Abstract
Although it is widely appreciated that the use of global translation inhibitors, such as cycloheximide, in protein degradation assays may result in artefacts, these inhibitors continue to be employed, owing to the absence of robust alternatives. We describe here the promoter reference technique (PRT), an assay for protein degradation with two advantageous features: a reference protein and a gene-specific inhibition of translation. In PRT assays, one measures, during a chase, the ratio of a test protein to a long-lived reference protein, a dihydrofolate reductase (DHFR). The test protein and DHFR are coexpressed, in the yeast Saccharomyces cerevisiae, on a low-copy plasmid from two identical PTDH3 promoters containing additional, previously developed DNA elements. Once transcribed, these elements form 5′-RNA aptamers that bind to the added tetracycline, which represses translation of aptamer-containing mRNAs. The selectivity of repression avoids a global inhibition of translation. This selectivity is particularly important if a component of a relevant proteolytic pathway (e.g. a specific ubiquitin ligase) is itself short-lived. We applied PRT to the Pro/N-end rule pathway, whose substrates include the short-lived Mdh2 malate dehydrogenase. Mdh2 is targeted for degradation by the Gid4 subunit of the GID ubiquitin ligase. Gid4 is also a metabolically unstable protein. Through analyses of short-lived Mdh2 as a target of short-lived Gid4, we illustrate the advantages of PRT over degradation assays that lack a reference and/or involve cycloheximide. In sum, PRT avoids the use of global translation inhibitors during a chase and also provides a “built-in” reference protein.
Keywords: aptamer, protein degradation, Saccharomyces cerevisiae, ubiquitin, yeast, assay, chase, degradation, reference, GID, Mdh2, Gid4, tetracycline
Introduction
In vivo half-lives of intracellular proteins range from less than a minute to many days (1–6). The term “half-life” is, at best, an approximate descriptor of a protein's degradation curve. The reasons for this include (i) a significant (and varying) probability of cotranslational degradation of a nascent (still being made) polypeptide chain (7–12); (ii) the process of conformational maturation, i.e. transitions from a newly formed, still partially unfolded (and often more vulnerable to proteolysis) monomer of a protein to its final state, either as a folded monomer or an oligomer of the homo or hetero kind (mature proteins are usually more resistant to proteolysis in part through steric shielding of their degradation signals (degrons)) (4, 5, 13–19); (iii) a variety of chemical (usually enzymatic) modifications of a protein that can change its metabolic stability either cotranslationally or posttranslationally (e.g. phosphorylation of a protein may either activate or inhibit its degron) (20–27); and (iv) posttranslational in vivo cleavage(s) of a protein by a nonprocessive protease, such as, for instance, a calpain or a caspase (28–30). The rates of subsequent degradation of the resulting protein fragments can be quite dissimilar. The in vivo destruction of a protein usually involves either its polyubiquitylation, followed by the proteasome-mediated degradation, or the entrapment of a protein in an autophagosome, followed by the lysosome-mediated proteolysis. Some proteins can be targeted through both of these routes concurrently, depending on the nature of a protein, its degron(s), and the physiological state of a cell (31).
These and other complexities of intracellular protein degradation make it particularly important to have the means for determining the in vivo decay curves of specific proteins with high accuracy and through methods that do not perturb the rates of proteolysis that these methods are designed to measure. A variety of protein degradation assays are currently in use, including pulse-chases as well as proteome-scale techniques based on mass spectrometry (19, 32–48).
One class of widely employed approaches, historically among the first to be introduced, are assays for in vivo protein degradation that involve the use of a global translation inhibitor to halt protein synthesis, followed by measurements of decreasing levels of a protein of interest (a “chase”) by SDS-PAGE and immunoblotting or by other means. The resulting decay curves are usually calibrated by measuring, in parallel, the levels of an abundant endogenous protein (e.g. tubulin or actin) that is presumed to be both long-lived and expressed at approximately equal levels under different physiological conditions and in varying genetic backgrounds.
A shortcoming, in the context of these assays, of a global translation inhibitor such as, for example, cycloheximide (CHX),3 is not only the inhibitor's cytotoxicity but also the fact that a proteolytic pathway under study may itself involve a short-lived protein(s). Naturally unstable components of specific proteolytic systems continue to be identified (49–53). In all such cases, a global halt to the synthesis of cellular proteins would perturb the very process that a degradation assay is meant to measure, because the activity of a relevant proteolytic pathway would start to decrease as soon as its short-lived component is no longer produced. In addition and independently, it can be problematic to calibrate chase-based degradation assays by measuring the relative amounts of endogenous “marker” proteins, inasmuch as their levels often change in response to altered physiological conditions or genetic backgrounds.
Two of our 2017 studies described and employed a degradation assay termed the promoter reference technique (PRT) (54, 55). Key features of PRT are (i) a coexpression, from identical transcriptional promoters, of a test protein and a long-lived reference protein; and (ii) a gene-specific (i.e. not global) inhibition of translation during a chase (Fig. 1). In the present work, this technique is described in detail (Figs. 1–4), was further optimized, and was also used, in specific PRT-based degradation assays, to demonstrate a significant downside of using a global translation inhibitor, in comparison with a gene-specific inhibition of translation (Fig. 4).
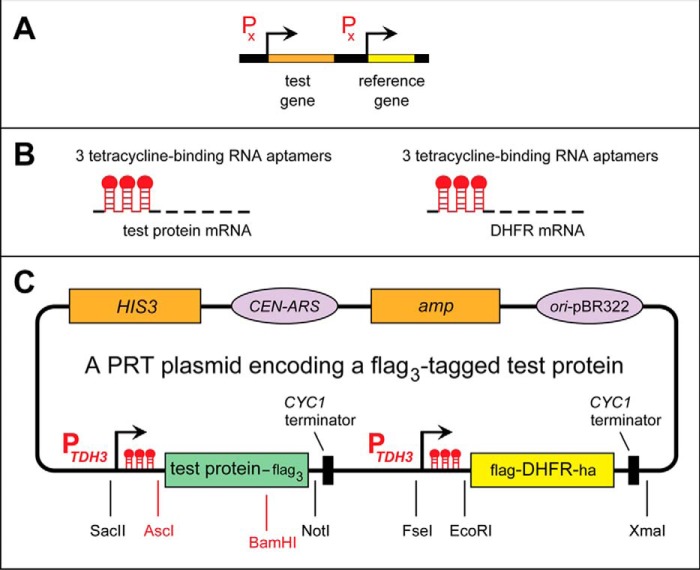
Figure 1.
