Abstract
The degenerin/epithelial sodium channel (DEG/ENaC) superfamily of ion channels contains subfamilies with diverse functions that are fundamental to many physiological and pathological processes, ranging from synaptic transmission to epileptogenesis. The absence in mammals of some DEG/ENaCs subfamily orthologues such as FMRFamide peptide–activated sodium channels (FaNaCs), which have been identified only in mollusks, indicates that the various subfamilies diverged early in evolution. We recently reported that the nonproton agonist 2-guanidine-4-methylquinazoline (GMQ) activates acid-sensing ion channels (ASICs), a DEG/ENaC subfamily mainly in mammals, in the absence of acidosis. Here, we show that GMQ also could directly activate the mollusk-specific FaNaCs. Differences in ion selectivity and unitary conductance and effects of substitutions at key residues revealed that GMQ and FMRFamide activate FaNaCs via distinct mechanisms. The presence of two activation mechanisms in the FaNaC subfamily diverging early in the evolution of DEG/ENaCs suggested that dual gating is an ancient feature in this superfamily. Notably, the GMQ-gating mode is still preserved in the mammalian ASIC subfamily, whereas FMRFamide-mediated channel gating was lost during evolution. This implied that GMQ activation may be essential for the functions of mammalian DEG/ENaCs. Our findings provide new insights into the evolution of DEG/ENaCs and may facilitate the discovery and characterization of their endogenous agonists.
Keywords: epithelial sodium channel (ENaC), ion channel, ligand-binding protein, neuropeptide, small molecule, 2-guanidine-4-methylquinazoline (GMQ), FMRFamide (Phe-Met-Arg-Phe-NH2) peptides, FMRFamide peptide-gated sodium channel (FaNaC), acid-sensing ion channels (ASIC)
Introduction
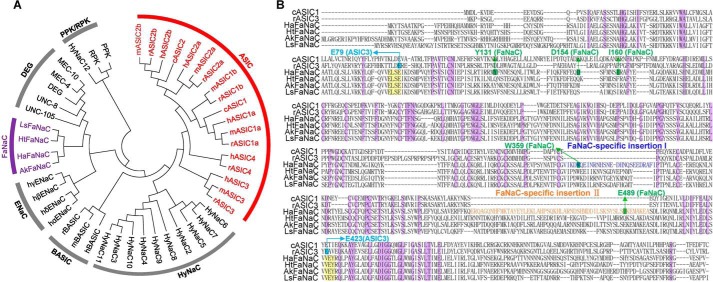
The degenerin/epithelial sodium channel (DEG/ENaC)4 superfamily consists of a diverse group of nonvoltage-gated, sodium-selective amiloride-sensitive cation channels (1, 2). According to channel properties and ligand sensitivities, they are classified into diverse subfamilies, including ENaC, acid-sensing ion channels (ASIC), bile acid–sensing ion channels (BASIC), DEG, FMRFamide peptide–activated sodium channels (FaNaC), Hydra sodium channels (HyNaCs), and PPK/RPK (Pickpocket/Ripped Pocket) (Fig. 1A). Different members of these subfamilies of DEG/ENaC are involved in diverse physiological and pathological processes (1, 2), such as synaptic transmission (3), learning and memory (4), fear conditioning (5, 6), anxiety (7), pain (8, 9), stroke (10, 11), Parkinson's disease (12, 13), and epilepsy (14). Unlike some classical channels that originated from remote antiquity and are present in almost all organisms, DEG/ENaC subfamilies are only expressed in tissues of metazoans, such as coelenterates, mollusks, arthropods, and mammals (1, 2). The sequence homology among DEG/ENaC subfamilies is quite low (around 15–20%), and some DEG/ENaC subfamilies do not have mammalian orthologues, indicating that DEG/ENaC subfamilies may be diverged early in evolution (1, 2, 15). FaNaC, a DEG/ENaC subfamily identified only in mollusks, can be directly activated by FMRFamide peptide (16), and may be an elder subfamily member among the entire DEG/ENaC superfamily (2). So far, four FaNaC orthologues have been identified, named HaFaNaC from Helix aspersa (16), HtFaNaC from Helisoma trivolvis (17), LsFaNaC from Lymnaea stagnalis (18), and AkFaNaC from Aplysia kurodai (19), sharing ∼65% sequence identity (Fig. 1B). Despite of their high sequence homology, FaNaC channels display different sensitivities to amiloride, a nonselective inhibitor of DEG/ENaCs, and native peptide FMRFamide (20). Purely based on sequence homology, ASICs have been considered to share a common ancestor with either the FaNaC (18) or HyNaC subfamily (21); FaNaC has also been deemed to be more closely related to ENaC (Fig. 1A) (15, 22).
Figure 1.
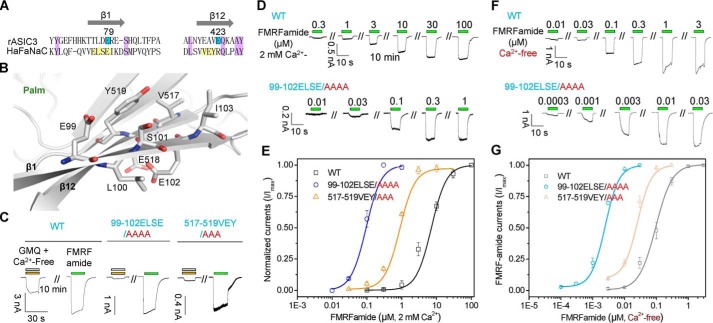
Phylogenetic and sequence analysis of DEG/ENaC superfamily of ion channels. A, phylogenetic tree generated from genes of ASIC, HyNaC, BASIC, ENaC, FaNaC, DEG, and PPK/RPK subfamilies. The genes of ASIC and FaNaC subfamilies are highlighted with red and purple. B, multiple sequence alignment, made by Clastal W with manual adjustments, from two ASICs and four FaNaCs. The conserved sequence and the key binding regions are indicated with different colors. Purple, conservative residues; cyan, key residues essential for GMQ's action in rASIC3 (Glu-79 (E79) and Glu-423 (E423)); yellow, FaNaC regions identical to key site of GMQ in rASIC3; green, residues essential for FMRFamide-mediated HaFaNaC activation; blue and orange, FaNaC-specific insertions I and II in comparison with other members of DEG/ENaCs.
Different DEG/ENaC subfamilies have been known to use different activation mechanisms (1, 2, 23). For example, ENaCs open spontaneously, DEGs respond upon mechanical stimulation, ASICs are sensitive to extracellular acidosis, whereas FaNaCs are directly activated by native FMRFamide peptide (1, 2, 23). Recently, there have been multiple breakthroughs in identifying new activation mechanisms of some DEG/ENaCs, for instance, the activation of ENaC by a synthetic compound S3969 (24) or shear stress (25), ASIC3 by a synthetic small molecule GMQ (26, 27), and ASIC1a by snake venom MitTx (28). Thus, it is reasonable to speculate that DEG/ENaCs may be activated or modulated by some endogenous agents via allosteric mechanisms similar to that used by the exogenous activators. Moreover, understanding the dual or polymodal mode of DEG/ENaC activation is pivotal for elucidating the functional significance of these channels. In the case of ASICs, although the channels are best known to sense extracellular acidosis (29), some of the physiological functions implicated to them cannot be fully explained by their responses to low pH (27, 30). The recent identification of nonproton ligands/toxins should help advance our understanding of ASIC-mediated functions (27, 28, 31, 32) and encourage further exploration of endogenous nonproton ligands for these channels (32–34). The identification of the synthetic activator of ENaC, S3969, also brings new opportunities for therapeutic development, such as treatment of false aldosterone deficiency (24). Therefore, more in-depth studies into polymodal gating mechanisms of the DEG/ENaC channels have important implications in both basic research and disease intervention.
Here, we demonstrated that GMQ, a previously described nonproton ligand of ASIC3, can activate all four FaNaC orthologues via a mechanism distinct from the native FMRFamide peptide (35), but similar to the one used by GMQ to activate ASIC3 (27). Our data not only indicate a closer evolutionary relationship between ASIC and FaNaC subfamilies, despite the low sequence homology (12–18%), but also a conservation of the “GMQ-activation mode” that was already present in the common ancestor of these two subfamilies. In addition, two distinct activation modes of FaNaCs also reflect that the dual gating may already exist in the ancestor of DEG/ENaC superfamily of ion channels.
