Abstract
Thrombolites are buildups of carbonate that exhibit a clotted internal structure formed through the interactions of microbial mats and their environment. Despite recent advances, we are only beginning to understand the microbial and molecular processes associated with their formation. In this study, a spatial profile of the microbial and metabolic diversity of thrombolite-forming mats of Highborne Cay, The Bahamas, was generated by using 16S rRNA gene sequencing and predictive metagenomic analyses. These molecular-based approaches were complemented with microelectrode profiling and in situ stable isotope analysis to examine the dominant taxa and metabolic activities within the thrombolite-forming communities. Analyses revealed three distinctive zones within the thrombolite-forming mats that exhibited stratified populations of bacteria and archaea. Predictive metagenomics also revealed vertical profiles of metabolic capabilities, such as photosynthesis and carboxylic and fatty acid synthesis within the mats that had not been previously observed. The carbonate precipitates within the thrombolite-forming mats exhibited isotopic geochemical signatures suggesting that the precipitation within the Bahamian thrombolites is photosynthetically induced. Together, this study provides the first look at the spatial organization of the microbial populations within Bahamian thrombolites and enables the distribution of microbes to be correlated with their activities within modern thrombolite systems.
Keywords: Thrombolites, Microbial diversity, Metagenome, Stable isotopes, Microbialites
1. Introduction
With their long evolutionary history, microbialites serve as important model systems to explore and understand the coevolutionary dynamics among lithifying microbial communities and their local environment. These carbonate structures are formed via the metabolic activity of microbes, which influence and drive biological processes associated with sediment capture and microbiologically induced organomineralization. Microbialites have been found in a wide range of habitats including brackish (e.g., Laval et al., 2000; Breitbart et al., 2009; White et al., 2015; Chagas et al., 2016), marine (e.g., Dravis, 1983; Reid et al., 2000; Stolz et al., 2009; Casaburi et al., 2016), and hypersaline (e.g., Logan, 1961; Glunk et al., 2011; Wong et al., 2015; Paul et al., 2016; Ruvindy et al., 2016; Suosaari et al., 2016) environments and are classified based on their internal micro-fabrics (Burne and Moore, 1987; Dupraz et al., 2009). Two of the most well-studied types of microbialites are stromatolites, which exhibit laminated internal fabrics (Walter, 1994; Reid et al., 2000), and thrombolites with irregular clotted fabrics (Aitken, 1967; Kennard and James, 1986).
Much of our understanding of microbialite formation comes from the study of modern systems (e.g., Reid et al., 2000; Breitbart et al., 2009; Petrash et al., 2012; Russell et al., 2014; Valdespino-Castillo et al., 2014; Saghaï et al., 2015; White et al., 2015, 2016; Casaburi et al., 2016; Chagas et al., 2016; Ruvindy et al., 2016; Warden et al., 2016). Microbialites in The Bahamas have been particularly important in expanding research in this area, as they are the only known modern open marine microbialite system and serve as potential analogues to ancient systems (Reid et al., 2000). In Bahamian stromatolites, processes underlying formation include iterative growth by cycling microbial mat communities and seasonal environmental controls; the resulting lamination represents a chronology of past surface communities (Visscher et al., 1998; Reid et al., 2000; Bowlin et al., 2012). In thrombolites, the processes that form the clotted fabrics are not well defined. In some Bahamian thrombolites, the clots appear to be products of calcified cyanobacterial filaments, which through their metabolism cause shifts in the carbonate saturation state and thereby drive precipitation (Dupraz et al., 2009; Planavsky et al., 2009; Myshrall et al., 2010). Alternatively, it has been suggested that the clotted textures in thrombolites are sometimes linked to disruption or modification of microbial fabrics (Planavsky and Ginsburg, 2009; Bernhard et al., 2013; Edgcomb et al., 2013).
To further explore the formation of clotted fabrics, the thrombolites of Highborne Cay, The Bahamas, were targeted, as they represent one of the few modern locations of actively accreting thrombolitic microbialites in open marine environments (Planavsky et al., 2009; Myshrall et al., 2010; Mobberley et al., 2012, 2013, 2015). These marine thrombolites form in the intertidal zone of a 2.5 km fringing reef complex that extends along the eastern margin of Highborne Cay (Fig. 1A; Reid et al., 1999). The thrombolites range in size from up to 1 m in height to several meters in length (Andres and Reid, 2006; Myshrall et al., 2010) and are covered with several distinct microbial mat types (Mobberley et al., 2012).
FIG. 1.
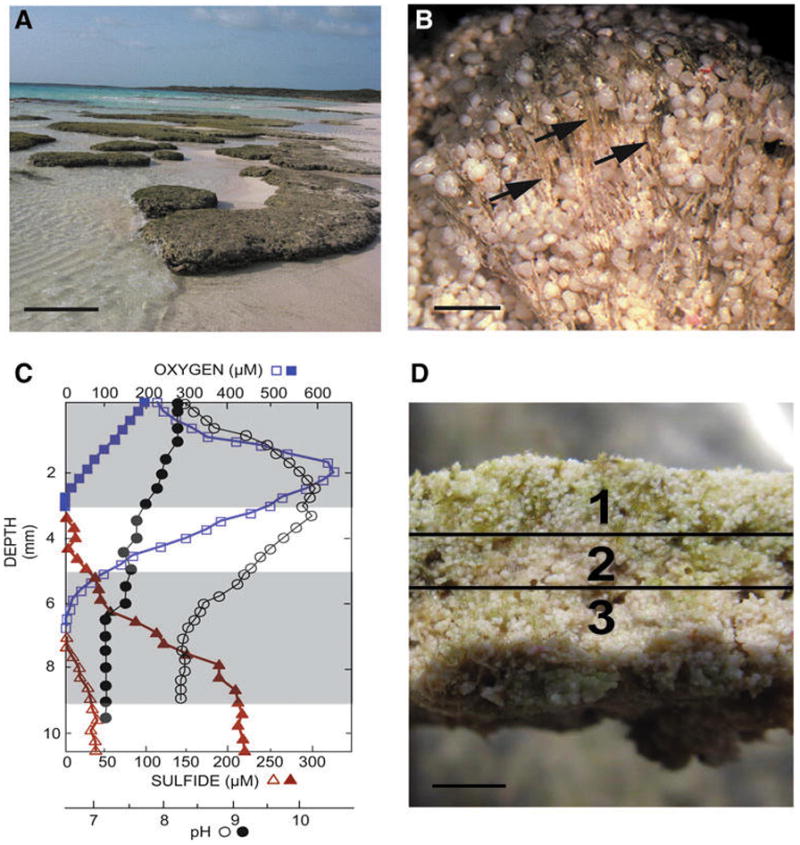
The thrombolites of Highborne Cay, The Bahamas. (A) Intertidal thrombolite platforms from Site 5. Bar, 1 m. (B) Light micrograph of a thrombolite-forming button mat revealing extensive vertical assemblages of calcified filaments (arrows). Bar, 500 μm. (C) In situ depth profiles of oxygen (square), sulfide (triangle), and pH (circle) collected at peak of photosynthesis (open symbols) or respiration (filled symbols). Shaded areas reflect the targeted areas collected for analysis. Depths below 9 cm were not sampled, as that region shared the same biochemical profile as in Zone 3. (D) Cross section of button mat depicting the three spatial regions including an oxic Zone 1 (0–3 mm), transitional Zone 2 (3–5 mm), and anoxic Zone 3 (5–9 mm). Bar, 3 mm.
The dominant mat type associated with the Bahamian thrombolites, referred to as “button” mat, harbors tufts of vertically orientated calcified cyanobacterial filaments (Fig. 1B; Myshrall et al., 2010; Mobberley et al., 2012). The dominant cyanobacterium identified within these tufts with both morphological and molecular tools is Dichothrix sp. (Planavsky et al., 2009; Mobberley et al., 2012). At the surface, these Dichothrix-enriched button mats are calcified with aragonite precipitates located in the exopolymeric sheath of the cell. With depth, precipitates undergo dissolution and filaments degrade (Planavsky et al., 2009). In addition to the tufts of calcified filaments, the thrombolite-forming button mats also harbor a genetically diverse and active microbial community that appears to form vertical gradients of metabolic activity (Myshrall et al., 2010; Mobberley et al., 2013, 2015).
Previous work in other microbialite systems, such as stromatolites, has shown that the relationship among active, distinct microbial guilds can alter the local physiochemical environment and generate discrete gradients of both solutes and redox conditions (e.g., Dupraz et al., 2009; Glunk et al., 2011; Wong et al., 2015). Within these microenvironments, the microbial activity can alter both the carbonate saturation index (i.e., carbonate alkalinity and availability of free calcium) and the cycling of exopolymeric substances (EPS; Braissant et al., 2009), which serve as important nucleation sites for precipitation (Dupraz and Visscher, 2005). Certain metabolisms, such as photosynthesis and some types of sulfate reduction, can lead to an increase in pH and thereby promote precipitation (Visscher et al., 1998; Dupraz et al., 2009; Gallagher et al., 2012). Contrastingly, some metabolisms, such as sulfide oxidation, aerobic respiration, and fermentation, can increase dissolved inorganic carbon concentrations but lower the pH and carbonate saturation state of the local environment and promote dissolution (Walter, 1994; Visscher et al., 1998; Dupraz et al., 2009). Together, it is the parity between categories of metabolisms that determines the extent and net precipitation potential within the lithifying mat community (Visscher and Stolz, 2005).
In addition to the precipitation potential, another component that is critical to the formation of microbialites is the availability of nucleation sites, which can be controlled by the production and degradation of EPS material. The EPS matrix serves several essential roles in the formation of microbialites, as it binds cations (e.g., Ca2+) critical for carbonate precipitation, serves as attachment sites for microbes to withstand the high-energy wave impacts, and protects microbes from environmental stresses, such as UV exposure and desiccation (Dupraz et al., 2009). Metagenomic analyses of both stromatolites and thrombolites across the globe have shown that Cyanobacteria and Proteobacteria are the two primary producers of EPS material (Khodadad and Foster, 2012; Mobberley et al., 2013, 2015; Casaburi et al., 2016; Ruvindy et al., 2016; Warden et al., 2016). Alteration or restructuring of the EPS through microbial degradation can reduce the cation-binding capability and thereby facilitate the precipitation of calcium carbonate on the EPS matrix (Dupraz et al., 2004, 2009; Dupraz and Visscher, 2005).
There have been major advances in understanding the processes controlling stromatolite formation; in contrast, the factors controlling carbonate precipitation in thrombolites are less understood. Several recent studies have begun to use metaomic approaches to understand thrombolite communities and how they may initiate precipitation. For example, metatranscriptomic sequencing of the Bahamian thrombolite-forming mats at midday revealed distinct profiles of gene expression within the thrombolites (Mobberley et al., 2015). This study, however, captured only those metabolically active communities and did not provide a comprehensive assessment of the total microbial population within the observed thrombolites’ zones. Additionally, previous shotgun meta-genomic studies have been used to examine the overall metabolic potential in thrombolite ecosystems, including those at Highborne Cay (Mobberley et al., 2013) and in hypersaline thrombolites of Lake Clifton (Warden et al., 2016); however, neither of these metagenomic studies provided spatial information of the thrombolite-forming communities.
In the present study, we build on this previous research by examining the spatial distribution of the bacterial and archaeal diversity associated with the button mats of Bahamian thrombolites using a targeted phylogenetic marker gene approach coupled with a predictive computational reconstruction of the metagenome to ascertain how thrombolite-forming communities change, both taxonomically and functionally, with depth. These molecular-based approaches are complemented by stable isotope analysis with secondary ion mass spectrometry (SIMS), a high-resolution technique that has not been previously used in any microbialite study, to provide additional constraints on carbonate precipitation in the Dichothrix calcified filaments. Together, these methodologies elucidate the juxtapositioning of the taxa and metabolic functions associated with the thrombolite-forming mats as well as provide key insight into the metabolic metabolisms that initiate precipitation within these lithifying ecosystems.