The promoter reference technique. A, PRT design: two identical transcriptional promoters expressing, respectively, a test protein and a reference protein. B, identical sets of DNA elements, developed by Kötter et al. (80), that are present at the two identical, tandem PTDH3 promoters in the current version of PRT. C, a “generic” PRT plasmid, derived from the previously described pJO629 plasmid (54, 55). It contains DNA elements that enable propagation of this plasmid in E. coli (including the ampicillin-encoding amp gene and the origin of DNA replication from the E. coli plasmid pBR322) and in S. cerevisiae (as a low-copy plasmid), including the HIS3 marker gene, the replication/segregation CEN-ARS DNA segment, and the CYC1-derived transcription-terminating DNA segment. This plasmid also encodes the flag/ha-tagged fDHFRha reference protein and a C-terminally triple-flag-tagged test protein. In the present study, the test proteins were X-Mdh23f bearing either N-terminal Pro or N-terminal Ser, as described under “Results and discussion” (Fig. 3 and Fig. S1). The test protein and the reference protein are expressed from two identical PTDH3 promoters containing additional DNA elements (80). Once transcribed into an mRNA, these elements form 5′-RNA aptamers that can bind to Tc, thereby repressing translation of that mRNA in cis.
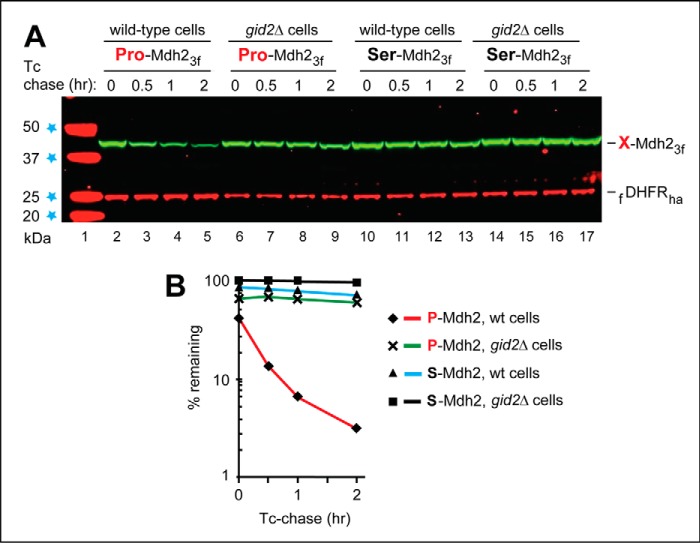
Figure 2.
PRT-based chase assays with Mdh2. A, S. cerevisiae Mdh2 is the cytosolic malate dehydrogenase and a substrate of the GID-mediated Pro/N-end rule pathway (54). Lane 1, kDa markers. Tc-based chases were performed at 30 °C during the transition from ethanol to glucose medium (see “Experimental procedures”) with wild-type (lanes 2–5 and 10–13) or gid2Δ S. cerevisiae (lanes 6–9 and 14–17) expressing the fDHFRha reference and either wild-type P-Mdh23f (bearing N-terminal Pro; lanes 2–9) or S-Mdh23f (bearing N-terminal Ser; lanes 10–17). At the indicated times of a chase, proteins in cell extracts were fractionated by SDS-PAGE, followed by immunoblotting with anti-flag antibody. The bands of X-Mdh23f test proteins and the fDHFRha protein are indicated on the right. B, quantification of the data in A. The time 0 (before-chase) level of S-Mdh23f in gid2Δ cells was taken as 100%. Note the much lower time 0 level (40%) of the short-lived, wild-type P-Mdh23f in wild-type cells. Its half-life (the time it took for the level of P-Mdh23f to decrease from the initial 40% to 20%) was ∼20 min. All chases in this study were performed at least twice and yielded results that differed by <10%.
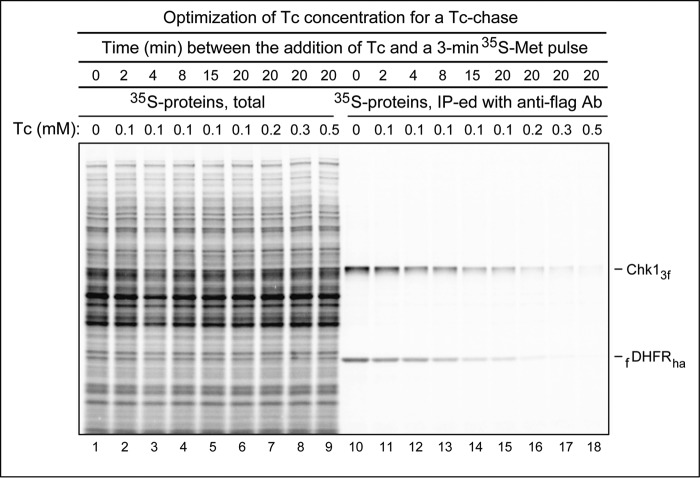
Figure 3.
Optimizing the level of tetracycline during a chase. ubr1Δ S. cerevisiae carried the previously described (55), PRT-based plasmid pJO630 that expressed the Chk13f protein and the fDHFRha reference protein (see “Experimental procedures” and “Results and discussion” for a brief introduction of the mitotic checkpoint kinase Chk1, a physiological substrate of the Arg/N-end rule pathway). Cells were labeled for 3 min at 30 °C with 35S-EXPRESS Met/Cys at the specified times after the addition of Tc to the indicated varying final concentrations. Lanes 1–9, total 35S-labeling patterns, before immunoprecipitation. Lanes 10–18, same as lanes 1–9 but after immunoprecipitation of labeled cell extracts with anti-flag antibody (Ab). Lane 1, no addition of Tc before a 3-min pulse with 35S-EXPRESS Met/Cys. Lanes 2–6 and 10–15, Tc was added to cells to a final concentration of 0.1 mm, followed by incubation for 2, 4, 8, 15, and 20 min, respectively, before a 3-min pulse with 35S-EXPRESS Met/Cys. Lanes 7–9 and 16–18, Tc was added to cells to the final concentrations of 0.2, 0.3, and 0.5 mm, respectively, followed by incubation for 20 min and a 3-min pulse with 35S-EXPRESS Met/Cys. See “Results and discussion” for additional details.
