Abstract
About one-quarter to nearly one-third of the proteins synthesized in the cytosol of eukaryotic cells are integrated into the plasma membrane or are secreted. Translocation of secretory proteins into the lumen of the endoplasmic reticulum or the periplasm of bacteria is mediated by a highly conserved heterotrimeric membrane protein complex denoted Sec61 in eukaryotes and SecYEG in bacteria. To evaluate a possible modulation of the translocation efficiency by secondary structures of the nascent peptide chain, we performed a comparative analysis in bacteria, yeast, and mammalian cells. Strikingly, neither the bacterial SecY nor the eukaryotic Sec61 translocon was able to efficiently transport proteins entirely composed of intrinsically disordered domains (IDDs) or β-strands. However, translocation could be restored by α-helical domains in a position- and organism-dependent manner. In bacteria, we found that the α-helical domains have to precede the IDD or β-strands, whereas in mammalian cells, C-terminally located α-helical domains are sufficient to promote translocation. Our study reveals an evolutionarily conserved deficiency of the Sec61/SecY complex to translocate IDDs and β-strands in the absence of α-helical domains. Moreover, our results may suggest that adaptive pathways co-evolved with the expansion of IDDs in the proteome of eukaryotic cells to increase the transport capacity of the Sec61 translocon.
Keywords: endoplasmic reticulum (ER), intrinsically disordered protein, prion, protein secretion, protein translocation, SecY, Sec61, ER import
Introduction
Around 25–30% of all proteins synthesized in the cytosol of eukaryotic cells are integrated into the plasma membrane or secreted. Membrane integration and import into the lumen of the endoplasmic reticulum (ER)3 is mainly mediated by a heterotrimeric membrane protein complex called the Sec61 complex, which is highly homologous to the SecY translocon in bacteria (reviewed in Refs. 1–5). A major fraction of secretory proteins contains N-terminal signal peptides, which mediate transport of the nascent chain to the Sec61 translocon and initiate translocation into the ER lumen (reviewed in Refs. 3 and 6–13). Failure to deliver secretory or membrane proteins to the ER can have fatal consequences. First, secretory proteins do not reach their final destination (loss of function), and second, these proteins may form cytosolic aggregates with toxic activities (gain of toxic function) because they often contain hydrophobic domains. For example, mislocalization of the cellular prion protein to the cytosol can induce neuronal cell death in mammalian cell culture models and transgenic mice (14–17). Thus, quality control pathways have evolved to rapidly eliminate mistargeted proteins (18–21).
Each secretory protein has an individual signal sequence, although the targeting peptide is usually removed during transmembrane transport and therefore is not part of the mature protein. However, there are differences in ER import efficiency due to the variability of ER signal sequences (reviewed in Refs. 22–25). Context-specific regulation of protein translocation might enable the cell to prevent an overload of protein folding and quality control pathways in the ER lumen (26–28). In addition, it might allow for dual targeting of secretory proteins to other cellular compartments to expand their physiological function (29, 30).
ER signal peptides have an intrinsic activity to mediate ER import. For example, they can be fused to heterologous non-secretory proteins to mediate their ER import. However, we and others demonstrated that intrinsically disordered proteins (IDPs) equipped with authentic N-terminal ER signal peptides failed to be productively imported into the ER lumen of neuronal cells (31–35). This might be related to the lack of structural elements, which have been shown to already form within the ribosomal tunnel and to modulate the translocation process (36–46).
We now present evidence for a deficiency of the Sec61/SecY complex of eukaryotes and bacteria to translocate substrates that are entirely unstructured or composed only of β-strands. Moreover, our study may indicate the evolution of auxiliary translocon components in eukaryotes to expand the transport capacity of the Sec61 complex for secretory proteins with extended unstructured domains.
Results
The Sec61 and SecY complex cannot efficiently translocate intrinsically disordered proteins
Previous studies revealed that productive ER import of IDPs equipped with authentic N-terminal ER signal peptides is greatly impaired in mammalian cells (31, 32, 34, 35). Because the Sec61 complex in eukaryotes is highly homologous to the SecY complex in bacteria (reviewed in Refs. 1–4), the question emerged of whether impaired translocation of IDPs is a general phenomenon of Sec61/SecY-mediated translocation. To address this question experimentally, we studied transport of IDPs through the Sec61 complex in mammalian and yeast cells and the SecY complex in Escherichia coli. We started with two sets of model substrates that are either composed of α-helical domains or entirely intrinsically disordered. These model substrates were equipped with authentic N-terminal signal peptides of the respective organisms. Mammalian constructs contain the signal peptide of mouse PrP, and for secretion in E. coli, the substrates contained the PelB or DsbA signal peptide to target the proteins via either the SecA (mainly post-translational) (47) or SRP-dependent pathway (mainly co-translational) (48) to the SecY complex. The sequences of all constructs used in this study are shown in Table S1. For the analysis of eukaryotic cells, all constructs contained acceptor sites for N-linked glycosylation to monitor productive import into the ER by the appearance of a glycosylated protein fraction (Fig. 1A). Transport of the constructs into the periplasm was analyzed by separating transformed bacteria into periplasmic and cytoplasmic fractions and subsequent Western blotting (Fig. 1B). To monitor the purity of the periplasmic fraction, the cytosolic chaperonin GroEL and the periplasmic maltose-binding protein (MBP) were analyzed in parallel.
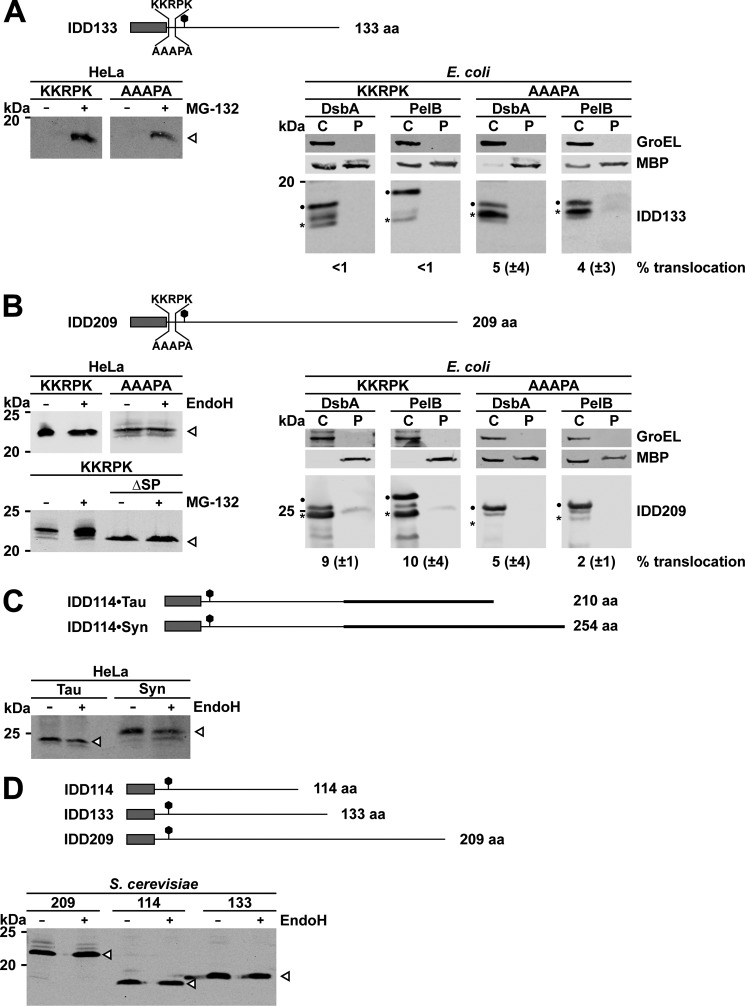
Figure 1.
α-Helical proteins are efficiently transported through the Sec61 or SecY translocon. A and B, experimental strategy to study Sec61- and SecY-mediated translocation in eukaryotic cells and E. coli. Rectangle, signal peptide; helices, α-helical structure; straight line, intrinsic disorder. A, Sec61-mediated translocation in eukaryotic cells. Left, schematic diagram to illustrate the subcellular localization and post-translational modification of the analyzed substrates (1). Proteins containing α-helical domains are translocated through the Sec61 translocon. During translocation, the signal peptide is cleaved, and N-linked glycans are added (2). Intrinsically disordered proteins are not imported into the ER and remain unmodified in the cytosol. Right, schematic diagram of a Western blot analysis. Upon Endo H or PNGase F treatment, the glycans of proteins imported into the ER (1) are cleaved off, leading to an increased electrophoretic mobility during SDS-PAGE (3). Proteins that have not been imported into the ER contain an uncleaved signal peptide, are unglycosylated (2), and migrate slightly more slowly than unglycosylated proteins after retrotranslocation from the ER. B, SecY-mediated translocation in E. coli. Left, schematic diagram to illustrate the subcellular localization of the analyzed substrates (1). Proteins dominated by α-helical domains are translocated through the SecY translocon into the periplasm. Upon translocation, the signal peptide is cleaved (2). Intrinsically disordered proteins are not imported into the periplasm and stay in the cytosol. Respective fractions were separated, and proteins were detected afterward by Western blotting (right). C and D, top, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; α-helical structure derived from PrP is indicated by helices; polygons, N-linked glycosylation acceptor site; gray rectangle, GPI anchor signal sequence. The total length of the proteins is indicated. C, α-helical proteins are efficiently translocated through the Sec61 complex in mammalian cells and the SecY complex in E. coli. Left, cell lysates of transiently transfected HeLa cells were analyzed by Western blotting. One set of cells was cultivated in the presence of the proteasomal inhibitor MG-132 (30 μm, 3 h). To analyze N-linked glycosylation, lysates were treated with PNGase F (+) before Western blotting. White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Right, transformed E. coli expressing the protein either with the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fractions were analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and the periplasmic MBP were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments. D, charged residues interfere with translocation in E. coli, but not in mammalian cells. The naturally occurring polybasic motif (amino acid sequence KKRPK) of PrPC was introduced next to the signal peptide. Expression and translocation in HeLa cells (left) and E. coli (right) was analyzed as described in C.