2. Methods
2.1. Sample collection
Thrombolite-forming button mats were collected from the island of Highborne Cay, The Bahamas (24°43′N, 76°49′W), in February 2010 and October 2013 from an intertidal thrombolitic platform from Site 5 (Andres and Reid, 2006). The 2010 mats were partitioned in the field into three distinct vertical sections (0–3, 3–5, and 5–9 mm depth horizons, respectively) with a sterile scalpel to cut the thrombolite-forming mats, and the sections were immediately placed into RNAlater (Life Technologies, Inc., Grand Island, NY). These samples were transported to Space Life Sciences Lab, Merritt Island, Florida, where they were stored at −80°C until processing. The 2013 mats were processed for isotope analyses as described below.
2.2. Microelectrode measurements
Depth profiles of oxygen, sulfide, and pH were determined in triplicate with needle microelectrodes (Visscher et al., 1991, 1998; Pages et al., 2014) either in situ or ex situ under ambient temperature and light intensity. Microelectrodes with a tip diameter between 60 and 150 μm were deployed in 250 μm depth increments with a manual micromanipulator (National Aperture, Salem, NH). Oxygen profiles were measured in submerged mats (in ca. 5–15 cm water) with a polarographic Clark-type needle electrode with an outer diameter of 0.4 mm, and readings were recorded with a picoammeter (PA2000; Unisense, Aarhus, Denmark). Polarographic sulfide electrodes (Unisense, Denmark) were used in combination with a Unisense PA2000 picoammeter, and pH and S2− electrodes (Diamond General, Ann Arbor, MI) were connected to a high-impedance millivolt meter (Microscale Measurements, The Netherlands). Both electrode types were encased in needles (outer diameter 0.5 mm). Sulfide electrodes were calibrated before and after each deployment with buffers of three different pH values that span the pH range observed in the thrombolite (i.e., pH 7, 8, and 9). Under an oxygen-free atmosphere, aliquots of a sulfide stock solution were added in increments to the buffer, and electrode signals were recorded. Subsamples of the buffer were taken to ascertain the actual concentration of sulfide in the calibration cocktail by using the methylene blue method. The pH electrodes were calibrated at pH 5, 7, and 10. The pH profiles were used to calculate the actual sulfide concentration at each depth.
2.3. Generation and sequencing of 16S rRNA gene libraries
DNA was extracted in triplicate from each vertical section with a modified MoBio PowerSoil DNA isolation kit that included a xanthogenate pretreatment, as previously described (Green et al., 2008). The DNA was then PCR amplified in triplicate with fusion 454-primers that included a unique eight-base-pair barcode on the 3′ end (Supplementary Table S1; supplementary material is available online at www.liebertonline.com/ast). The PCR reactions for the bacterial 16S rRNA libraries targeted the V1-2 region and included the following: 1 × Pfu reaction buffer (Stratagene, La Jolla, CA), 280 μM dNTPs, 2.5 μg bovine serum albumin (BSA), 600 nM of each primer, 1 ng of genomic mat DNA, 1.25 U of Pfu DNA polymerase (Stratagene, La Jolla, CA), and nuclease-free water (Sigma, St. Louis, MO) in a volume of 25 μL. The amplification parameters included a 95°C denaturation for 5 min, followed by 30 cycles of 95°C for 1 min, 64°C for 1 min, 75°C for 1 min, and a final extension at 75°C for 7 min.
The archaeal libraries required a nested PCR approach that included two rounds of amplification and targeted the V3-5 region. The reactions contained the same concentrations as the bacterial library with the exception of 400 nM of 23F and 958R primers (DeLong, 1992; Barns et al., 1994) and 10 ng of thrombolitic mat DNA in round one, whereas 400 nM of primers 334F and 915R (Casamayor et al., 2002) with 10 ng of round-one amplicon material as a template. The amplification parameters in round one included a denaturation step of 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 55°C for 1 min, 72°C for 2 min with an extension of 72°C for 10 min. In round two the parameters were similar except that the annealing temperature was changed to 61°C.
For each library, the PCR amplicons were purified with the Ultraclean PCR Clean-Up Kit (MoBio, Carlsbad, CA) and combined into equimolar ratios. Sequencing was performed per manufacturer’s protocol by a 454 GS-FLX platform with Titanium chemistry (Roche, Branford, CT) at the University of Florida’s Interdisciplinary Center for Biotechnology Research. The raw sequence data files were deposited into the NCBI sequencing read archive under number SRP068710 (bacteria) and SRP068710 (archaea) under project PRJNA305634.
2.4. Bioinformatic analysis of 16S rRNA gene libraries
The recovered bacterial and archaeal 16S rRNA gene sequences were analyzed by Quantitative Insights Into Microbial Ecology (QIIME; version 1.9.1; Caporaso et al., 2010). Preprocessing was completed to separate the replicate libraries by depth, remove barcode adaptors, and filter for quality by using default parameters including minimum sequence length of 200 bp, maximum sequence length of 1000 bp, minimum quality score of 25, maximum ambiguous bases of 6, and maximum homopolymer length of 6. Operational taxonomic units (OTUs) were assigned to the filtered reads at 97% identity against the Greengenes database (v13.8; DeSantis et al., 2006) using the UCLUST method within QIIME. Further filtering was completed including removal of unassigned reads and filtering for most abundant OTUs (> 0.005%). The generated OTU table was used for taxonomic comparison, filtering the OTUs at 0.005% and producing taxonomic trees with Meta Genome Analyzer (MEGAN5; Huson et al., 2007). OTU tables were filtered at 0.1%, and hierarchal taxonomic pie charts were created with the Krona tool (Ondov et al., 2011). The representative sequences were aligned with PyNAST (v1.2.2; Caporaso et al., 2010) to the Greengenes Core reference alignment, and a phylogenetic tree was built by FastTree (v2.1.3; Price et al., 2010). The phylogenetic tree was used for downstream community analyses. Diversity analyses were performed at a sequence depth of 3587 for archaea and 3691 for bacteria.
Alpha diversity indices were calculated by using observed species and Faith0s Phylogenetic Diversity measure (Faith, 1992), and the averaged results were used to generate rarefaction curves. Beta diversity comparisons were visualized by using Principal Coordinates Analyses and Emperor (Vázquez-Baeza et al., 2013) generated from unweighted UniFrac distance matrices (Lozupone and Knight, 2005). Statistical significance between the mat depths was calculated by adonis, a nonparametric, permutation-based metric.
2.5. Reconstruction of functional metagenome using the PICRUSt algorithm
Functional gene content from each of the three vertical sections was predicted from the recovered 16S rRNA gene sequences by using the algorithm Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt v.1 .0; Langille et al., 2013), as previously described (Casaburi et al., 2016). Results were collapsed at Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs (KOs) Level 3 within the pathway hierarchy of KEGG (Kanehisa and Goto, 2000). For comparison purposes, a shotgun meta-genomic data set of whole Bahamian thrombolite-associated mats previously collected from Highborne Cay (Mobberley et al., 2013) was downloaded from the MG-RAST database with accession number 4513715.3. Raw reads were filtered by SICKLE (v. 1.2; Joshi and Fass, 2011) with default parameters. Filtered reads were re-annotated for functionality at different KEGG levels by the Metagenome Composition Vector (MetaCV v. 2.3.0) with default parameters (Liu et al., 2012). Resulting hits were filtered at a correlation score > 30, collapsed at KO Level 3, and finally compared to the 16S rRNA gene-predicted functional profile.
2.6. Bulk stable isotope analysis
Samples of thrombolite-forming mats were collected for isotopic analysis during the same collection trip as the molecular samples from Site 5 (Andres and Reid, 2006) of Highborne Cay in October 2013. The mat samples were dried and examined by bulk isotope analysis for both inorganic and organic signatures. Calcified filaments were dissected from the button mats, dried, and ground to a fine powder in triplicate. Aliquots of the carbonate (i.e., aragonite; Planavsky et al., 2009) were measured for inorganic δ13C and δ18O with a Finnigan-MAT 252 isotope ratio mass spectrometer coupled with a Kiel III carbonate preparation device.
For isotopic analysis of organic matter, calcified filaments were dissected and treated with an acid solution (6 N HCl) at room temperature overnight until all CaCO3 was removed and rinsed with distilled water to remove HCl. Samples were loaded into tin capsules and placed in a 50-position automated Zero Blank sample carousel on a Carlo Erba NA1500 CNHS elemental analyzer. After flash combustion in a quartz column containing chromium oxide and silvered cobaltous/cobaltic oxide at 1000°C in an oxygen-rich atmosphere, the sample gas was transported in a He carrier stream and passed through a hot reduction column (650°C) consisting of reduced elemental copper to remove oxygen. The effluent stream then passed through a chemical (magnesium perchlorate) trap to remove water followed by a 3 m GC column at 45°C to separate N2 from CO2. The sample gas next passed into a ConFlo II preparation system and into the inlet of a Thermo Electron Delta V Advantage isotope ratio mass spectrometer running in continuous flow mode where the sample gas was measured relative to laboratory reference N2 and CO2 gases. All carbon and oxygen isotopic results are expressed in standard delta notation relative to Vienna Pee Dee Belemnite (VPDB), whereas nitrogen isotopic results are expressed in standard delta notation relative to air (AIR). The standard used for bulk C and O measurements was NBS-19, whereas USGS40 and USGS41 were used for N. Measurements were conducted in triplicate at the Light Stable Isotope Mass Spectrometry Laboratory in the Department of Geological Sciences at the University of Florida. Instrument precision was better than 0.10‰ for all bulk isotope measurements.
2.7. Stable isotope analysis using secondary ion mass spectrometry (SIMS)
Additional mat samples, collected in October 2013, were prepared as thin sections at the WiscSIMS laboratory, University of Wisconsin-Madison. Samples were cast with EpoxiCure resin in 25 mm epoxy rounds, cut with a Buehler IsoMet low speed to expose the most suitable section for analysis, and turned, together with two grains of UWC-3 WiscSIMS calcite standard (δ13C = −0.91 ± 0.04‰; δ18O = −17.87‰± 0.03‰ VPDB; Kozdon et al., 2009), into ~100 μm thick thin sections. An aragonite standard (UWArg-7, δ13C = 5.99‰; δ18O = −10.84‰ VPDB; Orland, 2012; Linzmeier et al., 2016) was also run at the beginning of each day of analysis to correct for the differences in instrumental mass fractionation between calcite and aragonite, which was 1.3‰ for δ18O and 1.5‰ for δ13C. The epoxy rounds were ground to expose features of interest for analysis. Petrographic microscopy was conducted with an Olympus BH-2 microscope with plane-polarized and cross-polarized transmitted light at various magnifications to identify potential sites suitable for SIMS analysis. The samples were then polished and sputter coated with palladium for scanning electron microscopy (SEM) at the University of Miami’s Center for Advanced Microscopy (UMCAM) to identify areas of precipitate for analysis and to screen for potential textural anomalies that might impede in situ δ13C and δ18O measurements. The SEM analysis was conducted on a FEI XL-30 field emission ESEM/SEM instrument with energy dispersive spectrometer. The SEM analysis was to ensure integrity of the sample and to identify specific target sites. After SEM analysis, the palladium coating was removed with 0.25 μm polish on a lapidary wheel, dried, and recoated with gold.