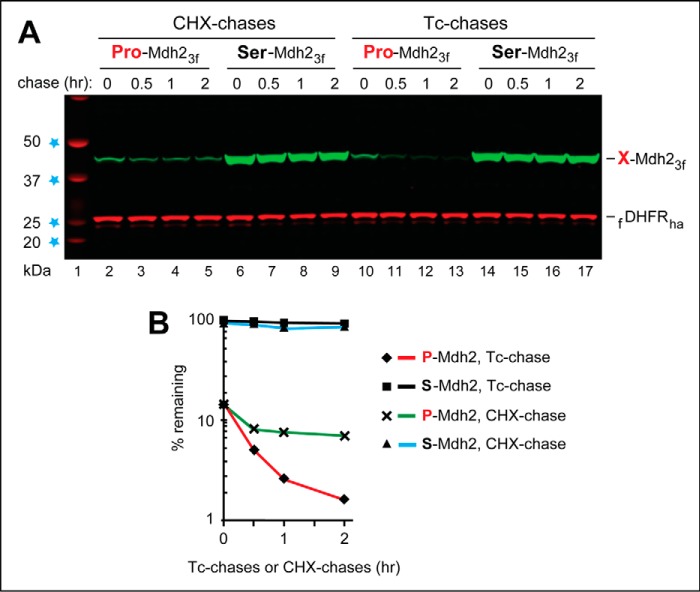
Figure 4.
Comparing a PRT-based tetracycline chase with a cycloheximide chase. PRT-based Tc chases and CHX chases were carried out with wild-type S. cerevisiae growing in SC medium and expressing either P-Mdh23f (bearing N-terminal Pro) or S-Mdh23f (bearing N-terminal Ser). A, lane 1, kDa markers. Lanes 2–5, CHX chase, for 0, 0.5, 1, and 2 h, with P-Mdh23f. Lanes 6–9, same as lanes 2–5 but with S-Mdh23f. Lanes 10–13, same as lanes 2–5 but a Tc chase. Lanes 14–17, same as lanes 6–9, but a Tc chase. The bands of X-Mdh23f test proteins and the fDHFRha protein are indicated on the right. B, quantification of the data in A. The time 0 (before-chase) level of S-Mdh23f in the Tc chase was taken as 100%. Note the time 0, before-chase degradation of ∼85% of P-Mdh23f, relative to S-Mdh23f, and the similar time 0 levels (in contrast to subsequent levels) of P-Mdh23f in the Tc chase versus the CHX chase. Note, also, the lower rate of P-Mdh23f decrease (and the subsequent flattening of the decay curve) in the CHX chase, in comparison with the Tc chase. All chases in this study were performed at least twice and yielded results that differed by <10%. Also see “Results and discussion.”
Results and discussion
PRT vis-à-vis the Pro/N-end rule pathway
PRT is described below through its applications to one of the N-end rule pathways (Fig. 1 and Fig. S1). These pathways are a set of proteolytic systems whose unifying feature is their ability to recognize proteins containing N-terminal degradation signals called N-degrons, thereby causing the degradation of these proteins, largely by the proteasome (and also by autophagy) in eukaryotes and by the proteasome-like ClpAP protease in bacteria (4, 54–64).
The main determinant of an N-degron is a destabilizing N-terminal residue of a protein. Initially, most N-degrons are pro-N-degrons. They are converted to active N-degrons either constitutively (e.g. during the emergence of a nascent protein from a ribosome) or conditionally, via regulated steps. A pro-N-degron can become an N-degron through a site-specific cleavage of a protein by a protease and/or through enzymatic acetylation, formylation, arginylation, or leucylation of specific proteins at the α-amino group of their N-terminal residues. Studies over the last 3 decades have shown that all 20 amino acids of the genetic code can act, in cognate sequence contexts, as destabilizing N-terminal residues (Fig. S1). Consequently, many, possibly most, proteins in a cell are conditionally short-lived N-end rule substrates, either as full-length proteins or as protease-generated natural protein fragments (54, 55, 65). Recognition components of N-end rule pathways are called N-recognins. Regulated degradation of proteins and their fragments by the N-end rule pathways has been shown to mediate a remarkably wide range of biological processes (Fig. S1) (4, 54–77).
Eukaryotes contain three N-end rule pathways (see Fig. S1 and its legend). One of these proteolytic systems, termed the Pro/N-end rule pathway, is mediated, in Saccharomyces cerevisiae, by the multisubunit GID Ub ligase. Gid4, a subunit of GID that functions as the Pro/N-recognin, targets proteins that bear the N-terminal Pro residue or a Pro at position 2, in addition to adjoining and also required sequence motifs (54) (Fig. S1C). Physiological substrates of the S. cerevisiae Pro/N-end rule pathway include the gluconeogenic enzymes Fbp1, Mdh2, Icl1, and Pck1, which are long-lived in cells deprived of glucose but are selectively destroyed upon return to glucose-replete conditions (Fig. S1C) (54, 78, 79). Degradation assays of the present study focus on Mdh2, a physiological substrate of the Pro/N-end rule pathway (Fig. S1C).
The promoter reference technique
Fig. 1 illustrates PRT. A plasmid, based on the low-copy S. cerevisiae–Escherichia coli pJO629 vector (see “Experimental procedures”), contains the required PRT elements, including an ORF encoding an epitope-tagged protein of interest (Fig. 1C, test protein-flag3). The test protein and a long-lived reference protein (an epitope-tagged mouse DHFR) are coexpressed, in S. cerevisiae, from a pJO629-based plasmid and its two identical PTDH3 promoters containing additional DNA elements, developed by Kötter et al. (80). Once transcribed into an mRNA, these elements form 5′-RNA aptamers that can bind to tetracycline (Tc), thereby repressing translation of that mRNA in cis (Fig. 1, B and C).
As a protein degradation assay, PRT has two advantageous features: a “built-in” reference protein and the possibility of using a Tc-mediated, gene-specific (i.e. non-global and therefore largely nontoxic) repression of translation (Fig. 1, B and C). The logic of PRT (Fig. 1A) is not confined to Tc as a gene-specific translation inhibitor. It should also be possible to develop PRT assays that employ, for example, other transcriptional promoters and/or aptamer-mediated regulatory elements specific for small compounds other than Tc.