To validate our experimental strategy, we first analyzed Sec61- and SecY-mediated translocation of the highly structured C-terminal domain (aa 122–254) of the mammalian prion protein (49–51) (Fig. 1C, top). In this context, it is important to note that in vitro experiments indicated that even a PrP fragment comprising only helix 2 and helix 3 shows complete structural autonomy (i.e. it independently adopts an α-helical conformation) (52). Corroborating previous results (31), the α-helical model substrate was modified in HeLa cells with N-linked glycans (i.e. imported into the ER), because it was sensitive to digestion with peptide:N-glycosidase F (PNGase F). The multiple glycosylation bands result from the formation of glycans of complex structure, specifically seen for membrane-anchored PrP constructs (53). After transient inhibition of the proteasome with MG-132, we did not observe stabilization of unglycosylated species, which would be indicative of an incomplete ER import of the α-helical substrate (Fig. 1C, left). Similarly, the α-helical protein was transported through the SecY complex in E. coli. Both the DsbA and PelB signal peptides were proficient in mediating secretion into the periplasm (Fig. 1C, right). Note that some MBP was always found in the cytosolic fraction due to incomplete permeabilization of the outer membrane. Importantly, GroEL was absent in the periplasmic fraction, indicating the purity of this fraction.
A highly conserved feature of PrP is a polybasic motif (KKRPK) located directly after the signal peptide cleavage site (aa 22 in human or mouse PrP). Because charged residues in close proximity to the signal peptide cleavage site may affect translocation (54–56), we were wondering whether such a motif would modulate the translocation efficiency of our substrates. Thus, we inserted the polybasic motif directly after the signal peptide and analyzed secretion. Interestingly, in E. coli, the polybasic motif abrogated translocation (Fig. 1D, right), whereas it had no obvious effect on the translocation into the ER of HeLa cells (Fig. 1D, left).
Next, we analyzed Sec61/SecY-mediated translocation of substrates that are entirely unstructured. We used two model substrates that are either 133 aa (IDD133; Fig. 2A) or 209 aa (IDD209; Fig. 2B) in length. In addition, we generated two versions of each construct, one containing the polybasic motif and the other containing a mutated version to eliminate the positively charged residues (AAAPA). In line with previous studies (31, 32, 35), all unstructured constructs were not imported into the ER of mammalian cells, because we could not detect any glycosylated variants (Fig. 2, A and B, left). The shorter constructs could only be detected after proteasomal inhibition (Fig. 2A, left, + MG-132). The longer constructs were more stable, but the endoglycosidase H (Endo H) digestion revealed that these proteins were not glycosylated (Fig. 2B, left, + EndoH). Similarly to the findings in mammalian cells, secretion of the intrinsically disordered proteins was greatly impaired in E. coli. All proteins were detectable in the cytosol of transformed E. coli, indicating that impaired secretion was not due to lack of expression or rapid degradation in the cytosol (Fig. 2, A and B, right). Interestingly, cleavage of the signal peptide seems to be more efficient in the KKRPK IDD209 constructs (Fig. 2B, right).
Figure 2.
Intrinsically disordered proteins are not efficiently transported through the Sec61 or SecY translocon. A–C, top panels, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; straight line, intrinsic disorder; polygons, N-linked glycosylation acceptor site. The total length of the proteins is indicated. A and B, completely unstructured substrates derived from PrP were expressed in Hela cells (left panels) or E. coli (right panels). One set of proteins contained the polybasic motif (KKRPK) next to the signal peptide. Left, cell lysates of transiently transfected HeLa cells were analyzed by Western blotting. One set of cells was cultivated in the presence of the proteasomal inhibitor MG-132 (30 μm, 5 h). To monitor ER import, lysates were treated with Endo H (+) before Western blotting. White arrowhead, unglycosylated protein fraction. To analyze processing by the signal peptidase in HeLa cells, expression of a mutant lacking the ER signal peptide (ΔSP) was analyzed in parallel. Right, transformed E. coli expressing the protein either with the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fraction was analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and the periplasmic MBP were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments. C, heterologous intrinsically disordered domains do not restore ER import. To study intrinsically disordered domains with different primary structure, we fused the intrinsically disordered domain of Tau (aa 103–197) and α-synuclein (aa 2–140) to the IDD114 derived from PrP. To monitor ER import, lysates were treated with EndoH (+) before Western blotting. White arrowhead, unglycosylated protein fraction. D, intrinsically disordered proteins are not imported into the ER of S. cerevisiae. Expression of three intrinsically disordered proteins derived from PrP S. cerevisiae was analyzed by Western blotting of total cell lysates after treatment with Endo H as described for HeLa cells in A.
In HeLa cells, the unstructured proteins could be stabilized by MG-132, revealing that they were subjected to proteasomal degradation in the cytosol. The proteasome is part of a quality control system designated ER-associated degradation (ERAD), which mediates post-translational degradation of non-native ER proteins after their retrotranslocation back into the cytosol (57–59). ERAD substrates are usually characterized by a cleaved N-terminal signal peptide. To analyze whether this was the case for IDD209, we generated a mutant lacking the N-terminal ER signal peptide (IDD209ΔSP). IDD209 migrated more slowly on SDS-PAGE than IDD209ΔSP, suggesting that IDD209 contained an uncleaved N-terminal ER signal peptide and had not been imported into the ER lumen (Fig. 2B, bottom left). Thus, degradation of IDD209 by the proteasome is obviously not linked to ERAD. To test for the possibility that heterologous intrinsically disordered domains could restore ER import of the unstructured domain of PrP, we fused the intrinsically disordered domains of Tau (Tau40/P301L aa 103–197) or α-synuclein (aa 2–114) to an IDD derived from PrP (IDD114·Tau and IDD114·Syn). Similarly to IDD133 and IDD209, the chimeric proteins were not imported into the ER (Fig. 2C).
Because the translocation efficiency in mammalian cells can be modulated in a signal peptide sequence–specific manner (22–25), we equipped the unstructured protein with the ER signal peptide of rat growth hormone (GH·209), which has previously been shown to be more efficient in promoting translocation than the PrP signal peptide (31, 60). However, productive ER import of the fully unstructured protein in HeLa cells was not restored (data not shown).
To study Sec61-mediated translocation in Saccharomyces cerevisiae, the unstructured model substrates were equipped with the ER signal peptide of the yeast protein Kre5p, which mediates co-translational targeting to the ER. Cell lysates of transformants were prepared and analyzed by Western blotting. In line with the findings in mammalian cells, the electrophoretic mobility of the three different intrinsically disordered proteins was not changed after Endo H digestion, indicating that they have not been imported into the ER of S. cerevisiae (Fig. 2D).
In summary, our studies in bacteria, yeast, and mammalian cells revealed a conserved impairment of the Sec61/SecY complex to translocate substrates that are entirely intrinsically disordered. Aborted translocation seems to be independent of the length of the constructs, the efficiency of the signal peptide, and the targeting mode.
Selective impairment of the SecY complex in E. coli to translocate substrates with extended unstructured N-terminal domains
To test whether translocation of an unstructured domain can be restored, we fused α-helical domains to the C terminus of an unstructured protein. Indeed, Endo H digestion revealed that in HeLa cells, ER import of the unstructured domain was restored by the α-helical domains (Fig. 3A, HeLa, black arrowhead). Note that these constructs are modified only with high-mannose glycans but are secreted (data not shown). The faster migrating bands represent unglycosylated molecules (white arrowhead) that have failed to be imported into the ER. Consequently, this fraction is stabilized in cells incubated with the proteasomal inhibitor MG-132 (data not shown). The same phenomenon was observed in our yeast model. The Western blot analysis of cell lysates prepared from transformed S. cerevisiae showed two prominent bands. After Endo H digestion, the upper band disappeared, indicating that a fraction of the substrate with an extended unstructured domain was imported into the ER (Fig. 3A, S. cerevisiae). In contrast, a C-terminally located α-helical domain was not able to promote SecY-mediated translocation of an extended intrinsically disordered domain in E. coli. Secretion was not improved by using a signal peptide for co-translational targeting, as neither the DsbA nor the PelB signal peptide allowed translocation of this construct into the periplasm (Fig. 3A, bottom, E. coli).