The thrombolite mat samples were then analyzed for δ13C and δ18O on a CAMECA ims-1280 secondary ion microprobe mass spectrometer (SIMS) using a 133Cs+ primary ion beam at the WiscSIMS Laboratory, Department of Geoscience, University of Wisconsin-Madison. A primary beam of 600 pA, with mean 0.77/‰ spot-to-spot precision (2SD), was used for δ13C, and 1.7 nA was used for δ18O with a 10 μm spot size (precision ~0.3‰). WiscSIMS carbonate analysis has been described in detail in previous publications (Orland et al., 2009; Valley and Kita, 2009; Kozdon et al., 2011; Williford et al., 2016).
Analysis of the thrombolitic mat sections (10–15 spot analyses per sample) was bracketed by 8–10 repeat measurements on the UWC-3 standard grain by using the same parameters as the samples to help determine instrumental mass fractionation corrections for each set of measurements. After completion of each analytical session, the samples were returned to the University of Miami for SEM inspection of the pits to evaluate any features that may have impacted accuracy (e.g., cracks or epoxy). Additionally, for those measurements that penetrated down to epoxy material (depths of 1–2 μm) and had high secondary ion count rates (i.e., >100% for 12C of the measured counts per second on the standard grain), the final three to six cycles (of 20) were excluded from computations, and the values for the spots were recalculated as in the work of Vetter et al. (2014). Visualization of the data was conducted in R (v.3.2.2; R Core Team, 2015) by using the package ggplot2 (Wickham, 2009).
3. Results
3.1. Microelectrode profiling of thrombolite button mats
The in situ concentrations of oxygen and sulfide were measured with microelectrodes during early afternoon representing peak photosynthesis (i.e., 12:30 pm and 2:00 pm) and at the end of the night, which marks the end of a prolonged anoxic period (i.e., 4:00–6:00 am) (Fig. 1C). The profiles reveal steep vertical gradients that fluctuated throughout the diel cycle. During the day, the oxic zone extended through the first 5 mm of the button mat with the peak of oxygen production (> 600 μM) occurring in the upper 3 mm (Fig. 1C). At night, however, oxygen levels decreased significantly and were detectable only in the upper 2 mm of the mat, suggesting rapid consumption at night and limited diffusion of O2 from the overlying water column. Contrastingly, sulfide levels were low during the day with levels detectable only below 6 mm. At night, sulfide levels built up and were detectable at 4 mm with a peak concentration occurring at a depth of 8–10 mm within the mat.
In addition to oxygen and sulfide, pH was also monitored throughout the vertical profile of the button mat, revealing a wide shift throughout the diel cycle. At peak photosynthesis, the localized pH ranged from 8.4 to 10.4 throughout the depth profile with the highest pH occurring at a depth of 3 mm (Fig. 1C). At night, however, the pH steadily decreased to as low as 7.1 at depths below 5 mm. Based on these oxygen, sulfide, and pH microelectrode profiles, three distinct spatial zones emerged. Zone 1 included the upper 3 mm of the button mat and contained a supersaturated oxic zone that was suggestive of high rates of oxygen production and consumption. Zone 2 represented a transitional area 3–5 mm beneath the surface where oxygen levels decreased and sulfide levels began to build. Finally Zone 3, which included depths below 5 mm, represented a primarily anoxic region of the thrombolite-forming mat.
3.2. Phylogenetic composition of bacteria in thrombolite communities with depth
Immediately after the microelectrode profiles were generated, the thrombolite mats were then sectioned based on these three observed zones (Zone 1, 0–3 mm; Zone 2, 3–5 mm; and Zone 3, 5–9 mm), and each of these spatial regions was subsequently examined for taxonomic diversity (Fig. 1D). Three replicate amplicon libraries were generated for each zone, targeting the 16S rRNA gene for both the Bacteria and Archaea. A summary of the data associated with the amplicon libraries is provided in Table 1. The overall bacterial diversity increased with depth (Supplementary Fig. S1A) with 2044 OTUs at 97% sequencing similarity in the upper oxic Zone 1 and 2947 and 3525 OTUs recovered from Zone 2 and 3, respectively. The number of recovered OTUs was much higher than previous diversity assessments of the Highborne Cay thrombolites (Myshrall et al., 2010; Mobberley et al., 2012) and likely reflects the increased sequencing coverage as determined by Good’s estimates (Table 1).
Table 1.
Summary Statistics for Thrombolite Samples by Zone for Bacterial and Archaeal Samples
| Bacteria
|
Archaeal
|
|||||
|---|---|---|---|---|---|---|
| Zone 1 | Zone 2 | Zone 3 | Zone 1 | Zone 2 | Zone 3 | |
| Depth | 0–3 mm | 3–5 mm | 5–9 mm | 0–3 mm | 3–5 mm | 5–9 mm |
| No. of reads | 25609 | 21535 | 31217 | 14253 | 22794 | 21646 |
| Normalized readsa | 3691 | 3691 | 3691 | 3587 | 3587 | 3587 |
| Total OTUsb | 2044 | 2947 | 3525 | 671 | 506 | 654 |
| OTUs > 0.005% | 729 | 949 | 956 | 178 | 169 | 172 |
| Shannon Indexc | 6.59 | 8.67 | 8.59 | 4.91 | 3.58 | 3.97 |
| ± sd | ± 0.026 | ± 0.026 | ± 0.016 | ± 0.008 | ± 0.020 | ± 0.024 |
| (confidence) | (0.029) | (0.029) | (0.019) | (0.009) | (0.023) | (0.027) |
| % coveraged | 94.5 | 87.8 | 92.3 | 97.9 | 98.2 | 98.2 |
| ± sd | 0.13 | 5.02 | 0.71 | 0.07 | 0.09 | 0.31 |
Randomized sequence count of each replicate for each zone used to measure diversity.
OTU identification used a 97% similarity threshold.
Shannon diversity index calculated over 10 iterations for three replicate samples.
Goods coverage estimate.
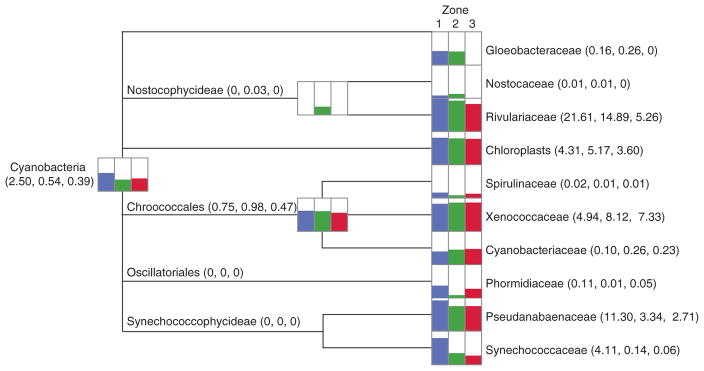
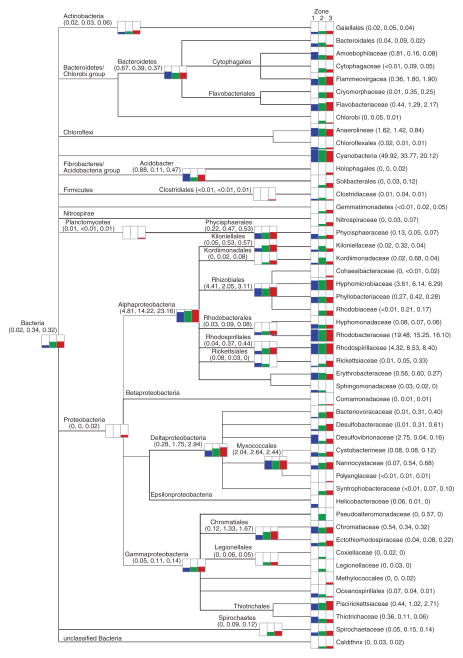
A total of 16 phyla were recovered from the three spatial zones within the thrombolite-forming mat, with the Proteobacteria, Cyanobacteria, Bacteroidetes, Chloroflexi, and Acidobacteria being highly represented in each zone (Supplementary Fig. S2). Distinct taxonomic differences, however, were observed across the three spatial regions of the thrombolite mat at the family level (Figs. 2 and 3; Supplementary Fig. S2). In the upper Zone 1, the most abundant family represented within the mat is the cyanobacterial family Rivulariaceae (Fig. 2; Supplementary Fig. S2). This taxon contains the genus Dichothrix, which was previously identified in the thrombolite mats as forming extensive tufts of calcified filaments (Fig. 1B) and has rarely been found in laminated stromatolites (Foster and Green, 2011). The Rivulariaceae dominated the oxic Zone 1, comprising 21% of annotated reads compared to 15% in the transitional Zone 2 and only 5% of the total recovered reads in Zone 3 (Fig. 2; Supplementary Fig. S2). In addition to Rivulariaceae, other prevalent Cyanobacteria in the oxic Zone 1 included Pseudanabaenaceae (11%), Xenococcaceae (5%), and Synechococcaceae (4%; Fig. 2; Supplementary Fig. S2).
FIG. 2.
Taxonomic distribution of cyanobacteria within the thrombolite-forming mats derived from MEGAN5 using the Greengenes database. Overall percentages based on read counts are presented logarithmically, depicting the distributions for Zone 1 (blue), Zone 2 (green), and Zone 3 (red). Read abundance data for each taxonomic level are included in parentheses.
FIG. 3.
Taxonomic distribution of Bacteria within the thrombolite-forming mats derived from MEGAN5 using the Greengenes database. Overall percentages based on read counts are presented logarithmically, depicting the distributions for Zone 1 (blue), Zone 2 (green), and Zone 3 (red). Read abundance data for each taxonomic level are included in parentheses.
Although Cyanobacteria was the dominant phylum recovered from Zone 1, there was also a diverse population of Proteobacteria, specifically, the class Alphaproteobacteria. Within the Alphaproteobacteria there was enrichment of the photoheterotrophic Rhodobacteraceae (19%) and Rhodospirillaceae (7%) families, and to a lesser extent the Rhizobiales (5%). These taxa were not only abundant in Zone 1 but were highly represented throughout the thrombolite vertical profile (Fig. 3; Supplementary Fig. S2). Other proteobacterial taxa that were abundant in Zone 1 compared to the other two zones included the sulfate-reducing Delta-proteobacteria family Desulfovibrionaceae (3%) and the Gammaproteobacteria family Thiotrichaceae (0.8%), which harbors several sulfide-oxidizing taxa (Fig. 3). A detailed Krona plot of the upper 3 mm of the thrombolite mat is provided in Supplementary Fig. S3.
Zone 2 represented a transitional phase in the thrombolite-forming mats, with several taxa first appearing in this 3–5 mm zone and gradually increasing in relative abundance in the anoxic Zone 3 (Fig. 3; Supplementary Figs. S2 and S4). For example, in the Deltaproteobacteria, the sulfate-reducing families Desulfobacteraceae and Syntrophobacteraceae were enriched in Zones 2 and 3 compared to Zone 1. Additionally, the purple sulfur bacterial Gammaproteobacteria family Ectothiorhodospiraceae (order Chromatiales) and the sulfide-oxidizing Piscirickettsiaceae (order Thiotrichales) also exhibited a gradual increase in relative abundance with depth (Fig. 3). In addition to the more prevalent taxa, there were several families that appeared to a lesser extent only at depth and included the photoheterotrophic Gemmatimonadetes, purple nonsulfur bacteria Rhodobiaceae, and nitrite-oxidizing Nitrospiraceae. Detailed taxonomic profiles of Zones 2 and 3 are depicted as Krona plots in Supplementary Figs. S4 and S5.