Fig. 2 illustrates the use of Tc-based PRT to characterize the degradation of Mdh2, a gluconeogenic enzyme. When glucose is low or absent, cells synthesize it through gluconeogenesis, an ATP-consuming process (81). When S. cerevisiae grow on ethanol as a carbon source, gluconeogenic enzymes, such as Mdh2, are expressed and long-lived. Transition to a medium containing glucose inhibits the synthesis of gluconeogenic enzymes and induces their degradation, which is mediated by the multisubunit GID Ub ligase (54, 78, 79). Gid4, a subunit of GID, was recently identified as the N-recognin of a distinct N-end rule pathway, termed the Pro/N-end rule pathway (Fig. S1C) (54). Gid4 recognizes the N-terminal Pro residue of Mdh2 in the context of an also required adjoining sequence motif (54). Previous work showed that Gid4, which is absent in yeast growing on ethanol, is an expressed but short-lived protein in cells that grow in a glucose-containing medium (82).
Our 2017 study of the Pro/N-end rule pathway (54) involved Tc chases with the gluconeogenic enzymes Mdh2, Fbp1, and Pck1. To avoid repetition, Fig. 2 shows a previously unpublished set of PRT-based Mdh2 results. These assays involved wild-type and gid2Δ S. cerevisiae carrying PRT-based plasmids (Fig. 1C) that expressed either the 377-residue, C-terminally flag-tagged P-Mdh23f (bearing N-terminal Pro) or its otherwise identical mutant S-Mdh23f, bearing N-terminal Ser. gid2Δ cells lack an essential subunit of the GID Ub ligase and therefore lack the Pro/N-end rule pathway (Fig. S1C). After 18 h in ethanol medium at 30 °C, cells were shifted to a glucose medium while the synthesis of both the fDHFRha reference protein and X-Mdh23f proteins was repressed by Tc, initiating a chase (Fig. 2). Under these conditions, P-Mdh23f (its initially present N-terminal Met is cotranslationally removed by Met-aminopeptidases (4)) was short-lived in wild-type cells but was much more stable in gid2Δ cells, which lacked the activity of GID (Fig. 2A, lanes 2–5; compare with lanes 6–9 and Fig. 2B). In contrast, the S-Mdh23f mutant, bearing N-terminal Ser instead of Pro, was long-lived even in wild-type cells (Fig. 2, A (lanes 10–17) and B).
Optimizing the level of tetracycline during a chase
The concentration of Tc used for gene-specific inhibition of translation in PRT-based chases of Fig. 2 was 0.2 mm. The same level of Tc was used in our 2017 studies that introduced and briefly described PRT (54, 55). In the present work, we wished to determine whether 0.2 mm Tc was an optimal concentration (i.e. whether it was low enough to cause no detectable cytotoxicity and high enough to result in a non-leaky gene-specific repression of translation). We found that 0.5 mm Tc was preferable to the also acceptable but slightly “leaky” 0.2 mm Tc (Fig. 3).
In contrast to the assays in Figs. 2 and 4, which involved Mdh2, the Tc-optimization experiments described in Fig. 3 involved S. cerevisiae Chk1, a mitotic checkpoint kinase. Chk1 and several other yeast proteins were recently identified as conditionally short-lived physiological substrates of the Ubr1-mediated Arg/N-end rule pathway (55). The Ubr1-containing targeting complex of this pathway (Fig. S1A) was shown to recognize a C terminus–proximal degron of the Chk1 protein (55). Our sole aim, in the experiments of Fig. 3, was to determine a level of Tc that would be optimal for PRT assays. Therefore, we used the ubr1Δ JOY379 S. cerevisiae strain (lacking the Arg/N-end rule pathway), in which Chk1 was long-lived. JOY379 cells carried a PRT-based plasmid (Fig. 1C) expressing the epitope-tagged Chk13f protein and the fDHFRha reference protein (55).
Tc was added, to different final levels (varying from 0.1 to 0.5 mm, as indicated in Fig. 3), to identical samples of JOY379 S. cerevisiae. After a time interval in the presence of Tc that varied from 0 to 20 min, cell suspensions were pulse-labeled for 3 min with [35S]Met/Cys (see “Experimental procedures”), followed by extraction of proteins, immunoprecipitation of Chk13f and fDHFRha with anti-flag antibody, SDS-PAGE, and autoradiography. Fig. 3, lanes 1–9 show the SDS-PAGE results with total 35S-pulse-labeled proteins, before anti-flag immunoprecipitation. Fig. 3 (lanes 10–18) shows the same samples from 35S-pulse-labeled cells but after anti-flag immunoprecipitation.
None of the examined concentrations of Tc, including 0.5 mm, caused significant changes in the levels (or patterns) of total 35S-pulse-labeled proteins, indicating that Tc did not significantly influence global cytosolic translation (Fig. 3, lanes 1–9). Fig. 3 (lanes 10–18) shows the bands of 35S-labeled (immunoprecipitated) Chk13f and fDHFRha, the two proteins expressed from mRNAs whose translation was repressed by Tc. In particular, lanes 10–15 show 35S-labeled Chk13f and fDHFRha in cells that were incubated with 0.1 mm Tc for either 0, 2, 4, 8, 15, or 20 min before the beginning of a 3-min [35S]Met/Cys pulse. The 35S-labeling of Chk13f and fDHFRha “flattened out” by 15 min after the addition of Tc (to a final concentration of 0.1 mm) and was much lower than the 35S-labeling immediately after the addition of Tc (Fig. 3, lane 10; compare with lane 15), indicating a gene-specific inhibition of translation by Tc.
The levels of 3-min 35S-labeling of Chk13f and fDHFRha 20 min after the addition of Tc decreased to essentially undetectable (at this and significantly longer autoradiographic exposures) at a Tc concentration of 0.5 mm (Fig. 3, lane 18) and to low but still detectable levels at a Tc concentration of 0.2 mm (Fig. 3, lane 16; compare with lane 18). We concluded that 0.5 mm Tc yielded an approximately optimal level of gene-specific inhibition of translation in PRT assays, as this concentration of Tc was sufficiently “non-leaky” while still without effects on either the level or pattern of total protein synthesis (Fig. 3, lane 9).
Comparing a PRT-based cycloheximide chase with a PRT-based tetracycline chase
Global translation inhibitors, such as CHX, continue to be widely used in studies that involve chase-based degradation assays. As mentioned in the Introduction, a major shortcoming of global translation inhibitors, in the context of chases, stems from the fact that a proteolytic pathway under study may itself involve a short-lived protein(s) (49–53). In such cases, a halt to the synthesis of cellular proteins would perturb the very process that a degradation assay is meant to measure, because the activity of a relevant proteolytic pathway would start to decrease as soon as its unstable component is no longer produced. Inasmuch as the Gid4 Pro/N-recognin of the Pro/N-end rule pathway that targets P-Mdh23f for degradation (Fig. 2 and Fig. S1C) is known to be a short-lived protein (82), we asked whether results of a PRT-based Tc chase with P-Mdh23f would significantly differ from those of an otherwise identical PRT-based CHX chase.
