Xanthine oxidase (XOD), also known as xanthine dehydrogenase, is a rate-limiting enzyme in purine nucleotide degradation, which produces uric acid. Uric acid concentrations in the blood and liver exhibit circadian oscillations in both humans and rodents; however, the underlying mechanisms remain unclear. Here, we demonstrate that XOD expression and enzymatic activity exhibit circadian oscillations in the mouse liver. We found that the orphan nuclear receptor peroxisome proliferator–activated receptor-α (PPARα) transcriptionally activated the mouse XOD gene and that bile acids suppressed XOD transactivation. The synthesis of bile acids is known to be under the control of the circadian clock, and we observed that the time-dependent accumulation of bile acids in hepatic cells interfered with the recruitment of the co-transcriptional activator p300 to PPARα, thereby repressing XOD expression. This time-dependent suppression of PPARα-mediated transactivation by bile acids caused an oscillation in the hepatic expression of XOD, which, in turn, led to circadian alterations in uric acid production. Finally, we also demonstrated that the anti-hyperuricemic effect of the XOD inhibitor febuxostat was enhanced by administering it at the time of day before hepatic XOD activity increased. These results suggest an underlying mechanism for the circadian alterations in uric acid production and also underscore the importance of selecting an appropriate time of day for administering XOD inhibitors.
Keywords: bile acid, circadian rhythm, nucleoside/nucleotide metabolism, peroxisome proliferator–activated receptor (PPAR), uric acid, xanthine oxidase
Introduction
Circadian rhythms are ∼24-h cycles that allow the adaptation of physiological and behavioral activities to environmental cues. Rhythmic changes in physiological functions have been suggested to help organisms anticipate daily changes in environmental conditions and feeding times (1). This control is achieved through a complex program of gene expression. In mammals, the molecular clock machinery consists of interconnected transcriptional–translational feedback loops that ultimately ensure the proper oscillation of a number of genes in a tissue-specific manner (2–4).
The circadian machinery also causes circadian alternations in the synthesis and degradation of small molecules in the body. The expression of genes responsible for nucleotide metabolism is known to be under the control of the circadian clock (5). Consequently, the contents of free purine and pyrimidine bases, nucleosides, and nucleotides in the livers of mice vary with the time of day (5). Because the liver is a site of active de novo nucleotide synthesis and controls the supply of free bases and nucleosides to other tissues for salvage (6–9), disturbances in the hepatic circadian clock have been implicated in nucleotide imbalance disorders.
Xanthine oxidase (XOD),2 also known as xanthine dehydrogenase, is a rate-limiting enzyme in the terminal step of purine nucleotide degradation that converts hypoxanthine to xanthine and xanthine to uric acid (10–12). Circulating uric acid is excreted mainly by the kidneys (13); however, a small amount of uric acid is also secreted into the intestines (14). The hepatic contents and blood concentrations of uric acid show significant circadian oscillations in male rats (15, 16), despite its efficient degradation by uricase (urate oxidase). Although uricase is a pseudogene in primates, significant circadian oscillations in blood uric acid levels are also observed in humans and monkey (17, 18), suggesting that the synthesis and/or excretion of uric acid in primates is also under the control of the circadian clock machinery. Despite the efficient mechanisms functioning to excrete uric acid, serum levels may readily increase in a manner that depends on dietary constituents, particularly the intake of meat and seafood rich in purine (19). Therefore, XOD inhibitors are more commonly used to prevent hyperuricemia than inhibitors of the tubular reabsorption of uric acid (20, 21). However, the role of XOD in the circadian regulation of uric acid metabolism remains largely unknown.
In the present study, we demonstrated that peroxisome proliferator–activated receptor-α (PPARα) acted as a transcriptional activator of the mouse XOD gene, and its hepatic expression exhibited circadian oscillation, which was generated by the time-dependent repression of PPARα activity by bile acids. The circadian expression of XOD affected its enzymatic activity, which appeared to cause rhythmic changes in serum uric acid levels. Because circadian variations in the expression and/or activity of drug target molecules often cause dosing time–dependent changes in the pharmacological efficacy of drugs, we also investigated how the rhythmic change in the XOD activity influences the anti-hyperuricemic effects of XOD inhibitors.
Results
PPARα-mediated transcriptional regulation of the mouse XOD gene
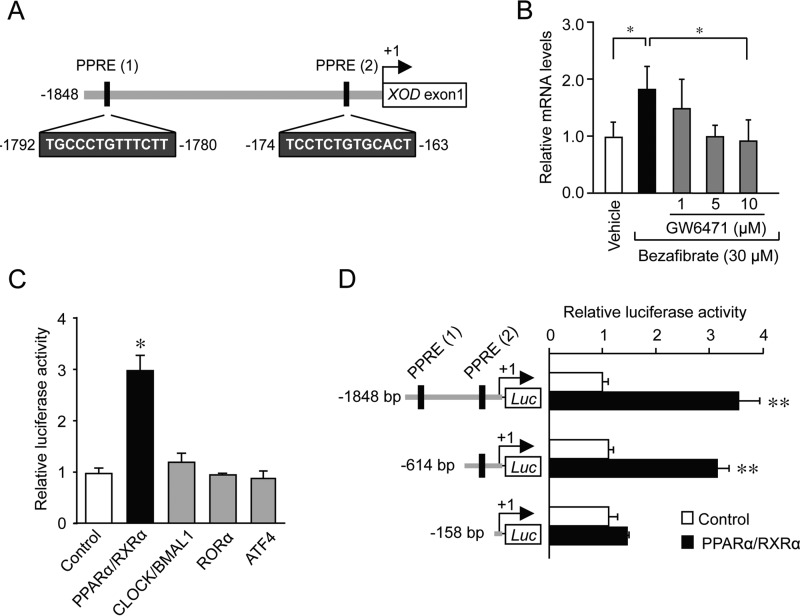
During the analysis of the upstream region of the mouse XOD gene, we noted that the nucleotide sequences located from bp −1792 to −1780 and from bp −174 to −163 (+1 indicates the transcription initiation site) showed homology with PPAR response elements (PPREs) (Fig. 1A). The sequences were also identified at a similar position in the XOD genes of rats and humans. Because the expression of some PPARα target genes shows circadian oscillations (22–24), we investigated whether the mouse XOD gene was expressed in a PPARα-dependent circadian manner. As shown in Fig. 1B, the treatment of Hepa1–6 cells with 30 μm bezafibrate, a typical PPARα agonist, significantly increased XOD mRNA levels (p < 0.05). However, the bezafibrate-induced expression of XOD mRNA was dose-dependently suppressed by the PPARα antagonist GW6471, suggesting that the hepatic expression of the XOD gene is under the control of PPARα.
Figure 1.