In addition to analysis of the bacterial composition, a beta diversity analysis was completed to assess whether these observed taxonomic differences were statistically significant. Unweighted UniFrac distance matrices were generated for the Bacteria amplicon libraries and visualized with a jackknifed principal coordinate analysis (PCoA; Fig. 4A). The statistical analyses revealed that each of the three spatial zones represented distinctive bacterial communities with low standard deviation among the library replicates (Fig. 4A). The R2 value showed an effect size of 0.402, indicating that approximately 40% of the variation in the bacterial populations could be explained by depth and potentially other environmental factors within the mats (p = 0.001; R2 = 0.402, adonis; Fig. 4A). Depth likely accounted for at least 27% (PC1) of the variation among the three zones based on the PCoA plots (Fig. 4A).
FIG. 4.
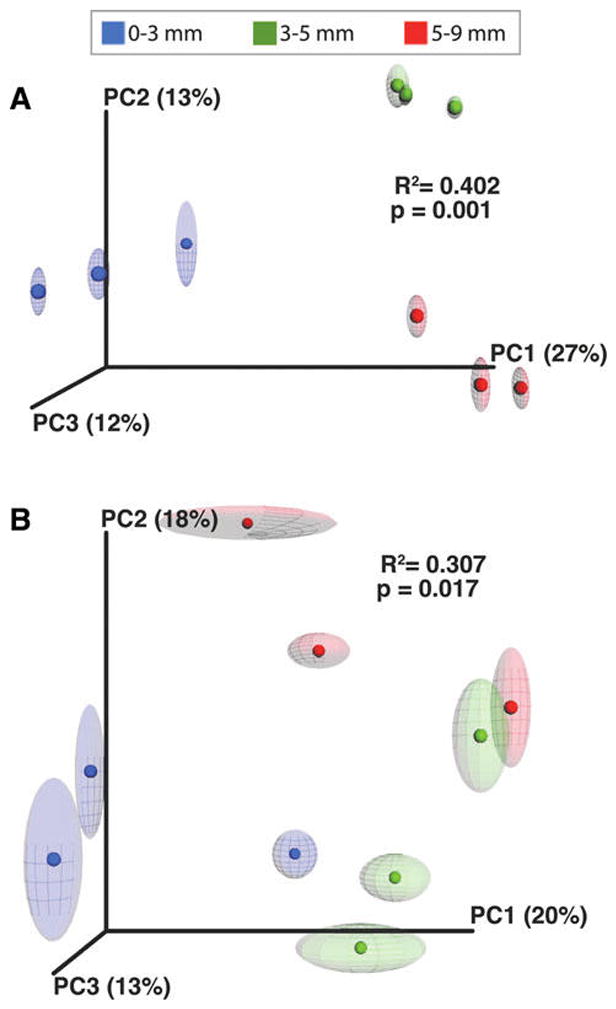
Comparison of diversity analyses of three spatial zones within the thrombolite-forming mats. Principal coordinate analysis of communities from unweighted UniFrac distance matrix of Zone 1 (0–3 mm, blue), Zone 2 (3–5 mm, green), and Zone 3 (5–9 mm, red) in (A) Bacteria and (B) Archaea populations. Ellipses represent standard deviation over 10 rarefaction samplings. Adonis tests suggest that depth is a significant predictor of community composition for both bacterial (R = 0.402, p = 0.001) and archaeal (R = 0.307, p = 0.017) communities.
3.3. Phylogenetic composition of archaea in thrombolite communities with depth
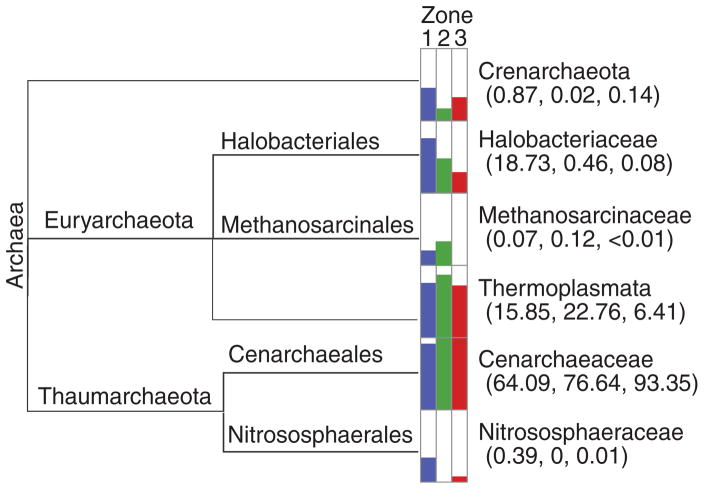
With regard to the overall archaeal diversity (e.g., Shannon Index), there was little difference across the three zones, with the recovered OTUs ranging from 506 to 671 (Table 1; Supplementary Fig. S1B). Of the three recovered phyla, the Thaumarchaeota were dominant in all three zones of the thrombolite-forming mats, with most of the reads sharing similarity to the ammonia-oxidizing family Cenarchaeaceae (Fig. 5), specifically the genus Nitrosopumilus. There were, however, some taxonomic differences between the different spatial regions in the thrombolites. For example, phototrophic Halobacteriales showed the highest abundance in the upper oxic Zone 1, as did the ammonia oxidizer Nitrososphaeraceae (Fig. 5). Although few methanogenic archaeal taxa were detected in each of the three zones, they had the highest representation in the transitional Zone 2 with most of the reads sharing similarity to the class Thermoplasmata and the family Methanosarcinaceae (Fig. 5). A beta diversity test was also completed for the archaeal libraries and showed increased statistical variation between replicates (Fig. 4B). Although the archaeal populations did not have as high an effect size (R2) as the bacterial population, 30% of variation within the archaea could be explained by environmental factors, such as depth. Based on the beta diversity analysis, just as in the bacterial population, the three zones did appear to have spatially distinct Archaea populations, with approximately 20% of the variation between the zones likely being associated with depth (p = 0.017; R2 = 0.307, adonis; Fig. 4B).
FIG. 5.
Taxonomic distribution of Archaea within the thrombolite-forming mats derived from MEGAN5 using the Greengenes database. Overall percentages based on read counts are presented logarithmically, depicting the distributions for Zone 1 (blue), Zone 2 (green), and Zone 3 (red). Read abundance data for each taxonomic level are included in parentheses.
3.4. Spatial profiling of functional gene complexity of thrombolite-forming mats using predictive sequencing analysis
In addition to profiling the microbial diversity within the thrombolite button mat, a reconstruction of the functional gene complexity was generated for each zone by using the 16S rRNA gene sequences and the PICRUSt algorithm (Langille et al., 2013). As the number of available reference genomes has steadily increased, PICRUSt has emerged as an effective tool to accurately predict the functional complexity of the metagenomes based on taxonomic information (Langille et al., 2013). The tool has successfully been used to reconstruct the metagenomes of a wide range of ecosystems including nonlithifying microbial mats and stromatolites (Langille et al., 2013; Casaburi et al., 2016). A predicted metagenome was generated for each spatial zone with the QIIME taxonomic output, which was then statistically compared to a previously published metagenome of the entire button mat (0–9 mm; Mobberley et al., 2013) to determine whether differences in the metabolic capabilities could be observed between zones. The previously sequenced thrombolite metagenome was re-annotated by using MetaCV to update the metagenomic data set and enabling comparable annotations to the PICRUSt predictive metagenomes.
A total of 272 KEGG functions were identified in the three zones corresponding to 328 Level 3 KO entries, which was consistent with the 268 KEGG functions observed in the re-annotated whole-mat metagenome (Supplementary Table S2). Additionally, there was a strong correlation between the PICRUSt predictive metagenomes and the whole mat metagenome (r = 0.93, Pearson), with most of the KOs (n = 222) showing little or no variation between zones (Supplementary Table S2). Of the 59 KOs that did show variation (> 0.1%), several of the differences occurred between the upper oxic Zone 1 and the two deeper Zones 2 and 3 (Fig. 6). In Zone 1, there was an increase in the relative abundance of KO pathways associated with photosynthesis, including the antennae proteins, porphyrin, and chlorophyll metabolism, whereas there was a lower abundance of genes associated with carboxylic acid metabolism (e.g., butanoate, benzoate, caprolactam metabolism; Fig. 6). Deeper within the mat in Zones 2 and 3 there was a higher relative abundance of genes associated with fatty acid metabolism and lipopolysaccharide (LPS) biosynthesis compared to Zone 1. Despite these few select differences, many highly represented pathways in the thrombolite-forming mats, such as DNA repair proteins, two-component signaling, and bacterial motility, showed no differences among the three spatial zones and likely reflect the central metabolisms associated with the thrombolite microbiome.
FIG. 6.
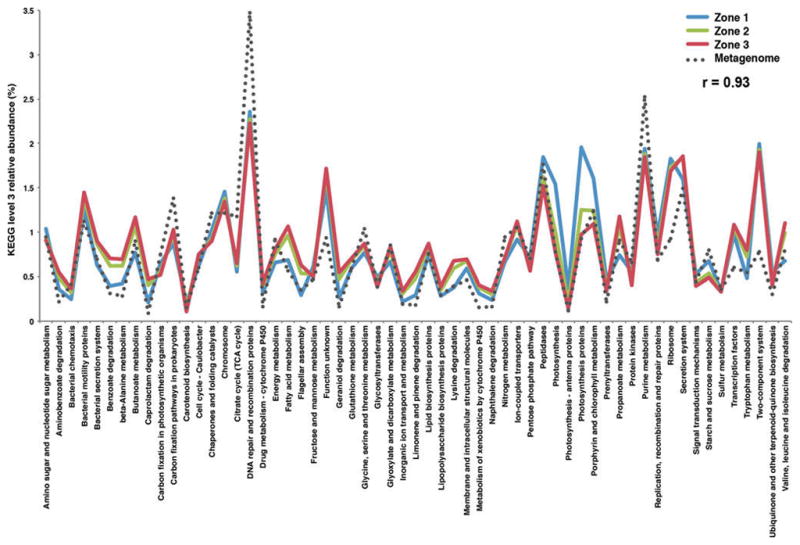
Functional gene comparison of the three thrombolitic mat spatial zones from 16S rRNA metabolic prediction (PICRUSt) and whole shotgun sequencing. Pearson correlation value (r) is shown for the comparison of metabolic predictions for Zone 1 (blue), Zone 2 (green), and Zone 3 (red) and the whole mat shotgun metagenome.
3.5. Stable isotope analyses of thrombolitic carbonates
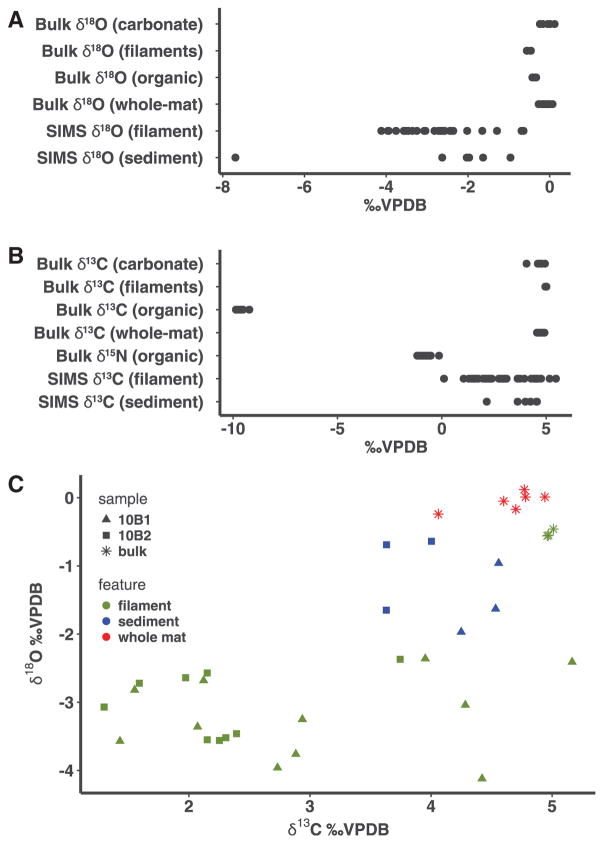
The calcified carbonate filaments associated with the Dichothrix cyanobacteria in the upper Zone 1 were examined by using a combined bulk isotopic analysis and targeted SIMS approach coupled, which enabled an in situ high-spatial resolution analysis (Kozdon et al., 2009; Valley and Kita 2009; Kita et al., 2011) (Fig. 7). Bulk samples of dissected calcified filaments had δ18O values with a mean of −0.5 ± 0.1‰ VPDB, suggesting that the precipitates associated with the filaments were not the result of evaporation, which would cause an enrichment in heavy isotopes. Bulk δ13Ccarb values of the dissected filaments had a mean of 5.0 ± 0.03‰, which was similar to the surrounding carbonate sediments within the thrombolite structure (+4.0‰ to +4.9‰; mean = 4.6 ± 0.3‰). The δ13C values for the organic matter associated with the filaments were depleted compared to the sediment, with values ranging −9.9‰ and −9.2‰ (mean = −9.6 ± 0.2‰), suggesting a relatively muted fractionation during organic matter uptake, similar to what has been produced in other modern microbial mats (Canfield and Des Marais, 1993). The δ15Norg values associated with the filaments ranged from −1.1‰ to −0.1‰ (mean = −0.8 ±0.3‰), suggesting that nitrogen fixation is a predominant means of N assimilation (Sigman et al., 2009) within the thrombolite-forming mats and correlates with the high number of recovered diazotrophic Cyanobacteria and Alphaproteobacteria from the mats.