These assays were carried out identically to the ones described in Fig. 2, except that the concentration of Tc during chase was 0.5 mm (instead of 0.2 mm Tc in the assays of Fig. 2), and cells were grown in a glucose-containing medium before and during chases. Both PRT-based Tc chase and PRT-based CHX chase assays with P-Mdh23f detected the in vivo degradation of P-Mdh23f, in contrast to the metabolic stability and much higher time 0 (before-chase) levels of S-Mdh23f (Fig. 4). Remarkably, however, the actual decay curves of P-Mdh23f were quite different when determined using the Tc chase, in comparison with the CHX chase.
Specifically, the time 0 levels of P-Mdh23f, at the beginning of both chases, were nearly identical to each other (Fig. 4, A (lane 2 versus lane 10) and B). However, whereas the rapid degradation of P-Mdh23f continued throughout 2 h of Tc chase, the degradation of P-Mdh23f during an otherwise identical CHX chase was not as fast. Moreover, the degradation curve of the CHX chase “flattened out” after 30 min, in contrast to the results with the Tc chase (Fig. 4, A (lanes 2–5 versus lanes 10–13) and B).
To the best of our knowledge, these results are the first direct and rigorously controlled evidence for a significant distortion of an in vivo decay curve of a protein in a PRT-based chase that involves a global translation inhibitor, in comparison with the otherwise identical PRT-based chase that utilizes Tc (Fig. 4). Because Gid4, the N-recognin of the Pro/N-end rule pathway (Fig. S1C), is a short-lived protein (82), the above results were in agreement with the a priori expectation of a difference between these PRT-based chase patterns (Fig. 4). Thus, at least in the setting of the Pro/N-end rule pathway, a chase that employs a global translation inhibitor, such as CHX, would underestimate the actual rate of degradation of a protein of interest, due to the pathway inhibition effect detected and described above (Fig. 4).
Concluding remarks
The Tc-based PRT (Figs. 1–4) has two independently advantageous features: a “built-in” reference protein and a gene-specific inhibition of translation during a chase. A long-lived reference protein, the epitope-tagged DHFR in the present version of PRT (Figs. 1C and 4A), addresses the problem of data scatter in reference-lacking pulse-chase assays and immunoblotting-based chase assays. The presence of a reference protein makes it possible to measure the level of a test protein during a chase as the ratio of its level to that of the reference at a given chase time. Consequently, in both immunoblotting-mediated chases and radioactive labeling–mediated pulse-chases, a reference-based and ratio-based measurement can compensate for the scatter of labeling efficiencies, scatter of immunoprecipitation (and/or immunoblotting) yields, imprecisions in sample volumes, and other sources of sample-to-sample variation. The resulting robustness and higher accuracy of reference-based degradation assays also makes it easier to detect and measure, at the start of a chase, the “early,” before-chase degradation of a test protein (Figs. 2 and 4).
In addition to optimizing Tc concentration in PRT (Fig. 3), we also demonstrated, through specific PRT-based degradation assays, the significant downside of using a global translation inhibitor, in comparison with a gene-specific inhibition of translation (Fig. 4). An extension of Tc-based PRT from its described use in S. cerevisiae (Figs. 1–4) to multicellular eukaryotes, including mammalian cells, is highly desirable and should be feasible but is not technically straightforward, because the set of RNA aptamers that works in yeast (Figs. 1B, 2, and 4) did not suffice, so far, with mammalian cells (80).
Experimental procedures
Antibodies and other reagents
The antibodies used were anti-flag mouse M2 monoclonal antibody (Sigma, F1804) and anti-ha rabbit monoclonal antibody (Sigma, H6908). Fluorescence of immunoblots was quantified using an Odyssey 9120 imaging system (LI-COR, Lincoln, NE). Secondary antibodies for immunoblotting were LI-COR IRDye-conjugated goat anti-mouse 800CW (LI-COR, C60405-05) or anti-rabbit 680RD (LI-COR, C51104-08). Other reagents included “complete protease inhibitor mixture” tablets (Roche Applied Science, 11697498001); protease inhibitor mixture “for use with fungal and yeast extracts” (Sigma, P8215); phenylmethylsulfonyl fluoride (Sigma, P7626); CHX (Sigma, C7698); and Tc (Sigma, T3383).
Genetic techniques, media, yeast strains, and plasmids
Standard techniques were used for strain construction and transformation (54, 55, 83, 84). S. cerevisiae media included YPD (1% yeast extract, 2% peptone, 2% glucose; only the most relevant components are cited); SD medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose); and synthetic complete (SC) medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose), plus a drop-out mixture of compounds required by specific auxotrophic strains. The S. cerevisiae strains used in this work were BY4741 (MATa his3-1 leu2-0 Met15-0 ura3-0), BY4742 (MATα his3-1 leu2-0 lys2-0 ura3-0 can1-100), and JOY379 (a ubr1Δ::HphNT1 derivative of the BY4742 strain) (54, 55). The plasmids pJO629 (a parent of the “generic” PRT plasmid encoding a C-terminally epitope-tagged test protein and the fDHFRha reference (Fig. 1C)), pCSJ125 (a PRT-based plasmid encoding P-Mdh23f and the fDHFRha reference), pCSJ126 (an otherwise identical plasmid encoding S-Mdh23f and the fDHFRha reference), and pJO630 (a PRT-based plasmid encoding Chk13f and the fDHFRha reference) were constructed and described previously (54, 55). pJO629 and plasmids derived from it contained two direct repeats of a relatively long nucleotide sequence (PTDH3 promoter and the adjoining aptamer-forming DNA segment) (Fig. 1), a recombinogenic configuration. Therefore, a small fraction of S. cerevisiae that received a pJO629-derived plasmid (about 5% of transformants, in our experience) contained recombination-produced derivatives of that plasmid. This minor complication could be dealt with by discarding transformants that failed to produce the correctly sized fDHFRha reference and/or a test protein.