Results
GMQ, the nonproton ligand of ASIC3, directly activates FaNaC channels
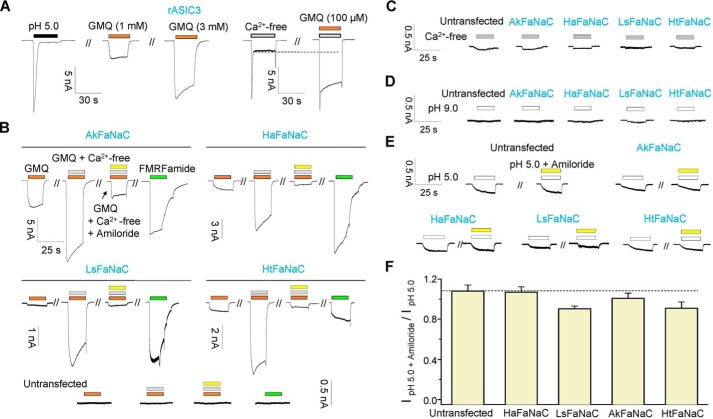
Previously, we identified GMQ as the first nonproton ligand of ASICs (Fig. 2A), which selectively activates ASIC3 but not ASIC1a, -1b, and -2a (27). Others, however, argued that the GMQ worked by altering pH dependence and steady-state inactivation of ASIC1a, -1b, -2a, and -3 differentially, and such shifts only allowed ASIC3 activation at the physiological pH 7.4 (36). To look for further evidence that GMQ acts as a direct agonist rather a modulator, we examined its ability to activate other DEG/ENaC subfamily members not known to be activated by extracellular protons. We tested FaNaCs because their native agonist, FMRFamide, has been shown to augment the sustained currents of ASIC3 and ASIC1a (37). Interestingly, GMQ directly activated all four FaNaC orthologues expressed in CHO cells (Fig. 2B), and there is no GMQ current observed in cells without FaNaC expression (Fig. 2B). GMQ-evoked currents in all FaNaCs are sensitive to amiloride (Fig. 2B), which is a common feature of DEG/ENaC superfamily of ion channels (1). GMQ also evoked amiloride-sensitive currents in Xenopus oocytes that were injected with HaFaNaC cRNA (see below), suggesting that the stimulatory effect of GMQ is independent of the host cell types.
Figure 2.
GMQ, a nonproton ligand of ASIC3, directly activates FaNaC channels. A, the representative current traces illustrating the activation of rASIC3 by pH 5.0, Ca2+o deprivations, and GMQ (n = 6–10). B, the representative current traces illustrating the activation of four FaNaC orthologues in CHO cells with or without expressed channels by GMQ (5 mm) and FMRFamide (0.1–1 mm) in the absence or presence of extracellular Ca2+. Most of those currents were inhibited by amiloride (100 μm) (n = 4–7). C and D, representative current traces in response to pH 9.0 (C) and Ca2+o deprivation (D) external solution in CHO cells without or with the expression of indicated FaNaC orthologues (n = 3–5). E and F, representative current traces (E) and pooled data (F, mean ± S.E., n = 4–10; p >0.05 versus control) for pH 5.0–induced currents in CHO cells without or with the expression of FaNaCs, and the lack of inhibition by amiloride.
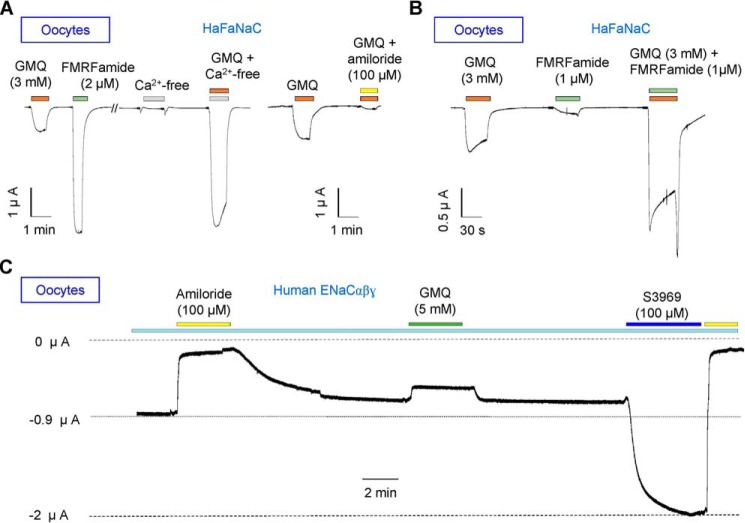
ASIC3 can be directly activated by deprivation of extracellular Ca2+ (Ca2+o) (Fig. 2A) (27, 38, 39), acidosis (lower pH than 7.2), or alkalization (pH 8.0) (34). However, neither Ca2+o deprivation (Fig. 2C), nor high (pH 9.0)/low (pH 5.0) pH (Fig. 2, D–F) activated those four FaNaC orthologues. A small endogenous response to pH 5.0 was observed in all cells (Fig. 2E), and it was not inhibited by amiloride (Fig. 2, E and F). Thus, it is unlikely that GMQ acted on FaNaCs through modulation of any intrinsic activation or inhibition of this DEG/ENaC subfamily by pH or Ca2+o. Furthermore, GMQ evoked amiloride-sensitive currents in Xenopus oocytes with HaFaNaC cRNA injection (Fig. 3, A and B), but caused only a slight inhibition of the spontaneous activity of human ENaCαβγ expressed (Fig. 3C). Thus, the agonistic effects of GMQ on FaNaCs and ASIC3 are specific, rather than universal, among the DEG/ENaC family of ion channels, which could imply a close evolutionary relationship between FaNaCs and ASICs.
Figure 3.
Characterization of the effects of extracellular Ca2+ deprivation, GMQ, S3969, and amiloride on HaFaNaC and hENaCαβγ expressed in Xenopus oocytes. A and B, representative current traces illustrating the effects of GMQ, FMRFamide, Ca2+-free bath, and amiloride in oocytes expressing HaFaNaC (n = 3). C, representative current traces illustrating the effects of amiloride, S3969 (a small molecule agonist of ENaCs), and GMQ, respectively, on oocytes expressing hENaCαβγ (n = 3).
Common features shared by GMQ-induced currents in FaNaC and ASIC3
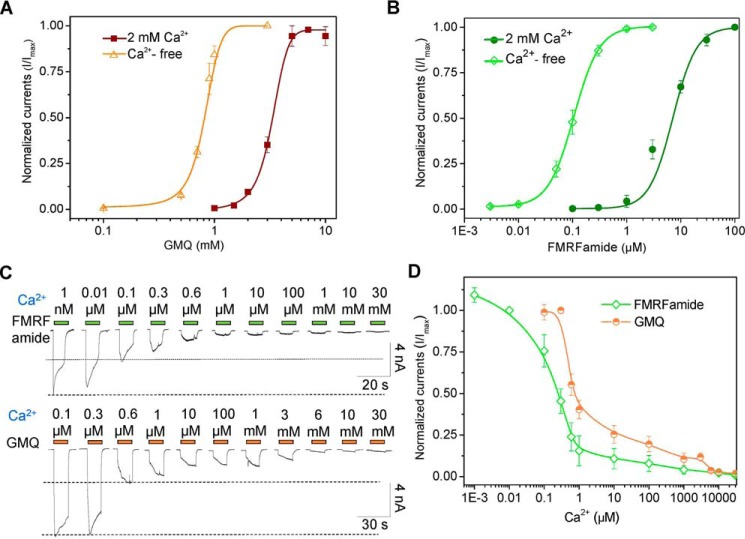
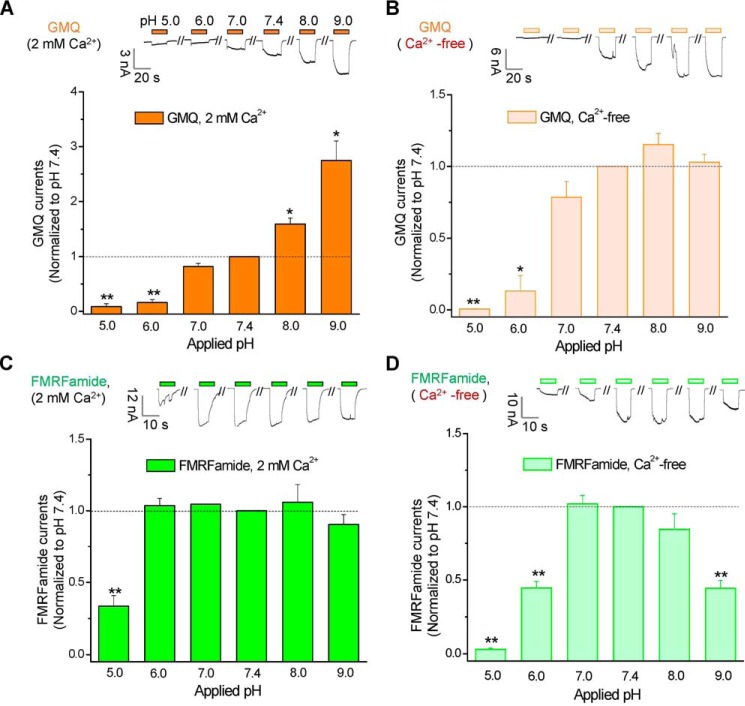
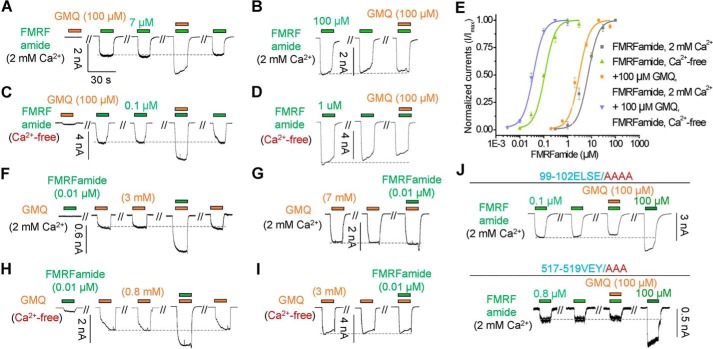
We recently identified the key sites essential for FMRFamide-mediated activation of HaFaNaC (35), here we continue using HaFaNaC as a representative to examine the mechanism by which GMQ activates FaNaCs, and compared it with GMQ's action on ASIC3. Previously, we have showed that GMQ could activate rat ASIC3 (rASIC3) at millimolar level (EC50 = 1.27 ± 0.13 mm) under normal physiological conditions (pH 7.4 and 2 mm Ca2+o) (27). Decrease or deprivation of Ca2+o significantly increased its apparent affinity by ∼20-fold (EC50 = 0.06 ± 0.01 mm, Ca2+-free) (27). In addition, GMQ also exhibited an increased potency on rASIC3 in mild acidosis (pH 7.0–6.9), whereas acidic (pH 6.5–5.0) or basic (pH 8.0–9.0) pH attenuated the GMQ-induced activation (27). Similarly, GMQ activated HaFaNaC at millimolar level (EC50 = 3.45 ± 0.24 mm, 2 mm Ca2+, pH 7.4) (Fig. 4A); depletion of Ca2+o also left shifted the GMQ's dose-response curve in HaFaNaC, but with a relative lower potentiation in efficacy when compared with rASIC3 (∼5-fold enhancement, EC50 = 0.8 ± 0.04 mm, Ca2+-free, pH 7.4) (Fig. 4A). Deprivation of Ca2+o also increased the GMQ's action on three other FaNaC orthologues (Fig. 2B), suggesting this is a common feature in the FaNaC subfamily. Although acidification (pH 5.0–6.0) inhibited the GMQ's action in HaFaNaC (Fig. 5A), just like what it does on rASIC3 (26, 27), GMQ-evoked currents were potentiated in alkaline solution (pH 8.0–9.0) (Fig. 5A), which was eliminated by extracellular Ca2+o deprivation (Fig. 5B), indicating that GMQ, proton, and Ca2+o interacted with each other on gating HaFaNaC via similar but more complicated mechanisms than they do on ASIC3 (27).