FIG. 7.
Stable isotope results for calcified filaments located in the upper 3 mm of thrombolite-forming button mat. (A) Oxygen isotope values of organic and inorganic fractions using both bulk and SIMS analysis. Analyses were completed for both background carbonate precipitates (sediment), calcified filaments (filaments), and untreated whole mat samples. (B) Carbon and nitrogen isotope values of both organic and inorganic fractions using both bulk and SIMS analysis. (C) Comparative plot of SIMS values collected for oxygen and carbon isotopes. All results are expressed in delta notation with respect to the carbon/oxygen Vienna Pee Dee Belemnite (VPDB) or nitrogen air (AIR) standard.
To complement the bulk stable isotope analyses, the calcified filaments were also analyzed in situ with SIMS to provide a higher spatial resolution (10 μm spot size) of the δ18O and δ13C compositions of the calcified filaments. Micrographs depicting the SIMS target sites along the filaments and associated carbonate precipitate are shown in Fig. 8. The δ18O value of the surrounding carbonate sediments ranged from −2.0‰ to −0.6‰ (mean = −1.3 ± 0.5‰), whereas the filaments exhibited a more depleted oxygen signature ranging from −7.7‰ to −2.0‰ (mean = −3.2 ± 1.1‰) (Fig. 7). The δ13C values of the surrounding sediments (i.e., ooids) in the thrombolite button mats had a narrow range of values (+3.6‰ to +4.6‰; mean = 4.1 ± 0.4‰) and matched values from previous studies of sediments that surround the thrombolite structures (e.g., ooids; Swart and Eberli, 2005), whereas the filaments had a much wider range (+0.1‰ to +5.5‰; mean = 2.7 ± 1.3‰). All stable isotope measurements are presented in order of analysis in supplementary Tables S4 and S5.
FIG. 8.
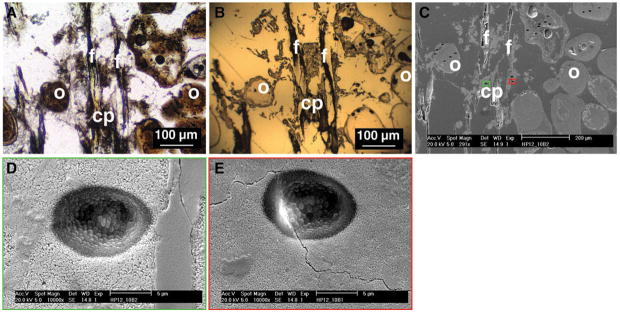
Overview of target areas for SIMS analyses within the thrombolite-forming mat. (A) Petrographic thin section of Dichothrix sp. filaments (f) and associated carbonate precipitate (cp) surrounded by sediments such as ooids (o). (B) Gold-coated reflected light image as viewed by the SIMS instrument. (C) Scanning electron micrograph showing the numerous 6–10 μm pits formed during the SIMS analysis. Boxes depict representative pits that show both high (green) and low (red) quality targets within the sample. (D) Higher-resolution scanning electron micrograph of representative high-quality pit (corresponding to green box in C) showing no textural anomalies or cracks. (E) Scanning electron micrograph of low-quality pit (corresponding to red box in C) showing crack within the targeted sample site. All low-quality target sites were removed from downstream analyses.
4. Discussion
4.1. Microbial diversity within thrombolite-forming mats is highly structured
The presence of discrete spatial zones of microbial and biochemical activity have been well documented in stromatolites (e.g., Visscher et al., 1998; Wong et al., 2015); however, the occurrence of similar zonation in mats that form clotted thrombolites has only been recently suggested (Mobberley et al., 2015). In this study, statistical analysis of the bacterial and archaeal communities revealed significantly different profiles of taxa with depth (Fig. 4), suggesting the microbes are not only active at different depths (Mobberley et al., 2015) but that there are also distinct populations that are forming discrete microenvironments within the thrombolite-forming mats.
In the upper oxic Zone 1, the dominance of cyanobacterial sequences with similarity to the filamentous Rivulariaceae reinforces the morphological observation that Dichothrix sp., a member of the Rivulariaceae, serves as a “hot spot” for photosynthetic activity and carbonate deposition within the thick EPS matrix associated with the filaments (Planavsky et al., 2009). Future sequencing of the Dichothrix sp. genome will help expand the relatively small database of filamentous, heterocystous cyanobacteria as well as delineate the specific pathways associated with EPS production in this keystone organism. In addition to the cyanobacteria, taxonomic analyses also revealed an enrichment of diazotrophic photoorganoheterotrophs primarily associated with the Rhodobacterales, Rhodospirillales, and Rhizobiales increasing with depth (Fig. 3). These metabolically flexible Alphaproteobacteria are ubiquitous in marine microbial communities including all previously characterized microbialites, coral symbioses, and sediments (e.g., Dang et al., 2013; Houghton et al., 2014; Wong et al., 2015; Casaburi et al., 2016; Hester et al., 2016; Suosaari et al., 2016) and may be contributing to the carbon fixation rates deeper within the thrombolitic mats where there are fewer cyanobacteria due to the reduced light levels and the presence of sulfide. Additionally, the diazotrophic photoheterotrophs may be helping to maintain the bioavailability of nitrogen in the thrombolite-forming communities.
Another key microbial functional group enriched within the thrombolite-forming communities was sulfate-reducing bacteria (SRB), whose activity has been directly correlated to deposition of carbonate in actively accreting stromatolites (Visscher et al., 2000; Decho et al., 2010). There was a pronounced vertical stratification of SRB in the thrombolite-forming communities. Taxa associated with Desulfovibrionaceae were enriched in the upper oxic Zone 1, whereas the Desulfobacteraceae increased in their relative abundance with depth. This vertical stratification of SRB has been seen in the nonlithifying hypersaline mats of Guerrero Negro, Mexico (Risatti et al., 1994) and Solar Lake, Egypt (Minz et al., 1999). Several species of sulfate-reducing Desulfovibrionaceae (e.g., Desulfovibrio spp. and Desulfomicrobium spp.) have been shown to be prevalent in the oxic zone of microbial mats (Krekeler et al., 1997), and high levels of sulfate reduction activity have been recorded in the upper oxic zone of nonlithifying and stromatolite-forming mats (e.g., Canfield and Des Marais, 1991; Visscher et al., 1992, 2000). The abundance of SRB in the oxic zone may be, in part, due to the presence of sulfide-oxidizing bacteria (SOB). There was an enrichment of the families Thiotrichaceae and Chromatiaceae in the upper Zone 1, which are known to harbor many sulfide-oxidizing taxa (Pfennig and Trüper, 1992; Lenk et al., 2011). The SOB may be removing the O2 and S2− generated by the cyanobacteria and SRB, both of which can be toxic to the SRB at high enough levels (Decho et al., 2010). Together, this enrichment of SOB, oxygen-tolerant SRB, and their vertical stratification in the thrombolite-forming mat suggest that, much like the stromatolites, these different phylogenetic groups may be playing distinctive community functions in response to variable carbon and electron donor availability at different depths as well as the diel flux of oxygen and sulfide.
The archaeal population also exhibited stratification of certain taxa within the thrombolite-forming mat. There was an enrichment of Halobacteriales in the upper oxic Zone 1 of the thrombolitic mats. Members of this order are typically chemoorganoheterotrophic and can grow on a wide range of sugars, carboxylic acids, alcohols, and amino acids. This aerobic taxon has been observed in both lithifying and nonlithifying microbial mat communities primarily in hypersaline environments (Burns et al., 2004; Arp et al., 2012; Schneider et al., 2013) and may be contributing to the heterotrophic degradation of EPS material associated with the calcified filaments. It should be noted that the salinity of the pore water in the upper part of the microbialites increases significantly (~135 PSU; Visscher, unpublished) upon exposure to the atmosphere during low tide, creating temporary hypersaline conditions.
Sequences were also recovered from methanogenic archaea in primarily Zone 2, and these were primarily associated with the Methanocarcinaceae and Thermoplasmata. These taxonomic results correspond to recovered methyltransferase-encoding genes in the thrombolite metagenome (Mobberley et al., 2013), and there was an enrichment of recovered sequences from Zone 2 (Fig. 5). Members of the Methanocarcinaceae can perform methanogenesis using CO2, acetate, and C1 compounds (Feist et al., 2006) and have been shown to elevate pH levels in mat communities via CO2 consumption (Kenward et al., 2009). More recently, methanogenic lineages of the Thermoplasmata have been identified in human and rumen gut microbiomes as well as wastewater sludge habitats that also can use methanogenic substrates (Dridi et al., 2012; Iino et al., 2013; Poulsen et al., 2013). Although not the dominant archaea within the thrombolite-forming mats, the recovered taxa in this study coupled with functional genes observed in the thrombolite metagenome (Mobberley et al., 2013) suggest that methanogenesis may have a potential, albeit minor, role in promoting an alkaline environment within these thrombolitic mats. Methane production has been observed within the thrombolites and adjacent stromatolites (Visscher, unpublished), and further work to more fully characterize methane levels within each zone may help elucidate the role of methanogenesis, if any, in the Bahamian thrombolite formation.
By far the largest component of the archaeal population within the thrombolite-forming mat was the Thaumarchaeota, specifically Cenarchaeaceae, which harbor many ammonia-oxidizing taxa (Fig. 5). Although the Cenarchaeaceae were found in all three zones, there was an enrichment in the lower two regions of the mat (Fig. 5). Thaumarchaeota have been found in a wide range of lithifying and nonlithifying microbial mat habitats (e.g., Ruvindy et al., 2016) and likely play a role in nitrogen cycling within the thrombolite-forming communities. Previous studies in which the metagenomics of lithifying systems have been examined found a paucity of bacterial nitrification genes (Breitbart et al., 2009; Mobberley et al., 2013; Ruvindy et al., 2016); and ammonia-oxidizing archaea, such as those taxa within the Thaumarchaeota, may be facilitating the metabolism of ammonia to nitrite.