Transcriptional regulation of the mouse XOD gene by PPARα. A, closed boxes indicate the sites that match the consensus PPRE in the mouse XOD gene. The number of nucleotide residues indicates the distance from the transcription start site. B, Hepa1–6 cells were treated with 30 μm bezafibrate for 4 h in the presence or absence of GW6471. In plots of intensity, the mean value of vehicle-treated cells (control) was set at 1.0. Each value represents the mean ± S.D. (n = 3). *, p < 0.05, significant difference between the groups (F4,10 = 10.404, p = 0.007; ANOVA with the Tukey-Kramer post hoc test). C, Hepa1–6 cells were transfected with XOD reporter constructs (XOD (−1848)::Luc) in the absence or presence of expression plasmids encoding CLOCK, BMAL1, RORα, ATF4, PPARα, and RXRα. Luciferase activity was measured 24 h after transfection. In plots of intensity, the mean value of the control (pcDNA3.1) was set at 1.0. Each value represents the mean ± S.D. (n = 3). *, p < 0.05, significant difference from the control group (F4,10 = 64.715, p < 0.001; ANOVA with the Dunnett's post hoc test). D, Hepa1–6 cells were transfected with luciferase reporter constructs containing various lengths of the 5′-flanking region of the XOD gene in the absence or presence of expression plasmids encoding PPARα and RXRα. In plots of intensity, the mean value of the control (pcDNA3.1) was set at 1.0. Each value represents the mean ± S.D. (n = 3). **, p < 0.01, significant difference from the control group (F5,12 = 113.652, p < 0.001; ANOVA with the Tukey-Kramer post hoc test).
The activity of the luciferase reporter under the control of the mouse XOD promoter containing these two PPREs (XOD (−1848)::Luc) were also enhanced by PPARα and its heterodimer partner retinoid X receptor-α (RXRα), but this reporter was unable to respond to the clock gene products CLOCK and BMAL1 or clock-controlled gene products RORα and ATF4 (Fig. 1C). Although the transactivation effect of PPARα/RXRα on XOD reporters was still observed with the elimination of the sequence up to bp −614, the deletion of the sequence from bp −614 to −158 caused a significant reduction in the transactivation effect of PPARα/RXRα by ∼60% (Fig. 1D). These results suggest that the PPARα and RXRα complexes positively regulate the transcription of the mouse XOD gene through the PPRE located within the upstream region between bp −174 and −163.
Disrupted circadian rhythm in the hepatic expression of XOD in PPARα-null mice
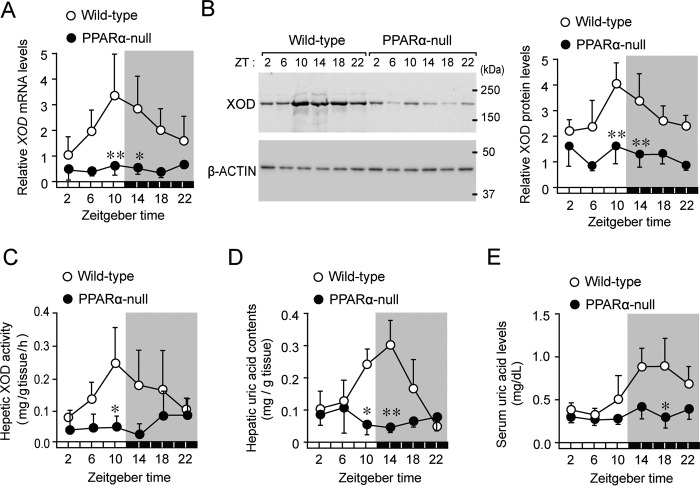
XOD mRNA and protein levels in the livers of wild-type mice exhibited significant circadian oscillations (p < 0.01 for mRNA, p < 0.05 for protein, Fig. 2, A and B). Their expression levels increased from the late light phase to the early dark phase. In the livers of wild-type mice, a similar circadian oscillation was also observed in the enzymatic activity of XOD, converting xanthine to uric acid (p < 0.01, Fig. 2C). Consistent with these results, the hepatic contents of uric acid and its serum concentrations fluctuated in a circadian time–dependent manner (p < 0.01 for hepatic contents; p < 0.05 for serum concentrations, Fig. 2, D and E); however, fluctuations in serum uric acid levels were delayed by ∼4–8 h with respect to the XOD circadian activity rhythm.
Figure 2.
Disrupted rhythms in the expression of XOD and its enzymatic activity in livers of PPARα-null mice. A, temporal expression profiles of XOD mRNA in the livers of wild-type and PPARα-null mice. Each value represents the mean ± S.D. (n = 3–6). There was a significant time-dependent variation in mRNA levels in wild-type mice (F5,30 = 3.743, p < 0.001; ANOVA). **, p < 0.01; *, p < 0.05, significant difference from wild-type mice at the corresponding time points (F11,42 = 5.932, p < 0.001; ANOVA with the Tukey-Kramer post hoc test). B, temporal expression profiles of the XOD protein in the livers of wild-type and PPARα-null mice. The left panel shows representative immunoblots of the XOD protein in the livers of mice. Each value represents the mean ± S.D. (n = 3). There was a significant time-dependent variation in protein levels in wild-type mice (F5,18 = 3.934, p = 0.014; ANOVA). **, p < 0.01, significant difference from wild-type mice at the corresponding time point (F11,35 = 9.234, p < 0.001; ANOVA with the Tukey-Kramer post hoc test). C, temporal profiles of XOD activity in the livers of wild-type and PPARα-null mice. Each value represents the mean ± S.D. (n = 3–4). *, p < 0.05, significant difference from wild-type mice at the corresponding time point (F11,29 = 3.006, p = 0.009; ANOVA with the Tukey-Kramer post hoc test). D, temporal profiles of uric acid contents in the livers of wild-type and PPARα-null mice. Each value represents the mean ± S.D. (n = 3). There was a significant time-dependent variation in the hepatic contents of uric acid in wild-type mice (F5,12 = 6.884, p = 0.003; ANOVA). **, p < 0.01; *, p < 0.05, significant difference from wild-type mice at the corresponding time point (F11,24 = 6.399, p < 0.001; ANOVA with the Tukey-Kramer post hoc test). E, temporal profiles of serum uric acid concentrations in wild-type and PPARα-null mice. Each value represents the mean ± S.D. (n = 3). There was a significant time-dependent variation in serum uric acid levels in wild-type mice (F5,12 = 3.748, p = 0.028; ANOVA). *, p < 0.05, significant difference from wild-type mice at the corresponding time point (F11,24 = 5.562, p < 0.001; ANOVA with the Tukey-Kramer post hoc test).
In contrast to findings in wild-type mice, circadian oscillations in XOD expression and its enzymatic activity were dampened in PPARα-null mice (Fig. 2, A–C), where its expression and activity decreased throughout the day. Similar disruptions were also detected in the circadian rhythms of the hepatic contents of uric acid and its serum concentrations (Fig. 2, D and E). These results suggest that PPARα acts as a positive regulator of XOD expression and is also required to generate circadian changes in its enzymatic activity.
