Abstract
Pectin methylesterases (PMEs) catalyze the demethylesterification of pectin, one of the main polysaccharides in the plant cell wall, and are of critical importance in plant development. PME activity generates highly negatively charged pectin and mutates the physiochemical properties of the plant cell wall such that remodeling of the plant cell can occur. PMEs are therefore tightly regulated by proteinaceous inhibitors (PMEIs), some of which become active upon changes in cellular pH. Nevertheless, a detailed picture of how this pH-dependent inhibition of PME occurs at the molecular level is missing. Herein, using an interdisciplinary approach that included homology modeling, MD simulations, and biophysical and biochemical characterizations, we investigated the molecular basis of PME3 inhibition by PMEI7 in Arabidopsis thaliana. Our complementary approach uncovered how changes in the protonation of amino acids at the complex interface shift the network of interacting residues between intermolecular and intramolecular. These shifts ultimately regulate the stability of the PME3–PMEI7 complex and the inhibition of the PME as a function of the pH. These findings suggest a general model of how pH-dependent proteinaceous inhibitors function. Moreover, they enhance our understanding of how PMEs may be regulated by pH and provide new insights into how this regulation may control the physical properties and structure of the plant cell wall.
Keywords: pH regulation, protein–protein interaction, plant biochemistry, molecular dynamics, molecular modeling, plant cell wall, plant physiology
Introduction
The cell wall of plants is an extremely organized, multifunctional, and dynamic compartment. Among many others, its functions include structural support of cells, the initiation of cellular diversification during the many life cycles of plants and protection from stress (1–4). The physicochemical structure of the plant cell wall regulates its functional properties and thus needs to be efficiently controlled through the chemical remodeling of its components, which is mainly performed through enzymatic activity (5, 6). The main constituent of plant primary cell walls is pectin, a complex polysaccharide mostly comprising of α-1–4-glycosidic-linked galacturonate monomers that can be methylesterified and form diversely methylesterified linear homogalacturonan polymers (HGs).4 The demethylesterification of HG chains, carried out by pectin methylesterase enzymes (PMEs) (7–9), is a key process for the development and defense of plants (10, 11). Upon the removal of methyl groups, PMEs create negatively charged monomeric units along the HG chains that can be further processed by other enzymes downstream in the enzymatic cascade (6). Thus, PMEs are responsible for regulating the physicochemical properties of the plant cell wall, and being the first enzymes along the cascade, their activity needs to be strongly controlled. Plants control PME activity by expressing highly specialized proteinaceous inhibitors (PMEIs), which bind PMEs through the formation of a 1:1 complex as reported by the crystallographic structure of a PME–PMEI pair (12). From a structural biology perspective, PMEs are carbohydrate-binding enzymes that show a triple β-helix featuring a groove that has the primary role of accommodating HG chains (13, 14). PMEIs, on the other hand, are highly similar to plant invertase inhibitors (15) and show a fully α-helical structure composed of four long helices (αI to αIV), with two of them (αII and αIII) creating the binding interface for the binding groove of PMEs and two others (αI and αIV) providing structural stability to the interface helices. Overall, the four helices of PMEIs form a four-helix bundle. A fifth, N-terminal segment, composed of three short-helices (αa, αb, and αc) has been suggested to promote the dimerization of the inhibitor (16). Nevertheless, inhibition of PMEs can efficiently be promoted by PMEI monomers as suggested by crystallographic evidence (12). The inhibition of PMEs is therefore achieved through a direct obstruction of the binding groove of the protein by helices αII and αIII of the inhibitor, which hinders the ability of the enzyme to bind its natural substrate.
Interestingly, because the chemical regulation of the plant cell wall is crucial for the survival and development of plants, PMEs and PMEIs are expressed with a strikingly high redundancy. A paradigmatic example of this can be found in Arabidopsis thaliana, where 66 PMEs and a similarly high number of PMEIs have so far been identified (17, 18). The reasons behind such a high redundancy are still unknown, although evidence suggests that PME activity strongly influences a wide variety of processes such as tissue development (19), mineral homeostasis, and exposure to toxicity (20), as well as resistance of the plant to pathogens (8). Thus, although showing a similar structure, different PME–PMEI pairs may promote a set of specialized functions that finely orchestrate the physiology of cells during the different life stages of plants. PMEIs have indeed different inhibitory capacities according to the PME that they target and often bind their target in response to particular microenvironmental conditions such as the pH. Recently, the pH-dependent activity of the kiwi PMEI has been tested, reporting on the inhibitory capacity of the protein as a function of its structural stability (21). Although some indications point toward a loss of secondary structure because of the reduction of the S–S bond covalently linking the αΙΙ and αΙΙΙ helices, this reduction can only occur when the pH rises well above neutrality. On the other hand, the changes in intrinsic fluorescence observed above pH 6 may reveal subtle but functionally important conformational variations affecting the stability of the investigated complex (21). Indeed, variations of PME–PMEI interaction at pH values around or above neutrality must occur via a mechanism aiming at destabilizing the binding between the partners.
If compared with the different inhibitory capacity in response to the change of few pH units, the high structural similarity of PMEIs suggests that the pH sensitivity must be related to the change in the protonation state of few residues that can modulate the PME–PMEIs complex stability. To elucidate the determinants of PME inhibition, it is therefore fundamental to employ the strategies able to resolve, at a high resolution, the structure and dynamics of PME–PMEI complexes. Understanding the mechanistic basis of PME inhibition has a strong significance for determining PME function in plants, as well as to finely control the activity of PMEs in industrial processes (22, 23).
To assess the structural determinants of the pH-dependent inhibition of PMEs, we employed a combination of computational and experimental approaches, which included homology modeling that allowed to overcome the lack of structures for the PME and PMEI investigated, molecular dynamics (MD) simulations that assessed the conformational dynamics of the complex in the ns time scale and in vitro characterization of the PME–PMEI complex at different pH values. We investigated the complex between PME3 and PMEI7 from A. thaliana (AtPME3 and AtPMEI7) at pH values of 5 and 7. Τhis PME–PMEI pair was shown to be highly sensitive to pH variations within such a range, and it has been proven to be fundamentally important for plant development (24). We further investigated the mechanism of the pH-dependence of the AtPME3–AtPMEI7 interaction by using several strategies. We identified protonatable residues responsible for the different responsivity of the PME–PMEI complex to changes in the pH. Moreover, MD simulations allowed the identification of a potential mechanism that causes changes in the protonation state of key PMEI protonatable amino acids across the binding interface, hinders their capacity to form intermolecular contacts and instead favors the formation of intramolecular contacts. Such contacts sequestration at the binding interface reduces the stability of the complex and may be a general strategy to achieve a finely tuned pH-dependent inhibition of the target. Overall, by interlacing the high predictive ability of computer simulations with the testing capability of experiments, this study adds an additional layer of comprehension to PME–PMEI association and reveals a mechanism that could be commonly found in other PME–PMEI pairs.