PRT-based degradation assays
These assays used low-copy pJO629-based plasmids that expressed a C-terminally triple-flag-tagged test protein and the long-lived, also tagged fDHFRha reference protein from a pair of identical PTDH3-based promoters. In this setting, the synthesis of both proteins could be selectively extinguished by the addition of Tc (Fig. 1). Initially, the final concentration of Tc was 0.2 mm (Fig. 2). It became 0.5 mm in later PRT assays (Figs. 3 and 4). When CHX, a global translation inhibitor, was used instead of Tc, it was at 0.36 mm (0.1 mg/ml). As described under “Results” (Fig. 3), 0.5 mm Tc was found to be preferable to the also acceptable but slightly “leaky” 0.2 mm Tc, the level used for Tc chases in our earlier work (54, 55) and also, initially, during the present study. Neither 0.2 nor 0.5 mm Tc detectably altered the growth rate of S. cerevisiae.
S. cerevisiae were grown to A600 of ∼1.0 in selective liquid media at 30 °C, followed by treatment with 0.2 mm Tc (and in later assays, with the optimized Tc concentration of 0.5 mm; see “Results and discussion”). When CHX was used instead of Tc, it was at a CHX concentration of 0.36 mm (0.1 mg/ml). At the indicated times, cell samples (corresponding to 1 ml of cell suspension at an A600 of ∼1.0) were harvested by centrifugation for 1 min at 3,000 rpm in a low-speed centrifuge. The pellet was resuspended in 0.8 ml of ice-cold water for 5 min, followed by centrifugation for 1 min at 3,000 rpm as above and resuspension in 0.8 ml of 0.2 m NaOH for 5 min on ice. The resulting suspension was centrifuged for 5 min at 21,130 × g. The pellet was resuspended in 50 μl of HU buffer (8 m urea, 5% SDS, 1 mm EDTA, 0.1 m DTT, 0.005% bromphenol blue, 0.2 m Tris-HCl, pH 6.8) containing 1× protease inhibitor mixture (Roche Applied Science) and 1× protease inhibitor mixture “for use with fungal and yeast extracts” (Sigma) and heated for 10 min at 70 °C. After centrifugation for 3 min at 21,130 × g, 15 μl of each supernatant was used to carry out SDS-4–12% NuPAGE (Invitrogen), followed by immunoblotting, performed as described previously (14, 54, 55, 75), using a mixture of anti-ha (1:2,000) and anti-flag (1:2,000) antibodies. Immunoblots were processed using secondary antibodies labeled with different fluorophores. Visualized protein bands were quantified using the Odyssey 9120 imaging system (LI-COR). The near-infrared fluorescence range and other features of the Odyssey scanner facilitate quantification of immunoblots.
Optimizing the level of tetracycline during chase
pJO630, a PRT-based plasmid (Fig. 1C) that expressed the flag epitope-tagged Chk13f protein and the fDHFRha reference protein, was described previously (55). See above for a brief introduction of Chk1, a kinase and a physiological substrate of the Arg/N-end rule pathway (55) (Fig. S1A). S. cerevisiae JOY379 cells (ubr1Δ::HphNT1 in the BY4742 strain background) that lacked the Arg/N-end rule pathway and carried pJO630 were grown at 30 °C to A600 of ∼1 in 50 ml of SC medium without histidine (SC−His). Nine 15-ml tubes were filled, each with 5 ml of cell suspension, followed by pelleting of cells by a low-speed centrifugation and washing of the pellets in 0.8 ml of SD medium (see above) supplemented with Lys, Trp, Leu, and Ura (83).
Cells were pelleted again and resuspended in 0.4 ml of fresh SD medium, followed by incubation at 30 °C for 20 min. Tc was added to cell suspensions, to varying final concentrations of 0.1, 0.2, 0.3, or 0.5 mm, either at the start of the 20-min incubation, during it, or immediately after it (see the legend to Fig. 3 for details). Thereafter (at the end of the 20-min incubation), cell suspensions were labeled for 3 min at 30 °C with 0.16 mCi of 35S-EXPRESS Met/Cys (PerkinElmer Life Sciences), followed by centrifugation to pellet the cells, their resuspension in 0.1 ml of amino acid–supplemented SD medium, and their freezing, immediately afterward, in liquid nitrogen. To process the samples further, 0.8 ml of lysis buffer (10% glycerol, 0.5% Nonidet P-40, 0.2 m KCl, 1 mm PMSF, 5 mm β-mercaptoethanol, 50 mm HEPES, pH 7.5) was added to each frozen sample, and extracts were prepared using Mini-Beadbeater-16 (BioSpec) (four times for 20 s each at maximum speed, with 5-min intervals on ice). The extracts were centrifuged at 21,000 × g for 15 min at 4 °C, and the supernatants were centrifuged (identically) again. 15 μl of each sample was taken and later analyzed as input samples. The rest of the supernatants were processed for immunoprecipitation, using 8 μl of a suspension of anti-flag antibody immobilized on magnetic beads (Sigma). Immunoprecipitates were analyzed by SDS-4–12% NuPAGE (Invitrogen) and autoradiography.
Author contributions
J.-H. O., S.-J. C., and A. V. designed the experiments. J.-H. O. and S.-J. C. performed the experiments. J.-H. O., S.-J. C., and A. V. wrote the paper. All authors discussed the results and commented on the manuscript.
Supplementary Material
Acknowledgments
We thank the present and former members of the Varshavsky laboratory for helpful discussions during this study. We also thank A. Melnykov, I. Printsev, and B. Wadas for their comments on the manuscript.
This work was supported by National Institutes of Health Grants 1R01GM031530 and 1R01DK039520 (to A. V.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- CHX
- cycloheximide
- DHFR
- dihydrofolate reductase
- PRT
- promoter reference technique
- Tc
- tetracycline
- Ub
- ubiquitin
- SC
- synthetic complete.