4.2. Predictive metagenome reconstruction shows strong correlation with taxa and function
The PICRUSt predictive metagenome strongly correlated (r = 0.93) with the previously published whole shotgun library, which had targeted the entire thrombolite-forming mat community and provided no spatial information regarding the metagenome (Mobberley et al., 2013). The PICRUSt reconstruction identified key differences between the different spatial zones, thereby providing further evidence that 16S rRNA gene libraries can provide useful insight into the metabolic capabilities of microbial ecosystems. For example, there was extensive overlap in the relative abundance of functional genes between the different depths in several pathways, such as nucleotide and amino acid metabolism, genetic information processing, and environmental information responses, with the shotgun sequence library suggesting there are key central metabolisms in the thrombolite-forming mat microbiome at all depths (Fig. 6). Additionally, genes associated with several key metabolisms associated with the promotion (e.g., photosynthesis, sulfate reduction) and dissolution (e.g., sulfide oxidation, fermentation, ammonia oxidation) of carbonate precipitation were observed within the thrombolite-forming mats.
Despite the extensive overlap between the core metagenome at each depth, differences were observed between the mat zones. The enrichment of genes associated with photosynthesis pathways in the upper Zone 1 and the increase of genes associated with different carboxylic, fatty acid, and LPS metabolisms deeper within the mat reveal distinctive metabolic transitions throughout the mat profile. The increase in LPS production at depth likely reflects the oxygen-limiting environment deeper in the thrombolite-forming mat. Previous studies with model organisms, such as Escherichia coli and Pseudomonas aeruginosa, have shown that anaerobic conditions can positively regulate production of LPS (Landini and Zehnder, 2002; Sabra et al., 2003). These spatial differences in metabolic capabilities are also reflected in the biochemical gradients observed within the mats (Fig. 1). These functional genes could serve as ideal targets to examine the potential regulation of these metabolisms within the thrombolite ecosystems, potentially providing insight into the molecular response to changing environmental variables, such as pH, oxygen, and sulfide. Additionally, by tracking these specific molecular pathways, it may be possible to elucidate the specific genes and taxa involved in the diagenetic alteration of organic material in the thrombolites over both spatial and temporal scales, which represents an important area of future microbialite research.
4.3. Stable isotope profiling suggests photosynthesis is the major inducer of precipitation in thrombolite-forming mats
In addition to the microbial and functional gene analyses, the stable isotope profiling provided new insights into the microbial nitrogen cycling and the mechanisms driving carbonate precipitation. Additionally, the SIMS approach enabled for one of the most highly spatially resolved carbonate oxygen and carbon isotopic data sets to date on modern thrombolites. Organic N isotope values approached 0‰, indicating nitrogen fixation was the dominant N source (Hoering and Ford, 1960; Minagawa and Wada, 1986; Sigman et al., 2009), which is consistent with the abundance of heterocystous cyanobacteria, such as Dichothrix sp., and numerous nitrogen-fixing anoxygenic phototrophs identified in Zone 1 (Fig. 8). These results are also consistent with the high number of nitrogen fixation genes (e.g., nifD, nifH, nifK) recovered from the metagenome and metatranscriptome of the thrombolites (Mobberley et al., 2015). Additionally, the enrichment of ammonia-oxidizing archaea and high number of recovered transcripts associated with ammonia mono-oxygenase (amoA; Mobberley et al., 2015) within the mat coupled with the low numbers of bacterial nitrification genes observed in both the predictive and whole shotgun libraries suggest that these archaeal chemolithotrophs may be playing an important role in controlling nitrification and the cycling of fixed nitrogen within the thrombolite-forming mats.
Analysis of δ18O values by using both bulk and SIMS analyses did not provide evidence of an evaporative signal, the results of which suggest biologically induced precipitation. The high rates of photosynthesis within the thrombolite-forming mats (Myshrall et al., 2010) coupled with the previously published observations that red algae distributed throughout the tufts of Dichothrix sp. filaments lack precipitates (Planavsky et al., 2009) make it unlikely that nonbiological processes, such as CO2 degassing, are driving the precipitation within the thrombolites. The SIMS δ18O values for filaments are highly depleted compared to the values associated with the sediments, and previous studies have shown that increased 18O depletion under elevated pH (Spero and Lea, 1996) is potentially suggestive of rapid rates of carbonate precipitation (McConnaughey, 1989). However, the offset between the bulk and SIMS δ18O values cannot yet be fully explained, as systematically lower SIMS values have been observed up to 2‰ (Orland et al., 2015) and may be the product of water or organics within the sample site. Despite this potential, there is low variability in the 16OH/16O values (Supplementary Table S4), which suggests that the zonation revealed by the SIMS data is accurate. The difference between SIMS and bulk measurements may, in part, reflect the extensive grinding during sample preparation for bulk isotope analysis. Previous studies in corals have shown that the friction generated during milling or drilling of the carbonate samples can cause inversion of aragonite to calcite (Waite and Swart, 2015). As a result of extensive processing (e.g., milling), the δ18O values cause correction errors from 0.2‰ per 1% of inversion from aragonite to calcite (Waite and Swart, 2015). Such differences between the two approaches reinforce the value of using a SIMS-based approach to capture the extensive variability that likely exists within the microenvironments of thrombolite-forming mats.
The bulk δ13C values of the organic matter associated within the thrombolites were heavy (mean −9.6 ± 0.2‰) relative to RuBisCO-mediated carbon fixation, which exhibits fractionations that typically span between −35‰ and −23‰ in both plant and microbial ecosystems and can be highly species-dependent (Farquhar et al., 1989; Falkowski, 1991). The values also appear heavier than other known microbialite systems. For example, unlaminated nodules of Pavilion Lake exhibit a mean bulk organic δ13C value of −26.8‰ (Brady et al., 2010), and microbialites in Cuatros Ciénegas range from −25‰ to −27‰ (Breitbart et al., 2009). These δ13C-enriched values in the Bahamian thrombolites may reflect diffusion limitations of CO2 into the intertidal microbialites, differences in light intensities (Cooper and DeNiro, 1989), and the relatively high rates of photosynthesis (Myshrall et al., 2010). Values of organic δ13C similar to the Bahamian thrombolites have been observed in microbial mats found in the hypersaline Solar Lake (−5.7 ± 1.4‰) and Gavish Sabkha (−10 ± 2.6‰) and have been attributed to EPS-rich materials on the surface of mats that impede transport of CO2 into the mats (Schidlowski et al., 1984). Previous studies have also shown that external factors, such as increased salinity and temperature, also decrease the solubility of CO2 (Mucci, 1983). Therefore, the abundance of EPS material within the thrombolite-forming mats coupled with high rates of productivity (Myshrall et al., 2010) may result in a potential shortage of CO2 that may reduce isotopic discrimination of 13C and is consistent with the idea of HCO3 dissociation driving a pH shift and inducing carbonate precipitation.
The overall carbon isotope profiles of the carbonate suggest that the thrombolites of Highborne Cay are primarily the result of photoautotrophic carbon fixation, which correlates to several lacustrine microbialite systems, such as Lake Clifton, Pavilion Lake, Great Salt Lake, Green Lake, and Bacalar (for review, see Chagas et al., 2016). The bulk isotope data for carbonates also correlates well with previous analyses on the Dichothrix calcified filaments (Planavsky et al., 2009), as well as several lacustrine microbialites, such as Pavilion Lake (−1.2–2.3‰; Brady et al., 2010; Russell et al., 2014), Kelly Lake (4–5‰; Ferris et al., 1997), Lake Van (6‰), Lake Alchichia (6.5‰), and Great Salt Lake (4.2‰) (Chagas et al., 2016). Interestingly, the thrombolite measurements are also higher than the δ13C values of the adjacent stromatolites located only a few meters away in the subtidal zone. The discrepancy may reflect the role of heterotrophic processes in carbonate precipitation in the Bahamian stromatolites (Andres et al., 2006), and similar results have been observed in the freshwater microbialites of Cuatros Ciénegas (Breitbart et al., 2009), suggesting that heterotrophic process may be also be influencing carbonate precipitation in the Mexican system.
Although SIMS data from Highborne Cay thrombolites show greater variability than bulk isotopes, the means are not statistically different. Some of the extensive variability in the SIMS δ13Ccarb values for filaments is tied to variations in the microenvironments along the vertically orientated cyanobacteria filaments. The lightest SIMS δ13C values in filaments may reflect the presence of localized organics (e.g., EPS material) associated with the calcified filaments, given that organic carbon has higher ionization efficiency than carbonate. However, as SIMS threshold cutoffs were applied to eliminate any spots that might include organics, the lower δ13C values likely accurately capture filament carbonate values. In contrast, the isotopically enriched samples, relative to values predicted from precipitation from local marine dissolved inorganic carbon, provide evidence for carbonate precipitation in a microenvironment influenced by carbon dioxide uptake, which increases the pH (Visscher et al., 1991, 1998; Visscher and Stolz, 2005; Planavsky et al., 2009). The highest SIMS δ13C values are more isotopically enriched than any previously reported Highborne Cay bulk thrombolite or filament δ13C values (Planavsky et al., 2009). Planavsky et al. (2009) used an offset between Dichothrix filament and detrital sediment δ13C values to argue for photosynthetic carbon dioxide consumption as the initiation factor for carbonate precipitation within the filament sheaths. The observed markedly enriched filament δ13C values strengthen the case for a photosynthetic carbonate precipitation trigger in the Bahamian thrombolites.
5. Conclusions
The integrated approaches of microbial diversity, meta-genome prediction, microelectrode, and stable isotope analysis address several important gaps in our previous understanding of modern thrombolite-forming communities. This study provides a comprehensive spatial portrait of thrombolite-forming communities revealing that, despite having un-laminated, clotted microstructures, these thrombolitic communities form distinct taxonomic and metabolic stratifications. Additionally, the SIMS results, the first ever generated for a microbialite-forming ecosystem, reveal SIMS δ13C values that are more isotopically enriched than any previously reported bulk thrombolite values (Planavsky et al., 2009; Warden et al., 2016), providing direct evidence of a photosynthetic trigger for carbonate precipitation in the thrombolite-forming communities, which differs from stromatolites. Even within the same environment, where thrombolites are juxtaposed to stromatolites under similar environmental conditions (e.g., pH, salinity, temperature, UV flux), these differences between their taxa and metabolic activities appear to generate very distinct carbonate microstructures. Elucidating how these disparate structural fabrics arise will require a more detailed look into the networking and connectivity of the microbial interactions and metabolisms. Regulation of these processes on both diel and seasonal timescales will help assess the patterns associated with microbial activities and their response to their changing environment. Together, these analyses help elucidate the pathways associated with microbialite formation and represent a valuable tool to help reconstruct the microbiological and environmental conditions of the past.
Supplementary Material
Acknowledgments
The authors would like to thank Jennifer Larimore for her technical assistance. A.S.L was supported by the NSF Graduate Research Fellowship Program, and J.M.M was supported by the NASA Graduate Student Research Program fellowship (NNX10AO18H). The research efforts were supported by the NASA Exobiology and Evolutionary Biology program element (NNX12AD64G). WiscSIMS is funded by NSF (EAR1355590).
Abbreviations Used
- EPS
exopolymeric substances
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOs
Kyoto Encyclopedia of Genes and Genomes Orthologs
- LPS
lipopolysaccharide
- MEGAN5
Meta Genome Analyzer
- MetaCV
Metagenome Composition Vector
- OTUs
operational taxonomic units
- PCoA
principal coordinate analysis
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- QIIME
Quantitative Insights Into Microbial Ecology
- SEM
scanning electron microscopy
- SIMS
secondary ion mass spectrometry, secondary ion microprobe mass spectrometer
- SOB
sulfide-oxidizing bacteria
- SRB
sulfate-reducing bacteria
- VPDB
Vienna Pee Dee Belemnite
Footnotes
Author Disclosure Statement
No competing financial interests exist.
Author Contributions
A.L., J.M., R.R., P.V., and J.F. conceived the experiments. J.M., P.H., J.F., and P.V. collected the samples. All authors contributed to the performance and analysis of the experiments. All authors reviewed and approved the final manuscript.