The time-dependent repression of PPARα-mediated transactivation by bile acids underlies the circadian expression of the XOD gene
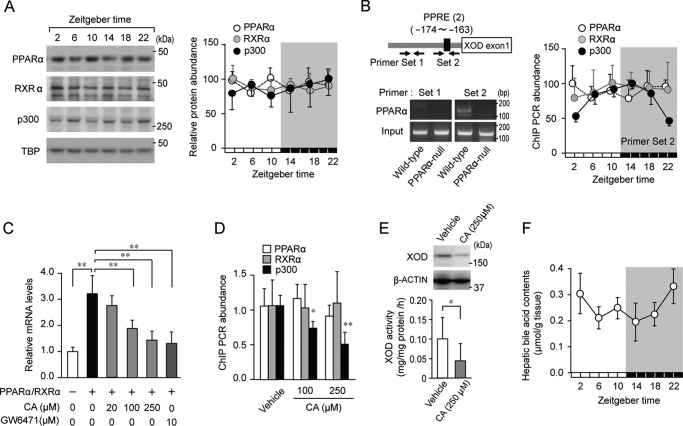
Although the transcription of PPARα was shown to be under the control of the molecular circadian clock (25), the strong expression of the PPARα protein was detected in the hepatic nuclear fraction of wild-type mice at all time points examined (Fig. 3A). In addition to PPARα, the protein levels of RXRα and the co-activator p300 did not appear to oscillate in the livers of wild-type mice (Fig. 3A). The results of the chromatin immunoprecipitation (ChIP) analysis for wild-type livers also revealed that PPARα and RXRα bound consistently to PPRE (from bp −174 to −163) in the XOD gene at both light and dark phases (Fig. 3B), whereas the recruitment of p300 on PPRE in the XOD gene varied in a circadian time–dependent manner. The oscillation observed in the recruitment of p300 showed a similar phase to the XOD mRNA rhythm (Fig. 2A).
Figure 3.
Repression of PPARα-mediated transactivation of the mouse XOD gene by bile acids. A, temporal expression profiles for PPARα, RXRα, and the p300 protein in the livers of wild-type mice. The left panel shows representative immunoblots of PPARα, RXRα, and the p300 protein in the livers of mice. Protein band intensity was normalized to TATA-binding protein (TBP) and converted to a ratio to maximal levels (set as 1.00). Each value represents the mean ± S.D. (n = 3). B, cross-linked chromatin was prepared from wild-type mice and subjected to PCR using primers surrounding the PPRE (from bp −174 to −163) in the 5′-flanking region of the mouse XOD gene. Solid arrows indicate the primer sets for the amplification area by PCR. In the right panel, the peak amounts of binding of PPARα, RXRα, and p300 were set at 100. Values shown are the mean ± S.D. (n = 4). There was a significant time-dependent variation in the amount of p300 binding (F5,12 = 10.916, p < 0.001; ANOVA). C, influence of CA on PPARα/RXRα-mediated expression of XOD mRNA in Hepa1–6 cells. Twenty-four hours after the co-transfection of PPARα and PXRα expression vectors, Hepa1–6 cells were treated with CA or GW6471 (applied as a positive control) for 4 h. Each value represents the mean ± S.D. (n = 4). **, p < 0.01, significant difference between the two groups (F5,23 = 20.252, p < 0.001; ANOVA with the Tukey-Kramer post hoc test). D, suppression effects of CA on the recruitment of p300 on PPRE in the mouse XOD gene. Hepa1–6 cells were treated with 100 and 250 μm CA or vehicle for 4 h. Cross-linked chromatin was subjected to PCR using primers surrounding the PPRE of the mouse XOD gene. Values shown are the mean ± S.D. (n = 4). **, p < 0.01; *, p < 0.05 significant decrease in the amount of p300 binding from that in the vehicle-treated group (F2,9 = 16.459, p < 0.001; ANOVA with the Tukey-Kramer post hoc test). E, suppressive effects of CA on protein expression of XOD and its activity in Hepa1–6 cells. The upper panel shows representative immunoblots of the XOD protein in Hepa1–6 cells treated with 250 μm CA or vehicle. Data were confirmed in more than three experiments in each group. The lower panel shows XOD activity in Hepa1–6 cells treated with 250 μm CA or vehicle. Each value represents the mean ± S.D. (n = 6). *, p < 0.05, significant difference between the two groups (t8 = 2.919, p = 0.015, unpaired t test, two-sided). F, temporal profile of the accumulation of bile acids in the livers of wild-type mice. Each value represents the mean ± S.D. (n = 4). There was a significant time-dependent variation in bile acid accumulation (F5,18 = 3.519, p = 0.022; ANOVA).
Because PPARα is a ligand-activated transcription factor, its transcription activity is modulated by endogenous and exogenous compounds (26). We demonstrated previously that the expression of PPARα target genes, in which intestinal expression exhibits circadian oscillations, was repressed by bile acid (25). Consistent with our previous findings (25), the PPARα/RXRα-mediated transactivation of XOD was dose-dependently repressed by cholic acid (CA) (Fig. 3C), a major component of bile acid in rodents (27). Furthermore, the treatment of Hepa1–6 cells with cholic acid interfered with the recruitment of p300 to PPRE without changing the amounts of PPARα and RXRα binding (Fig. 3D). Treatment with CA also decreased the protein levels of XOD and its enzymatic activity in hepatic cells (Fig. 3E). Because the rhythmic phase of bile acid accumulation in the livers of wild-type mice was nearly opposite that of the XOD mRNA rhythm (Fig. 3F), the time-dependent accumulation of bile acid in hepatic cells may cause circadian oscillations in XOD expression through the periodic repression of PPARα/RXRα-mediated transactivation.
Dosing time–dependent changes in the anti-hyperuricemic effect of the XOD inhibitor febuxostat in hyperuricemia mice
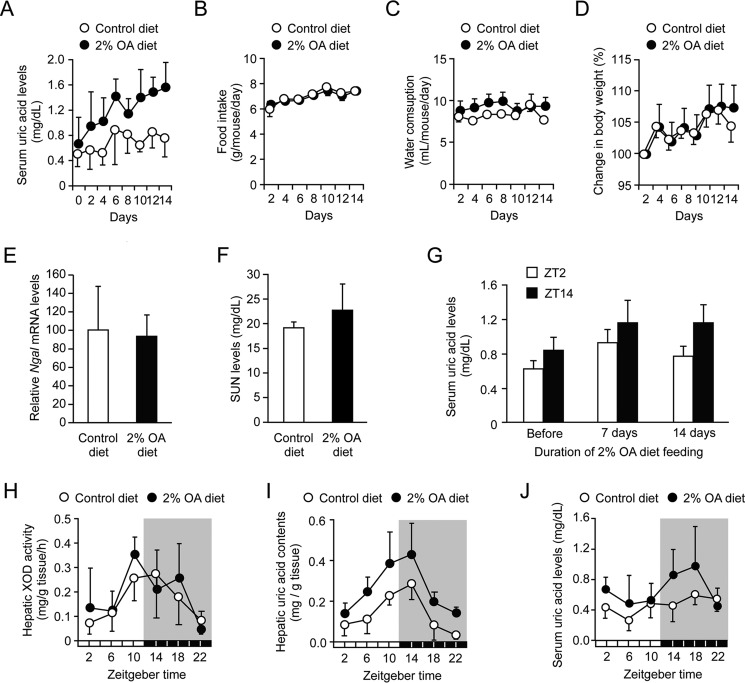
Because the activity of XOD showed circadian oscillations in the livers of wild-type mice, we investigated whether the anti-hyperuricemic effects of XOD inhibitors change in a manner that is dependent on the administration time. To achieve this goal, we prepared a hyperuricemia mouse model using the uricase inhibitor oxonic acid (OA). In rodents, uric acid is efficiently degraded by uricase. Therefore, the inhibition of this oxidase leads to the accumulation of uric acid in the blood and tissues (28, 29). Indeed, serum uric acid concentrations in mice fed a 2% OA-containing diet gradually increased during the duration of the experiment (Fig. 4A), whereas the feeding of this diet had negligible effects on food intake and water consumption (Fig. 4, B and C). Furthermore, body weight gains in mice fed the 2% OA-containing diet were similar to those observed in control mice (Fig. 4D).