References
- 1. Varshavsky A. (2005) Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 [DOI] [PubMed] [Google Scholar]
- 2. Inobe T., and Matouschek A. (2014) Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 24, 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dikic I. (2017) Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 [DOI] [PubMed] [Google Scholar]
- 4. Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Prot. Sci. 20, 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McShane E., Sin C., Zauber H., Wells J. N., Donnelly N., Wang X., Hou J., Chen W., Storchova Z., Marsh J. A., Valleriani A., and Selbach M. (2016) Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167, 803–815.e21 [DOI] [PubMed] [Google Scholar]
- 6. Toyama B. H., Savas J. N., Park S. K., Harris M. S., Ingolia N. T., Yates J. R., and Hetzer M. W. (2013) Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner G. C., and Varshavsky A. (2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120 [DOI] [PubMed] [Google Scholar]
- 8. Ha S. W., Ju D., Hao W., and Xie Y. (2016) Rapidly translated polypeptides are preferred substrates for cotranslational protein degradation. J. Biol. Chem. 291, 9827–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lykke-Andersen J., and Bennett E. J. (2014) Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 204, 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gandin V., and Topisirovic I. (2014) Co-translational mechanisms of quality control of newly synthesized polypeptides. Translation 2, e28109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duttler S., Pechmann S., and Frydman J. (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F., Durfee L. A., and Huibregtse J. M. (2013) A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell 50, 368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baker R. T., and Varshavsky A. (1991) Inhibition of the N-end rule pathway in living cells. Proc. Natl. Acad. Sci. U.S.A. 88, 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shemorry A., Hwang C. S., and Varshavsky A. (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buchler N. E., Gerland U., and Hwa T. (2005) Nonlinear protein degradation and the function of genetic circuits. Proc. Natl. Acad. Sci. U.S.A. 102, 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schrader E. K., Harstad K. G., and Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nie M., and Boddy M. N. (2015) Pli1(PIAS1) SUMO ligase protected by the nuclear pore-associated SUMO protease Ulp1SENP1/2. J. Biol. Chem. 290, 22678–22685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bursać S., Brdovčak M. C., Pfannkuchen M., Orsolić I., Golomb L., Zhu Y., Katz C., Daftuar L., Grabušić K., Vukelić I., Filić V., Oren M., Prives C., and Volarevic S. (2012) Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl. Acad. Sci. U.S.A. 109, 20467–20472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu H., Singh Gautam A. K., Wilmington S. R., Wylie D., Martinez-Fonts K., Kago G., Warburton M., Chavali S., Inobe T., Finkelstein I. J., Babu M. M., and Matouschek A. (2016) Conserved sequence preferences contribute to substrate recognition by the proteasome. J. Biol. Chem. 291, 14526–14539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng N., and Shabek N. (2017) Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 [DOI] [PubMed] [Google Scholar]
- 21. Vittal V., Stewart M. D., Brzovic P. S., and Klevit R. E. (2015) Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 290, 21244–21251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meszaros B., Kumar M., Gibson T. J., Uyar B., and Dosztanyi Z. (2017) Degrons in cancer. Sci. Signal. 10, eaak9982. [DOI] [PubMed] [Google Scholar]
- 23. Lucas X., and Ciulli A. (2017) Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small-molecule mimicry strategies. Curr. Opin. Struct. Biol. 44, 101–110 [DOI] [PubMed] [Google Scholar]
- 24. Ravid T., and Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Omnus D. J., Pfirrmann T., Andréasson C., and Ljungdahl P. O. (2011) A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol. Biol. Cell 22, 2754–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ye X., Nalepa G., Welcker M., Kessler B. M., Spooner E., Qin J., Elledge S. J., Clurman B. E., and Harper J. W. (2004) Recognition of phosphodegron motifs in human cyclin E by the SCF (Fbw7) ubiquitin ligase. J. Biol. Chem. 279, 50110–50119 [DOI] [PubMed] [Google Scholar]
- 27. Davey N. E., and Morgan D. O. (2016) Building a regulatory network with short linear sequence motifs: lessons from the degrons of the anaphase-promoting complex. Mol. Cell 64, 12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green D. R. (2011) Means to an End: Apoptosis and Other Cell Death Mechanisms, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Piatkov K. I., Brower C. S., and Varshavsky A. (2012) The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. U.S.A. 109, E1839–E1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piatkov K. I., Oh J.-H., Liu Y., and Varshavsky A. (2014) Calpain-generated natural protein pragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E817–E826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cha-Molstad H., Yu J. E., Feng Z., Lee S. H., Kim J. G., Yang P., Han B., Sung K. W., Yoo Y. D., Hwang J., McGuire T., Shim S. M., Song H. D., Ganipisetti S., Wang N., et al. (2017) p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 8, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belle A., Tanay A., Bitincka L., Shamir R., and O'Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arnold L., Höckner S., and Seufert W. (2015) Insights into the cellular mechanism of the yeast ubiquitin ligase APC/C-Cdh1 from the analysis of in vivo degrons. Mol. Biol. Cell 26, 843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim I., Miller C. R., Young D. L., and Fields S. (2013) High-throughput analysis of in vivo protein stability. Mol. Cell. Proteomics 12, 3370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holm L., O'Rourke B., Ebenstein D., Toth M. J., Bechshoeft R., Holstein-Rathlou N. H., Kjaer M., and Matthews D. E. (2013) Determination of steady-state protein breakdown rate in vivo by the disappearance of protein-bound tracer-labeled amino acids: a method applicable in humans. Am. J. Physiol. Endocrinol. Metab. 304, E895–E907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L., Nelson C. J., Solheim C., Whelan J., and Millar A. H. (2012) Determining degradation and synthesis rates of Arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol. Cell. Proteomics 11, M111.010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terweij M., van Welsem T., van Deventer S., Verzijlbergen K. F., Menendez-Benito V., Ontoso D., San-Segundo P., Neefjes J., and van Leeuwen F. (2013) Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae. G3 (Bethesda) 3, 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Butko M. T., Yang J., Geng Y., Kim H. J., Jeon N. L., Shu X., Mackey M. R., Ellisman M. H., Tsien R. Y., and Lin M. Z. (2012) Fluorescent and photo-oxidizing TimeSTAMP tags track protein fates in light and electron microscopy. Nat. Neurosci. 15, 1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan K. T., Rendahl A. K., Chen W. P., Freund D. M., Gray W. M., Cohen J. D., and Hegeman A. D. (2016) Proteome-scale protein turnover analysis using high resolution mass spectrometric data from stable-isotope labeled plants. J. Proteome Res. 15, 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Christiano R., Nagaraj N., Fröhlich F., and Walther T. C. (2014) Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep. 9, 1959–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cambridge S. B., Gnad F., Nguyen C., Bermejo J. L., Krüger M., and Mann M. (2011) Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res. 10, 5275–5284 [DOI] [PubMed] [Google Scholar]