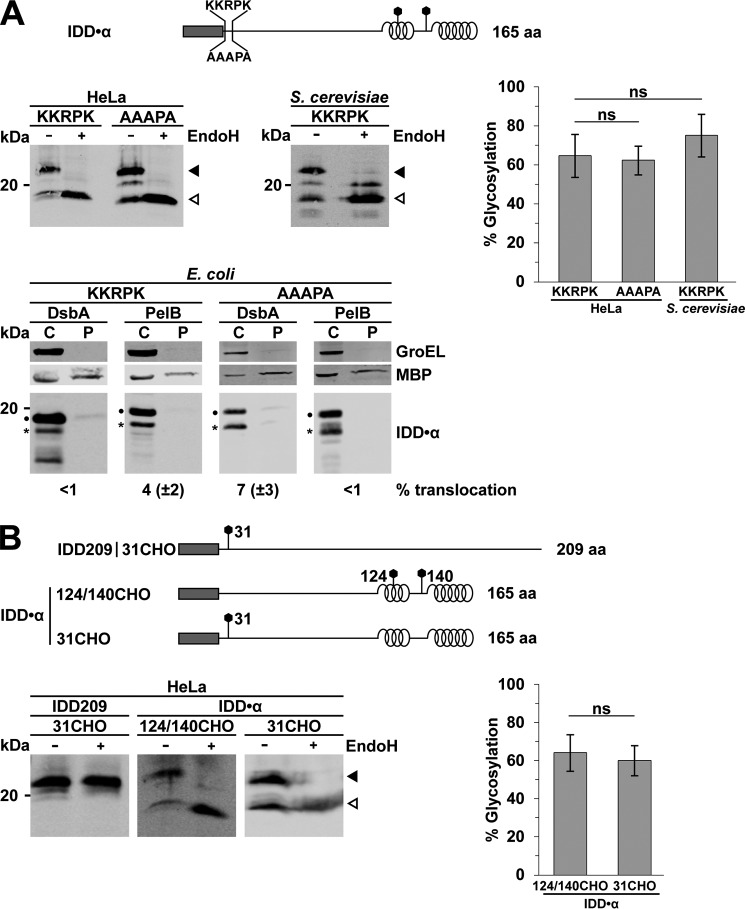
Figure 3.
Secretory proteins with extended N-terminal unstructured domains are not transported through the SecY translocon in E. coli. A and B, top, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; helices, α-helical structure derived from PrP; straight line, intrinsic disorder derived from PrP; polygons, N-linked glycosylation acceptor site. The total length of the proteins is indicated. A, C-terminally located α-helical domains restore translocation of unstructured proteins in eukaryotic cells. A protein composed of an intrinsically disordered domain followed by an α-helical domain was expressed in HeLa cells and S. cerevisiae (top) and in E. coli (bottom). One set of the proteins contained the polybasic motif (KKRPK) next to the signal peptide. To analyze ER import in eukaryotic cells, lysates were treated with Endo H (+) before Western blotting. White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Right, the percentage of the ER-translocated fraction was determined as the ratio between the signal of the glycosylated fractions and the total protein signal. Data represent mean ± S.E. (error bars) of three independent experiments. ns, not significant. Bottom, transformed E. coli expressing the proteins either with the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fractions were analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and the periplasmic maltose-binding protein (MBP) were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments. B, the position of the N-glycosylation site does not modulate glycosylation efficiency. Top, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; helices, α-helical structure derived from PrP; straight line, intrinsic disorder derived from PrP. The N-glycosylation sites (polygons) are either at aa 31 (31CHO) or at aa 124 and 140 (124/140CHO). The total length of the proteins is indicated. Bottom left, the indicated substrates, containing the glycosylation site at different positions, were expressed in transiently transfected HeLa cells. Cell lysates were treated with Endo H (+) before Western blotting to monitor ER import. White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Bottom right, the percentage of the ER-translocated fraction was determined as the ratio between the signal of the glycosylated fractions and the total protein signal. Data represent mean ± S.E. of three independent experiments. ns, not significant.
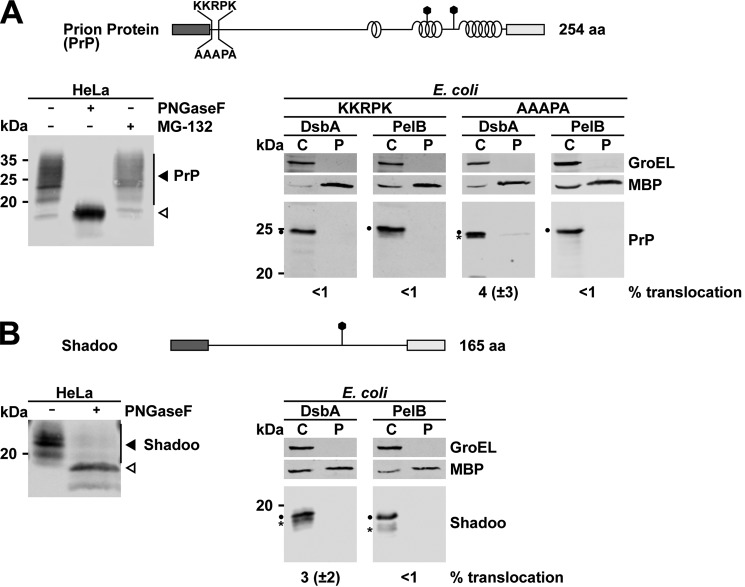
To address the possibility that the localization of the N-glycosylation site within a construct affects the glycosylation efficiency, we analyzed glycosylation of IDD·α constructs with the N-glycosylation site located either at aa 31 (31CHO) or at aa 124 and 140 (124/140CHO) (Fig. 3B). However, the position of the N-glycosylation site has no significant influence on the glycosylation efficiency (Fig. 3B, 124/140CHO versus 31CHO). Based on the impaired secretion of the artificial IDD·α constructs in E. coli, we analyzed secretion of two naturally occurring mammalian proteins in E. coli that are characterized by extended unstructured domains: wild-type prion protein (49, 51) and Shadoo. Shadoo is a highly conserved neuronal glycoprotein present in all vertebrates with a stress-protective activity (35, 61, 62). Of note, full-length PrP is efficiently imported into the ER of mammalian (Fig. 4A, left) and yeast cells (63, 64). In contrast, wild-type PrP equipped with the DsbA or PelB signal peptide was not secreted in E. coli, not even in the absence of the positively charged residues next to the signal peptide (Fig. 4A, right). Similarly, full-length Shadoo, a protein that is efficiently imported into the ER of HeLa cells, is not secreted into the periplasm of bacteria (Fig. 4B).
Figure 4.
Naturally occurring secretory proteins with extended unstructured domains are not secreted in E. coli. A and B, top panels, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; helices, α-helical structure; straight line, intrinsic disorder; polygons, N-linked glycosylation acceptor site; gray rectangle, GPI anchor signal sequence. The total length of the proteins is indicated. The cellular prion protein PrPC (A) and Shadoo (B) were expressed in HeLa cells (left panels) or E. coli (right panels). To analyze ER import in eukaryotic cells, lysates were treated with PNGase F (+) before Western blotting. White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Transformed E. coli expressing the proteins either with the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fractions were analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and periplasmic MBP were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments.