Figure 4.
Extracellular Ca2+ inhibits FMRFamide- and GMQ-induced currents in cells expressing HaFaNaC. A and B, concentration responses to GMQ (A) and FMRFamide (B) in cells expressing HaFaNaC in the absence or presence of extracellular Ca2+. Data points are mean ± S.E. (n = 4–6). C and D, representative current traces (C) and summary data (D) to illustrate the inhibition by extracellular Ca2+ on FMRFamide (0.1 μm)- and GMQ (1 mm)-induced currents in cells expressing HaFaNaC (mean ± S.E., n = 3–10).
Figure 5.
Differential effects of extracellular pH on GMQ- and FMRFamide-evoked activation of HaFaNaC in the presence and absence of extracellular Ca2+. A–D, representative current traces (upper) and summary data (lower, mean ± S.E.) for currents induced by GMQ (A, 3 mm in 2 mm Ca2+o; B, 1 mm in Ca2+-free), and FMRFamide (C, 7 μm in 2 mm Ca2+o; D, 0.01 μm in Ca2+-free) under different pH conditions. n = 3; *, p < 0.05, **, p < 0.001 versus control (pH 7.4, broken line).
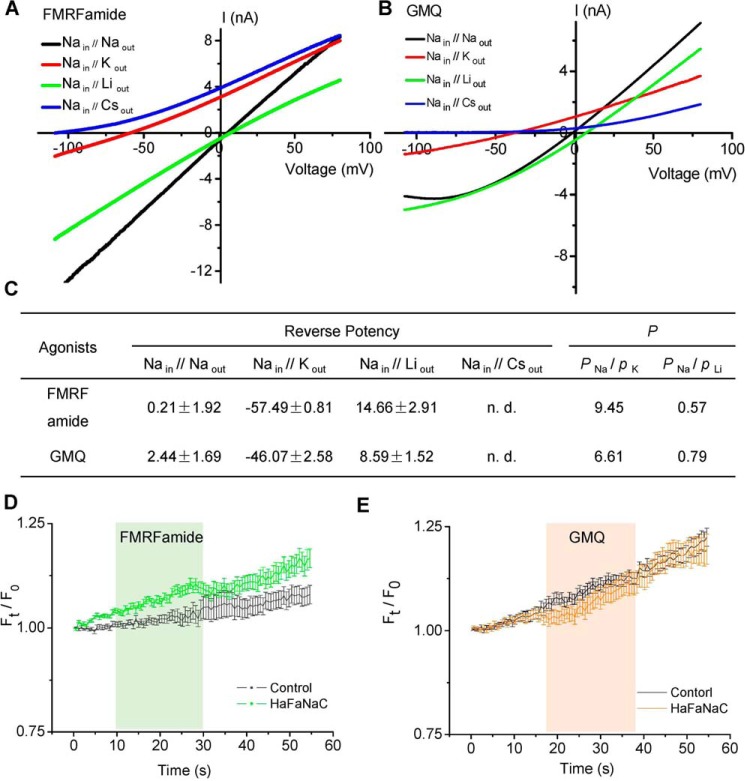
The selectivity ratio of Na+/Li+ over K+ (pNa+/pK+) in GMQ-evoked ASIC3 current (3.32) was lower than that induced by low pH (5.57) (27). Similarly, the pNa+/pK+ value of GMQ-evoked HaFaNaC current (6.6) was also markedly lower than that evoked by FMRFamide (9.45) (Fig. 6, A–C). Moreover, neither FMRFamide nor GMQ elicited Cs+ or Ca2+ conductance through HaFaNaC (Fig. 6, D and E), similar to GMQ's action on rASIC3 (27). Most importantly, key residues located within the palm domain of HaFaNaC mediated the action of GMQ (see below), further demonstrating that GMQ uses a similar mechanism to activate both ASIC3 (27) and FaNaC channels. The fact that FaNaCs are only identified in invertebrates suggests that this activation mode may evolve earlier before the mammalian subfamilies of DEG/ENaCs.
Figure 6.
Ion selectivity of FMRFamide- and GMQ-induced currents in cells expressing HaFaNaC. A and B, I–V relationships of HaFaNaC currents induced by 100 μm FMRFamide (A) and 5 mm GMQ (B) under different bi-ionic conditions. The internal solution contained 150 mm Na+; external solutions contained 150 mm Na+, Li+, K+, or Cs+. C, summary data of measured reversal potentials (mean ± S.E., n = 4–8) and derived ion permeability (P). The reversal potentials were recorded by a double voltage-ramp protocol from −110 to +80 mV in 800 ms, whereas P was calculated using the modified Goldman-Hodgkin-Katz equation (see “Experimental Procedures”). n.d., not determined. D and E, changes in fluorescent intensity induced by 100 μm FMRFamide (D) and 5 mm GMQ (E) in Fluo-4-loaded CHO cells with or without transfected cDNA for HaFaNaC. Data points are mean ± S.E. (n = 15–17), expressed as Ft/F0 (a ratio of fluorescent intensities at time (t) and time zero (0)). Green and orange shadows indicate the treatments of FMRFamide and GMQ, respectively. Saturating concentrations of FMRFamide (100 μm) or GMQ (5 mm) did not induce Ca2+ influx through HaFaNaC (p > 0.05 versus control).
Differences existed between GMQ- and peptide-mediated FaNaC activations
The above data revealed that the FaNaC currents activated by GMQ exhibited differences in sensitivity to Ca2+o and selectivity for Na+ versus K+ as compared with that evoked by FMRFamide (Figs. 5 and 6). Deprivation of Ca2+o dramatically enhanced the apparent affinity of FMRFamide of HaFaNaC by ∼60- to 70-fold (EC50 = 7.33 ± 0.82 and 0.11 ± 0.02 μm, for 2 mm Ca2+o and Ca2+-free, respectively) (Fig. 4B), but was much less effective on GMQ, reducing the EC50 only by ∼5-fold (Fig. 4A). In addition, the FMRFamide-induced HaFaNaC currents appeared to be more sensitive to Ca2+o than the GMQ-evoked ones, with the former being attenuated at 1–3 nm Ca2+o whereas the latter suppressed only by at least 100–300 nm Ca2+o (Fig. 4, C and D). Meanwhile, in the presence of 2 mm Ca2+o, FMRFamide-induced FaNaC activation (Fig. 5C) is much less sensitive to pH variations than GMQ (Fig. 5A). Changing the extracellular pH from 6.0 to 9.0 only slightly influenced the activation of FaNaC by FMRFamide, although pH 5.0 inhibited the activation (Fig. 5, C and D); by contrast, increasing the pH from 6.0 to 9.0 continuously potentiated the GMQ-evoked HaFaNaC current (Fig. 5A).