Figure 4.
Characterization of hyperuricemia model mice. A–D, time course of serum uric acid levels (A), food intake (B), water consumption (C), and body weight (D) in mice during the feeding of a 2% OA-containing diet. Each value represents the mean ± S.D. (n = 6–7). E and F, the renal expression of neutrophil gelatinase-associated lipocalin-2 (Ngal) mRNA (E) and SUN concentrations (F) in mice after feeding of the 2% OA-containing diet for 14 days. Each value represents the mean ± S.D. (n = 6–7). G, temporal profiles of serum uric acid levels in mice during feeding of the 2% OA-containing diet. Blood samples were collected at ZT2 and ZT14 on the indicated day. Each value represents the mean ± S.D. (n = 3). H, temporal profiles of XOD activity in the livers of mice after feeding the 2% OA-containing diet for 14 days. Each value represents the mean ± S.D. (n = 3). I, temporal profiles of uric acid contents in the livers of mice after feeding the 2% OA-containing diet for 14 days. Each value represents the mean ± S.D. (n = 3). There was a significant time-dependent variation in the hepatic contents of uric acid in mice fed the control diet (F5,12 = 6.884, p = 0.003; ANOVA) and 2% OA-containing diet (F5,12 = 4.638, p = 0.014; ANOVA). J, temporal profiles of serum uric acid levels in mice after feeding of the 2% OA-containing diet for 14 days. There were significant time-dependent variations in serum uric acid levels in mice fed the control diet (F5,30 = 2.994, p = 0.026; ANOVA) and those fed the 2% OA-containing diet (F5,36 = 2.907, p = 0.026; ANOVA).
It is reported that the chronic intake of OA sometimes causes renal dysfunction (30, 31); however, neither the renal expression of neutrophil gelatinase-associated lipocalin-2 (Ngal) mRNA, a marker for tubular damage, nor serum urea nitrogen (SUN), a marker for renal dysfunction, was affected by feeding the 2% OA-containing diet for 2 weeks (Fig. 4, E and F). Feeding of the 2% OA-containing diet revealed a trend toward increased serum uric acid concentrations while still exhibiting time-dependent variations (Fig. 4G). Although circadian variations in hepatic XOD activity were not affected by feeding with the 2% OA-containing diet (Fig. 4H), the chronic feeding of the uricase inhibitor increased the amount of uric acid in the mouse liver at all time points examined (Fig. 4I). The amplitude of the rhythm in hepatic uric acid contents was not significantly increased by feeding with the 2% OA-containing diet, whereas the serum uric acid oscillation in mice fed the 2% OA-containing diet was enhanced from that in mice fed the control diet (Fig. 4J).
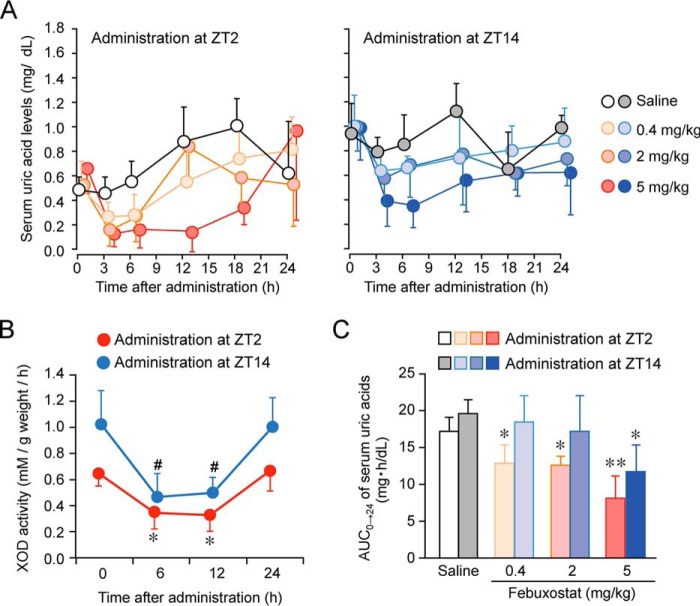
On day 14 after the initiation of 2% OA-containing diet feeding, hyperuricemic mice were intraperitoneally (i.p.) administered 0.4, 2, or 5 mg/kg body weight of febuxostat at zeitgeber time (ZT) 2 or ZT14, times at which serum uric acid levels in 2% OA-fed mice increased and declined, respectively (Fig. 4J). Serum uric acid concentrations in hyperuricemic mice transiently decreased after the single intraperitoneal administration of febuxostat at both dosing times (Fig. 5A) but returned to the basal level within 24 h of its administration. When hyperuricemic mice were administered 5 mg/kg febuxostat i.p. at ZT2 or ZT14, hepatic XOD activity was suppressed for at least 12 h, and the inhibitory effects of febuxostat wore off within 24 h of its administration at both dosing times (Fig. 5B). The time course of serum uric acid concentrations after the administration of febuxostat paralleled its inhibitory effect on XOD activity in the liver.
Figure 5.
Dosing time–dependent differences in the ability of febuxostat to decrease serum uric acid levels in chronic hyperuricemia model mice. A, time course of serum uric acid levels in chronic hyperuricemia mice after the intraperitoneal administration of 0.4, 2, or 5 mg/kg febuxostat at ZT2 or ZT14. Each value represents the mean ± S.D. (n = 4–5). B, time course of the inhibitory effects of febuxostat (5 mg/kg, i.p.) on XOD activity in the livers of mice after its administration at ZT2 and ZT14. Each value represents the mean ± S.D. (n = 4). *, p < 0.05; #, p < 0.05, significant difference from the basal level (time = 0) in the corresponding group (F3,12 = 7.904, p = 0.003 for the ZT2 administration group; F3,11 = 9.203, p = 0.002 for the ZT14 administration group, ANOVA with Tukey-Kramer post hoc test). C, the value of AUC0→24 in chronic hyperuricemia mice after the intraperitoneal administration of 0.4, 2, or 5 mg/kg febuxostat at ZT2 or ZT14. Each value represents the mean ± S.D. (n = 4–5). **, p < 0.01; *, p < 0.05, significant difference from the saline-treated groups at the corresponding dosing time (F3,13 = 15.888, p < 0.001 for the ZT2 administration group; F3,13 = 3.803, p = 0.037 for the ZT14 administration group, ANOVA with Tukey-Kramer post hoc test).
To compare the anti-hyperuricemic effects of febuxostat, we calculated the area under the curve of serum uric acid concentrations from 0 to 24 h after the febuxostat injection (AUC0→24). Dose-dependent reductions in the AUC0→24 of serum uric acid concentrations were observed in hyperuricemic mice after the administration of febuxostat at both dosing times; however, AUC0→24 values after the administration of the drug at ZT2 were slightly lower than those in mice given the drug at ZT14 (Fig. 5C). These results suggest that the XOD inhibitor febuxostat effectively decreased serum uric acid concentrations when administered at the time of day prior to elevations in hepatic XOD activity.