Results
AtPME3 and AtPMEI7 form a pH-dependent reversible complex
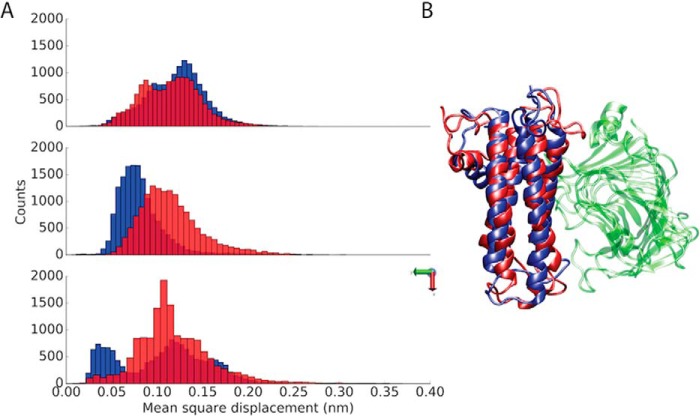
The analysis of the collected MD trajectories was firstly focused on the capacity of the investigated PME and PMEI to stably interact (Fig. S1). To elucidate the stability of AtPME3–AtPMEI7 complex at different pH values, the mean square displacement (MSD) of AtPMEI7 with respect to AtPME3 was monitored in all three dimensions (Fig. 1). A higher MSD of one of the partners, after the removal of the rotational and translational motions of the other molecule involved in the complex, would reveal how the interactions taking place at the complex interface contribute to reduce the diffusive motions of one partner with respect to the other and keep the complex stable. The MSD of AtPMEI7 shows wide oscillations at pH 7 when compared with the motions at acidic pH (Fig. 1), pointing toward a reduced ability of the inhibitor to bind the target at neutral pH. The lower capacity of AtPMEI7 to bind at pH 7 was also investigated as a function of the structural stability of the inhibitor while bound to AtPME3. An analysis of the conformational dynamics of the PMEI helices revealed a consistent twist of helix αII, which is mostly involved in the binding of AtPME3 (Fig. S2). Additionally, the dihedral space explored by helix αII revealed the presence of several populations at pH 7, with values of ψ dihedral angles shifting toward regions of the Ramachandran plot that do not characterize α-helices (Fig. S3). The Ramachandran plot of the residues composing helix αΙ also reports two distinct dihedral populations at the two investigated pH values, even though such a helix does not appear at the binding interface. The root mean square fluctuations (RMSF) of AtPME3 and AtPMEI7 at different pH in the apo and bound states revealed key differences in the dynamics of the two partners (Fig. S4). Compared with the inhibitor, AtPME3 shows lower fluctuations. AtPMEI7, on the other hand, not only shows higher fluctuations but a remarkably diverse behavior at the two different pH values investigated. Although at pH 5 the RMSF profile of the bound AtPMEI7 suggests higher dynamics for the loops connecting helices αI to αII and αII to αIII, upon binding with AtPME3 this trend is reversed and at pH 7 a higher RMSF can be seen for the loops that interact with AtPME3.
Figure 1.
Stability of the AtPME3–AtPMEI7 complex at acidic and neutral pH. A, distributions of the MSD for AtPMEI7 bound to AtPME3 at pH 5 (blue) and pH 7 (red). The MSD is shown in the three dimensions. B, the average conformations of the AtPMEI7 bound to AtPMEI3 at pH 5 (blue) and pH 7 (red) is shown, with the proteins depicted using a cartoon representation.
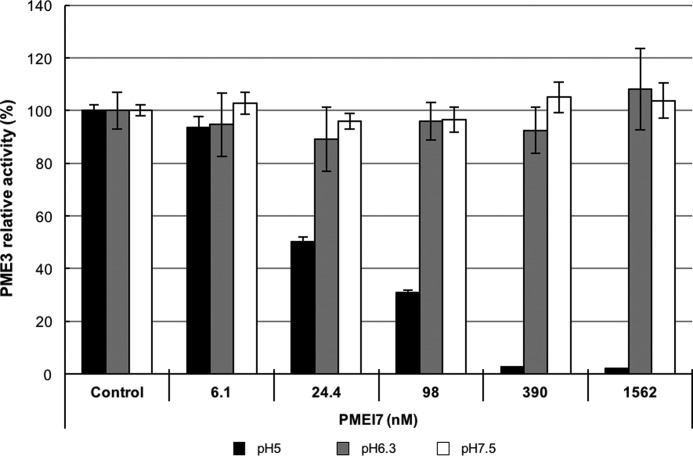
Following the discovery that AtPME3 and AtPMEI7 interact in a pH-dependent manner (21, 24, 25) and the observations made by MD simulations, we then experimentally tested the reversibility of the complex formed at pH 5. For this purpose, we incubated AtPME3 (333 nm) and AtPMEI7 at various concentrations ranging from 1.56 μm to 6.10 nm, for 30 min and at pH 5 to allow complex formation as described by Sénéchal et al. (24). The complex was subsequently transferred into buffers at different pH levels (pH 5, 6.3, and 7.5). As illustrated in Fig. 2, the inhibition of the AtPME3 activity at pH 5 is still total when using concentrations of AtPMEI7 above 390 nm, which corresponds to the formation of a complex at a 1:1 ratio. In contrast, when the complex is transferred at pH 6.3 and 7.5, AtPME3 activity levels were similar to that of the control, showing that the AtPME3–AtPMEI7 complex formed at pH 5 can be reversibly broken when the pH is not in favor of complex stability, ultimately suggesting that the pH-dependent formation of the complex is coupled to the establishment of key interactions between protonatable chemical groups.
Figure 2.
Reversibility of the AtPME3–AtPMEI7 complex. AtPME3 (333 nm) and increasing concentrations of AtPMEI7 were mixed for 30 min at pH 5 to induce complex formation, before being transferred to gel diffusions at pH 5 (black), pH 6.3 (gray), and pH 7.5 (white). AtPME3 activity was measured as the halo diameter revealed by ruthenium red staining after 16 h of incubation time, and the results are presented as the percentages of control activity (AtPME3 incubated without AtPMEI7). The results are the means ± standard deviation of two independent replicates.