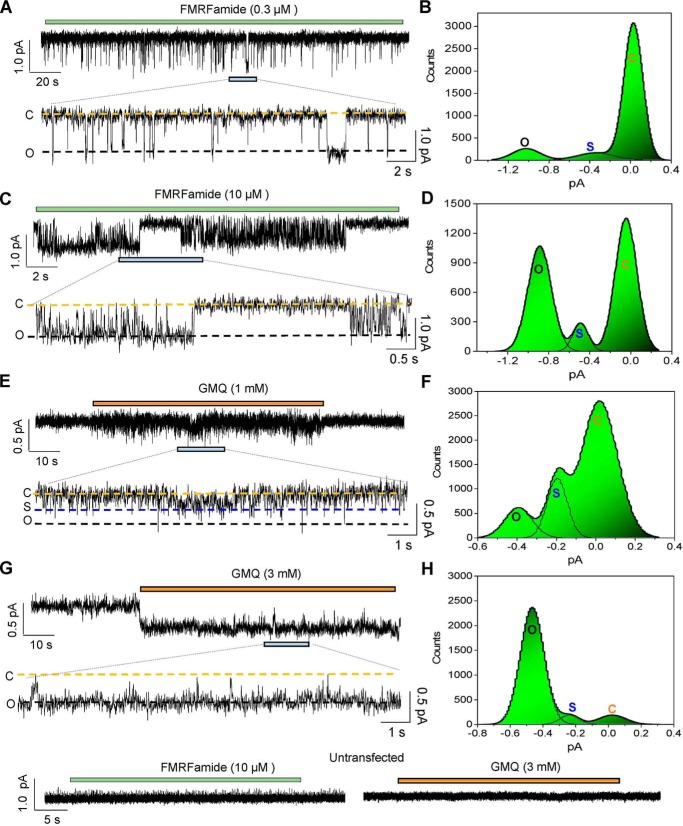
To further characterize the GMQ-evoked activation of FaNaC channels, we performed single channel recordings in an outside-out configuration from CHO cells transiently transfected with HaFaNaC. As a control, FMRFamide (0.1–10 μm) evoked single channel activities with the main conductance generating ∼0.9–1.1 pA of currents at −80 mV (Fig. 7, A–D) was similar to previous reports (16, 40, 41), and there was no single channel activity observed in cells without FaNaC expression (Fig. 7I). Interestingly, the unitary currents of FaNaC induced by 1 mm GMQ were very flickery and only reached ∼0.4–0.5 pA at −80 mV (Fig. 7, E and F). The channels were mostly closed or existed in subconductance states (Fig. 7F). At a higher concentration of GMQ, more fully opened channels were observed (Fig. 7, G and H), but the unitary chord conductance remained to be lower (∼4 picosiemens at −80 mV) than that evoked by FMRFamide (∼10–12 picosiemens), indicating distinct channel gating mechanisms between GMQ and FMRFamide.
Figure 7.
Distinct unitary currents evoked by GMQ and FMRFamide in HaFaNaC. A–H, The single channel currents recorded from outside-out patches held at −80 mV in responses to FMRFamide (A, 0.3 μm, n = 16; C, 10 μm, n = 6) and GMQ (E, 1 mm, n = 8; G, 3 mm, n = 10) and their all-points histograms (FMRFamide, (B) 0.3 μm, (D) 10 μm; GMQ, (F) 1 mm; GMQ, (H) 3 mm) fitted to the sum of three Gaussians. Full opening (O), closing (C), and sub-open states (S) are indicated by black, yellow, and blue lines, respectively. y axis (count) denotes the ratio of the number of events to the number of bins (the bin number is set to 320). I, representative single channel currents recorded in responses to FMRFamide (10 μm) and GMQ (3 mm) in untransfected CHO cells (n = 4–7).
FMRFamide and GMQ target distinct regions of FaNaC for channel activation
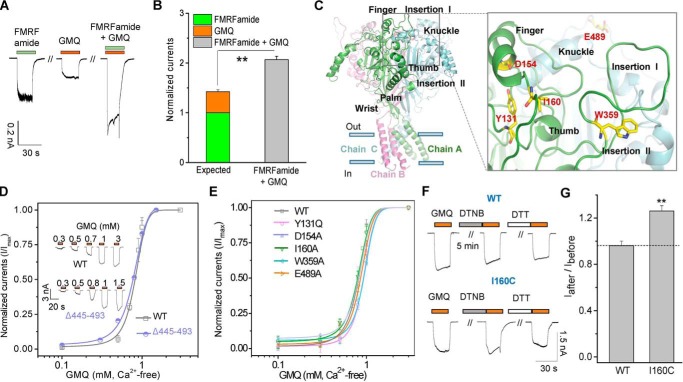
Given that the key residues essential for the low pH-induced activation of ASIC3 are not involved in the GMQ-mediated channel gating (26, 27), it is possible that GMQ and FMRFamide also act on different sites of HaFaNaC. Coapplication of unsaturated GMQ (3 mm) and FMRFamide (1 μm) to cells expressing HaFaNaC (pH 7.4, 2 mm Ca2+o) caused a larger current than the sum of the responses from individual applications of GMQ and FMRFamide (Fig. 8, A and B), indicating a likelihood of synergy between the two agonists. For the mutation HaFaNaCΔGln-445–Ala-493 (with an ∼50-residue truncation at the specific insertion II of the knuckle domain) (Figs. 1B and 8C), its apparent affinity (∼80-fold decrease), and the maximal currents of FMRFamide decreased profoundly (35). In contrast, the EC50 of GMQ and the maximal currents were not changed by a truncation of Gln-445–Ala-493 (Fig. 8D). Meanwhile, mutations at the residues of other three domains, including I160A, D154A, Y131Q, W359A, and E489A, which significantly reduced (∼10- to 40-folds) the apparent affinity of FMRFamide (35) showed little effect on the dose-response curves of GMQ on HaFaNaC (EC50 = 0.8 ± 0.04, 0.78 ± 0.05, 0.81 ± 0.03, 0.92 ± 0.02, 0.93 ± 0.03, 0.82 ± 0.04 mm, for HaFaNaC WT, I160A, D154A, Y131Q, W359A and E489A, respectively, Ca2+-free/pH 7.4) (Fig. 8E). In addition, a covalent modification of I160C decreased the apparent affinity for at least 700-fold and significantly reduced the maximal current of FMRFamide in HaFaNaC (EC50 = 7.33 ± 0.82 μm versus over 5 mm, before versus after the DTNB treatment of HaFaNaCI160C, respectively), and DTT treatment can reverse those effects (35). However, unlike the FMRFamide-evoked activation, DTNB treatment of cells that expressed HaFaNaCI160C or HaFaNaCWT, only a slight potentiation or no apparent difference was detected between the GMQ-induced currents before and after DTNB modifications (Fig. 8, F and G). Collectively, these results demonstrate that the channel regions and critical residues targeted by GMQ and FMRFamide are distinct.
Figure 8.
Key sites essential for peptide-mediated HaFaNaC activation are not required for the action of GMQ on HaFaNaC. A and B, representative current traces (A) and pooled data (B) (mean ± S.E., n = 4, normalized to FMRFamide-induced maximal currents) to illustrate the synergistic effect between FMRFamide (1 μm) and GMQ (3 mm). **, p < 0.001 versus expected ratio. C, a three-dimensional homology model of HaFaNaC to show key domains and residues (Tyr-131, Asp-154, Ile-160, Trp-359, and Glu-489, displayed in sticks for emphasis) essential for FMRFamide-mediated HaFaNaC activation. Insertion I: Lys-357–Ala-379; Insertion II: Glu-445–Ala-493. D and E, representative current traces and dose-response curves of GMQ for wild type (WT) HaFaNaC and its deletion mutant ΔGln-445–Ala-493 (D), and point mutations (E). Each data point represents the mean ± S.E. of four to five measurements. The solid lines illustrate the fits to the Hill equation. F and G, representative traces (F) and pooled data (G) (mean ± S.E., n = 10–11) for the effect of DTNB treatment (4 mm) on GMQ (3 mm, 2 mm Ca2+o/pH 7.4)-induced currents of WT HaFaNaC and its I160C mutant. **, p < 0.001 versus control.
Moreover, although a low concentration of GMQ (100 μm) failed to evoke any current in cells that expressed HaFaNaC (Fig. 9A), it significantly potentiated the activation by an unsaturated concentration of FMRFamide (7 μm for 2 mm Ca2+o, 0.1 μm for Ca2+-free) (Fig. 9, A and C), and left-shifted the dose-response curves of FMRFamide both in the absence (5 mm EGTA) and the presence of Ca2+o (2 mm) (Fig. 9E). Similar potentiation was produced by a low concentration of FMRFamide (0.01 μm) on HaFaNaC activation by unsaturated concentration of GMQ (3 mm for 2 mm Ca2+o, 0.8 mm for Ca2+-free) (Fig. 9, F and H), indicating that the key sites for FMRFamide and GMQ interact with each other. Interestingly, the potentiation effects of the low concentrations of GMQ (100 μm) and FMRFamide (0.01 μm) on HaFaNaC channels diminished with the use of saturating concentrations of, respectively, FMRFamide (100 μm for 2 mm Ca2+o, 1 μm for Ca2+o –free) (Fig. 9, B and D) and GMQ (7 mm for 2 mm Ca2+o, 3 mm for Ca2+o –free) (Fig. 9, G and I), implying that the two agonists opened a common channel pore or ion permeation pathway, despite the key sites for their actions being different.
Figure 9.