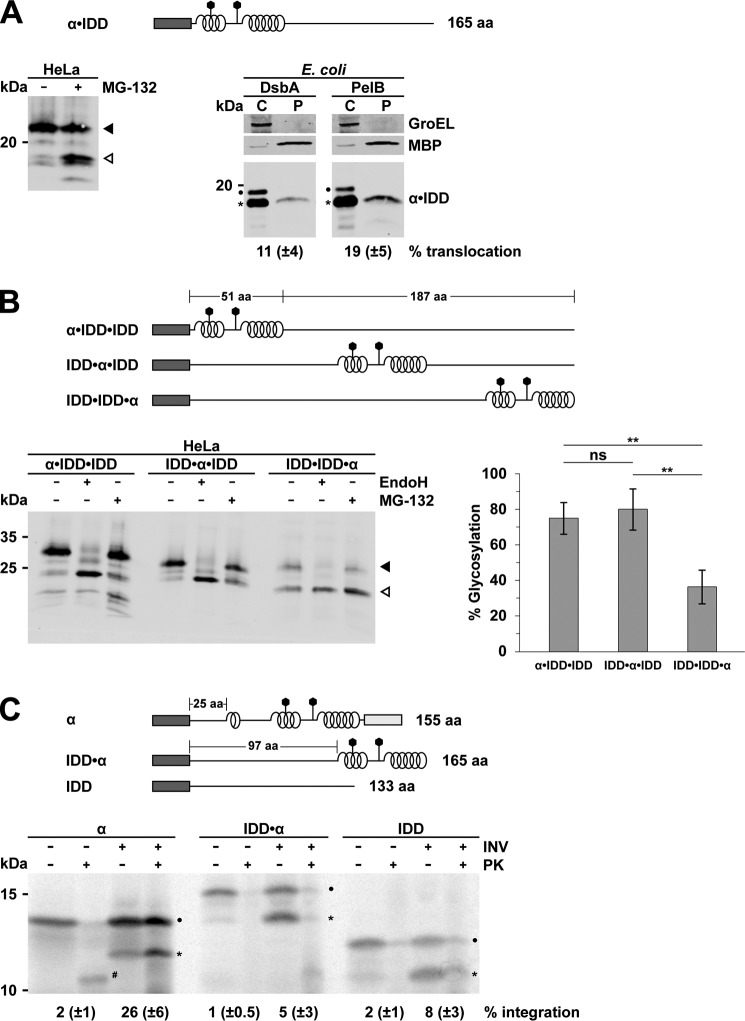
To address the possibility that the localization of an unstructured domain within the protein has an effect on the translocation efficiency, we cloned the domains in reversed order (i.e. the α-helical domains were placed directly after the signal peptide followed by a C-terminally unstructured domain). In mammalian cells, import efficiency was increased by positioning the α-helical domain N-terminal to the unstructured domain. Moreover, this construct was also secreted in E. coli, revealing that extended unstructured domains can be transported through the SecY complex if they are preceded by an α-helical domain (Fig. 5A). However, compared with mammalian cells, the translocation of the α·IDD construct in E. coli was less efficient. Next, we evaluated the role of the position of the α-helical domain within an intrinsically disordered protein in more detail. To this end, we placed the α-helical domains at three different positions into a long unstructured protein: either at the C or N termini or in the middle (Fig. 5B). Remarkably, the analysis in HeLa cells revealed that α-helical domains located 187 aa downstream to the signal peptide are still able to restore ER import of an intrinsically disordered domain (Fig. 5B). To further study impaired translocation of proteins with an unstructured N-terminal domain in E. coli, we employed an in vitro system for SecY-mediated translocation (65). As expected, the α-helical protein was imported into SecY-containing inverted inner membrane vesicles (INVs) (Fig. 5C, α), as indicated by signal sequence cleavage and proteinase K protection. A typical feature of bacterial in vitro systems is that protease K protection is also observed for the non-processed substrate in the presence of INVs. This is related to the low activity of the signal peptidase in the isolated INVs (65, 66). An additional band of low molecular weight (Fig. 5C (#)) was present after proteinase K treatment in the absence of INVs, which most likely corresponds to partially protease-resistant aggregates, but this was not further analyzed. Consistent with our in vivo results observed in E. coli, SecY-mediated translocation in vitro was also significantly impaired for both the completely unstructured protein (IDD) and the construct with an extended N-terminal unstructured domain (IDD·α) (Fig. 5C). For both constructs, only background protease protection was observed. It should be noted that complete membrane-free in vitro systems are almost impossible to generate, as the sucrose-gradient purified ribosomes usually contain some minor membrane contamination (65). This explains the weak protease resistance even in the absence of added INVs. Interestingly, cleavage of the signal peptide seemed to occur for all three proteins, suggesting that the secondary structure did not impair targeting and the initial insertion of the proteins into the translocon but rather their productive translocation. Processing without translocation has been observed before for native E. coli secretory proteins when the translocation activity of the SecYEG translocon is impaired (e.g. by deleting the SecG subunit of the SecYEG translocon) (66). The notion that targeting of the IDDs to the translocon occurs is in line with previous in vitro studies employing stalled ribosome-nascent chain complexes and canine pancreatic microsomal membranes (34).
Figure 5.
α-Helical domains restore translocation of unstructured proteins in a position- and organism-specific manner. A–C, top, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; helices, α-helical structure derived from PrP; straight line, intrinsic disorder derived from PrP; polygons, N-linked glycosylation acceptor site; gray rectangle, GPI anchor signal sequence. The total length of the proteins or individual parts is indicated. A, N-terminally located α-helical domains restore secretion of intrinsically disordered domains in both mammalian cells and E. coli. A protein consisting of an α-helical domain N-terminal to an intrinsically disordered domain was expressed in transiently transfected HeLa cells (left) or in E. coli (right). Cell lysates of HeLa cells (left) were analyzed by Western blotting. One set of cells was cultivated in the presence of the proteasomal inhibitor MG-132 (30 μm, 3 h). White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Transformed E. coli cells expressing the protein either with the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fractions were analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and periplasmic MBP were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments. B, C-terminally located α-helical domains restore secretion of intrinsically disordered domains in mammalian cells. The three proteins depicted were expressed in transiently transfected HeLa cells, and expression was analyzed as described in A. To analyze glycosylation of the proteins, one set of lysates was treated with EndoH (+) before Western blotting. Right, the percentage of the ER-translocated fraction was determined as the ratio between the signal of the glycosylated fractions and the total protein signal. Data represent mean ± S.E. (error bars) of three independent experiments. **, p < 0.005; ns, not significant. C, intrinsically disordered domains interfere with import into E. coli inner membrane vesicles in vitro. The indicated substrates containing a DsbA signal peptide were translated and labeled with l-[35S]methionine in vitro in the presence (+) or absence (−) of E. coli INVs. After translation, samples were treated with proteinase K (+PK) or left untreated (−PK) and analyzed by SDS-PAGE and phosphorimaging. Indicated are the precursor forms of the substrates (dots) and the mature form after cleavage of the signal peptide (asterisks). For the calculation of percentage of integration, the radioactive signals in the −PK and +PK lanes were quantified using the ImageQuant software. The obtained value for −PK was set to 100%, and the value +PK was calculated. This latter value was then corrected for the amount of material that was loaded (i.e. 80% of the total in vitro synthesized material). The mean of at least three independent experiments and the S.D. values are indicated.
These findings indicate that α-helical domains are required in addition to N-terminal signal peptides to mediate translocation of unstructured domains through the Sec61 or SecY translocon. The α-helical domains must be close to the signal peptide in the case of SecY-mediated translocation in E. coli. In eukaryotic cells, α-helical domains promote translocation even if they are located more than 180 aa distal to the signal peptide, although translocation efficiency decreases with increasing distance.
β-Strand proteins are poor substrates for Sec61- and SecY-mediated translocation
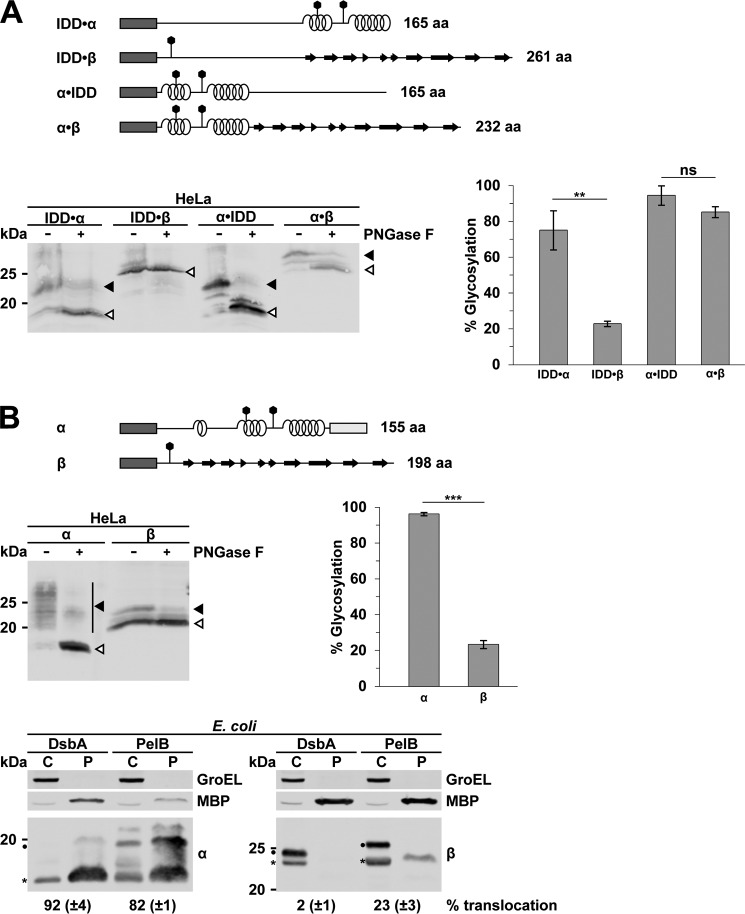
In a next step, we extended our analysis to assess a possible impact of β-strands on ER translocation. We therefore fused a domain composed of β-strands C-terminally to the unstructured domain (IDD·β) and compared ER import efficiency to an IDD·α protein in mammalian cells. Remarkably, the β-strands were markedly less potent in restoring ER import of the IDD compared with α-helical domains (Fig. 6A, IDD·α versus IDD·β). Located C-terminally to α-helical domains, neither β-strands nor an unstructured domain significantly impaired ER import (Fig. 6A, α·IDD versus α·β). To follow up on this observation, we analyzed ER import of two substrates that are dominated by either α-helical domains or β-strands without discernible α-helical domains. Indeed, in eukaryotic cells, ER import efficiency of a protein composed only of β-strands was significantly decreased compared with an α-helical protein (Fig. 6B, α versus β). Similarly, SecY-mediated translocation of the β-strand protein in E. coli was also reduced (bottom) and only seen when the β-strand protein was equipped with a signal peptide for a SecA-dependent targeting (PelB) (Fig. 6B, bottom right). These findings are in line with the observation that β-barrel proteins in E. coli are targeted via the SecA-dependent pathway to the SecY complex. Moreover, when we inspected the secondary structure of the outer membrane porins OmpC, OmpE, and OmpF, we noticed that these proteins contain several short α-helical domains in addition to the β-strands (UniProtKB P06996, P02932, and P02931).