Discussion
Several orphan nuclear receptors have been identified, and some have been characterized as lipid sensors that respond to elevated cellular lipid levels to regulate gene expression (32, 33). Although a number of polyunsaturated fatty acids serve as ligands of PPARα (34), transcriptional activity is also modulated by endogenous as well as exogenous compounds (30). In this study, we demonstrated that PPARα acted as a transcriptional activator of the mouse XOD gene and the PPARα-mediated transactivation of XOD was repressed by bile acids. As previous and our present studies have demonstrated that the hepatic contents of bile acids vary in a circadian time–dependent manner (35, 36), the expression of XOD may have oscillated due to the time-dependent suppression of PPARα-mediated transactivation by bile acids.
PPARα and its heterodimer partner RXRα activate the transcription of the XOD gene by binding to PPRE; however, their binding on PPRE in the XOD gene was unaffected by CA. Consistent with previous findings (22), CA interfered with the recruitment of p300 on PPRE in mouse XOD genes, thereby repressing its transcription. Assembled or preassembled coactivator complexes facilitate the liganded PPARα to achieve the transcriptional activation of its target genes (37). Once p300 and also the CREB-binding protein are recruited to the liganded PPARα, they remodel the chromatin structure by intrinsic histone acetyltransferase activities. p300 interacts directly with the ligand-binding domain of PPARα (38). The C-terminal region of PPARα (residues 448–468) is required for interaction with the N terminus of p300 spanning amino acids 39–117. Therefore, CA appears to interfere with the interaction between PPARα and p300, thereby suppressing XOD expression.
In wild-type mice, hepatic bile acid contents fluctuated between 0.2 and 0.3 μmol/g tissue. Because the mean volume of the mouse liver was 1.33 ± 0.05 ml/g tissue (mean ± S.D.), hepatic concentrations of bile acid appear to oscillate between 154 and 231 μm. CA is a major component of bile acid in rodents. Its content is ∼42% of total bile acids (27). Therefore, hepatic concentrations of CA appear to fluctuate between 65 and 97 μm. This fluctuation is sufficient to affect XOD expression in the livers of mice because the PPARα/RXRα-induced expression of XOD mRNA in Hepa1–6 cells was significantly suppressed by CA at concentrations greater than 100 μm.
A previous study reports that the expression of Pparα is under the control of the circadian clock (25), and the expression of Pparα mRNA exhibits significant circadian oscillations. Despite demonstrating clear circadian expression of Pparα mRNA, there are no unified views on the circadian rhythmicity in the expression of PPARα protein (39–41). In the present study, we were also unable to detect significant circadian expression of the PPARα protein in the hepatic nucleus of ICR mice. Furthermore, the binding of PPARα to the XOD gene promoter region was constant throughout the day. These results suggest that PPARα and RXRα constantly bind to PPRE in the XOD gene to activate its expression. The temporal accumulation of CA in hepatic cells interferes with the association of PPARα with p300, resulting in the rhythmic expression of XOD mRNA and its protein.
Although the XOD gene is expressed ubiquitously in the body (42), the liver, which is the largest internal organ in humans and rodents, serves as the major site for the production of uric acid. Because the expression of XOD and its enzymatic activity in the livers of mice peaked around the late light phase, the hepatic synthesis of uric acid may be enhanced during this time window. After its synthesis in hepatic cells, uric acid is exported from the liver to the circulation. Therefore, oscillations in serum uric acid concentrations appeared to be delayed by ∼4–8 h relative to the hepatic rhythm of XOD activity.
In rodents, uric acid is efficiently degraded by uricase (28, 29); thus, the inhibition of this oxidase enzyme by oxonic acid is often employed to prepare a hyperuricemic model in rodents. In the present study, no marked changes were observed in the rhythmicity of XOD activity in the livers of mice fed the 2% OA-containing diet, whereas hepatic and serum uric acid concentrations were elevated throughout the day and still appeared to exhibit circadian oscillations. These results suggest that the feeding of mice with this dosage of oxonic acid effectively suppresses uricase activity without changing the rhythm of uric acid production. Although no significant change was observed in the amplitude of the rhythm in hepatic uric acid contents, serum uric acid oscillations were enhanced by feeding with the 2% OA-containing diet. Elevated serum uric acid concentrations in mice fed the 2% OA-containing diet were detected mainly during their daily feeding time (the dark phase). Therefore, the dietary consumption of uric acid may also contribute to enhancing serum uric acid oscillations.
A previous study demonstrated that the hepatic contents of uric acid increase in mice deficient in Bmal1, a major component of the circadian clock (5). This finding supports the present results and also suggests that the production and/or degradation of uric acid is under the control of the circadian clock. We found an elevation in XOD mRNA levels in the livers of Bmal1−/− mice, despite no significant alteration in the expression of uricase (Fig. S1). The mRNA levels of PPARα and RXRα were also not significantly increased in Bmal1−/− mice (Fig. S2). However, BMAL1 has been suggested to act as a positive regulator of bile acid synthesis (36, 43, 44). Therefore, the transcriptional activity of PPARα in the livers of Bmal1−/− mice appears to be enhanced due to a decrease in bile acid production. The alleviation of the bile acid–mediated repression of PPARα activity may induce the expression of XOD, thereby facilitating uric acid production in Bmal1−/− mice.
Because uricase is a pseudogene in primates (45), the excess production of uric acid induces hyperuricemia and gout in humans. XOD inhibitors are often administered to these patients after meals during the daytime. Febuxostat has an apparent elimination half-life of ∼5–8 h (46); therefore, this drug is generally taken once a day, mainly in the morning. In hyperuricemia model mice, the anti-hyperuricemic effect of febuxostat was enhanced by its administration in the early light phase (ZT2), during which nocturnally active mice begin to fall asleep. The inhibitory effect of febuxostat on XOD activity in the livers of mice continued for at least 12 h after its administration (5 mg/kg, i.p.) at both dosing times. However, XOD activity, namely the production of uric acid, in the livers of mice was elevated from the late light phase to the early dark phase. Therefore, pretreatment of hyperuricemic animals with febuxostat prior to elevations in hepatic XOD activity would more effectively suppress the production of uric acid.
The present results obtained from animal model reveal that XOD is a PPARα-targeted gene that is expressed in a bile acid–dependent circadian manner. The time-dependent suppression of PPARα-mediated transactivation by bile acid appeared to generate circadian oscillations in XOD expression and its enzymatic activity in the livers of mice. Thus, our present findings also indicate a molecular link connecting the circadian machinery to uric acid metabolism. Because dosing time–dependent differences in drug efficacy or side effects are caused not only by circadian changes in the sensitivity of living organisms to drugs but also by drug pharmacokinetics, selecting the most appropriate time of day for the administration of febuxostat will contribute to achieving rational chronopharmacotherapy for the treatment of patients with hyperuricemia and gout.