Synergistic effects between FMRFamide and GMQ on HaFaNaC. A–D, a concentration of GMQ (100 μm) that could not evoke currents in cells expressing HaFaNaC significantly potentiated the currents evoked by unsaturating concentrations of FMRFamide (A, 7 μm for 2 mm Ca2+o; C, 0.1 μm for Ca2+-free), but not that by saturating concentrations of FMRFamide (B, 100 μm in 2 mm Ca2+o; D, 1 μm in Ca2+-free) (n = 3–5). E, concentration-response curves to FMRFamide without or with GMQ (100 μm) and in the absence or presence of extracellular Ca2+ (2 mm) in CHO cells that expressed HaFaNaC. Data points are mean ± S.E. (n = 3–5). The low concentration of GMQ (100 μm) left-shifted the concentration responses of FMRFamide under both normal Ca2+o (EC50 = 7.33 ± 0.82 and 2.30 ± 0.23 μm, without and with GMQ, respectively) and Ca2+-free (EC50 = 0.10 ± 0.003 and 0.03 ± 0.002 μm, without and with GMQ, respectively) conditions. F–I, a concentration of FMRFamide (0.1 μm) that could not evoke currents in cells expressing HaFaNaC significantly potentiated the currents evoked by unsaturating concentrations of GMQ (F, 3 mm in 2 mm Ca2+o; H, 0.8 mm in Ca2+-free), but not those by saturating concentrations of GMQ (G, 7 mm in 2 mm Ca2+o; I, 3 mm in Ca2+-free) (n = 4). Similar results were obtained in at least three other independent recordings. J, GMQ (100 μm) could not potentiate unsaturated FMRFamide-induced currents in HaFaNaC99–102ELSE/AAAA and HaFaNaC517–519VEY/AAA (2 mm Ca2+o) (n = 3–4).
Residues in the palm domain serve opposite roles in the FaNaC activations by FMRFamide and GMQ
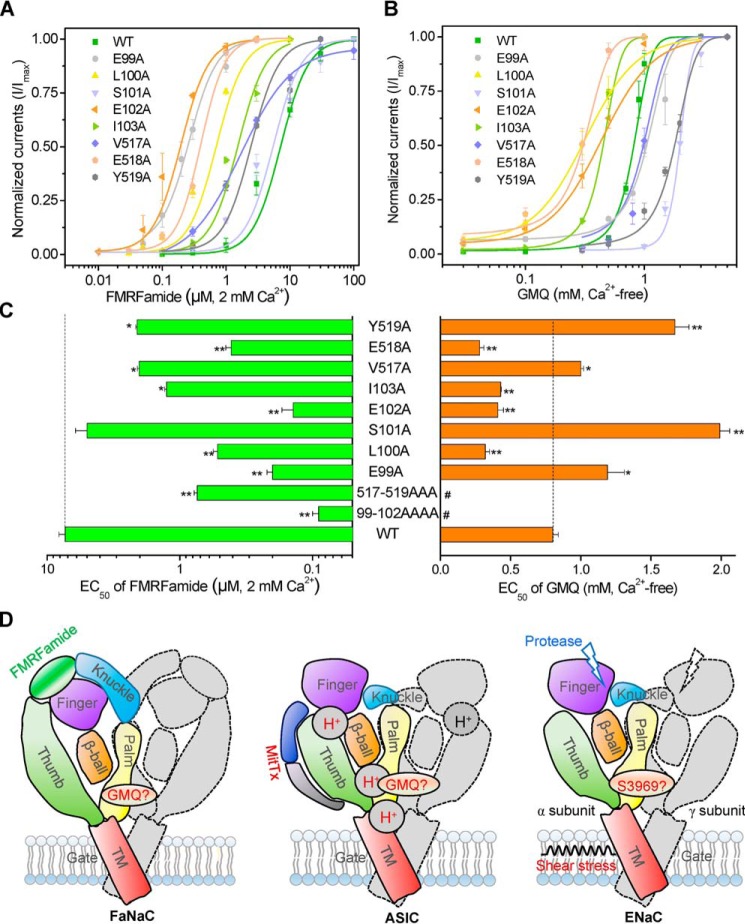
To further understand the mechanism by which GMQ activates the FaNaC subfamily channels, we searched for the channel regions and key amino acid residues involved in the GMQ-mediated HaFaNaC activation. Previously, we demonstrated that residues located at the β1- and β12-sheets of the palm domain, namely Glu-79 and Glu-423, are essential for the GMQ-evoked activation of ASIC3 (Figs. 1B and 10A, shown in cyan) (27), which prompts us to perform mutations on the corresponding residues in HaFaNaC. However, because of the low sequence homology between the HaFaNaC and rASIC3 at this region (Figs. 1B and 10A, shown in yellow), we replaced three to four amino acids of β1- and β12-sheets (Fig. 10, A and B) with alanine simultaneously to ensure that the residues corresponding to Glu-79 or Glu-423 in ASIC3 were mutated, yielding two mutants HaFaNaC99–102ELSE/AAAA and HaFaNaC517–519VEY/AAA. Similar to GMQ's action on rASIC3, these two mutations5 nearly fully abolished GMQ-induced direct activations (Fig. 10C) and allosteric potentiation of HaFaNaC (Fig. 9J), indicating that residues located in the palm domain are also essential for GMQ-mediated HaFaNaC stimulus. Surprisingly, those two mutants led a profound increase in the apparent affinity of FMRFamide in HaFaNaC both in 2 mm Ca2+o (EC50 = 7.33 ± 0.82, 0.09 ± 0.01, and 0.78 ± 0.03 μm for WT, 99–102ELSE/AAAA, and 517–519VEY/AAA, respectively) (Fig. 10, D and E) and Ca2+-free solutions (EC50 = 110 ± 15, 2.03 ± 0.11, and 21.5 ± 1.97 nm for WT, 99–102ELSE/AAAA, and 517–519VEY/AAA, respectively) (Fig. 10, F and G), indicating that the site located in the palm domain of HaFaNaC may be an allosteric site of FMRFamide both in the presence and absence of extracellular Ca2+, further supporting the idea that GMQ and FMRFamide use distinct mechanisms to gate FaNaC channels.
Figure 10.
Residues in the palm domains have opposite roles in FMRFamide- and GMQ-mediated FaNaC activations. A, sequence alignment of rASIC3 and HaFaNaC. Purple, conservative residues; cyan, key residues essential for GMQ's action in rASIC3 (Glu-79 and Glu-423); yellow, FaNaC regions (99–102ELSE and 517–519VEY) identical to GMQ sites in rASIC3. B, putative residues (displayed in sticks for emphasis) essential for GMQ-mediated activation of FaNaC, shown in the homology model of HaFaNaC. C, representative current traces illustrating the effects of GMQ (3 mm, Ca2+-free/pH 7.4) and FMRFamide (100 μm) on WT, HaFaNaC99–102ELSE/AAAA (in β1-sheet of the palm domain) and HaFaNaC517–519VEY/AAA (in β12-sheet) (n = 3–6). D–G, representative traces (D and F) and summarized data (E and G) to illustrate the altered dose-dependent responses of HaFaNaC99–102ELSE/AAAA and HaFaNaC517–519VEY/AAA (D and E, 2 mm Ca2+o/pH 7.4; F and G, Ca2+-free/pH 7.4) (mean ± S.E., n = 3–5).
Echoing the findings from the compound mutations, substitutions of the residues in the β1- and β12-sheets profoundly decreased the EC50 of FMRFamide for HaFaNaC (EC50 = 7.33 ± 0.82, 5.01 ± 1.01, 0.2 ± 0.02, 0.52 ± 0.04, 0.14 ± 0.03, 1.26 ± 0.04, 2.03 ± 0.06, 0.41 ± 0.04, and 2.1 ± 0.05 μm, for WT, S101A, E99A, L100A, E102A, and I103A in β1-sheet, and V517A, E518A, and Y519A in β12-sheet, respectively) (Fig. 11, A and C). E99A, S101A, E517A, and Y519A decreased the apparent affinity of GMQ for HaFaNaC (∼2- to 3-fold, EC50 = 0.8 ± 0.04, 1.19 ± 0.07, 1.99 ± 0.07, 1.0 ± 0.02, and 1.67 ± 0.1 mm, for WT, E99A, S101A, E517A, and Y519A, respectively; Ca2+-free, pH 7.4) (Fig. 11, B and C), which is similar to effects of ASIC3E79A and ASIC3E423A on the apparent affinity of GMQ in rASIC3 (27). Because of the extreme difficulty in obtaining the crystal structure of FaNaC-GMQ complex, we cannot conclude that GMQ activates FaNaC by directly interacting with residues in the β1- and β12-sheets. Likewise, it has not been firmly established that GMQ activates ASIC3 by binding to Glu-79 and Glu-423 at the palm domain either, although covalently linking GMQ or TNB to Cys-79 of ASIC3E79C was sufficient to activate the channel in the absence of acidosis (27). Even so, based on the different ion selectivity, unitary conductance, and the distinct key sites determining their actions, it is reasonable to conclude that GMQ activates FaNaC via a mechanism distinct to that of native FMRFamide peptide.
Figure 11.
A comparison of effects of the palm domain mutations in HaFaNaC on the concentration responses to FMRFamide and GMQ. A and B, concentration-response curves to FMRFamide (A, 2 mm Ca2+, pH 7.4) and GMQ (B, Ca2+-free, pH 7.4) for WT HaFaNaC and its point mutations located in β1- and β12-sheets of the palm domain. Data points are mean ± S.E. (n = 3–5). Nearly all the alanine substitutions of the residues in β1- and in β12-sheets caused left-shifts in the concentration-response curve of FMRFamide as compared with that of WT (green line). C, summary of EC50 values for FMRFamide and GMQ in WT and the mutant HaFaNaC (mean ± S.E., n = 3–5). *, p < 0.05; **, p < 0.001, versus WT. #, EC50 could not be determined because of the very small response. D, illustration of dual or polymodal gating of FaNaC, ASIC, and ENaC subfamilies. Key sites for GMQ's and FMRFamide actions on FaNaC are shown on the left; key sites/binding modes for proton, toxin (MitTx), and GMQ in ASIC are illustrated in the middle; shear stress–, protease-, and S3969-induced activations in ENaC are shown in right. The question mark indicates that whether or not these sites directly interact with GMQ or S3969 needs further extensive studies.