Figure 6.
Impaired translocation of β-strands through the Sec61 and SecY translocon. A and B, top panels, schematic presentation of the constructs analyzed. Dark rectangle, signal peptide; helices, α-helical structure derived from PrP; straight line, intrinsic disorder derived from PrP; arrows, β-strands derived from GFP; polygons, N-linked glycosylation acceptor site; gray rectangle, GPI anchor signal sequence. The total length of the proteins is indicated. A, impaired activity of β-strands to promote ER import of intrinsically disordered domains. HeLa cells were transiently transfected with the constructs indicated and analyzed by Western blotting. To analyze N-linked glycosylation, lysates were treated with PNGase F (+) before Western blotting. White arrowhead, unglycosylated protein fraction; black arrowhead, glycosylated protein fraction. Right, the percentage of the ER-translocated fraction was determined as the ratio between the signal of the glycosylated fractions and the total protein signal. Data represent mean ± S.E. (error bars) of three independent experiments. **, p < 0.005; ns, not significant. B, proteins composed of β-strand are inefficiently transported through the Sec61 and the SecY translocon. The indicated proteins were transiently expressed and analyzed in HeLa cells as described in A. Right, translocation efficiency of constructs was measured densitometrically as described in A. ***, p < 0.0005; NS, not significant. Bottom panels, transformed E. coli cells expressing the proteins with either the DsbA or PelB signal peptide were fractionated, and the cytoplasmic (C) and periplasmic (P) fractions were analyzed by Western blotting. Unprocessed full-length constructs (dots) and constructs after signal peptide cleavage (asterisks) are marked. The cytosolic chaperonin GroEL and the periplasmic MBP were analyzed in parallel. Translocation efficiency is indicated. Data represent mean ± S.E. of at least three independent experiments.
Discussion
In all forms of life, protein secretion is mediated by a highly conserved complex designated Sec61 in eukaryotes and SecY in prokaryotes and archaea (reviewed in Refs. 1–5). In this study, we show that the Sec61/SecY complex is designed for the translocation of client proteins with α-helical structures. Even if equipped with highly potent signal peptides, proteins with β-strands are only less efficiently translocated, whereas intrinsically disordered proteins cannot be productively translocated at all. As a consequence, α-helical domains are required in addition to the N-terminal signal peptide to ensure effective translocation. Furthermore, our study reveals adaptive pathways to enable the transport of substrates with extended N-terminal unstructured domains through the Sec61 translocon in eukaryotic cells.
Impaired translocation of intrinsically disordered and β-strand domains is an evolutionarily conserved feature
In addition to their role to target substrates and initiate transport through the translocon, signal peptides can modulate translocation efficiency in a sequence- and context-specific manner (22–28). Our study was designed to address the question of whether secondary structures within the nascent chain serve as additional regulatory elements. The experimental approach was based on the comparative analysis of a set of substrates of similar length but different secondary structure. Importantly, all proteins were equipped with well-studied signal peptides derived from authentic secretory proteins of the respective organisms. Indeed, our results revealed that secondary structure can significantly modulate translocation efficiency. Specifically, neither the Sec61 complex in mammalian and yeast cells nor the SecY complex in E. coli can efficiently transport client proteins that are completely intrinsically disordered or exclusively composed of β-strands. Increasing the length of these substrates or the use of more efficient signal peptides did not significantly improve ER import or secretion.
The unstructured substrates that failed to be imported contained an uncleaved signal peptide and were subjected to proteasomal degradation in mammalian cells. Thus, we disfavor an interpretation that the proteins were first imported into the ER lumen and then subjected to the ERAD pathway. However, it seems unlikely that the targeting of unstructured proteins to the translocon was impaired. First, in vitro studies employing stalled ribosome nascent chain complexes indicated that also unstructured nascent chains are productively targeted to the Sec61 translocon (34). Second, we used model substrates longer than 200 aa to exclude the possibility that reduced SRP binding to shorter substrates might account for the observed translocation defects. Based on these findings, it seems plausible that unstructured substrates are targeted to the translocon, where the signal peptide initiates early translocation steps. However, translocation is aborted before the signal peptide is cleaved. As a consequence, the unstructured substrate is released from the translocon and subjected to proteasomal degradation. Alternatively, translocon-associated and/or cytosolic components might recognize unstructured domains and β-strands and actively interfere with their translocation. For such a model, however, one has to consider that such components have to be present in bacteria, yeast, and mammalian cells and, at least in eukaryotic cells, spare proteins with an extended (≥180-aa) unstructured N-terminal domain followed by C-terminal α-helical domains (see below). In E. coli, the signal peptides of a fraction of the unstructured proteins are processed before translocation is aborted. This possibility is based on the in vivo experiments in which two protein species are often detected in the cytoplasmic fractions and on the observed cleavage of the signal peptide of all constructs in the in vitro import assays after the addition of inverted inner membrane vesicles.
Another interesting finding of our study was the apparent advantage of the SecA-dependent pathway in E. coli for the secretion of β-strand proteins. In support of this concept, naturally occurring outer membrane proteins, such as OmpC, OmpD, OmpF, OmpE, and OmpN, which are nearly entirely composed of β-strands, are targeted to the SecY complex via SecA.
Efficient translocation requires α-helical domains in addition to the signal peptide
Our study emphasized the importance of α-helical domains in the mature part of the protein to achieve efficient translocation through both the Sec61 and the SecY translocon. This activity was not only seen for model proteins designed for this study but also for naturally occurring secretory proteins with extended unstructured domains in mammalian cells, such as the GPI-anchored proteins PrPC and Shadoo (this study) and the neuropeptide hormones somatostatin and gonadotropin-releasing hormone (34, 35). As mentioned above, targeting of the ribosome-nascent chain complex of unstructured proteins to the translocon was apparently not impaired (34), indicating a role of α-helical domains in a post-targeting event. Which step may be affected? After targeting of the ribosome-nascent chain complex to the translocon, the signal peptide also contributes to the opening of the translocation channel (67–70). Thus, it is conceivable that after the initial gating by the signal peptide, additional α-helical domains are required to keep the pore open and to ensure efficient translocation into the ER lumen. Once the N-terminal domain of a protein is in the ER lumen, the secondary structure of the remaining part is of minor importance because folding of the mature part and interaction with lumenal proteins, such as BiP, provide a driving force for complete translocation. This concept would also explain why α-helical domains are most potent when they are located next to the signal peptide, because then the pore is already in an open conformation. Located further downstream, gating may already have ceased, making it more difficult for the C-terminally located α-helical domain to re-initiate gating.
Notably, the activity of α-helical domains to promote secretion may also be of physiological relevance to SecY-mediated translocation in E. coli. By inspecting the secondary structure of naturally occurring β-barrel proteins, such as OmpF, OmpC, and OmpE, we noticed that in addition to β-strands, short α-helical domains are present at conserved positions.
Adaptive pathways in eukaryotic cells enable Sec61-mediated translocation of secretory proteins with extended N-terminal unstructured domains
Another important finding of our study was that abortive ER import of unstructured domains can be restored by α-helical domains, arguing against a dominant negative activity of these domains during translocation. Moreover, our comparative analysis in bacteria and eukaryotic cells revealed organism-specific differences. Located N-terminally to an unstructured domain, α-helical domains promoted translocation through both the Sec61 complex in mammalian cells and the SecY complex in bacteria. Interestingly, however, the activity of N-terminally located α-helical domains to rescue IDD translocation was rather poor in E. coli compared with the activity in mammalian cells. Located at the C terminus of the unstructured protein, α-helical domains were still proficient in promoting translocation in eukaryotic cells, but not in bacteria. An attractive model to explain the activity of C-terminal α-helical domains in mammalian cells is based on the findings that nascent polypeptides can pause on the cytosolic side of the translocon before productive translocation (43, 45, 71). Indeed, it was shown before that translocation of the unstructured N-terminal domain of the prion protein was delayed until the α-helical domains in the C terminus were synthesized (45). Moreover, delayed translocation induced recruitment of translocon accessory factors, such as Sec62 and Sec63, components that are missing in E. coli. Thus, one could envisage a pathway in mammalian cells allowing secretory proteins with extended unstructured domains to remain at the cytosolic side of the translocon until α-helical domains are synthesized to promote translocation. Bacteria seem to lack such a pathway to handle secretory proteins with extended N-terminal unstructured domains. Interestingly, structural disorder within the proteome increased with the transition from prokaryotic to eukaryotic cells (72), consistent with the notion that intrinsically disordered proteins play a major role in several eukaryotic protein classes including signaling molecules (reviewed in Refs. 73–78). Hence, the apparent translocation deficiency is probably of little relevance to bacteria, because secretory proteins with extended N-terminal unstructured domains have not been described so far in prokaryotes. In eukaryotic cells, adaptive mechanisms may have evolved in parallel with the expansion of intrinsically disordered domains in the proteome to compensate for the inherent translocation deficiency. It will now be interesting to identify these putative translocon-associated pathways in eukaryotic cells and study their role in the regulation of translocation efficiency in a context- and substrate-dependent manner.