Experimental procedures
Animals and treatment
Male PPARα-null mice (129S4-Svjae- PPARαmuGonzN12) with a JcI:ICR background and wild-type mice of the strain (Kyudo Co., Ltd., Tosu, Japan) were housed with ad libitum access to food and water in a temperature-controlled (24 ± 1 °C) room under a 12-h light/dark cycle. Under the light/dark cycle, ZT0 was designated as “lights on” and ZT12 as “lights off.” To prepare a hyperuricemia model, mice were fed a 2% (w/w) potassium oxonate (Wako Pure Chemical Industries, Osaka, Japan)-containing diet (28, 29). All animal experiments followed the Law for the Human Treatment and Management of Animals and other related laws and regulations. All of the experiments were conducted under a protocol approved by the internal committee for animal experiments at Kyushu University.
Cell culture and treatment
The mouse hepatoma cell line Hepa1–6 (Cell Resource Center for Biomedical Research, Sendai, Japan) was cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin. Cells were grown in monolayer cultures at 37 °C in 5% CO2. After growing to semiconfluency, cells were treated with a PPARα agonist, bezafibrate (Wako Pure Chemical Industries), a PPARα antagonist, GW6471 (Sigma Aldrich), or cholic acid (Wako Pure Chemical Industries).
Construction of reporter and expression vectors
The upstream region of the mouse XOD gene (from bp −1848 to +131, bp −614 to +131, or bp −158 to +131, where +1 indicates the transcription start site) was amplified using Platinum PCR SuperMix High Fidelity (Life Technologies). PCR products were purified and ligated into the pGL4.12 promoter vector (Promega, Madison, WI). The primer sequences used in the construction of reporter vectors are listed in Table 1. The expression vectors of CLOCK, BMAL1, RORα, ATF4, PPARα, and RXRα were made previously in our laboratory (22, 47). In brief, the coding regions of each gene were obtained by RT-PCR and used after their sequences were confirmed. All coding regions were ligated into the pcDNA3.1 vector (Life Technologies).
Table 1.
Primer sets for the construction of reporter vectors
| Mouse XOD upstream region | Primers |
|---|---|
| Forward for −1848 bp | 5′-CTAGGTACCGGTTTCACATTCCTATCTC-3′ |
| Forward for −614 bp | 5′-CTCGAGCATTCTCTGCAAGTTTATAAG-3′ |
| Forward for −158 bp | 5′-TTTCTCGAGGACTAGGAGGACAGAGTA-3′ |
| Reverse | 5′-TAATCTAGAACCAAAGGTCAACTGCCA-3′ |
Luciferase reporter assay
Hepa1–6 cells were seeded into 6-well culture plates (BD, Franklin Lakes, NJ). Cells were incubated to semiconfluency and transfected with reporter vectors (200 ng/well) and expression vectors (3000 ng/well) using Lipofectamine LTX reagent (Life Technologies). As an internal control reporter, the phRL-TK vector (Promega) was co-transfected in all experiments. The total amount of DNA in each well was adjusted by the addition of empty pcDNA3.1 vectors (Life Technologies). Twenty-four hours after transfection, cells were harvested, and the lysate was analyzed using a Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized by Renilla luciferase activity in each sample.
Real-time PCR analysis
Total RNA was extracted from mouse liver or cultured Hepa1–6 cells using RNAiso Plus (Takara Bio Inc., Shiga, Japan). cDNA was synthesized from total RNA using a ReverTra Ace® qPCR RT kit (Toyobo, Osaka, Japan). Diluted cDNA samples were analyzed using THUNDERBIRDTM SYBR® qPCR mix (Toyobo) and the 7500 real-time PCR system (Applied Biosystems, Foster City, CA). The ΔCt (delta threshold cycle) method was used for quantification, and transcript levels were normalized to β-actin. The primer sequences used in the PCR analysis are listed in Table 2.
Table 2.
Primer sets for RT-PCR
| Mouse gene | Primers |
|---|---|
| XOD | |
| Forward | 5′-AGGAGAGAATTGCCAAAAGCC-3′ |
| Reverse | 5′-AAGGCATTCTCGATCTCCTCG-3′ |
| Ngal | |
| Forward | 5′-GCTGTCGCTACTGGATCAGAACATT-3′ |
| Reverse | 5′-AAATACCATGGCGAACTGGTTGTAGT-3′ |
| β-Actin | |
| Forward | 5′-GGCTGTATTCCCCTCCATCG-3′ |
| Reverse | 5′-CCAGTTGGTAACAATGCCATGT-3′ |
Western blot analysis
Hepatic tissues were homogenized in CelLyticTM cell lysis reagent (Sigma Aldrich), and nuclear fractions were prepared using a LysoPureTM nuclear and cytoplasmic extractor kit (Wako). Protein samples (10 μg) were denatured and separated on SDS-polyacrylamide gels. The separated proteins were transferred to polyvinylidene difluoride membranes. The membranes were reacted with antibodies against XOD (sc-20991, Santa Cruz Biotechnology, Santa Cruz, CA), PPARα (sc-9000, Santa Cruz Biotechnology), RXRα (sc-553, Santa Cruz Biotechnology), p300 (sc-584, Santa Cruz Biotechnology), β-actin (sc-1616, Santa Cruz Biotechnology), or TATA-binding protein (TBP, 1:1000; ab51841, Abcam). Specific antigen–antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibodies and Chemi-Lumi One (Nacalai Tesque, Kyoto, Japan). Signals from the membrane were detected and imaged using LAS3000 (Fuji Film, Tokyo, Japan).
Measurement of XOD activity
Approximately 30 mg of liver tissue was homogenized with 1.0 ml of 0.25 m sucrose solution. The liver suspension was centrifuged at 3000 rpm for 10 min, and the supernatants were collected. A total of 0.05 ml of the supernatant was incubated with 0.7 ml of 0.5 m phosphate buffer (pH 7.4) containing 7.5 μm potassium oxonate and 62.5 nmol of xanthine at 37 °C for 2 h. After deproteinization of the reacted solution by 1% trichloroacetic acid, the concentration of uric acid was assessed by high performance liquid chromatography (HPLC), which is described as follows. An increase in uric acid after the 2-h incubation was assessed as XOD activity. XOD activity was normalized by the weight of the liver.
ChIP analysis
Cross-linked chromatin in the livers were sonicated on ice, and nuclear fractions were obtained by centrifugation at 10,000 × g for 5 min. Supernatants were incubated with antibodies against PPARα (sc-9000, Santa Cruz Biotechnology), RXRα (sc-553, Santa Cruz Biotechnology), p300 (sc-584, Santa Cruz Biotechnology), or rabbit IgG (sc66931, Santa Cruz Biotechnology). DNA was purified using a DNA purification kit (Promega) and amplified by PCR for the surrounding PPRE in the 5′-flanking region of the XOD gene. The primer sequences used in the amplification of the surrounding or outside PPRE are listed in Table 3. The quantitative reliability of PCR was evaluated by the kinetics of the amplified products to ensure that signals were derived only from the exponential phase of amplification. This analysis also proceeded in the absence of an antibody or in the presence of rabbit IgG as negative controls. Ethidium bromide staining did not detect any PCR products in these samples.
Table 3.