- 42. Bojkowska K., Santoni de Sio F., Barde I., Offner S., Verp S., Heinis C., Johnsson K., and Trono D. (2011) Measuring in vivo protein half-life. Chem. Biol. 18, 805–815 [DOI] [PubMed] [Google Scholar]
- 43. Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., Cohen L., Mayo A., and Alon U. (2011) Proteome half-life dynamics in living human cells. Science 331, 764–768 [DOI] [PubMed] [Google Scholar]
- 44. Cohen I., Geffen Y., Ravid G., and Ravid T. (2014) Reporter-based growth assay for systematic analysis of protein degradation. J. Vis. Exp. e52021 25406949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lévy F., Johnsson N., Rümenapf T., and Varshavsky A. (1996) Using ubiquitin to follow the metabolic fate of a protein. Proc. Natl. Acad. Sci. U.S.A. 93, 4907–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu D., Hsiao J. Y., Davey N. E., Van Voorhis V. A., Foster S. A., Tang C., and Morgan D. O. (2014) Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 207, 23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maurer M. J., Spear E. D., Yu A. T., Lee E. J., Shahzad S., and Michaelis S. (2016) Degradation signals for ubiquitin-proteasome dependent cytosolic protein quality control (CytoQC) in yeast. G3 (Bethesda) 6, 1853–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Piatkov K., Graciet E., and Varshavsky A. (2013) Ubiquitin reference technique and its use in ubiquitin-lacking prokaryotes. PLoS One 8, e67952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X. S., Trojer P., Matsumura T., Treisman J. E., and Tanese N. (2010) Mammalian SWI/SNF—a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 30, 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu X., Yen L., Irwin L., Sweeney C., and Carraway K. L. 3rd (2004) Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 24, 7748–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Galan J. M., and Peter M. (1999) Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 96, 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi Q., Gu S., Yu X. S., White T. W., Banks E. A., and Jiang J. X. (2015) Connexin controls cell-cycle exit and cell differentiation by directly promoting cytosolic localization and degradation of E3 ligase Skp2. Dev. Cell 35, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cui Y., He S., Xing C., Lu K., Wang J., Xing G., Meng A., Jia S., He F., and Zhang L. (2011) SCFFBXL(1)(5) regulates BMP signalling by directing the degradation of HECT-type ubiquitin ligase Smurf1. EMBO J. 30, 2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen S. J., Wu X., Wadas B., Oh J.-H., and Varshavsky A. (2017) An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oh J. H., Hyun J. Y., and Varshavsky A. (2017) Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 114, E4370–E4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bachmair A., Finley D., and Varshavsky A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 [DOI] [PubMed] [Google Scholar]
- 57. Tasaki T., Sriram S. M., Park K. S., and Kwon Y. T. (2012) The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Finley D., Ulrich H. D., Sommer T., and Kaiser P. (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hwang C. S., Shemorry A., and Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gibbs D. J., Bacardit J., Bachmair A., and Holdsworth M. J. (2014) The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611 [DOI] [PubMed] [Google Scholar]
- 61. Dissmeyer N., Rivas S., and Graciet E. (2017) Life and death of proteins after protease cleavage: protein degradation by the N-end rule pathway. New Phytol. 10.1111/nph.14619 [DOI] [PubMed] [Google Scholar]
- 62. Dougan D. A., Micevski D., and Truscott K. N. (2012) The N-end rule pathway: from recognition by N-recognins to destruction by AAA+ proteases. Biochim. Biophys. Acta 1823, 83–91 [DOI] [PubMed] [Google Scholar]
- 63. Cha-Molstad H., Sung K. S., Hwang J., Kim K. A., Yu J. E., Yoo Y. D., Jang J. M., Han D. H., Molstad M., Kim J. G., Lee Y. J., Zakrzewska A., Kim S. H., Kim S. T., Kim S. Y., et al. (2015) Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 17, 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stein B. J., Grant R. A., Sauer R. T., and Baker T. A. (2016) Structural basis of an N-degron adaptor with more stringent specificity. Structure 24, 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim H. K., Kim R. R., Oh J. H., Cho H., Varshavsky A., and Hwang C. S. (2014) The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim M. K., Oh S. J., Lee B. G., and Song H. K. (2016) Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 113, 12438–12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brower C. S., Piatkov K. I., and Varshavsky A. (2013) Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol. Cell 50, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamano K., and Youle R. J. (2013) PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choi W. S., Jeong B.-C., Joo Y. J., Lee M.-R., Kim J., Eck M. J., and Song H. K. (2010) Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 17, 1175–1181 [DOI] [PubMed] [Google Scholar]
- 70. Matta-Camacho E., Kozlov G., Li F. F., and Gehring K. (2010) Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 17, 1182–1187 [DOI] [PubMed] [Google Scholar]
- 71. Wadas B., Piatkov K. I., Brower C. S., and Varshavsky A. (2016) Analyzing N-terminal arginylation through the use of peptide arrays and degradation assays. J. Biol. Chem. 291, 20976–20992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kwon Y. T., Kashina A. S., Davydov I. V., Hu R.-G., An J. Y., Seo J. W., Du F., and Varshavsky A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 [DOI] [PubMed] [Google Scholar]
- 73. Hu R.-G., Sheng J., Qi X., Xu Z., Takahashi T. T., and Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 74. Brower C. S., and Varshavsky A. (2009) Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Park S. E., Kim J. M., Seok O. H., Cho H., Wadas B., Kim S. Y., Varshavsky A., and Hwang C. S. (2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347, 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Aksnes H., Drazic A., Marie M., and Arnesen T. (2016) First things first: vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 41, 746–760 [DOI] [PubMed] [Google Scholar]
- 77. Scott D. C., Monda J. K., Bennett E. J., Harper J. W., and Schulman B. A. (2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Menssen R., Schweiggert J., Schreiner J., Kusevic D., Reuther J., Braun B., and Wolf D. H. (2012) Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J. Biol. Chem. 287, 25602–25614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hung G.-C., Brown C. R., Wolfe A. B., Liu J., and Chiang H.-L. (2004) Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J. Biol. Chem. 279, 49138–49150 [DOI] [PubMed] [Google Scholar]
- 80. Kötter P., Weigand J. E., Meyer B., Entian K.-D., and Suess B. (2009) A fast and efficient translational control system for conditional expression of yeast genes. Nucl. Acids Res. 37, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fraenkel D. G. (2011) Yeast Intermediary Metabolism, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 82. Santt O., Pfirrmann T., Braun B., Juretschke J., Kimmig P., Scheel H., Hofmann K., Thumm M., and Wolf D. H. (2008) The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol. Biol. Cell 19, 3323–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Smith J. A., Seidman J. G., and Struhl K. (2010) Current Protocols in Molecular Biology, Wiley-Interscience, New York [Google Scholar]
- 84. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.