Discussion
This study demonstrated here that the “primitive” member of DEG/ENaC, FaNaCs, including all four orthologues AkFaNaC, LsFaNaC, HaFaNaC, and HtFaNaC, could all be directly activated by GMQ (Figs. 2B and 11D), the nonproton ligand of mammalian ASICs. GMQ was the first nonproton ligand ever identified for mammalian ASICs and shown to activate ASIC3 in the absence of acidosis (27). A recent report, however, suggested that GMQ might not directly activate ASIC3, but instead work by altering the pH dependence and steady-state inactivation of ASICs in an isoform-specific manner so that the pH dependence of ASIC3 was shifted to a higher pH range (36). Here, because FaNaC cannot be activated by low pH, high pH or Ca2+o deprivation (Fig. 2, C–F), the activation of FaNaC by GMQ in the absence of FMRFamide cannot be attributed to a GMQ-induced change in pH or Ca2+o sensitivity. Thus, we believe that the GMQ, may act as an activator of FaNaCs, rather than a modulator. These results also suggested that GMQ may directly activate ASIC3 channels rather than change its pH sensitivity. This point is supported by the following evidence. First, the residues essential for the low pH-induced channel gating of rASIC3 are not essential for GMQ's action on ASIC3 (27), indicating that distinct channel regions are involved in ASIC3 gating by GMQ and protons. Second, GMQ-evoked inward currents are sustained whereas low-pH–induced currents are transient. If GMQ's activation is merely attributed to shifting the pH sensitivity of ASIC3, the currents should also be transient. Furthermore, it has been demonstrated that the activation of ASIC3 by pH 6.9 and pH 6.0 produced currents with different desensitization rates (2, 42). If GMQ were to affect the pH sensitivity of ASIC3, the currents evoked by different concentrations of GMQ should also exhibit distinct desensitization rates. However, both low and high concentrations of GMQ evoke persistent currents (27). Third, the sites of action for GMQ in HaFaNaC and ASIC3 are both located within the lower palm domain, along with the conformational transition pathway of DEG/ENaCs (43–45). Although the agonistic function of GMQ does not exclude the capability of this compound to potentiate proton-induced activation of ASIC3, just like that GMQ also potentiated the FMRFamide-evoked currents of HaFaNaC (Fig. 9), these potentiation effects, occurring normally between agonists that act on separate pathways, should not be taken as evidence that GMQ acted just as a modulator rather than a direct agonist on the channel. As we have demonstrated here, GMQ and FMRFamide are both direct agonists (Figs. 2 and 9) of the FaNaC subfamily and modulators to each other's actions.
Additionally, according to the evolutionary tree of DEG/ENaC superfamily of ion channels, HyNaC is closely related with ASIC (Fig. 1A) (21), and FaNaC to ENaC (15, 22). Unexpectedly, GMQ could activate FaNaC, but not ENaC (Fig. 3C), suggesting a closer relationship between FaNaC and ASICs. The fact that FMRFamide can interact with both ASICs and FaNaC further supports this hypothesis (16, 37, 46), although FMRFamide only regulates, rather than directly activates ASIC3 (37). As a newly discovered nonproton ligand of ASIC3 (27), GMQ is able to activate DRG neurons and induce pain sensation, indicating that nonproton ligand-mediated activation may be an activation mode with physiological functions (27). Some endogenous ligands of ASIC3 have been identified, such as agmatine, although its potency is weak (27, 33). Recently, lipids have also been demonstrated to activate ASIC3 in neutral environments (32). Therefore, it is possible that there are potent endogenous nonproton ligands of ASICs awaiting identification. FaNaCs are expressed only in invertebrates whereas ASICs are mainly found in mammalian (1, 46). These two kinds of subfamilies are distant in the evolution although they share common agonists. Therefore, this activation mode of GMQ maybe already exist in the common ancestor of those two subfamilies of ion channels, rather than appear during the later evolution. However, we could not exclude the possibility that other members of DEG/ENaC belonging to the superfamily can also be activated by other small molecules. For instance, the discovery of the small agonist of ENaC, namely S3969, demonstrates that besides spontaneous activation, ENaC could also be activated by small molecules (24). Furthermore, the key site of ENaC activation for S3969 is also located in the palm domain (24), which is similar to GMQ-induced activation of ASIC3 and FaNaC, suggesting that palm domain is possibly the key site of DEG/ENaC superfamily of ion channels activated by small molecules. However, whether GMQ's actions on FaNaC or ASIC3 are through a direct interaction with the residues of the palm domain or not still require extensive experiments and additional structural determinants of GMQ-ASIC3 and GMQ-FaNaCs, and a similar situation for S3969-mediated ENaC activation. Nevertheless, our observations together with previous reports (23, 24) suggest the presence of a distinct gating mode in some members of DEG/ENaC besides activation modes identified previously, for example, the FMRFamide-mediated FaNaC gating, low-pH–evoked ASIC gating, and the spontaneous opening of ENaCs. Some of these gating modes might have disappeared or become unused during evolution of DEG/ENaCs, like the direct activation by FMRFamide peptides, which is only found in FaNaCs, or they have not been experimentally identified. Others, such as the gating possibly through interaction with the palm domain residues, via the binding of small molecules such as GMQ in the case of ASIC3 (26, 27) and FaNaC, and S3969 in the case of ENaC (24) are still preserved (Fig. 11D), indicating that this novel gating mode uncovered by GMQ and S3969 may be essential for the functions of mammalian DEG/ENaC channels.
Additionally, our study unveils a dual gating of FaNaC, induced by FMRFamide and GMQ, (Fig. 11D). Polymodal activation modes exist in other members of DEG/ENaC superfamily. For ENaC, not only the channel is spontaneously open (23), but also it can be activated by proteases (47, 48), small molecules, e.g. S3969 stimulation (24), or shearing stress stimulations (25). For ASICs, the activation can be achieved by low pH (29), GMQ (27, 31), toxins (28), lipid (32), alkali stimulation (34), and mechanical force (49, 50). Therefore, we speculate that DEG/ENaC superfamily is another type of ion channels, similar to transient receptor potential (TRP) channels (51) and K2P channels (52), responding to multiple stimulations. Although it remains to be determined whether the palm domain and the thumb-finger interface located in the extracellular loop represents the only or at least the major areas of gating in the DEG/ENaC superfamily, it is quite clear from the studies on ASIC3 and FaNaCs that these two domains represent distinct gating modes that result in different open states. The polymodal activation of FaNaC indicates that this mode, at least dual gating, is a characteristic of DEG/ENaC superfamily of ion channels existed in remote antiquity before the evolution of this superfamily. It should be interesting to explore further these two distinct gating modes among different members of the DEG/ENaC superfamily and with the use of different activation methods, as well as the interplay between them, which will shed more light on the mechanisms of regulation of these channels and help identify new strategies to perturb their function for clinical therapies.
Experimental procedures
cDNAs and drugs
Full-length cDNAs for HaFaNaC, AkFaNaC, and human α-, β-, and γ-ENaC were kind gifts of Drs. Eric Lingueglia, Yasuo Furukawa, and Michael Welsh. That for LsFaNaC (GenBankTM ID: AF335548) and HtFaNaC (GenBankTM ID: AF254118) were synthesized by GENEWIZ (Shanghai) and YouBio (Changsha), respectively. All four FaNaC orthologues were subcloned into the pRc/CMV vector. FMRFamide was synthesized by GL Biochem Ltd. (Shanghai) with a purity >98%. GMQ, amiloride, DTNB, DTT, and other salts were purchased from Sigma. S3969 was a kind gift from Dr. Wang-Sheng Sun.
Site-directed mutagenesis, cell culture, and transfection
Mutations were generated by the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) and verified by DNA sequencing as we described previously (27, 35). FaNaCs and rASIC3 were individually transfected into CHO cells by Hilymax (DOJINDO Laboratories) as we described previously (27, 35). Briefly, CHO cells were cultured in F12 medium (Gibco) supplemented with 10% fetal bovine serum (Gibco), 1% l-glutamine (Gibco), and 1% penicillin/streptomycin (HyClone) at 37 °C with 5% CO2 in humidified atmosphere.
Whole-cell and single-channel recordings
As we described previously (27, 35), whole-cell recordings were carried out with CHO cells 24–48 h post transfection. Patch pipettes (3–5 megaohms) were filled with internal solution, which contains (in mm): 30 NaCl, 120 KCl, 1 MgCl2, 10 HEPES, and 5 EGTA, pH 7.4. The standard external solution (SS) contains (in mm): 150 NaCl, 10 glucose, 5 KCl, 1 MgCl2, 2 CaCl2, and 10 HEPES (pH 6.0–7.4). HEPES was replaced by equimolar MES when pH <6.0. CaCl2 and MgCl2 were substituted with 5 mm EGTA in the Ca2+-free solution. During electrophysiological recordings, 80–90% of the series resistance was compensated. All currents were measured at a holding potential of −60 mV by Axon Axopatch 200B connected to a DIGIDATA 1440A A/D converter, and analyzed using Clampex 10.2. Single-channel recordings using outside-out configuration were carried out in CHO cells at room temperature (23 ± 2 °C) 24–48 h after transfection. Recording pipettes were pulled for borosilicate glass (World Precision Instruments, Inc.) and fired polished to yield resistance of 13–18 megaohms. The holding potential was set at −80 mV. The external solution and internal solutions are the same as those of whole-cell recordings. Currents were sampled at 50 kHz with a 2-kHz filter, and a low-pass filtered at 300 Hz, using an AxonPatch 200B amplifier in conjunction with pClamp 10 software (Molecular Device).