Key acidic residues modulate the pH-dependence of the AtPME3–AtPMEI7 association through a competitive establishment of intramolecular and intermolecular contacts
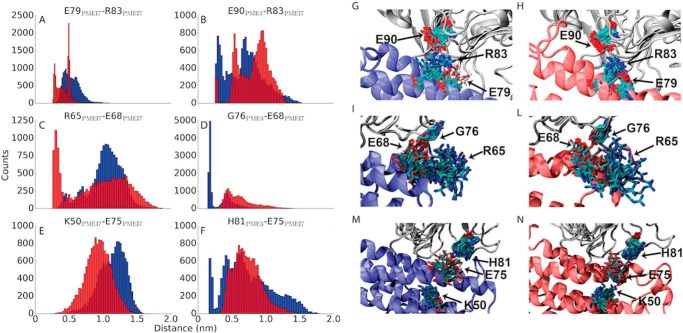
The analysis of MD simulations was then performed to understand the role of protonatable residues at the binding interface. To achieve a pH-dependent inhibition of the target, AtPMEI7 and/or AtPME3 must undergo a protonation switch for one or more residues across the pH range at which inhibition takes place. Importantly, re-incubating the sample at pH values where the inhibitory capacity of AtPMEI7 is not expected, fully restores AtPME3 activity (Fig. 2), suggesting the reversibility of AtPME3–AtPMEI7 complex formation. We found that three acidic residues on the αΙΙ helix of AtPMEI7 are able to considerably shift their contact space at the two different pH investigated: these residues are Glu-79AtPMEI7, Glu-68AtPMEI7, and Glu-75AtPMEI7 (Fig. 3). Although Glu-79AtPMEI7 does not directly establish contacts with AtPME3, its deprotonation at pH 7 favors the establishment of a stable intramolecular salt bridge with the vicinal Arg-83AtPMEI7. Upon raising the pH, Arg-83AtPMEI7 loses the intermolecular contacts established at pH 5 with Glu-90AtPME3. The formation of the intramolecular salt bridge Glu-79AtPMEI7–Arg-83AtPMEI7 at pH 7 hinders the establishment of contacts with the binding partner, hence partially destabilizing complex formation with the rise in pH. This can be evinced from the analysis of intramolecular versus intermolecular distance distributions (Fig. 3). At pH 5 the distance between Glu-90AtPME3 and Arg-83AtPMEI7 is lower than at pH 7. This trend is reversed by raising the pH, because at neutral pH, the distance between Glu-79PMEI7 and Arg-83AtPMEI7 becomes lower (Fig. 3, A and B) with conformer populations shifted toward the establishment of a salt bridge rather than a hydrogen bond (Fig. 3, G and H). Importantly, this mechanism of competition between intramolecular and intermolecular contacts is not isolated to a single pair of residues but can also be observed for the other two acidic residues that, conversely to Glu-79AtPMEI7, directly establish contacts with AtPME3. At pH 5 Glu-68AtPMEI7 establishes a strong hydrogen bond with the backbone carbonyl oxygen of Gly-76AtPME3, whereas the change in protonation at pH 7 favors the establishment of an intramolecular salt bridge with Arg-65AtPMEI7. The distance distributions defining the contact between these two residues are also swapped at the two different pH values investigated (Fig. 3, C and D). Similarly, Glu-75AtPMEI7 establishes a strong (distance between interacting residues of ∼0.2 nm) hydrogen bond with His-81AtPME3. Such a contact is lost because the carboxylate group created at pH 7 upon Glu-75AtPMEI7 deprotonation interacts with Lys-50AtPMEI7 located on the αΙ helix (Fig. 3, E and F).
Figure 3.
Competition between intramolecular and intermolecular contacts in titratable residues of AtPMEI7. A–F, distributions of intramolecular (within AtPMEI7) (A, C, and E) and intermolecular (B, D, and F) distances for residues involved in modulating AtPME3–AtPMEI7 complex stability at pH 5 (blue) and pH 7 (red). G–N, conformer populations of the residues involved in the intramolecular contacts as sampled in MD simulations. Conformers are shown every ns of the collected trajectories. AtPME3 is shown in gray, and AtPMEI7 is shown in blue for pH 5 and in red for pH 7, respectively.
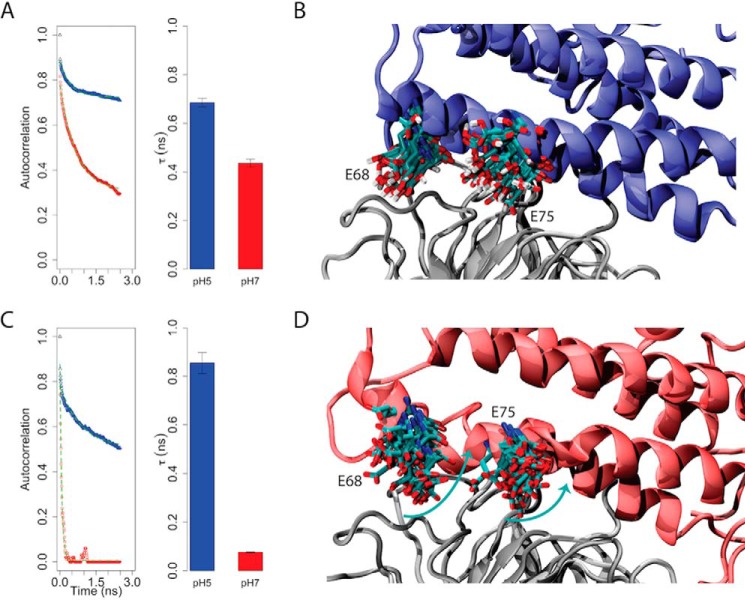
A subsequent analysis of the ability of Glu-68AtPMEI7 and Glu-75AtPMEI7 to establish hydrogen bonds with AtPME3 revealed that these residues may play an important role in defining complex stability and formation. The autocorrelation function calculated for the monitored hydrogen bonds made by Glu-68AtPMEI7 and Glu-75AtPMEI7 reveals a faster decay at pH 7 with different magnitudes observed for the two residues. Although involvement of Glu-68AtPMEI7 in establishing hydrogen bonds drops only slightly with the raise of pH, for Glu-75AtPMEI7 the same autocorrelation is fully lost within the early nanosecond time scale, suggesting a stronger contribution of Glu-75AtPMEI7 to the stability of the complex (Fig. 4). Overall, these observations more generally suggest that the competition between intramolecular and intermolecular contacts of the partners may destabilize, at higher pH values, the binding interface changing the half-life of the complex.
Figure 4.
Role of AtPMEI7 acidic residues for pH-dependent interaction with AtPME3. A and C, left panel, autocorrelation functions of the hydrogen bonds made between Glu-68 (A) and Glu-75 (C) of AtPMEI7 and AtPME3 at pH 5 and 7 during MD simulations. Right panel, half-life (τ) of the contacts made by Glu-68 at pH 5 (blue) and pH 7 (red). The half-life has been calculated by fitting each autocorrelation function shown in the left panel by using a single exponential function y = e−x/τ. The fit is shown in each panel by the dashed green line. B and D, representative conformers for the acidic residues (Glu-68 and Glu-75) involved in contacts with AtPME3 (gray) at pH 5 (B) and pH 7 (D). The turquoise arrows in D resemble the change in conformer population of Glu-68 and Glu-75 at pH 7.