Experimental procedures
Constructs/plasmids
Plasmid amplification and maintenance was carried out in E. coli TOP10® (Thermo Fisher Scientific). All PrP mutants are based on the coding region of mouse PRNP (GenBankTM accession number M18070), modified to express PrP-L108M/V111M, allowing detection by the monoclonal antibody 3F4. The following constructs for mammalian expression were described previously: wild-type PrPC, IDD114 (previously designated PrP-A115X/31CHO), IDD209 (previously designated PrP-115/31CHO+115), IDD·α (previously designated PrP·115α2α3), α·IDD (previously designated PrP·α2α3–115), IDD114·Tau (previously designated 115/31CHO+Tau), and IDD114·Syn (previously designated 115/31CHO+αsyn) (32). All other constructs used in this study were generated by standard PCR cloning techniques. In PrP mutants lacking the C-terminal glycosylation sites of PrP, the aa Trp and Asn at position 31/32 were replaced by Asn and Phe to generate an additional glycosylation acceptor site (NFT). If indicated, the polybasic motif KKRPK of PrP (aa 23–27) was changed to AAAPA. Constructs α and α(PB) were created by deleting aa 23–121 and aa 34–121 of PrP, respectively. Construct IDD133 was created by introducing a stop codon (TAA) at aa position 134 of PrP. Constructs α·IDD·IDD, IDD·α·IDD, and IDD·IDD·α were created by fusing unstructured/structured parts of PrP using overlap extension PCR as listed: α·IDD·IDD (aa 1–22 + 171–221 + 23–114 + 23–114), IDD·α·IDD (aa 1–114 + 171–221 + 23–114), and IDD·IDD·α (aa 1–114 + 23–114 + 171–221). The β-sheet–rich construct β was created by fusing aa 1–39 of PrP with aa 92–238 of GFP. Construct α·β was created by fusing aa 1–22 and 171–221 of PrP with aa 92–238 of GFP. Both constructs β and α·β contain C-terminal HA tag for Western blot detection. IDD·β was created by fusing aa 1–114 of PrP (containing glycosylation acceptor site NFT, as described above) with aa 92–238 of GFP. All constructs described above were inserted into pcDNA3.1/Neo(+) vector (Invitrogen). For translocation studies in E. coli, the constructs described above were cloned into pET27b(+) expression vector (Novagen), and the PrP signal peptide was exchanged with the PelB signal peptide (MKYLLPTAAAGLLLLAAQPAMA; GenBankTM reference sequence M17364.1) for SecA-dependent (e.g. PelB·IDD133 = PelB signal peptide + aa 23–133 of PrP) or DsbA signal peptide (MKKIWLALAGLVLAFSASA; NCBI reference sequence NP_418297.1) for SRP-dependent translocation, respectively. For translocation studies in yeast, constructs were cloned into pYES2 expression vector (Thermo Fisher Scientific), and the PrP signal peptide was exchanged with the Kre5p signal peptide (MRLLALVLLLLCAPLRA; NCBI reference sequence NM_001183756.1). The wild-type human Shadoo construct has been described previously (35). For translocation studies in E. coli, the signal peptide was exchanged with DsbA and PelB as described for PrP. All Shadoo constructs are equipped with a V5 tag (5′-GGT AAA CCG ATA CCG AAC CCG CTC CTC GGT CTC GAT TCG ACG-3′) inserted between amino acids 124 and 125. All amino acid numbers refer to the human Shadoo sequence (NCBI reference sequence NP_001012526.2). The primary and secondary structures of all substrates analyzed in this study are listed in Table S1.
Antibodies and reagents
The following antibodies were used: anti-PrP 3F4 monoclonal antibody (79), mouse monoclonal anti-V5 antibody (mAb, R960CUS; Thermo Fisher Scientific), anti-HA (mAb, MMS-101R; Covance), anti-MBP-probe (mAb, sc-32747; Santa Cruz Biotechnology, Inc.), anti-Hsp60 (polyclonal antibody, SC-1052; Santa Cruz Biotechnology), and IR-Dye–conjugated secondary antibody (IR-Dye 800CW donkey anti-mouse/anti-goat; LI-COR). All standard chemicals and reagents were purchased from Sigma-Aldrich if not otherwise noted. The following reagents were used: Endo H (New England Biolabs), PNGase F (New England Biolabs), cOmplete® Mini EDTA-free protease inhibitor mixture (Roche Applied Science), aproptin, benzamidine, pepstatine, and leupeptin (all from MP Biomedicals).
Studies in mammalian cells
Human HeLa cells were cultured in DMEM (Thermo Fisher Scientific) with the addition of 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. They were grown in a humidified 5% CO2 atmosphere at 37 °C. Cells cultivated on a 3.5-cm cell culture dish (Nunc, Roskilde, Denmark) were transfected with a total of 1 μg of DNA by a liposome-mediated method, using Lipofectamine Plus® reagent (Life Technologies) according to the manufacturer's instructions. For proteasomal inhibition cells were treated with 30 μm MG-132 (Sigma-Aldrich) for 3–5 h at 37 °C before cell lysis. To deglycosylate proteins, cell lysates were treated with PNGase F (New England Biolabs) or endoglycosidase H (New England Biolabs) for 1 h at 37 °C according to the manufacturer's instructions. For Western blot analysis, cells were washed twice with cold saline buffer (PBS), scraped off the plate, pelleted by centrifugation (5,000 × g, 5 min), and lysed in detergent buffer (0.5% Triton X-100, 0.5% sodium deoxycholate in PBS). The cell lysates were either analyzed directly or centrifuged (20,000 × g, 10 min) to analyze the postnuclear supernatant.
Studies in S. cerevisiae
For expression in S. cerevisiae, constructs bearing the Kre5p signal peptide were cloned into pYES2 expression vector and transformed into K39 wild-type yeast cells according to established protocols (80). A single clone of transformed K39 wild-type yeast cells was picked and inoculated in 20 ml of −Ura+Glu medium. Cells were grown overnight at 30 °C and 120 rpm in the presence of glucose. The next day, cells were centrifuged (2,000 × g, 5 min), and the pellet was resuspended in −Ura+Gal medium (0.4% (w/v) galactose, 0.1% (w/v) yeast extract, 0.5% (w/v) ammonium sulfate, 0.17% (w/v) YNB-amino acid mix (pH 6.0)) to induce gene expression. Cells were harvested (2,000 × g, 5 min) and lysed using glass beads (Alekto Malzperle, Willy A. Backofen GmbH) in 3 times the cell weight of PBS lysis buffer (0.5% (w/v) Triton X-100, 0.5% (w/v) deoxycholate in PBS (−/−) (Mg/Ca), 10 μg/ml aproptin, 1 mm benzamidine, 10 μg/ml pepstatin, 1 μm leupeptin) by vortexing 10 times for 1 min with 1-min intervals on ice in between. The soluble fraction was aliquoted into one untreated sample and one digested with endoglycosidase H (New England Biolabs) for 1 h at 37 °C according to the manufacturer's instructions before Western blot analysis.
Studies in E. coli
Constructs bearing DsbA or PelB signal peptide were cloned into pET27b(+) (Novagen) and transformed in E. coli BL21(DE3) (Thermo Fisher Scientific). Main expression cultures were inoculated from overnight culture to an optical density of A600 nm = 0.05 and grown in LB substituted with 50 mg/ml kanamycin at 30 °C at 120 rpm. Protein expression was induced at A600 nm = 0.5 for 2 h by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. The isolation of the periplasmic fraction was described previously (81). In brief, 10 ml of main culture were harvested by centrifugation at 4,000 × g for 1 min at 4 °C. The cell pellet was carefully resuspended in TSE periplasmic extraction buffer (200 mm Tris-HCl, pH 8.0, 500 mm sucrose, 1 mm EDTA, cOmplete® Mini EDTA-free protease inhibitor mixture (Roche Applied Science)) using a wire loop. Respective volume of TSE for resuspension was calculated according to A600 nm of the main culture at the time of harvesting (A600 nm = 1.0 ≈ 1.000 μl of TSE buffer). After a 30-min incubation on ice, cells were centrifuged for 30 min at 21,000 × g and 4 °C. Supernatant containing periplasmic fraction was isolated for Western blotting. The cell pellet was resuspended in TE buffer (same volume as TSE buffer, 10 mm Tris-HCl, 1 mm EDTA, pH 8, cOmplete® Mini EDTA-free protease inhibitor mixture (Roche Applied Science)), and cells were opened by sonification and again centrifuged for 30 min at 21,000 × g and 4 °C to obtain soluble cytoplasmic fraction for Western blotting.