Primer sets for the ChIP analysis
| Amplification | Primers |
|---|---|
| Primer Set 1 | |
| Surrounding PPRE | Forward: 5′-GCAAGTTTATAAGGCACGC-3′ |
| Reverse: 5′-TCAGAGACACAGACCCTTAT-3′ | |
| Primer Set 2 | |
| Outside PPRE | Forward: 5′-GCTTCCTTAATCTACGGCT-3′ |
| Reverse: 5′-CAGCCAATCCACAGATCCTA-3′ | |
Measurement of bile acid contents in the liver
Approximately 30 mg of liver tissue was homogenized with 1.0 ml of 70% ethanol and then incubated at 55 °C for 4 h. The liver suspension was centrifuged at 3000 rpm for 10 min, and the supernatant was collected. The supernatant was evaporated to dryness and resuspended in 300 μl of 0.5 m phosphate buffer (pH 7.0). The concentrations of total bile acids were measured using the Total Bile Acids assay kit (Diazyme Laboratories). The amount of bile acids was normalized by the weight of the liver.
Measurement of serum urea nitrogen
To assess renal function, SUN was measured using a urea nitrogen kit, UN3 (Wako Pure Chemical Industries) according to the manufacturer's protocol.
Measurement of serum uric acid levels
The concentration of uric acids was measured as described previously (48). In brief, 10 μl of mouse serum, urine, or liver homogenates was mixed with 0.4 μl of 0.6 m sodium perchlorate. The mixture was centrifuged at 12,000 rpm for 10 min, and the supernatant was collected. The supernatant was added to an equal volume of 0.2 m sodium hydrogen phosphate solution, including 1 mg/dl adenine as an internal control, and subjected to HPLC. The mobile phase of phosphate buffer (pH 2.2, 74 mm)-methanol (96:4, v/v) was eluted at 1.0 ml/min through a 5C18-MS-II column (4.6 × 150 mm, Nacalai Tesque) using an LC-20AD pump (Shimadzu Corp., Kyoto, Japan). The separated analyte was detected using an SPD-20A detector (284 nm, Shimadzu Corp.).
Statistical analysis
The significance of the 24-h variations in each parameter was tested by ANOVA. The significance of differences among groups was analyzed by ANOVA followed by the Tukey-Kramer or Dunnett's post hoc test. The Student's t test was applied for comparisons between two groups. Equal variances were not formally tested. p < 0.05 was considered significant.
Author contributions
T. K., N. K., S. K., and S. O. designed the study and wrote the paper. T. K., Y. T., M. O., and S. K. performed and analyzed the experiments shown in Figs. 1–5 and contributed to the preparation of these figures. M. O., and N. M. provided technical assistance. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We are grateful for the technical support provided by the Research Support Center, Graduate School of Medical Sciences, Kyushu University.
This work was supported in part by KAKENHI Grant-in-aid for Scientific Research 16H02636 and 17H06262 from the Japan Society for the Promotion of Science (JSPS) (to S. O.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- XOD
- xanthine oxidase
- PPARα
- peroxisome proliferator–activated receptor-α
- PPRE
- PPAR response element
- RXRα
- retinoid X receptor-α
- CA
- cholic acid
- OA
- oxonic acid
- SUN
- serum urea nitrogen
- AUC
- area under the curve
- ZT
- zeitgeber time
- ANOVA
- analysis of variance.
References
- 1. Panda S., Hogenesch J. B., and Kay S. A. (2002) Circadian rhythms from flies to human. Nature 417, 329–335 [DOI] [PubMed] [Google Scholar]
- 2. Ueda H. R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., and Hashimoto S. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 [DOI] [PubMed] [Google Scholar]
- 3. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 4. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., and Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 5. Fustin J. M., Doi M., Yamada H., Komatsu R., Shimba S., and Okamura H. (2012) Rhythmic nucleotide synthesis in the liver: Temporal segregation of metabolites. Cell Reports 1, 341–349 [DOI] [PubMed] [Google Scholar]
- 6. Barsotti C., Tozzi M. G., and Ipata P. L. (2002) Purine and pyrimidine salvage in whole rat brain. utilization of ATP-derived ribose-1-phosphate and 5-phosphoribosyl-1-pyrophosphate generated in experiments with dialyzed cell-free extracts. J. Biol. Chem. 277, 9865–9869 [DOI] [PubMed] [Google Scholar]
- 7. Cansev M. (2006) Uridine and cytidine in the brain: Their transport and utilization. Brain Res. Rev. 52, 389–397 [DOI] [PubMed] [Google Scholar]
- 8. Cao D., Leffert J. J., McCabe J., Kim B., and Pizzorno G. (2005) Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J. Biol. Chem. 280, 21169–21175 [DOI] [PubMed] [Google Scholar]
- 9. Gasser T., Moyer J. D., and Handschumacher R. E. (1981) Novel single-pass exchange of circulating uridine in rat liver. Science 213, 777–778 [DOI] [PubMed] [Google Scholar]
- 10. Harris C. M., and Massey V. (1997) The oxidative half-reaction of xanthine dehydrogenase with NAD: Reaction kinetics and steady-state mechanism. J. Biol. Chem. 272, 28335–28341 [DOI] [PubMed] [Google Scholar]
- 11. Krenitsky T. A., Neil S. M., Elion G. B., and Hitchings G. H. (1972) A comparison of the specificities of xanthine oxidase and aldehyde oxidase. Arch. Biochem. Biophys. 150, 585–599 [DOI] [PubMed] [Google Scholar]
- 12. Rosemeyer H., and Seela F. (1983) Methylated 7-Deazahypoxanthines as regiochemical probes of xanthine oxidase. Eur. J. Biochem. 134, 513–515 [DOI] [PubMed] [Google Scholar]
- 13. Lipkowitz M. S. (2012) Regulation of uric acid excretion by the kidney. Curr. Rheumatol. Rep. 14, 179–188 [DOI] [PubMed] [Google Scholar]
- 14. Xu X., Li C., Zhou P., and Jiang T. (2016) Uric acid transporters hiding in the intestine. Pharm. Biol. 54, 3151–3155 [DOI] [PubMed] [Google Scholar]
- 15. O'Neill R. D. (1990) Uric acid levels and dopamine transmission in rat striatum: Diurnal changes and effects of drugs. Brain Res. 507, 267–272 [DOI] [PubMed] [Google Scholar]
- 16. Minematsu S., Watanabe M., Tsuchiya N., Watanabe M., and Amagaya S. (1995) Diurnal variations in blood chemical items in Sprague-Dawley rats. Exp. Anim. 44, 223–232 [DOI] [PubMed] [Google Scholar]
- 17. Singh R., Singh R. K., Tripathi A. K., Cornélissen G., Schwartzkopff O., Otsuka K., and Halberg F. (2005) Chronomics of circulating plasma lipid peroxides and anti-oxidant enzymes and other related molecules in cirrhosis of liver. Biomed. Pharmacother. 59, S229–S235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shinosaki T., Inagaki H., Nakai T., Yamashita T., and Yonetani Y. (1992) Circadian rhythm of plasma uric acid and handling stress-induced hyperuricemia in conscious Cebus monkeys. Jpn. J. Pharmacol. 58, 443–450 [DOI] [PubMed] [Google Scholar]
- 19. Choi H. K., Liu S. and Curhan G. (2005) Intake of purine-rich foods, protein, and dairy products and relationship to serum levels of uric acid: The Third National Health and Nutrition Examination Survey. Arthritis Rheumatol. 52, 283–289 [DOI] [PubMed] [Google Scholar]