Mutated forms of AtPMEI7 are altered in their inhibiting capacity toward orange PME and PME from root cell wall extracts
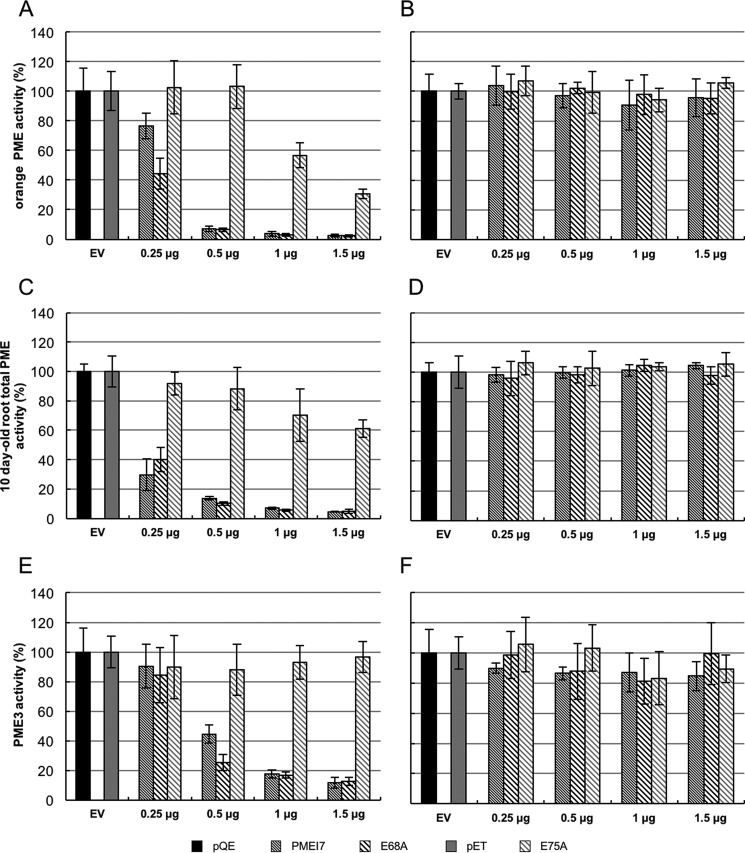
To investigate the role of residues predicted by MD simulation as potentially involved in the pH-dependence inhibition of AtPME3, either wild-type AtPMEI7 or their mutated forms E68A and E75A were expressed in Escherichia coli. Purification was achieved through His-tag affinity chromatography of recombinant wild-type and mutant proteins followed by Western blot analysis using antibodies raised against the His tag. This showed a clear band at molecular mass ranging from ∼20 to 33 kDa, visible for wild-type and mutated forms of AtPMEI7 and absent in the empty vector (pQE and pET) (Fig. S5, A and B). Excision of clear bands on resolving SDS-PAGE, followed by trypsin digestion and peptides identification by nano-LC-ESI-MS/MS, confirmed the production and purification of recombinant wild-type and mutated forms of AtPMEI7. Moreover, substitution of a Glu residue to an Ala residue in E68A and E75A was validated by the identification of alanine in the corresponding peptides (Fig. S5C). Purified fractions were used for PME activity inhibition tests on orange PME and on total PME activity from 10-day-old roots cell wall–enriched protein extract. Increasing quantities of recombinant wild-type and mutated forms of AtPMEI7 were incubated for 30 min in the presence of 1 milliunit of PME activity and then loaded into the wells of gel diffusion at pH 5 and 7 for orange PME (Fig. 5, A and B) and root PMEs (Fig. 5, C and D). At pH 7, neither orange nor root PMEs were inhibited with whichever AtPMEI7 form was used (Fig. 5, B and D). On the contrary, at pH 5, PMEI7 mutated version E68A (as the wild-type protein) is able to inhibit orange PME activity, whereas E75A differs in this ability as shown in Fig. 5A. These differences were even more striking when considering the inhibition of PME activity from root extracts (Fig. 5C). When using 1 μg of wild-type or mutated forms of AtPMEI7, the inhibition of PME activity from roots was ∼30, 90, and 90% for E75A, E68A, and AtPMEI7, respectively. The same figures were ∼50, 90, and 90% when considering orange PME (Fig. 5, A and C). Overall, this suggests that AtPMEI7–PME interactions are likely to depend on the PME considered but at the same time suggests that Glu-75 plays an important role in defining the stability of different AtPMEI7–PME complexes.
Figure 5.
Inhibition of PME activity by wild-type or mutated forms of AtPMEI7. Shown are AtPMEI7, Glu-68, and Glu-75 on orange PME activity at pH 5 (A) and pH 7 (B), on total PME activity from 10 day-old root at pH 5 (C) and pH 7 (D), and on purified AtPME3 activity at pH 5 (E) and pH 7 (F). For each of the experiments, 1 milliunit of PME activity was used with a range of 0.5–1.5 μg of recombinant wild-type or mutated AtPMEI7s. The results are the means ± standard deviation. The same variations are shown for three replicates. EV, empty vector.
Glu-75 plays a fundamental role in the pH-dependence of the AtPMEI7–AtPME3 interaction.
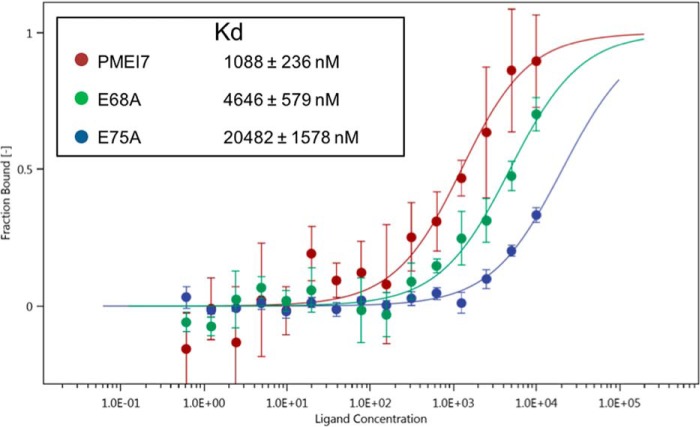
AtPME3 protein was used to assess whether recombinant wild-type or mutated forms of AtPMEI7 differ in their inhibiting capacity toward a purified isoform of the protein expressed by A. thaliana. Increasing quantities of wild-type and mutated AtPMEI7 were incubated for 30 min in presence of 1 milliunit of AtPME3 and loaded into the wells for gel diffusion at pH 5 and pH 7 (Fig. 5, E and F). No inhibition of AtPME3 activity was measured at pH 7, whatever the forms of AtPMEI7 used (Fig. 5F). In contrast, in acidic conditions, E75A appeared inefficient in inhibiting AtPME3 whatever the protein quantities used, thus fully validating the predictions of MD simulations (Fig. 5E and Fig. S6). To overcome cloning difficulties, the E75A construct was cloned in a distinct vector, which led to the production of a protein with a longer N-terminal extension and thus a higher molecular mass (Fig. S5A). To rule out the possibility that the lack of inhibition by E75A could be related to the N-terminal extension of the protein, E75A was digested by enterokinase and, after purification and validation comparing molecular mass and sequence (Fig. S7, A and B), was incubated with AtPME3. As observed for the non-cleaved E75A AtPMEI7, no inhibition of AtPME3 activity was measured at pH 5 (Fig. S7C and Fig. 5E), therefore dismissing a potential conformational issue in the E75A form of AtPMEI7. When using E68A, no differences were observed with the wild-type form of AtPMEI7, thus questioning the real importance of residue Glu-68 in defining the stability of the AtPME3–AtPMEI7 complex. To assess the thermodynamics of the complex between AtPME3 and wild-type or mutated AtPMEI7, we employed microscale thermophoresis (MST) to monitor the changes in the dissociation constant (Kd) at pH 5, because at this pH the maximal inhibition of AtPME3 is observed. Labeled AtPME3 at a constant concentration (333 nm) was titrated with recombinant wild-type or mutated forms of AtPMEI7 (from 10 mm to 0.6 nm), and the in vitro binding affinities were determined. To have an interaction as close to that which was used for the gel diffusion assays, purified proteins in acidic buffer were mixed with 0.1% pectin. From the determined fit curves, the Kd was estimated for each AtPME3–AtPMEI7 pairs (Fig. 6). The obtained results for MST overall corroborated the inhibition tests. Wild-type AtPMEI7 indeed showed the highest binding affinity with AtPME3, with a Kd of ∼1 μm, whereas E75A has the lowest affinity with a Kd larger than 20 μm, which approximately accounts for a 20-fold decrease in affinity. Although the Kd determined for E68A was 4.6 μm, representing a 4-fold increase with respect to the wild-type AtPMEI7 (Fig. 6), no significant change in the inhibiting ability of AtPME3 was measured. This could reflect differences in the sensitivity of the methods and notably the fact that changes in PME activity were measured over a large time course.