Western blotting
For Western blot analysis, lysates were boiled in Laemmli sample buffer with β-mercaptoethanol (4% (v/v)). Following SDS-PAGE, proteins were transferred to nitrocellulose by electroblotting. Membranes were blocked by incubation in TBS-T (TBS with 0.1% (w/v) Tween 20) containing 5% skimmed milk for 1 h at room temperature and incubated with primary antibody in TBS-T + 5% skimmed milk overnight at 4 °C. After washing with TBS-T, blots were incubated with respective secondary antibody (IRDye-Infrared Technology, LI-COR) in TBS-T for 1 h at room temperature. Protein signals were visualized and quantified using an ODYSSEY® 9120 scanner and Image Studio Light software (LI-COR).
Quantification of translocation efficiency
To quantify Sec61-mediated translocation, the respective bands of the constructs were measured densitometrically, and the percentage of glycosylated protein relative to total protein amount was determined. For E. coli/SecY, translocation efficiency was calculated as the ratio of substrate present in the periplasmic fraction normalized to the relative amount of the periplasmic protein MBP in the periplasmic fraction. Specifically, the translocation efficiency of MBP was set as 100% and assumed that the fraction of MBP present in the cytoplasmic fraction is due to an incomplete lysis of bacteria by cold osmotic shock. For example, when 30% of MBP was detected in the cytosolic fraction, the amount of our substrate present in the cytoplasmic fraction was reduced by 30% for the calculation of the ratio between cytoplasmic and periplasmic localization. For quantification, both the species representing the full-length unprocessed protein (marked by a dot) and the protein with the signal peptide cleaved off (marked by an asterisk) were included. Note that only the soluble fractions of the proteins were analyzed. Data represent mean ± S.E. of at least three independent experiments for each construct. The significance was determined by using Student's t test (**, p < 0.005; ***, p < 0.0005).
In vitro protein synthesis and transport
The components of the in vitro system, salt-washed ribosomes, initiation factors, and the cytosolic translation factors were prepared as described previously (65, 82). In vitro protein synthesis was performed in 50-μl aliquots in the presence of 1 μl of E. coli inverted inner membrane vesicles (∼50 μg of protein). The reaction was started by the addition of 5 units of T7 RNA polymerase and 8 units of placental RNase inhibitor. Incubation time was 30 min at 37 °C. Samples were subsequently divided into two aliquots of 10 and 40 μl each. The 10-μl aliquot was directly precipitated with ice-cold TCA (final concentration 10%), whereas the 40-μl aliquot was incubated with 40 μl of proteinase K (PK) (0.3 mg/ml, incubation on ice for 15 min). TCA was subsequently added (10% final concentration), and the sample was incubated at 56 °C for 10 min, followed by a 30-min incubation at 4 °C. The samples were then centrifuged for 20 min at 13,000 rpm, and the pellet was resuspended in 20 μl of SDS protein loading dye. Samples were separated by 15% SDS-PAGE and analyzed by phosphorimaging and Image Quant software (Storm PhosphorImager, GE Healthcare). For the calculation of percentage of integration, the radioactive signals in the −PK and +PK lanes were quantified using the ImageQuant software. The obtained value for −PK was set to 100%, and the value +PK was calculated. This latter value was then corrected for the amount of material that was loaded (i.e. 80% of the total in vitro synthesized material).
Author contributions
A. G., S. J., S. U., A. O., A. Z. acquired data; M. B. contributed essential reagents; H.-G. K. and R. Z. analyzed and interpreted data; K. F. W. and J. T. conceived and designed the study, analyzed and interpreted data, and drafted the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Jessica Klümper, Anna Chan, Kathrin Günnewig, Kristoffer Klewe, Jan Albrecht, and Jan Schwichtenberg for experimental support with S. cerevisiae and to Petra Goldmann, Barbara Kachholz, and Andrea Roth-Sturm for technical support.
This work was supported by a short-term fellowship of the International Max Planck Research School in Chemical and Molecular Biology (to A. G.) and Deutsche Forschungsgemeinschaft Grants TA 167/6 (to J. T.), GRK 1326 (to A. Z.), ZI 234/13-1 (to R. Z.), and Ko2184/8-1 and RTG 2202 (to H. G. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1.
- ER
- endoplasmic reticulum
- IDP
- intrinsically disordered protein
- MBP
- maltose-binding protein
- aa
- amino acid(s)
- PNGase F
- peptide:N-glycosidase F
- Endo H
- endoglycosidase H
- ERAD
- ER-associated degradation
- IDD
- intrinsically disordered domain
- INV
- inverted inner membrane vesicle
- SRP
- signal recognition particle
- GPI
- glycosylphosphatidylinositol
- PK
- proteinase K.
References
- 1. Driessen A. J., and Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 2. Park E., and Rapoport T. A. (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 [DOI] [PubMed] [Google Scholar]
- 3. Zimmermann R., Eyrisch S., Ahmad M., and Helms V. (2011) Protein translocation across the ER membrane. Biochim. Biophys. Acta 1808, 912–924 [DOI] [PubMed] [Google Scholar]
- 4. Walter P., Gilmore R., and Blobel G. (1984) Protein translocation across the endoplasmic reticulum. Cell 38, 5–8 [DOI] [PubMed] [Google Scholar]
- 5. Mandon E. C., Trueman S. F., and Gilmore R. (2009) Translocation of proteins through the Sec61 and SecYEG channels. Curr. Opin. Cell Biol. 21, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilmore R., Blobel G., and Walter P. (1982) Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 95, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilmore R., Walter P., and Blobel G. (1982) Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 95, 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer D. I., and Dobberstein B. (1980) Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. J. Cell Biol. 87, 503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walter P., and Blobel G. (1980) Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 77, 7112–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walter P., and Blobel G. (1982) Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299, 691–698 [DOI] [PubMed] [Google Scholar]
- 11. Rapoport T. A. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 12. Keenan R. J., Freymann D. M., Stroud R. M., and Walter P. (2001) The signal recognition particle. Annu. Rev. Biochem. 70, 755–775 [DOI] [PubMed] [Google Scholar]
- 13. Schatz G., and Dobberstein B. (1996) Common principles of protein translocation across membranes. Science 271, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 14. Wang X., Bowers S. L., Wang F., Pu X. A., Nelson R. J., and Ma J. (2009) Cytoplasmic prion protein induces forebrain neurotoxicity. Biochim. Biophys. Acta 1792, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma J., Wollmann R., and Lindquist S. (2002) Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298, 1781–1785 [DOI] [PubMed] [Google Scholar]
- 16. Rane N. S., Yonkovich J. L., and Hegde R. S. (2004) Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 23, 4550–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rambold A. S., Miesbauer M., Rapaport D., Bartke T., Baier M., Winklhofer K. F., and Tatzelt J. (2006) Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol. Biol. Cell 17, 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodrigo-Brenni M. C., and Hegde R. S. (2012) Design principles of protein biosynthesis-coupled quality control. Dev. Cell 23, 896–907 [DOI] [PubMed] [Google Scholar]
- 19. Rodrigo-Brenni M. C., Gutierrez E., and Hegde R. S. (2014) Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell 55, 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drisaldi B., Stewart R. S., Adles C., Stewart L. R., Quaglio E., Biasini E., Fioriti L., Chiesa R., and Harris D. A. (2003) Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 278, 21732–21743 [DOI] [PubMed] [Google Scholar]
- 21. Hessa T., Sharma A., Mariappan M., Eshleman H. D., Gutierrez E., and Hegde R. S. (2011) Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475, 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. von Heijne G. (1985) Signal sequences: the limits of variation. J. Mol. Biol. 184, 99–105 [DOI] [PubMed] [Google Scholar]
- 23. Martoglio B., and Dobberstein B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol. 8, 410–415 [DOI] [PubMed] [Google Scholar]
- 24. Hegde R. S., and Bernstein H. D. (2006) The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571 [DOI] [PubMed] [Google Scholar]
- 25. Hegde R. S., and Kang S. W. (2008) The concept of translocational regulation. J. Cell Biol. 182, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oyadomari S., Yun C., Fisher E. A., Kreglinger N., Kreibich G., Oyadomari M., Harding H. P., Goodman A. G., Harant H., Garrison J. L., Taunton J., Katze M. G., and Ron D. (2006) Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126, 727–739 [DOI] [PubMed] [Google Scholar]
- 27. Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., and Hegde R. S. (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hegde R. S., and Ploegh H. L. (2010) Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yogev O., Naamati A., and Pines O. (2011) Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278, 4230–4242 [DOI] [PubMed] [Google Scholar]
- 30. Shaffer K. L., Sharma A., Snapp E. L., and Hegde R. S. (2005) Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell 9, 545–554 [DOI] [PubMed] [Google Scholar]
- 31. Heske J., Heller U., Winklhofer K. F., and Tatzelt J. (2004) The C-terminal domain of the prion protein is necessary and sufficient for import into the endoplasmic reticulum. J. Biol. Chem. 279, 5435–5443 [DOI] [PubMed] [Google Scholar]