Two-electrode voltage clamp in Xenopus laevis oocytes
HaFaNaC and human ENaC (αβγ) were subcloned into pcDNA3.1 with poly (A) for oocyte expression. Using linearized cDNAs as templates, cRNAs were synthetized by T7 mMESSAGE mMACHINE Kit (Ambion, Austin, TX). Oocytes (stage V–VI) of X. laevis were injected with 5 ng HaFaNaC or hENaC (αβγ) cRNAs, and incubated in OR-2 medium (HaFaNaC) or low-Na+ OR-2 medium (hENaC) at 19 °C. OR-2 medium contained (in mm): 82.5 NaCl, 2.5 KCl, 1 Na2HPO4, 5 HEPES, 1 MgCl2, 1 CaCl2, and 1% penicillin/streptomycin. 77.5 NaCl was substituted with equimolar N-methyl-d-glucamine (NMDG) in low-Na+ OR-2 medium. Oocytes were clamped at a holding potential of −80 mV and recorded using OC-725C amplifier (AD Instruments, Australia) connected to an Axon 1550 Digitizer (Molecular Devices) at 24–48 h after cRNA injection, and analyzed using Clampex 10.2. The standard bath solution ND96 contained (in mm): 96 NaCl, 1.8 CaCl2, 1 MgCl2, 2 KCl, and 5 HEPES. NaCl was substituted with equimolar N-methyl-d-glucamine in Na+-free experiments. CaCl2 and MgCl2 were substituted with 5 mm EGTA in Ca2+-free solution.
Calcium imaging
CHO cells expressing HaFaNaC seeded on glass coverslips were incubated with Fluo-4-AM (1 μm, Molecular Probes) and Pluronic F127 (0.02% (w/v), Sigma) for 30 min at room temperature (23 ± 2 °C) in the dark. After washing the cells three times in standard external solution, the coverslip was mounted to a recording chamber placed on the stage of a Nikon N-SIM microscope. Fluorescent intensities in response to 488 nm laser excitation were monitored and recorded using NIS-Elements Soft in the dark. FMRFamide (100 μm) or GMQ (5 mm) were added when desired in the presence of 5 μm nimodipine (Aladdin) to block voltage-gated calcium channels. Changes in intracellular calcium concentrations were expressed as Ft/F0, a ratio of fluorescent intensities at time (t) and time zero (0).
Homology modeling
Homology model of HaFaNaC was created based on the cASIC1 structure (PDB ID: 4NYK) as we described previously (35). The alignment of HaFaNaC (GenBankTM ID: CAA63084) and cASIC1 (GenBankTM ID: AAY28986) were made by Modeler 9.9 (53) and manually adjusted by published alignments (54). The obtained model was checked and validated by ProCheck (55).
Data analysis
Data were expressed as the mean ± S.E.; statistical comparisons were made using Student's t test (*, p < 0.05; **, p < 0.001). The data were fit to the Hill or Boltzmann equation. Hill equation: I/Imax = 1/{1 + (EC50/[FMRFamide])n}, where I is the normalized current at a given concentration of FMRFamide, Imax is the maximum current, n is the Hill coefficient. Boltzmann equation: I/Imax = 1/{1 + exps [(EC50 − [GMQ])/k]}, where I is the normalized current at a given concentration of GMQ, Imax is the maximum current, k is the slope factor. As we described previously (27, 56), the permeability ratios of PNa/PLi and PNa/PK were calculated using the modified Goldman-Hodgkin-Katz equation: PX/PNa = exp (ΔVrevF/RT) because of the equimolar cations in the external and internal solution, where X represents the test cations, ΔVrev is the change in reversal potential when Na+ was replaced by the tested cation, F is the Faraday constant, R is the gas constant, and T is the absolute temperature.
Author contributions
Y. Yu designed the project. X.-N. Y., Y.-Y. N., Y. L., Y.-M. H., and Y. Yang performed cell culture, patch-clamp recording, mutagenesis, and calcium image. Y. Yang and Y. L. made mutations. Y. L., X.-N. Y., and J. W. performed single channel recordings. Y. Yu and Y.-Y. N. performed homology modeling. Y. T. and X.-Y. L. supervised X.-N. Y. and Y.-Y. N. Y. Yu, Y.-Y. N., Y. Yang, X.-N. Y., X.-Y. C., H.-S. W., H. L., T.-L. X., and M. X. Z. wrote the manuscript.
Acknowledgments
We thank Drs. Eric Lingueglia, Yasuo Furukawa, and Michael Welsh for their kind gifts of full-length cDNAs for HaFaNaC, AkFaNaC, and human α-, β-, and γ-ENaC.
This study was supported by National Program on Key Basic Research Project of China Grant 2014CB910300/02, National Natural Science Foundation of China Grants 31570832, 21431001, 31170787, 81760626, and 31400707, Guangxi Foundation for Distinguished Experts (2017), National Excellent Young Scientist Foundation of China Grant 31222018, Innovation Program of Shanghai Municipal Education Commission Grant 14YZ034, and Research Foundation of Education Bureau of Hunan Province, China Grant 17A153. The authors declare that they have no conflicts of interest with the contents of this article.
Throughout this manuscript, the following designations were used: ELSE/AAAA, E99A/L100A/S101A/E102A; VEY/AAA, V517A/E518A/Y519A.
- DEG/ENaC
- degenerin/epithelial sodium channel
- FaNaCs
- FMRFamide peptide–activated sodium channels
- GMQ
- 2-guanidine-4-methylquinazoline
- ASICs
- acid-sensing ion channels
- HyNaC
- Hydra sodium channel
- DTNB
- 5,5′-dithiobis(nitrobenzoic acid).
References
- 1. Kellenberger S., and Schild L. (2002) Epithelial sodium channel/degenerin family of ion channels: A variety of functions for a shared structure. Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 2. Kellenberger S., and Schild L. (2015) International Union of Basic and Clinical Pharmacology. XCI. Structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol. Rev. 67, 1–35 [DOI] [PubMed] [Google Scholar]
- 3. Kreple C. J., Lu Y., Taugher R. J., Schwager-Gutman A. L., Du J., Stump M., Wang Y., Ghobbeh A., Fan R., Cosme C. V., Sowers L. P., Welsh M. J., Radley J. J., LaLumiere R. T., and Wemmie J. A. (2014) Acid-sensing ion channels contribute to synaptic transmission and inhibit cocaine-evoked plasticity. Nat. Neurosci. 17, 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wemmie J. A., Chen J., Askwith C. C., Hruska-Hageman A. M., Price M. P., Nolan B. C., Yoder P. G., Lamani E., Hoshi T., Freeman J. H. Jr., and Welsh M. J. (2002) The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron 34, 463–477 [DOI] [PubMed] [Google Scholar]
- 5. Wemmie J. A., Askwith C. C., Lamani E., Cassell M. D., Freeman J. H. Jr., and Welsh M. J. (2003) Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J. Neurosci. 23, 5496–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziemann A. E., Allen J. E., Dahdaleh N. S., Drebot I. I., Coryell M. W., Wunsch A. M., Lynch C. M., Faraci F. M., Howard M. A. 3rd, Welsh M. J., and Wemmie J. A. (2009) The amygdala is a chemosensor that detects carbon dioxide and acidosis to elicit fear behavior. Cell 139, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wemmie J. A., Coryell M. W., Askwith C. C., Lamani E., Leonard A. S., Sigmund C. D., and Welsh M. J. (2004) Overexpression of acid-sensing ion channel 1a in transgenic mice increases acquired fear-related behavior. Proc. Natl. Acad. Sci. U.S.A. 101, 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ugawa S., Ueda T., Ishida Y., Nishigaki M., Shibata Y., and Shimada S. (2002) Amiloride-blockable acid-sensing ion channels are leading acid sensors expressed in human nociceptors. J. Clin. Invest. 110, 1185–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., and Lingueglia E. (2008) ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao J., Duan B., Wang D. G., Deng X. H., Zhang G. Y., Xu L., and Xu T. L. (2005) Coupling between NMDA receptor and acid-sensing ion channel contributes to ischemic neuronal death. Neuron 48, 635–646 [DOI] [PubMed] [Google Scholar]
- 11. Xiong Z. G., Zhu X. M., Chu X. P., Minami M., Hey J., Wei W. L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., and Simon R. P. (2004) Neuroprotection in ischemia: Blocking calcium-permeable acid-sensing ion channels. Cell 118, 687–698 [DOI] [PubMed] [Google Scholar]
- 12. Pidoplichko V. I., and Dani J. A. (2006) Acid-sensitive ionic channels in midbrain dopamine neurons are sensitive to ammonium, which may contribute to hyperammonemia damage. Proc. Natl. Acad. Sci. U.S.A. 103, 11376–11380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arias R. L., Sung M. L., Vasylyev D., Zhang M. Y., Albinson K., Kubek K., Kagan N., Beyer C., Lin Q., Dwyer J. M., Zaleska M. M., Bowlby M. R., Dunlop J., and Monaghan M. (2008) Amiloride is neuroprotective in an MPTP model of Parkinson's disease. Neurobiol. Dis. 31, 334–341 [DOI] [PubMed] [Google Scholar]
- 14. Ziemann A. E., Schnizler M. K., Albert G. W., Severson M. A., Howard M. A. 3rd, Welsh M. J., and Wemmie J. A. (2008) Seizure termination by acidosis depends on ASIC1a. Nat. Neurosci. 11, 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Assmann M., Kuhn A., Dürrnagel S., Holstein T. W., and Gründer S. (2014) The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol. 12, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lingueglia E., Champigny G., Lazdunski M., and Barbry P. (1995) Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature 378, 730–733 [DOI] [PubMed] [Google Scholar]