Figure 6.
Determination of interaction constants between AtPME3 and wild-type or mutated forms of AtPMEI7 at pH 5. The binding affinities between AtPME3 and wild-type or mutated forms of AtPMEI7 were determined by microscale thermophoresis. For the binding assay, AtPME3 was labeled and was titrated by decreasing concentrations of AtPMEI7s. The fit curves and the resulting dissociation constant (Kd) values were calculated by averaging replicates assimilated using NT analysis software. Kd values represent the means ± standard deviation from six replicates. Concentrations on the x axis are plotted (nm).
Discussion
PME enzymes are critically involved in physiologically relevant remodeling of the plant cell wall and, at the same time, are of industrial interest to extract pectin polysaccharides with a controlled degree of methylesterification (26, 27). Understanding the mechanism leading to their inhibition would both provide opportunities to comprehend how plants develop and engineer the inhibition of PMEs in industrial applications. We have studied the inhibition of PME3 by PMEI7 in A. thaliana, previously described as being pH-dependent, by employing a multidisciplinary approach that involved both computations and experiments. The study pointed toward a mechanism that encompasses the competition between intramolecular and intermolecular contacts in the PMEI. The deprotonation of acidic amino acids would shift the conformer populations of such residues so that intramolecular salt bridges are formed with positively charged residues sitting in the vicinity of titratable residue. The competition of intramolecular and intermolecular interactions has been established to be an important mechanism for regulating protein– binding (28–30) and protein aggregation at low pH (31) and to regulate protein post-translational modifications crucial in cellular signaling (32). This could potentially be a mechanism commonly used by PMEIs that work in a pH-dependent manner and will be the subject of further investigations. Previous investigations on a plant cell-wall invertase inhibitor showed how the protonation shift of a network of acidic residues is majorly responsible for the pH-dependent inhibitory activity of the protein (33). Interestingly, a comparison of the sequences between AtPMEI7 and a recently characterized pH-independent PMEI from A. thaliana (AtPMEI9) reveals that residues Glu-68AtPMEI7 and Glu-75AtPMEI7 involved in the formation of intramolecular contacts at acidic pH are not conserved (25). It is important to mention that the consideration of these two residues as solely responsible for the pH-dependent inhibition of AtPME3 by AtPMEI7 may be too simplistic, especially considering that protonatable residues widely span the binding interface. Nevertheless, simulations were able to pinpoint Glu-75AtPMEI7 as crucial to the pH-dependence of the association. Experiments confirmed lack of inhibition in activity assays and the lower thermodynamic stability of the AtPME3–AtPMEI7E75A complex with an overall 20-fold reduction of affinity between the partners. Interestingly, the thermodynamic stability of the E68A mutant in complex with AtPME3 is in apparent contradiction to what the activity profiles of AtPME3 show because AtPMEI7E86A did not show a significantly different inhibitory capability compared with the wild type, even though van't Hoff plots report on a reduced Kd of ∼4-fold. This could be related to the time scale of the experiments (15 min to obtain the Kd and overnight to determine PME activity). On the other hand, the RMSF profiles observed for the apo and unbound AtPMEI7 states, with the peculiar lowering of dynamics observed at pH 7, are most likely dictated by the establishment of a series of interactions that enhance the binding at lower pH, whereas the fluctuations of such loops eventually increase instability also having an effect at the binding interface. This finding may point toward a more generalized and less specific strategy to destabilize PME–PMEI complexes, because it could involve tuning, upon mutations, the dynamics of the loops connecting the α-helices of the inhibitor that participate in the complex. These outlined findings highlight, on one side, the importance of approaching the study of PME–PMEI complexes using a multidisciplinary approach that can unveil different biochemical, structural, and dynamical features of the investigated systems at once and on the other side to elucidate potential strategies adopted by PMEIs to inhibit their partners. Ultimately, the detailed comprehension of the molecular determinants that finely regulate the activity of PMEIs in different microenvironments will allow us to understand how protein–protein interactions can be so tightly regulated within a highly redundant proteome and will improve the comprehension of the physiological processes of plants in doing so, providing the means for the use of PMEIs in industrial processes of economic interest.
Experimental procedures
Homology modeling and molecular dynamics simulations of AtPME3, AtPMEI7, and AtPME3–AtPMEI7 complexes
To perform MD simulations of the AtPME3–AtPMEI7 complex, models were created through homology modeling by using as a template the PME from tomato (Solanum lycopersicum) and the PMEI from kiwi fruit (Actinidia chinensis), which have been previously co-crystallized into a 1:1 complex (12). MD simulations were performed at pH 5 and 7 by adjusting the protonation state of protonatable residues for both partners. The pKa for all the protonatable side chains were calculated through an empirical approach using the software PROPKA version 3 (34, 35), and hydrogen atoms were added according to the calculated protonation states. Molecular topologies were obtained using the Amber99-sb*-ILDN force field (36), and the complex was then placed into a cubic box of dimensions 15 nm3. The box was then solvated using TIP3P water molecules (37). At each pH, adding Na+ or Cl− counter ions neutralized the net charge of the system. Additionally, sodium and chloride ions were added to reach a final salt concentration of 0.15 m. The complex was subsequently minimized by using a steep descent algorithm with a tolerance of 10−6 kJ mol−1 nm−1 until convergence within the set tolerance was reached. Equilibration was then achieved in two steps. First, in a NVT ensemble, the temperature of the system was coupled to a value of 300 K using the V-rescale thermostat (38). In this step the velocity of each particle composing the system was assigned randomly and followed a Boltzmann distribution. Second, a NpT equilibration step in which pressure and temperature were kept constant at the values of 1 atm and 300 K, respectively was performed. The pressure was kept constant by using the Parrinello–Rahman barostat (39) and coupled every 0.5 ps. Both NVT and NpT steps had a duration of 500 ps, in which protein atoms were subjected to positional restraint through the application of a restraining harmonic potential of 1000 kJ mol−1 in each dimension. Subsequently to reach the equilibration of the complex, 50-ns-long production MD runs were performed in three replicates at each investigated pH using a 2-fs time step. A cutoff of 1 nm was used to sample 12-6 Lennard–Jones and electrostatic interactions in direct space. The latter were computed using the particle mesh Ewald summation method in Fourier space over the above mentioned cutoff.