References
- Aitken JD. Classification and environmental significance of cryptalgal limestones and dolomites, with illustrations from the Cambrian and Ordovician of southwestern Alberta. J Sediment Petrol. 1967;37:1163–1178. [Google Scholar]
- Andres MS, Reid RP. Growth morphologies of modern marine stromatolites: a case study from Highborne Cay, Bahamas. Sediment Geol. 2006;185:319–328. [Google Scholar]
- Andres M, Sumner D, Reid RP, Swart PK. Isotopic fingerprints of microbial respiration in aragonite from Bahamian stromatolites. Geology. 2006;34:973–976. [Google Scholar]
- Arp G, Helms G, Kalinska K, Schumann G, Reimer A, Reitner J, Trichet J. Photosynthesis versus exopolymer degradation in the formation of microbialites on the atoll of Kiritimati, Republic of Kiribati, Central Pacific. Geomicrobiol J. 2012;29:29–65. [Google Scholar]
- Barns SM, Fundyga RE, Jeffries MW, Pace NR. Remarkable archaeal diversity detected in a Yellow-stone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard JM, Edgcomb VP, Visscher PT, McIntyre-Wressnig A, Summons RE, Bouxsein ML, Louis L, Jeglinski M. Insights into foraminiferal influences on microfabrics of microbialites at Highborne Cay, Bahamas. Proc Natl Acad Sci USA. 2013;110:9830–9834. doi: 10.1073/pnas.1221721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlin EM, Klaus J, Foster JS, Andres M, Custals L, Reid RP. Environmental controls on microbial community cycling in modern marine stromatolites. Sediment Geol. 2012;263–264:45–55. [Google Scholar]
- Brady AL, Slater GF, Omelon CR, Southam G, Druschel G, Andersen DT, Lim DSS. Photosynthetic isotope biosignatures in laminated micro-stromatolitic and non-laminated nodules associated with modern, freshwater microbialites in Pavilion Lake, BC. Chem Geol. 2010;274:56–67. [Google Scholar]
- Braissant O, Decho AW, Przekop KM, Gallagher KL, Glunk C, Dupraz C, Visscher PT. Characteristics and turnover of exopolymeric substances in a hypersaline microbial mat. FEMS Microbiol Ecol. 2009;67:293–307. doi: 10.1111/j.1574-6941.2008.00614.x. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E, Edwards R, Souza V, Rohwer F, Hollander D. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Ciénegas, Mexico. Environ Microbiol. 2009;11:16–34. doi: 10.1111/j.1462-2920.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- Burne RV, Moore LS. Microbialites: organosedimentary deposits of benthic microbial communities. Palaios. 1987;2:241–254. [Google Scholar]
- Burns BP, Goh F, Allen M, Neilan BA. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ Microbiol. 2004;6:1096–1101. doi: 10.1111/j.1462-2920.2004.00651.x. [DOI] [PubMed] [Google Scholar]
- Canfield DE, Des Marais DJ. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- Canfield DE, Des Marais DJ. Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochim Cosmochim Acta. 1993;57:3971–3984. doi: 10.1016/0016-7037(93)90347-y. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaburi G, Duscher AA, Reid RP, Foster JS. Characterization of the stromatolite microbiome from Little Darby Island, The Bahamas using predictive and whole shotgun metagenomic analysis. Environ Microbiol. 2016;18:1452–1469. doi: 10.1111/1462-2920.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor EO, Massana R, Benlloch S, Ovreas L, Diez B, Goddard VJ, Gasol JM, Joint I, Rodriguez-Valera F, Pedros-Alio C. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- Chagas AAP, Webb GE, Burne RA, Southam G. Modern lacustrine mcirobialites; towards a synthesis of aquous and carbonate geochemistry and minerology. Earth-Science Reviews. 2016;162:338–363. [Google Scholar]
- Cooper LW, DeNiro MJ. Stable carbon isotope variability in the seagrass Posidonia oceanica: evidence for light intensity effects. Mar Ecol Prog Ser. 1989;50:225–229. [Google Scholar]
- Dang H, Yang J, Li J, Luan X, Zhang Y, Gu G, Xue R, Zong M, Klotz MG. Environment-dependent distribution of the sediment nifH-harboring microbiota in the Northern South China Sea. Appl Environ Microbiol. 2013;79:121–132. doi: 10.1128/AEM.01889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decho AW, Norman RS, Visscher PT. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 2010;18:73–80. doi: 10.1016/j.tim.2009.12.008. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis JJ. Hardened subtidal stromatolites, Bahamas. Science. 1983;219:385–386. doi: 10.1126/science.219.4583.385. [DOI] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 2012;62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- Dupraz C, Visscher PT. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005;13:429–438. doi: 10.1016/j.tim.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Dupraz C, Visscher PT, Baumgartner LK, Reid RP. Microbe-mineral interactions: early carbonate precipitation in a hypersaline lake (Eleuthera Island, Bahamas) Sedimentology. 2004;51:1–21. [Google Scholar]
- Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. Processes of carbonate precipitation in modern microbial mats. Earth-Science Reviews. 2009;96:141–162. [Google Scholar]
- Edgcomb VP, Bernhard JM, Beaudoin D, Pruss S, Welander PV, Schubotz F, Mehay S, Gillespie AL, Summons RE. Molecular indicators of microbial diversity in oolitic sands of Highborne Cay, Bahamas. Geobiology. 2013;11:234–251. doi: 10.1111/gbi.12029. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- Falkowski PG. Species variability in the fractionation of 13C and 12C by marine phytoplankton. J Plankton Res. 1991;13:21–28. [Google Scholar]
- Farquhar GD, Ehleringer JR, Hubick KT. Carbon isotope discrimination and photosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:503–537. [Google Scholar]
- Feist AM, Scholten JC, Palsson BO, Brockman FJ, Ideker T. Modeling methanogenesis with a genomescale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006:2. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris FG, Thompson JB, Beveridge TJ. Modern freshwater microbialites from Kelly Lake, British Columbia, Canada. Palaios. 1997;12:213–219. [Google Scholar]
- Foster JS, Green SJ. Microbial diversity in modern stromatolites. In: Seckbach J, Tewaris V, editors. Cellular Origin, Life in Extreme Habitats and Astrobiology: Interactions with Sediments. Springer; Dordrecht, The Netherlands: 2011. pp. 385–405. [Google Scholar]
- Gallagher KL, Kading TJ, Braissant O, Dupraz C, Visscher PT. Inside the alkalinity engine: the role of electron donors in the organomineralization potential of sulfate-reducing bacteria. Geobiology. 2012;10:518–530. doi: 10.1111/j.1472-4669.2012.00342.x. [DOI] [PubMed] [Google Scholar]
- Glunk C, Dupraz C, Braissant O, Gallagher KL, Verrecchia EP, Visscher PT. Microbially mediated carbonate precipitation in a hypersaline lake, Big Pond (Elutera, Bahamas) Sedimentology. 2011;58:720–738. [Google Scholar]
- Green SJ, Blackford C, Bucki P, Jahnke LL, PrufertBebout L. A salinity and sulfate manipulation of hypersaline microbial mats reveals stasis in the cyanobacterial community structure. ISME J. 2008;2:457–470. doi: 10.1038/ismej.2008.6. [DOI] [PubMed] [Google Scholar]
- Hester ER, Barott KL, Nulton J, Vermeij NJA, Rohwer FL. Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 2016;10:1157–1169. doi: 10.1038/ismej.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoering T, Ford HT. The isotope effect in the fixation of nitrogen by Azotobacter. J Am Chem Soc. 1960;82:376–378. [Google Scholar]
- Houghton J, Fike D, Druschel G, Orphan V, Hoehler TM, Des Marais DJ. Spatial variability in photosynthetic and heterotrophic activity drives localized delta13C org fluctuations and carbonate precipitation in hypersaline microbial mats. Geobiology. 2014;12:557–574. doi: 10.1111/gbi.12113. [DOI] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K, Igarashi Y, Haruta S. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ. 2013;28:244–250. doi: 10.1264/jsme2.ME12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NA, Fass JN. [accessed May 3, 2017];Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. (Version 1.21) 2011 Available from: github.com/najoshi/sickle.
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard JM, James NP. Thrombolites and stromatolites: two distinct types of microbial structures. Palaios. 1986;1:492–503. [Google Scholar]
- Kenward PA, Goldstein RH, Gonzalez LA, Roberts JA. Precipitation of low-temperature dolomite from an anaerobic microbial consortium: the role of methanogenic Archaea. Geobiology. 2009;7:556–565. doi: 10.1111/j.1472-4669.2009.00210.x. [DOI] [PubMed] [Google Scholar]
- Khodadad CL, Foster JS. Metagenomic and metabolic profiling of nonlithifying and lithifying stromatolitic mats of Highborne Cay, The Bahamas. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita NT, Huberty JM, Kozdon R, Beard BL, Valley JW. High-precision SIMS oxygen, sulfur and iron stable isotope analyses of geological materials: accuracy, surface topography and crystal orientation. Surf Interface Anal. 2011;43:427–431. [Google Scholar]
- Kozdon R, Ushikubo T, Kita NT, Spicuzza M, Valley JW. Intratest oxygen isotope cariability in the planktonic foraminifer N. pachyderma: real versus apparent vital effects by ion microprobe. Chem Geol. 2009;258:327–337. [Google Scholar]
- Kozdon R, Kelly DC, Kita NT, Fournelle JH, Valley JW. Planktonic foraminiferal oxygen isotope analysis by ion microprobe technique suggests warm tropical sea surface temperatures during the Early Paleogene. Paleoceanography. 2011:26. doi: 10.1029/2010PA002056. [DOI] [Google Scholar]
- Krekeler D, Teske AP, Cypionka H. Strategies of sulfate-reducing bacteria to escape oxygen stress in a cyanobacterial mat. FEMS Microbiol Ecol. 1997;25:89–96. [Google Scholar]
- Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval B, Cady SL, Pollack JC, McKay CP, Bird JS, Grotzinger JP, Ford DC, Bohm HR. Modern freshwater microbialite analogues for ancient dendritic reef structures. Nature. 2000;407:626–629. doi: 10.1038/35036579. [DOI] [PubMed] [Google Scholar]
- Lenk S, Arnds J, Zerjatke K, Musat N, Amann R, Mussmann M. Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ Microbiol. 2011;13:758–774. doi: 10.1111/j.1462-2920.2010.02380.x. [DOI] [PubMed] [Google Scholar]
- Linzmeier B, Kozdon R, Peters S, Valley JW. Oxygen isotope variability within Nautilus shell growth bands. PLoS One. 2016:11. doi: 10.1371/journal.pone.0153890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Yang H, Zhang Y, Wang J, Zhao F, Qi J. Composition-based classification of short metagenomic sequences elucidates the landscapes of taxonomic and functional enrichment of microorganisms. Nucleic Acids Res (2013) 2012;41(1):e3. doi: 10.1093/nar/gks828. doi: https://doi.org/10.1093/nar/gks828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan BW. Crytpzoan and associated stromatolties from the Recent, Shark Bay, Western Australia. J Geol. 1961;69:517–533. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnaughey T. 13C and 18O isotopic disequilibrium in biological carbonates: I. Patterns. Geochim Cosmochim Acta. 1989;53:151–162. [Google Scholar]