- 20. Gliozzi M., Malara N., Muscoli S., and Mollace V. (2016) The treatment of hyperuricemia. Int. J. Cardiol. 213, 23–27 [DOI] [PubMed] [Google Scholar]
- 21. Abhishek A., Roddy E., and Doherty M. (2017) Gout: A guide for the general and acute physicians. Clin. Med. 17, 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okamura A., Koyanagi S., Dilxiat A., Kusunose N., Chen J. J., Matsunaga N., Shibata S., and Ohdo S. (2014) Bile acid-regulated peroxisome proliferator-activated receptor-α (PPARα) activity underlies circadian expression of intestinal peptide absorption transporter PepT1/Slc15a1. J. Biol. Chem. 289, 25296–25305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wada E., Koyanagi S., Kusunose N., Akamine T., Masui H., Hashimoto H., Matsunaga N., and Ohdo S. (2015) Modulation of peroxisome proliferator-activated receptor-α activity by bile acids causes circadian changes in the intestinal expression of Octn1/Slc22a4 in mice. Mol. Pharmacol. 87, 314–322 [DOI] [PubMed] [Google Scholar]
- 24. Yang X., Downes M., Yu R. T., Bookout A. L., He W., Straume M., Mangelsdorf D. J., and Evans R. M. (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126, 801–810 [DOI] [PubMed] [Google Scholar]
- 25. Oishi K., Shirai H., and Ishida N. (2005) CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor-α (PPARα) in mice. Biochem. J. 386, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Forman B. M., Chen J., and Evans R. M. (1997) Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. U.S.A. 94, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Aguiar Vallim T. Q., Tarling E. J., and Edwards P. A. (2013) Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osada Y., Tsuchimoto M., Fukushima H., Takahashi K., Kondo S., Hasegawa M., and Komoriya K. (1993) Hypouricemic effect of the novel xanthine oxidase inhibitor, TEI-6720, in rodents. Eur. J. Pharmacol. 241, 183–188 [DOI] [PubMed] [Google Scholar]
- 29. Tapia E., Cristóbal M., García-Arroyo F. E., Soto V., Monroy-Sánchez F., Pacheco U., Lanaspa M. A., Roncal-Jiménez C. A., Cruz-Robles D., Ishimoto T., Madero M., Johnson R. J., and Sánchez-Lozada L. G. (2013) Synergistic effect of uricase blockade plus physiological amounts of fructose-glucose on glomerular hypertension and oxidative stress in rats. Am. J. Physiol. Renal Physiol. 304, F727–F736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mazzali M., Kanellis J., Han L., Feng L., Xia Y. Y., Chen Q., Kang D. H., Gordon K. L., Watanabe S., Nakagawa T., Lan H. Y., and Johnson R. J. (2002) Hyperuricemia induces a primary renal arteriolopathy in rats by a blood pressure-independent mechanism. Am. J. Physiol. Renal Physiol. 282, F991–F997 [DOI] [PubMed] [Google Scholar]
- 31. Sánchez-Lozada L. G., Soto V., Tapia E., Avila-Casado C., Sautin Y. Y., Nakagawa T., Franco M., Rodríguez-Iturbe B., and Johnson R. J. (2008) Role of oxidative stress in the renal abnormalities induced by experimental hyperuricemia. Am. J. Physiol. Renal Physiol. 295, F1134–F1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kliewer S. A., Lehmann J. M., and Willson T. M. (1999) Orphan nuclear receptors: Shifting endocrinology into reverse. Science 284, 757–760 [DOI] [PubMed] [Google Scholar]
- 33. Bensinger S. J., and Tontonoz P. (2008) Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454, 470–477 [DOI] [PubMed] [Google Scholar]
- 34. Jump D. B. (2002) The biochemistry of n-3 polyunsaturated fatty acids. J. Biol. Chem. 277, 8755–8758 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y. K., Guo G. L., and Klaassen C. D. (2011) Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 6, e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han S., Zhang R., Jain R., Shi H., Zhang L., Zhou G., Sangwung P., Tugal D., Atkins G. B., Prosdocimo D. A., Lu Y., Han X., Tso P., Liao X., Epstein J. A., and Jain M. K. (2015) Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat. Commun. 6, 7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKenna N. J., and O'Malley B. W. (2002) Minireview: Nuclear receptor coactivators–an update. Endocrinology 143, 2461–2465 [DOI] [PubMed] [Google Scholar]
- 38. Mochizuki K., Suruga K., Sakaguchi N., Takase S., and Goda T. (2002) Major intestinal coactivator p300 strongly activates peroxisome proliferator-activated receptor in intestinal cell line, Caco-2. Gene 291, 271–277 [DOI] [PubMed] [Google Scholar]
- 39. Lemberger T., Saladin R., Vázquez M., Assimacopoulos F., Staels B., Desvergne B., Wahli W., and Auwerx J. (1996) Expression of the peroxisome proliferator-activated receptor α gene is stimulated by stress and follows a diurnal rhythm. J. Biol. Chem. 271, 1764–1769 [DOI] [PubMed] [Google Scholar]
- 40. Lu Y. F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y. K., and Liu J. (2013) Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 30, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gachon F., Leuenberger N., Claudel T., Gos P., Jouffe C., Fleury Olela F., de Mollerat du Jeu X., Wahli W., and Schibler U. (2011) Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor α (PPARα) activity. Proc. Natl. Acad. Sci. U.S.A. 108, 4794–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moriwaki Y., Yamamoto T., Yamaguchi K., Takahashi S., and Higashino K. (1996) Immunohistochemical localization of aldehyde and xanthine oxidase in rat tissues using polyclonal antibodies. Histochem. Cell Biol. 105, 71–79 [DOI] [PubMed] [Google Scholar]
- 43. Lavery D. J., and Schibler U. (1993) Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 7, 1871–1884 [DOI] [PubMed] [Google Scholar]
- 44. Ripperger J. A., Shearman L. P., Reppert S. M., and Schibler U. (2000) CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 [PMC free article] [PubMed] [Google Scholar]
- 45. Wu X. W., Muzny D. M., Lee C. C., and Caskey C. T. (1992) Two independent mutational events in the loss of urate oxidase during hominoid evolution. J. Mol. Evol. 34, 78–84 [DOI] [PubMed] [Google Scholar]
- 46. Pandey A., Chaturvedi M., Prakash H., and Meena D. (2012) Febuxostat, a new treatment for hyperuricemia in gout: A review article. Natl. J. Physiol. Pharm. Pharmacol. 2, 23–28 [Google Scholar]
- 47. Hamdan A. M., Koyanagi S., Wada E., Kusunose N., Murakami Y., Matsunaga N., and Ohdo S. (2012) Intestinal expression of mouse Abcg2/breast cancer resistance protein (BCRP) gene is under control of circadian clock-activating transcription factor-4 pathway. J. Biol. Chem. 287, 17224–17231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Japan Society of Clinical Chemistry (1993) Recommended method for determination of uric acid in serum by HPLC. Jpn. J. Clin. Chem. 22, 300–307 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.