Non-bonded interactions were sampled by building a neighbor list every 10 ps, and the Verlet cutoff scheme was used to compute interparticle interactions (40). During production runs, temperature and pressure were coupled every 0.5 ps to the values of 300 K and 1 atm using a V-rescale thermostat and a Parrinello–Rahman barostat, respectively. The analysis of the collected trajectories was performed on the last 30 ns as the first 20 ns were considered as equilibration time by using tools available in the GROMACS suite or built in-house. Images of the proteins were created using the Visual Molecular Dynamics (41) and the Chimera (42) visualization packages.
Cloning, expression, and purification of recombinant proteins
Cloning, expression, and purification of AtPME3–6×His from a transgenic tobacco line was performed as described by Sénéchal et al. (43). In the text, purified AtPME3–6×His are referred to as either “AtPME3” or “recombinant AtPME3.” The wild-type AtPMEI7 construct used for bacterial expression (pQE-30 vector; Qiagen) has been described previously (24). The codons for Glu-68 and Glu-75 in the cDNA of AtPMEI7 were mutated into Ala to obtain two simple mutants (E68A and E75A). The E68A mutant sequence was cloned into pQE-30 vector, and E. coli strain Rosetta-gami was transformed. To obtain E75A mutant protein, the E75A mutated sequence was subcloned in pMK vector (Invitrogen). The recombinant pMK vector was digested with BamHI and SalI, and the E75A mutant sequence was subsequently inserted into the same sites of pET-30a(+) vector (Novagen). This was necessary to overcome cloning difficulties in pQE vector. For the E75A mutant, E. coli strain Rosetta-gami 2 was used as an expression strain. E. coli bacteria were grown in LB medium at 37 °C to reach an A600 of 0.6 with 100 μg/ml ampicillin, 12.5 μg/ml tetracyclin, 50 μg/ml streptomycin, and 15 μg/ml kanamycin for Rosetta-gami strain carrying 6×His-AtPMEI7 and 6×His-E68A constructs or with 50 μg/ml kanamycin, 12.5 μg/ml tetracyclin, and 50 μg/ml streptomycin for Rosetta-gami 2 strain carrying the 6×His-E75A construct. The expressions of the fusion proteins were induced by the addition of 0.2 mm isopropylthio-β-galactoside and subsequently grown at room temperature for 17 h. The bacterial pellet was harvested by centrifugation. Protein extraction was performed by resuspension and incubation of this pellet in a lysozyme solution (1 mg/ml), and this step was followed by sonication. The lysate was collected by centrifugation and used to perform purification using nickel-nitrilotriacetic acid-agarose resin (Qiagen, catalog no. 30210). Purification was performed by elution at pH 8 using a buffer at containing 50 mm Na2HPO4, 300 mm NaCl, and increasing concentrations of imidazole from 40 to 500 mm. Clear purified fractions were used for all experiments. In the text, purified 6×His-AtPMEI7, 6×His-E68A, and 6×His-E75A are referred to as AtPMEI7, E68A, and E75A, respectively or, more generally, as recombinant AtPMEI7.
Protein analysis
For Western blot and proteomic analyses, the proteins were resolved using a 15% acrylamide/bisacrylamide SDS-PAGE, using a running buffer composed of 25 mm Tris-base, 192 mm glycine, and 0.1% SDS at pH 8.7. After electrophoresis and washes, proteins were stained with PageBlue protein staining solution (Thermo Scientific, catalog no. 24620) and destained with distilled water for proteomic analysis or directly transferred on Hybond-P PVDF transfer membrane (GE Healthcare, catalog no. RPN303F) for Western blot analysis. To identify recombinant wild-type and mutated forms of 6×His-AtPMEI7, proteins were transferred from resolving gel to PVDF blotting membrane using appropriate cathode and anode buffers and a Trans-Blot TURBO transfer system (Bio-Rad, catalog no. 170-4155) at 25 V and 1 A for 30 min. Transferred proteins were incubated for 1 h at room temperature under shaking with 1:3000 dilution of commercial primary antibody raised against His tag and directly coupled with peroxidase (Sigma, catalog no. A7058). The immunoblot analysis was performed using the DAB substrate for peroxidase (Thermo Scientific, catalog no. 34002) following the supplier's manual. To dismiss the hypothesis that the occurrence of the N-terminal extension of E75A construction could influence the property of the inhibitor, a digestion by enterokinase was realized. After fixation of the crude cell lysate onto nickel-nitrilotriacetic acid resin as described previously, a buffer composed of 25 mm Tris-HCl, 50 mm NaCl buffer, 2 mm CaCl2 at pH 7.5 with 20 units of enterokinase (Bio Basic Inc., catalog no. RC572) was added to the resin. After overnight incubation, at room temperature, the resin was separated by centrifugation, and the eluate was analyzed by SDS-PAGE, immunodetection, and LC-ESI-MS/MS. Inhibition test with E75A with and without the N-terminal extension was also undertaken. For peptide identification by LC-ESI-MS/MS, excision of bands from the gel and trypsin hydrolysis were performed, and all digested peptide mixtures were separated online using nano-LC and analyzed by nano-electrospray tandem mass spectrometry according to the method previously described (43). The experiments were performed on an Ultimate 3000 RSLC system coupled with an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific).
Inhibition test of PME activity by gel diffusion assay
Inhibition of PME activities by recombinant wild-type and mutated forms of AtPMEI7 were quantified by a gel diffusion assay (43) at various pH levels with some modifications (44). For assays on total PME activity, protein extraction from Arabidopsis 10-day-old roots was performed according to a previously published method (45): 50 mg of fine powder of frozen material was mixed with 50 mm sodium phosphate dibasic buffer, containing 20 mm citric acid, 1 m NaCl, and 0.01% Tween 20, at pH 7.0, for 1 h at 4 °C with shaking. The extracts were clarified by centrifugation at 20,000 × g for 30 min at 4 °C. For the assays on purified PMEs, commercial orange PME (Sigma, catalog no. P5400) and Arabidopsis AtPME3 were used (24). Protein concentrations were quantified by the Bradford method (46), using a protein assay dye reagent concentrate kit (Bio-Rad, catalog no. 500-0006). To load identical activities (milliunits or nmol/min) in each well, quantification of PME activities were performed as described in Baldwin et al. (47). For each experiment, 1 milliunit of orange PME, of AtPME3, and of total PME activity from 10-day-old root cell wall–enriched protein extracts in a volume of 5 μl were preincubated for 30 min at room temperature with 5 μl of wild-type or mutated forms of AtPMEI7s at different concentrations. Then the reaction mixtures were loaded into each well in the gels containing 0.1% of pectin from citrus at degree of methylesterification > 85% (Sigma, catalog no. P9561) and prepared at pH 5, 6.3, or 7. To assess the reversibility of the complex, AtPME3 and wild-type AtPMEI7 were mixed for 30 min at pH 5 in the same concentration that previously described (24) to induce complex formation. After incubation, the mixes were loaded on the gel diffusion at three different pH levels: 5, 6.3, and 7.5. After incubation at 37 °C for 16 h, gel was stained with a 0.02% (w/v) ruthenium red solution for 1 h and washed with distilled water to reveal PME activity. Diameters of the red halos around each well were measured using ImageJ software (48–50), and PME activity based on red-stain diameter was quantified using a standard curve, derived from commercial orange PME (Sigma, catalog no. P5400), as previously described (51).