- 17. Jeziorski M. C., Green K. A., Sommerville J., and Cottrell G. A. (2000) Cloning and expression of a FMRFamide-gated Na(+) channel from Helisoma trivolvis and comparison with the native neuronal channel. J. Physiol. 526, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perry S. J., Straub V. A., Schofield M. G., Burke J. F., and Benjamin P. R. (2001) Neuronal expression of an FMRFamide-gated Na+ channel and its modulation by acid pH. J. Neurosci. 21, 5559–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Furukawa Y., Miyawaki Y., and Abe G. (2006) Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na+ channel. Pflugers Arch. 451, 646–656 [DOI] [PubMed] [Google Scholar]
- 20. Cottrell G. A. (2005) Domain near TM1 influences agonist and antagonist responses of peptide-gated Na+ channels. Pflugers Arch. 450, 168–177 [DOI] [PubMed] [Google Scholar]
- 21. Golubovic A., Kuhn A., Williamson M., Kalbacher H., Holstein T. W., Grimmelikhuijzen C. J., and Gründer S. (2007) A peptide-gated ion channel from the freshwater polyp Hydra. J. Biol. Chem. 282, 35098–35103 [DOI] [PubMed] [Google Scholar]
- 22. Gründer S., and Assmann M. (2015) Peptide-gated ion channels and the simple nervous system of Hydra. J. Exp. Biol. 218, 551–561 [DOI] [PubMed] [Google Scholar]
- 23. Eastwood A. L., and Goodman M. B. (2012) Insight into DEG/ENaC channel gating from genetics and structure. Physiology 27, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu M., Echeverri F., Kalabat D., Laita B., Dahan D. S., Smith R. D., Xu H., Staszewski L., Yamamoto J., Ling J., Hwang N., Kimmich R., Li P., Patron E., Keung W., Patron A., and Moyer B. D. (2008) Small molecule activator of the human epithelial sodium channel. J. Biol. Chem. 283, 11981–11994 [DOI] [PubMed] [Google Scholar]
- 25. Carattino M. D., Sheng S., and Kleyman T. R. (2004) Epithelial Na+ channels are activated by laminar shear stress. J. Biol. Chem. 279, 4120–4126 [DOI] [PubMed] [Google Scholar]
- 26. Yu Y., Li W. G., Chen Z., Cao H., Yang H., Jiang H., and Xu T. L. (2011) Atomic level characterization of the nonproton ligand-sensing domain of ASIC3 channels. J. Biol. Chem. 286, 24996–25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Y., Chen Z., Li W. G., Cao H., Feng E. G., Yu F., Liu H., Jiang H., and Xu T. L. (2010) A nonproton ligand sensor in the acid-sensing ion channel. Neuron 68, 61–72 [DOI] [PubMed] [Google Scholar]
- 28. Bohlen C. J., Chesler A. T., Sharif-Naeini R., Medzihradszky K. F., Zhou S., King D., Sánchez E. E., Burlingame A. L., Basbaum A. I., and Julius D. (2011) A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 479, 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waldmann R., Champigny G., Bassilana F., Heurteaux C., and Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 30. Wemmie J. A., Price M. P., and Welsh M. J. (2006) Acid-sensing ion channels: advances, questions and therapeutic opportunities. Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 31. Smith R. N., and Gonzales E. B. (2014) Protons and Psalmotoxin-1 reveal nonproton ligand stimulatory sites in chicken acid-sensing ion channel: Implication for simultaneous modulation in ASICs. Channels 8, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marra S., Ferru-Clément R., Breuil V., Delaunay A., Christin M., Friend V., Sebille S., Cognard C., Ferreira T., Roux C., Euller-Ziegler L., Noel J., Lingueglia E., and Deval E. (2016) Non-acidic activation of pain-related acid-sensing ion channel 3 by lipids. EMBO J. 35, 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li W. G., Yu Y., Zhang Z. D., Cao H., and Xu T. L. (2010) ASIC3 channels integrate agmatine and multiple inflammatory signals through the nonproton ligand sensing domain. Mol. Pain 6, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Delaunay A., Gasull X., Salinas M., Noël J., Friend V., Lingueglia E., and Deval E. (2012) Human ASIC3 channel dynamically adapts its activity to sense the extracellular pH in both acidic and alkaline directions. Proc. Natl. Acad. Sci. U.S.A. 109, 13124–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niu Y. Y., Yang Y., Liu Y., Huang L. D., Yang X. N., Fan Y. Z., Cheng X. Y., Cao P., Hu Y. M., Li L., Lu X. Y., Tian Y., and Yu Y. (2016) Exploration of the peptide recognition of an amiloride-sensitive FMRFamide peptide-gated sodium channel. J. Biol. Chem. 291, 7571–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alijevic O., and Kellenberger S. (2012) Subtype-specific modulation of acid-sensing ion channel (ASIC) function by 2-guanidine-4-methylquinazoline. J. Biol. Chem. 287, 36059–36070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Askwith C. C., Cheng C., Ikuma M., Benson C., Price M. P., and Welsh M. J. (2000) Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron 26, 133–141 [DOI] [PubMed] [Google Scholar]
- 38. Immke D. C., and McCleskey E. W. (2003) Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron 37, 75–84 [DOI] [PubMed] [Google Scholar]
- 39. Yang Y., Yu Y., Cheng J., Liu Y., Liu D. S., Wang J., Zhu M. X., Wang R., and Xu T. L. (2012) Highly conserved salt bridge stabilizes rigid signal patch at extracellular loop critical for surface expression of acid-sensing ion channels. J. Biol. Chem. 287, 14443–14455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhainazarov A. B., and Cottrell G. A. (1998) Single-channel currents of a peptide-gated sodium channel expressed in Xenopus oocytes. J. Physiol. 513, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Green K. A., and Cottrell G. A. (2002) Activity modes and modulation of the peptide-gated Na(+) channel of Helix neurones. Pflugers Arch. 443, 813–821 [DOI] [PubMed] [Google Scholar]
- 42. Gründer S., and Pusch M. (2015) Biophysical properties of acid-sensing ion channels (ASICs). Neuropharmacology 94, 9–18 [DOI] [PubMed] [Google Scholar]
- 43. Della Vecchia M. C., Rued A. C., and Carattino M. D. (2013) Gating transitions in the palm domain of ASIC1a. J. Biol. Chem. 288, 5487–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frey E. N., Pavlovicz R. E., Wegman C. J., Li C., and Askwith C. C. (2013) Conformational changes in the lower palm domain of ASIC1a contribute to desensitization and RFamide modulation. PloS One 8, e71733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baconguis I., and Gouaux E. (2012) Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lingueglia E., Deval E., and Lazdunski M. (2006) FMRFamide-gated sodium channel and ASIC channels: A new class of ionotropic receptors for FMRFamide and related peptides. Peptides 27, 1138–1152 [DOI] [PubMed] [Google Scholar]
- 47. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., and Kleyman T. R. (2004) Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 48. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., and Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ-subunit. J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 49. Cabo R., Alonso P., Viña E., Vázquez G., Gago A., Feito J., Pérez-Moltó F. J., García-Suárez O., and Vega J. A. (2015) ASIC2 is present in human mechanosensory neurons of the dorsal root ganglia and in mechanoreceptors of the glabrous skin. Histochem. Cell Biol. 143, 267–276 [DOI] [PubMed] [Google Scholar]
- 50. Omerbašić D., Schuhmacher L. N., Bernal Sierra Y. A., Smith E. S., and Lewin G. R. (2014) ASICs and mammalian mechanoreceptor function. Neuropharmacology 94, 80–86 [DOI] [PubMed] [Google Scholar]
- 51. Tominaga M., Caterina M. J., Malmberg A. B., Rosen T. A., Gilbert H., Skinner K., Raumann B. E., Basbaum A. I., and Julius D. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543 [DOI] [PubMed] [Google Scholar]
- 52. McClenaghan C., Schewe M., Aryal P., Carpenter E. P., Baukrowitz T., and Tucker S. J. (2016) Polymodal activation of the TREK-2 K2P channel produces structurally distinct open states. J. Gen. Physiol. 147, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 54. Jasti J., Furukawa H., Gonzales E. B., and Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 55. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., and Thornton J. M. (1996) AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 56. Yang H., Yu Y., Li W. G., Yu F., Cao H., Xu T. L., and Jiang H. (2009) Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLos Biol. 7, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]