- 32. Miesbauer M., Pfeiffer N. V., Rambold A. S., Müller V., Kiachopoulos S., Winklhofer K. F., and Tatzelt J. (2009) α-Helical domains promote translocation of intrinsically disordered polypeptides into the endoplasmic reticulum. J. Biol. Chem. 284, 24384–24393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zanusso G., Petersen R. B., Jin T., Jing Y., Kanoush R., Ferrari S., Gambetti P., and Singh N. (1999) Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J. Biol. Chem. 274, 23396–23404 [DOI] [PubMed] [Google Scholar]
- 34. Dirndorfer D., Seidel R. P., Nimrod G., Miesbauer M., Ben-Tal N., Engelhard M., Zimmermann R., Winklhofer K. F., and Tatzelt J. (2013) The α-helical structure of prodomains promotes translocation of intrinsically disordered neuropeptide hormones into the endoplasmic reticulum. J. Biol. Chem. 288, 13961–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfeiffer N. V., Dirndorfer D., Lang S., Resenberger U. K., Restelli L. M., Hemion C., Miesbauer M., Frank S., Neutzner A., Zimmermann R., Winklhofer K. F., and Tatzelt J. (2013) Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. EMBO J. 32, 1036–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holtkamp W., Kokic G., Jäger M., Mittelstaet J., Komar A. A., and Rodnina M. V. (2015) Cotranslational protein folding on the ribosome monitored in real time. Science 350, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 37. Whitley P., Nilsson I. M., and von Heijne G. (1996) A nascent secretory protein may traverse the ribosome endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271, 6241–6244 [DOI] [PubMed] [Google Scholar]
- 38. Woolhead C. A., McCormick P. J., and Johnson A. E. (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 39. Lu J., and Deutsch C. (2005) Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 44, 8230–8243 [DOI] [PubMed] [Google Scholar]
- 40. Mingarro I., Nilsson I., Whitley P., and von Heijne G. (2000) Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woolhead C. A., Johnson A. E., and Bernstein H. D. (2006) Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell 22, 587–598 [DOI] [PubMed] [Google Scholar]
- 42. Bhushan S., Gartmann M., Halic M., Armache J. P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., and Beckmann R. (2010) α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 43. Conti B. J., Elferich J., Yang Z., Shinde U., and Skach W. R. (2014) Cotranslational folding inhibits translocation from within the ribosome-Sec61 translocon complex. Nat. Struct. Mol. Biol. 21, 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Daniel C. J., Conti B., Johnson A. E., and Skach W. R. (2008) Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J. Biol. Chem. 283, 20864–20873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conti B. J., Devaraneni P. K., Yang Z., David L. L., and Skach W. R. (2015) Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell 58, 269–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin P. J., Jongsma C. G., Pool M. R., and Johnson A. E. (2011) Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J. Cell Biol. 195, 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steiner D., Forrer P., Stumpp M. T., and Plückthun A. (2006) Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat. Biotechnol. 24, 823–831 [DOI] [PubMed] [Google Scholar]
- 48. Schierle C. F., Berkmen M., Huber D., Kumamoto C., Boyd D., and Beckwith J. (2003) The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185, 5706–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Donne D. G., Viles J. H., Groth D., Mehlhorn I., James T. L., Cohen F. E., Prusiner S. B., Wright P. E., and Dyson H. J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. U.S.A. 94, 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., and Wüthrich K. (1996) NMR structure of the mouse prion protein domain PrP(121–321). Nature 382, 180–182 [DOI] [PubMed] [Google Scholar]
- 51. Riek R., Hornemann S., Wider G., Glockshuber R., and Wüthrich K. (1997) NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett. 413, 282–288 [DOI] [PubMed] [Google Scholar]
- 52. Eberl H., and Glockshuber R. (2002) Folding and intrinsic stability of deletion variants of PrP(121–231), the folded C-terminal domain of the prion protein. Biophys. Chem. 96, 293–303 [DOI] [PubMed] [Google Scholar]
- 53. Winklhofer K. F., Heske J., Heller U., Reintjes A., Muranyi W., Moarefi I., and Tatzelt J. (2003) Determinants of the in vivo-folding of the prion protein: a bipartite function of helix 1 in folding and aggregation. J. Biol. Chem. 278, 14961–14970 [DOI] [PubMed] [Google Scholar]
- 54. Nouwen N., Berrelkamp G., and Driessen A. J. (2009) Charged amino acids in a preprotein inhibit SecA-dependent protein translocation. J. Mol. Biol. 386, 1000–1010 [DOI] [PubMed] [Google Scholar]
- 55. Liang F. C., Bageshwar U. K., and Musser S. M. (2012) Position-dependent effects of polylysine on Sec protein transport. J. Biol. Chem. 287, 12703–12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fujita H., Yamagishi M., Kida Y., and Sakaguchi M. (2011) Positive charges on the translocating polypeptide chain arrest movement through the translocon. J. Cell Sci. 124, 4184–4193 [DOI] [PubMed] [Google Scholar]
- 57. Nakatsukasa K., and Brodsky J. L. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ellgaard L., and Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 59. Meusser B., Hirsch C., Jarosch E., and Sommer T. (2005) ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 60. Rutkowski D. T., Ott C. M., Polansky J. R., and Lingappa V. R. (2003) Signal sequences initiate the pathway of maturation in the endoplasmatic reticulum lumen. J. Biol. Chem. 278, 30365–30372 [DOI] [PubMed] [Google Scholar]
- 61. Daude N., Ng V., Watts J. C., Genovesi S., Glaves J. P., Wohlgemuth S., Schmitt-Ulms G., Young H., McLaurin J., Fraser P. E., and Westaway D. (2010) Wild-type Shadoo proteins convert to amyloid-like forms under native conditions. J. Neurochem. 113, 92–104 [DOI] [PubMed] [Google Scholar]
- 62. Watts J. C., Drisaldi B., Ng V., Yang J., Strome B., Horne P., Sy M. S., Yoong L., Young R., Mastrangelo P., Bergeron C., Fraser P. E., Carlson G. A., Mount H. T., Schmitt-Ulms G., and Westaway D. (2007) The CNS glycoprotein Shadoo has PrP(C)-like protective properties and displays reduced levels in prion infections. EMBO J. 26, 4038–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heller U., Winklhofer K. F., Heske J., Reintjes A., and Tatzelt J. (2003) Post-translational import of the prion protein into the endoplasmic reticulum interferes with cell viability: a critical role for the putative transmembrane domain. J. Biol. Chem. 278, 36139–36147 [DOI] [PubMed] [Google Scholar]
- 64. Li A., Dong J., and Harris D. A. (2004) Cell surface expression of the prion protein in yeast does not alter copper utilization phenotypes. J. Biol. Chem. 279, 29469–29477 [DOI] [PubMed] [Google Scholar]
- 65. Koch H. G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H. K., Schimz K. L., Mechler B., and Müller M. (1999) In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koch H. G., and Müller M. (2000) Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J. Cell Biol. 150, 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Crowley K. S., Liao S., Worrell V. E., Reinhart G. D., and Johnson A. E. (1994) Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461–471 [DOI] [PubMed] [Google Scholar]
- 68. Jungnickel B., and Rapoport T. A. (1995) A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 82, 261–270 [DOI] [PubMed] [Google Scholar]
- 69. Voorhees R. M., and Hegde R. S. (2016) Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li L., Park E., Ling J., Ingram J., Ploegh H., and Rapoport T. A. (2016) Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531, 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Patterson M. A., Bandyopadhyay A., Devaraneni P. K., Woodward J., Rooney L., Yang Z., and Skach W. R. (2015) The ribosome-Sec61 translocon complex forms a cytosolically restricted environment for early polytopic membrane protein folding. J. Biol. Chem. 290, 28944–28952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xue B., Dunker A. K., and Uversky V. N. (2012) Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30, 137–149 [DOI] [PubMed] [Google Scholar]
- 73. Tompa P., Fuxreiter M., Oldfield C. J., Simon I., Dunker A. K., and Uversky V. N. (2009) Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays 31, 328–335 [DOI] [PubMed] [Google Scholar]
- 74. Fuxreiter M., and Tompa P. (2012) Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 725, 1–14 [DOI] [PubMed] [Google Scholar]
- 75. Fuxreiter M., Tompa P., Simon I., Uversky V. N., Hansen J. C., and Asturias F. J. (2008) Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 4, 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Uversky V. N., and Dunker A. K. (2010) Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Longhi S. (2012) The measles virus N(TAIL)-XD complex: an illustrative example of fuzziness. Adv. Exp. Med. Biol. 725, 126–141 [DOI] [PubMed] [Google Scholar]
- 78. Wright P. E., and Dyson H. J. (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., and Diringer H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gietz R. D., and Woods R. A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 81. Quan S., Hiniker A., Collet J. F., and Bardwell J. C. (2013) Isolation of bacteria envelope proteins. Methods Mol. Biol. 966, 359–366 [DOI] [PubMed] [Google Scholar]
- 82. Hoffschulte H. K., Drees B., and Müller M. (1994) Identification of a soluble SecA/SecB complex by means of a subfractionated cell-free export system. J. Biol. Chem. 269, 12833–12839 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.