Molecular interaction assay and determination of thermodynamic parameters
Molecular interaction between purified AtPME3 and wild-type or mutated forms of AtPMEI7s, as well as thermodynamic parameters, were analyzed using MST, according to the method previously described (52–55). For each experiment, lysine residues of AtPME3 were labeled, according to manufacturer's recommendations, with the Monolith protein labeling kit blue NHS amine reactive (NanoTemper, catalog no. MO-L003) and conserved in a 50 mm sodium phosphate buffer at pH 7.5 containing BSA 0.1%. Wild-type or mutated forms of AtPMEI7 were transferred in a 50 mm sodium acetate buffer at pH 5 with 300 mm NaCl using a PD SpinTrap G-25 column (GE Healthcare, catalog no. 28-9180-04) following the manufacturer's protocol. For all experiments, a constant concentration of labeled AtPME3 (333 nm) was titrated with decreasing concentrations of non-labeled recombinant AtPMEI7 from 10000 nm to 0.6 nm and mixed with pectins from citrus (degree of methylesterification > 85%, Sigma, catalog no. P9561) at a 0.1% final concentration. The resulting mixtures were incubated 15 min at room temperature and then loaded into a Monolith NT.115 series capillaries standard treated (NanoTemper, catalog no. MO-K002). Thermophoresis experiments were performed with 40% of MST power and 80% of LED power for fluorescence acquisition, according to the method previously described by Sénéchal et al. (24). Room temperature was kept constant at 22 °C for comparisons of the molecular interactions between AtPME3 and various forms of AtPMEI7.
Author contributions
F. S., O. H., L. H., J.-M. D., P. M., V. L., J. P., and D. M. data curation; F. S., O. H., L. H., J.-M. D., P. M., V. L., J. P., and D. M. formal analysis; F. S., O. H., L. H., J.-M. D., P. M., V. L., J. P., and D. M. investigation; F. S., O. H., L. H., J.-M. D., P. M., V. L., J. P., and D. M. methodology; F. S., O. H., L. H., J.-M. D., P. M., V. L., J. P., and D. M. writing-original draft; J. P. and D. M. conceptualization; J. P. supervision; J. P. funding acquisition; J. P. and D. M. project administration; J. P. and D. M. writing-review and editing.
Supplementary Material
This work was supported by Agence Nationale de la Recherche Projects ANR-12-BSV5-0001 GALAPAGOS and ANR-14-CE34-0010-03 PECTOSIGN and by grants from the Conseil Régional de Picardie (to F. S.) and the Institut Universitaire de France (to J. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7.
- HG
- homogalacturonan polymer
- PME
- pectin methylesterase
- PMEI
- pectin methylesterase inhibitor
- MD
- molecular dynamics
- MSD
- mean square displacement
- RMSF
- root mean square fluctuation(s)
- ESI
- electrospray ionization
- MST
- microscale thermophoresis.
References
- 1. Le Gall H., Philippe F., Domon J.-M., Gillet F., Pelloux J., and Rayon C. (2015) Cell wall metabolism in response to abiotic stress. Plants 4, 112–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hamann T. (2012) Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tucker M. R., and Koltunow A. M. (2014) Traffic monitors at the cell periphery: the role of cell walls during early female reproductive cell differentiation in plants. Curr. Opin. Plant Biol. 17, 137–145 [DOI] [PubMed] [Google Scholar]
- 4. Houston K., Tucker M. R., Chowdhury J., Shirley N., and Little A. (2016) The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levesque-Tremblay G., Pelloux J., Braybrook S. A., and Müller K. (2015) Tuning of pectin methylesterification: consequences for cell wall biomechanics and development. Planta 242, 791–811 [DOI] [PubMed] [Google Scholar]
- 6. Sénéchal F., Wattier C., Rustérucci C., and Pelloux J. (2014) Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J. Exp. Bot. 65, 5125–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pelletier S., Van Orden J., Wolf S., Vissenberg K., Delacourt J., Ndong Y. A., Pelloux J., Bischoff V., Urbain A., Mouille G., Lemonnier G., Renou J. P., and Höfte H. (2010) A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 188, 726–739 [DOI] [PubMed] [Google Scholar]
- 8. Lionetti V., Cervone F., and Bellincampi D. (2012) Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 169, 1623–1630 [DOI] [PubMed] [Google Scholar]
- 9. Zhao C., Zhao S., Hou L., Xia H., Wang J., Li C., Li A., Li T., Zhang X., and Wang X. (2015) Proteomics analysis reveals differentially activated pathways that operate in peanut gynophores at different developmental stages. BMC Plant Biol. 15, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson C. T. (2016) We be jammin': an update on pectin biosynthesis, trafficking and dynamics. J. Exp. Bot. 67, 495–502 [DOI] [PubMed] [Google Scholar]
- 11. Sun C., Lu L., Yu Y., Liu L., Hu Y., Ye Y., Jin C., and Lin X. (2016) Decreasing methylation of pectin caused by nitric oxide leads to higher aluminium binding in cell walls and greater aluminium sensitivity of wheat roots. J. Exp. Bot. 67, 979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Matteo A., Giovane A., Raiola A., Camardella L., Bonivento D., De Lorenzo G., Cervone F., Bellincampi D., and Tsernoglou D. (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fries M., Ihrig J., Brocklehurst K., Shevchik V. E., and Pickersgill R. W. (2007) Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J. 26, 3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johansson K., El-Ahmad M., Friemann R., Jörnvall H., Markovic O., and Eklund H. (2002) Crystal structure of plant pectin methylesterase. FEBS Lett. 514, 243–249 [DOI] [PubMed] [Google Scholar]
- 15. Scognamiglio M. A., Ciardiello M. A., Tamburrini M., Carratore V., Rausch T., and Camardella L. (2003) The plant invertase inhibitor shares structural properties and disulfide bridges arrangement with the pectin methylesterase inhibitor. J. Protein Chem. 22, 363–369 [DOI] [PubMed] [Google Scholar]
- 16. Hothorn M., Wolf S., Aloy P., Greiner S., and Scheffzek K. (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16, 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pelloux J., Rustérucci C., and Mellerowicz E. J. (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277 [DOI] [PubMed] [Google Scholar]
- 18. Louvet R., Cavel E., Gutierrez L., Guénin S., Roger D., Gillet F., Guerineau F., and Pelloux J. (2006) Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 224, 782–791 [DOI] [PubMed] [Google Scholar]