- Minagawa M, Wada E. Nitrogen isotope ratios of red tide organisms in the East China Sea: a characterization of biological nitrogen fixation. Mar Chem. 1986;19:245–259. [Google Scholar]
- Minz D, Fishbain S, Green SJ, Muyzer G, Cohen Y, Rittmann BE, Stahl DA. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl Environ Microbiol. 1999;65:4659–4665. doi: 10.1128/aem.65.10.4659-4665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobberley JM, Ortega MC, Foster JS. Comparative microbial diversity analyses of modern marine thrombolitic mats by barcoded pyrosequencing. Environ Microbiol. 2012;14:82–100. doi: 10.1111/j.1462-2920.2011.02509.x. [DOI] [PubMed] [Google Scholar]
- Mobberley JM, Khodadad CL, Foster JS. Metabolic potential of lithifying cyanobacteria-dominated thrombolitic mats. Photosynth Res. 2013;118:125–140. doi: 10.1007/s11120-013-9890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobberley JM, Khodadad CL, Visscher PT, Reid RP, Hagan P, Foster JS. Inner workings of thrombolites: spatial gradients of metabolic activity as revealed by metatranscriptome profiling. Sci Rep. 2015:5. doi: 10.1038/srep12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucci A. The solubility of calcite and aragonite in seawater at various salinities, temperatures, and one atmosphere total pressure. Am J Sci. 1983;283:780–799. [Google Scholar]
- Myshrall K, Mobberley JM, Green SJ, Visscher PT, Havemann SA, Reid RP, Foster JS. Biogeochemical cycling and microbial diversity in the modern marine thrombolites of Highborne Cay, Bahamas. Geobiology. 2010;8:337–354. doi: 10.1111/j.1472-4669.2010.00245.x. [DOI] [PubMed] [Google Scholar]
- Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011:12. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orland IJ. PhD dissertation. University of Wisconsin-Madison; Madison, WI: 2012. Seasonality from spleothems: high-resolution ion microprobe studies at Soreq Cave, Israel. [Google Scholar]
- Orland IJ, Bar-Matthews M, Kita NT, Ayalon A, Matthews A, Valley JW. Climate deterioration in the Eastern Mediterranean as revealed by ion microprobe analysis of a speleothem that grew from 2.2 to 0.9 ka in Soreq Cave, Israel. Quat Res. 2009;71:27–35. [Google Scholar]
- Orland IJ, Edwards RL, Cheng H, Kozdon R, Cross M, Valley JW. Direct measurements of deglacial monsoon strength in a Chinese stalagmite. Geology. 2015;43:555–558. [Google Scholar]
- Pages A, Welsh DT, Teasdale PR, Grice K, Vacher M, Bennett WW, Visscher PT. Diel fluctuations in solute distributions and biogeochemical cycling in a hyper-saline microbial mat from Shark Bay, WA. Mar Chem. 2014;167:102–112. [Google Scholar]
- Paul VG, Wronkiewicz DJ, Mormile MR, Foster JS. Mineralogy and microbial diversity of the microbialites in the hypersaline Storr’s Lake, The Bahamas. Astrobiology. 2016;16:282–300. doi: 10.1089/ast.2015.1326. [DOI] [PubMed] [Google Scholar]
- Petrash DA, Gingras MK, Lalonde SV, Orange F, Pecoits E, Konhauser KO. Dynamic controls on accretion and lithification on modern gypsum-dominated thrombolites, Los Roques, Venezuela. Sediment Geol. 2012;245–246:29–47. [Google Scholar]
- Pfennig N, Trüper HG. The Prokaryotes. Springer; New York: 1992. The family Chromatiaceae; pp. 3200–3221. [Google Scholar]
- Planavsky N, Ginsburg RN. Taphonomy of modern marine Bahamian microbialites. Palaios. 2009;24:5–17. [Google Scholar]
- Planavsky N, Reid RP, Andres M, Visscher PT, Myshrall KL, Lyons TW. Formation and diagenesis of modern marine calcified cyanobacteria. Geobiology. 2009;7:566–576. doi: 10.1111/j.1472-4669.2009.00216.x. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Schwab C, Jensen BB, Engberg RM, Spang A, Canibe N, Hojberg O, Milinovich G, Fragner L, Schleper C, Weckwerth W, Lund P, Schramm A, Urich T. Methylotrophic methanogenic Thermo-plasmata implicated in reduced methane emissions from bovine rumen. Nat Commun. 2013:4. doi: 10.1038/ncomms2432. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010:5. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2015. [Google Scholar]
- Reid RP, Macintyre IG, Steneck RS. A microbialite/algal-ridge fringing reef complex, Highborne Cay, Bahamas. Atoll Res Bull. 1999;465:459–465. [Google Scholar]
- Reid RP, Visscher PT, Decho AW, Stolz JF, Bebout BM, Dupraz C, Macintyre IG, Paerl HW, Pinckney JL, PrufertBebout L, Visscher PT, Decho AW, Stolz JF, Bebout BM, Dupraz C, Macintyre IG, Paerl HW, Pinckney JL, PrufertBebout L, Steppe TF, Des Marais DJ. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature. 2000;406:989–992. doi: 10.1038/35023158. [DOI] [PubMed] [Google Scholar]
- Risatti JB, Capman WC, Stahl DA. Community structure of a microbial mat: the phylogenetic dimension. Proc Natl Acad Sci USA. 1994;91:10173–10177. doi: 10.1073/pnas.91.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Brady AL, Cardman Z, Slater GF, Lim DS, Biddle JF. Prokaryote populations of extant microbialites along a depth gradient in Pavilion Lake, British Columbia, Canada. Geobiology. 2014;12:250–264. doi: 10.1111/gbi.12082. [DOI] [PubMed] [Google Scholar]
- Ruvindy R, White RA, III, Neilan BA, Burns BP. Unravelling core microbial metabolisms in the hypersaline microbial mats of Shark Bay using high-throughput metagenomics. ISME J. 2016;10:183–196. doi: 10.1038/ismej.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W, Lunsdorf H, Zeng AP. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology. 2003;149:2789–2795. doi: 10.1099/mic.0.26443-0. [DOI] [PubMed] [Google Scholar]
- Saghaï A, Zivanovic Y, Zeyen N, Moreira D, Benzerara K, Deschamps P, Bertolino P, Ragon M, Tavera R, LopezArchilla AI, López-García P. Metagenome-based diversity analyses suggest a significant contribution of non-cyanobacterial lineages to carbonate precipitation in modern microbialites. Front Microbiol. 2015:6. doi: 10.3389/fmicb.2015.00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schidlowski M, Matzigkeit U, Krumbein WE. Superheavy organic carbon from hypersaline microbial mats. Naturwissenschaften. 1984;71:303–308. [Google Scholar]
- Schneider D, Arp G, Reimer A, Reitner J, Daniel R. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, Central Pacific. PLoS One. 2013:8. doi: 10.1371/journal.pone.0066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman DM, Karsh KL, Casciotti KL. Ocean process tracers: nitrogen isotopes in the ocean. Encyclopedia of Ocean Sciences. 2009;2009:4138–4153. [Google Scholar]
- Spero HJ, Lea DW. Experimental determination of stable isotope variability in Globigerina bulloides: implications for paleoceanographic reconstructions. Mar Micropaleontol. 1996;28:231–246. [Google Scholar]
- Stolz JF, Reid RP, Visscher PT, Decho AW, Norman RS, Aspden RJ, Bowlin EM, Franks J, Foster JS, Paterson DM, Przekop KM, Underwood GJC, PrufertBebout L. The microbial communities of the modern marine stromatolites at Highborne Cay, Bahamas. Atoll Res Bull. 2009;567:1–29. [Google Scholar]
- Suosaari EP, Reid RP, Playford PE, Foster JS, Stolz JF, Casaburi G, Hagan PD, Chirayath V, Macintyre IG, Planavsky NJ, Eberli GP. New multi-scale perspectives on the stromatolites of Shark Bay, Western Australia. Sci Rep. 2016:6. doi: 10.1038/srep20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart PK, Eberli GP. The nature of the δ13C of periplatform sediments: implications for stratigraphy and the global carbon cycle. Sediment Geol. 2005;175:115–129. [Google Scholar]
- Valdespino-Castillo PM, Alcantara-Hernandez RJ, Alcocer J, Merino-Ibarra M, Macek M, Falcon LI. Alkaline phosphatases in microbialites and bacterioplankton from Alchichica soda lake, Mexico. FEMS Microbiol Ecol. 2014;90:504–519. doi: 10.1111/1574-6941.12411. [DOI] [PubMed] [Google Scholar]
- Valley JW, Kita NT. In situ oxygen isotope geochemistry by ion microprobe. In: Fayek M, editor. Secondary Ion Mass Spectrometry in the Earth Sciences: Gleaning the Big Picture from a Small Spot. Mineralogical Association of Canada; Toronto: 2009. pp. 19–63. [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013:2. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter L, Kozdon R, Valley JW, Mora CI, Spero HJ. SIMS measurements of intrashell δ13C in the cultured planktic foraminifer Orbulina universa. Geochim Cosmochim Acta. 2014;139:527–539. [Google Scholar]
- Visscher PT, Stolz JF. Microbial mats as bioreactors: populations, processes and products. Palaeogeogr Palaeoclimatol Palaeoecol. 2005;219:87–100. [Google Scholar]
- Visscher PT, Beukema J, van Gemerden H. In situ characterization of sediments: measurement of oxygen and sulfide profiles with a novel combined needle electrode. Limnol Oceanogr. 1991;36:1476–1480. [Google Scholar]
- Visscher PT, Prins RA, van Gemerden H. Rates of sulfate reduction and thiosulfate consumption in a marine microbial mat. FEMS Microbiol Lett. 1992;86:283–293. [Google Scholar]
- Visscher PT, Reid RP, Bebout BM, Hoeft SE, Macintyre IG, Thompson JA. Formation of lithified micritic laminae in modern marine stromatolites (Bahamas): the role of sulfur cycling. Am Mineral. 1998;83:1482–1493. [Google Scholar]
- Visscher PT, Reid RP, Bebout BM. Microscale observations of sulfate reduction: correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology. 2000;28:919–922. [Google Scholar]
- Waite AJ, Swart PK. The inversion of aragonite to calcite during the sampling of skeletal archives: implications for proxy interpretation. Rapid Commun Mass Spectrom. 2015;29:955–964. doi: 10.1002/rcm.7180. [DOI] [PubMed] [Google Scholar]
- Walter MR. Stromatolites: the main geological source of information on the evolution of the early benthos. Early Life on Earth. 1994;84:270–286. [Google Scholar]
- Warden JG, Casaburi G, Omelon CR, Bennett PC, Breecker DO, Foster JS. Characterization of microbial mat microbiomes in the modern thrombolite ecosystem of Lake Clifton, Western Australia using shotgun metagenomics. Front Microbiol. 2016:7. doi: 10.3389/fmicb.2016.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, III, Power IM, Dipple GM, Southam G, Suttle CA. Metagenomic analysis reveals that modern microbialites and polar microbial mats have similar taxonomic and functional potential. Front Microbiol. 2015:6. doi: 10.3389/fmicb.2015.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, III, Chan AM, Gavelis GS, Leander BS, Brady AL, Slater GF, Lim DS, Suttle CA. Metagenomic analysis suggests modern freshwater microbialites harbor a distinct core microbial community. Front Microbiol. 2016:6. doi: 10.3389/fmicb.2015.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer Science & Business Media; New York: 2009. [Google Scholar]
- Williford KH, Ushikubo T, Lepot K, Kitajima K, Hallmann C, Spicuzza MJ, Kozdon R, Eigenbrode JL, Summons RE, Valley JW. Carbon and sulfur isotopic signatures of ancient life and environment at the microbial scale: Neoarchean shales and carbonates. Geobiology. 2016;14:105–128. doi: 10.1111/gbi.12163. [DOI] [PubMed] [Google Scholar]
- Wong HL, Smith DL, Visscher PT, Burns BP. Niche differentiation of bacterial communities at a millimeter scale in Shark Bay microbial mats. Sci Rep. 2015:5. doi: 10.1038/srep15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.