- 19. Sexton T. R., Henry R. J., Harwood C. E., Thomas D. S., McManus L. J., Raymond C., Henson M., and Shepherd M. (2012) Pectin methylesterase genes influence solid wood properties of Eucalyptus pilularis. Plant Physiol. 158, 531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weber M., Deinlein U., Fischer S., Rogowski M., Geimer S., Tenhaken R., and Clemens S. (2013) A mutation in the Arabidopsis thaliana cell wall biosynthesis gene pectin methylesterase 3 as well as its aberrant expression cause hypersensitivity specifically to Zn. Plant J. 76, 151–164 [DOI] [PubMed] [Google Scholar]
- 21. Bonavita A., Carratore V., Ciardiello M. A., Giovane A., Servillo L., and D'Avino R. (2016) Influence of pH on the structure and function of kiwi pectin methylesterase inhibitor. J. Agric. Food Chem. 64, 5866–5876 [DOI] [PubMed] [Google Scholar]
- 22. Jayani R. S., Saxena S., and Gupta R. (2005) Microbial pectinolytic enzymes: a review. Process Biochem. 40, 2931–2944 [Google Scholar]
- 23. Kashyap D. R., Vohra P. K., Chopra S., and Tewari R. (2001) Applications of pectinases in the commercial sector: a review. Bioresour. Technol. 77, 215–227 [DOI] [PubMed] [Google Scholar]
- 24. Sénéchal F., L'Enfant M., Domon J. M., Rosiau E., Crépeau M. J., Surcouf O., Esquivel-Rodriguez J., Marcelo P., Mareck A., Guérineau F., Kim H. R., Mravec J., Bonnin E., Jamet E., Kihara D., et al. (2015) Tuning of pectin methylesterification: pectin methylesterase inhibitor 7 modulates the processive activity of co-expressed pectin methylesterase 3 in a ph-dependent manner. J. Biol. Chem. 290, 23320–23335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hocq L., Sénéchal F., Lefebvre V., Lehner A., Domon J.-M., Mollet J.-C., Dehors J., Pageau K., Marcelo P., Guérineau F., Kolšek K., Mercadante D., and Pelloux J. (2017) Combined experimental and computational approaches reveal distinct pH dependence of pectin methylesterase inhibitors. Plant Physiol. 173, 1075–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Degraeve P., Saurel R., and Coutel Y. (2003) Vacuum impregnation pretreatment with pectinmethylesterase to improve firmness of pasteurized fruits. J. Food Sci. 68, 716–721 [Google Scholar]
- 27. Demir N., Acar J., Sarıoğlu K., and Mutlu M. (2001) The use of commercial pectinase in fruit juice industry: Part 3. Immobilized pectinase for mash treatment. J. Food Eng. 47, 275–280 [Google Scholar]
- 28. Amrein K. E., Panholzer B., Flint N. A., Bannwarth W., and Burn P. (1993) The Src homology 2 domain of the protein-tyrosine kinase p56lck mediates both intermolecular and intramolecular interactions. Proc. Natl. Acad. Sci. U.S.A. 90, 10285–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greenall A., Willingham N., Cheung E., Boam D. S., and Sharrocks A. D. (2001) DNA binding by the ETS-domain transcription factor PEA3 is regulated by intramolecular and intermolecular protein·protein interactions. J. Biol. Chem. 276, 16207–16215 [DOI] [PubMed] [Google Scholar]
- 30. Griffith L. C. (2004) Regulation of calcium/calmodulin-dependent protein kinase II activation by intramolecular and intermolecular interactions. J. Neurosci. 24, 8394–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Routledge K. E., Tartaglia G. G., Platt G. W., Vendruscolo M., and Radford S. E. (2009) Competition between intramolecular and intermolecular interactions in an amyloid-forming protein. J. Mol. Biol. 389, 776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Calleja V., Alcor D., Laguerre M., Park J., Vojnovic B., Hemmings B. A., Downward J., Parker P. J., and Larijani B. (2007) Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 5, e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hothorn M., Van den Ende W., Lammens W., Rybin V., and Scheffzek K. (2010) Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc. Natl. Acad. Sci. U.S.A. 107, 17427–17432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Søndergaard C. R., Olsson M. H., Rostkowski M., and Jensen J. H. (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 35. Olsson M. H., Søndergaard C. R., Rostkowski M., and Jensen J. H. (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 36. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., and Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berendsen H. J. C., Grigera J. R., and Straatsma T. P. (1987) The missing term in effective pair potentials. J. Phys. Chem. 91, 6269–6271 [Google Scholar]
- 38. Bussi G., Donadio D., and Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 39. Parrinello M., and Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 [Google Scholar]
- 40. Páll S., and Hess B. (2013) A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 184, 2641–2650 [Google Scholar]
- 41. Humphrey W., Dalke A., and Schulten K. (1996) VMD: Visual Molecular Dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 42. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 43. Sénéchal F., Mareck A., Marcelo P., Lerouge P., and Pelloux J. (2015) Arabidopsis PME17 activity can be controlled by pectin methylesterase inhibitor 4. Plant Signal. Behav. 10, e983351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ren C., and Kermode A. R. (2000) An increase in pectin methyl esterase activity accompanies dormancy breakage and germination of yellow cedar seeds. Plant Physiol. 124, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pilling J., Willmitzer L., and Fisahn J. (2000) Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 210, 391–399 [DOI] [PubMed] [Google Scholar]
- 46. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 47. Baldwin L., Domon J.-M., Klimek J. F., Fournet F., Sellier H., Gillet F., Pelloux J., Lejeune-Hénaut I., Carpita N. C., and Rayon C. (2014) Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 104, 37–47 [DOI] [PubMed] [Google Scholar]
- 48. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schindelin J., Rueden C. T., Hiner M. C., and Eliceiri K. W. (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bourgault R., and Bewley J. D. (2002) Gel diffusion assays for endo-β-mannanase and pectin methylesterase can underestimate enzyme activity due to proteolytic degradation: a remedy. Anal. Biochem. 300, 87–93 [DOI] [PubMed] [Google Scholar]
- 52. André T., and Breitsprecher D. (2013) Thermodynamic characterization of DNA hybridization. DNA-DNA Interaction Analysis, NanoTemper Technologies GmbH, Munich, Germany [Google Scholar]
- 53. Griesbach J. (2013) Determination of thermodynamic parameters DG and DH of a small molecule binding to p38 using MST. Protein-Small Molecule Interaction Analysis, NanoTemper Technologies GmbH, Munich, Germany [Google Scholar]
- 54. Jerabek-Willemsen M., André T., Wanner R., Roth H. M., Duhr S., Baaske P., and Breitsprecher D. (2014) MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 1077, 101–113 [Google Scholar]
- 55. Wienken C. J., Baaske P., Rothbauer U., Braun D., and Duhr S. (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.