SUMMARY
Gut microbiota are linked to chronic inflammation and carcinogenesis. Chemotherapy failure is the major cause of recurrence and poor prognosis in colorectal cancer patients. Here, we investigated the contribution of gut microbiota to chemoresistance in patients with colorectal cancer. We found that Fusobacterium (F.) nucleatum was abundant in colorectal cancer tissues in patients with recurrence post chemotherapy, and was associated with patient clinicopathological characterisitcs. Furthermore, our bioinformatic and functional studies demonstrated that F. nucleatum promoted colorectal cancer resistance to chemotherapy. Mechanistically, F. nucleatum targeted TLR4 and MYD88 innate immune signaling and specific microRNAs to activate the autophagy pathway and alter colorectal cancer chemotherapeutic response. Thus, F. nucleatum orchestrates a molecular network of the Toll-like receptor, micro-RNAs, and autophagy to clinically, biologically, and mechanistically control colorectal cancer chemoresistance. Measuring and targeting F. nucleatum and its associated pathway will yield valuable insight into clinical management and may ameliorate colorectal cancer patient outcomes.
In Brief
Reducing a specific gut microbe in colorectal cancer patients may improve their response to chemotherapy and reduce cancer recurrence.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death worldwide (Cartwright, 2012; Siegel et al., 2013). In advanced CRC patients, the purpose of chemotherapy is to shrink tumor size, reduce tumor growth, and inhibit tumor metastasis. In general, active cytotoxic drugs, including 5-fluorouracil (5-FU) and capecitabine, inhibit the enzyme activity of thymidylate synthase during DNA replication (Walko and Lindley, 2005). Oxaliplatin, another chemotherapy drug, inhibits tumor cell growth and causes cell G2 phase arrest by covalently binding DNA and forming platinum-DNA adducts (Kelland, 2007). The combination of these chemotherapeutic agents is widely used in the treatment of CRCs (Cartwright, 2012). The majority of patients with advanced CRC are initially responsive to the combined chemotherapy. However, the patients eventually experience tumor recurrence due to drug resistance, and the 5 year survival rate is lower than 10% in advanced CRC patients (Dahan et al., 2009). Unfortunately, colon cancer patients are generally not responsive to novel immune checkpoint therapy (Zou et al., 2016). Thus, it is of paramount importance to elucidate the mechanism of chemotherapy resistance in CRC patients.
Cancer chemoresistance results from a complex interplay between gene regulation and the environment. The microbiota is linked to CRC initiation and progression via affecting intestinal inflammation (Arthur et al., 2012; Garrett, 2015; Man et al., 2015; Zitvogel et al., 2015) and tumor-related signaling pathways (Schwabe and Jobin, 2013). Recent mouse studies have shown that the gut microbiota may modulate local immune responses and in turn affect chemotherapy (Iida et al., 2013; Viaud et al., 2013) and immunotherapy (Sivan et al., 2015; Vétizou et al., 2015). Human gut microbiota are linked to inflammatory cytokine production (Schirmer et al., 2016). Two groups have recently shown that the abundance of F. nucleatum is gradually increased from normal tissues to adenoma tissues and to adenocarcinoma tissues in colorectal carcinogenesis (Castellarin et al., 2012; Kostic et al., 2012). Moreover, the amount of F. nucleatum in CRC tissues is associated with shorter survival (Mima et al., 2016). F. nucleatum adhesin FadA may bind to the E-cadherin protein and promote colorectal carcinogenesis (Rubinstein et al., 2013). In addition, F. nucleatum lectin Fap2 may recognize the host Gal-GalNAc and help this bacterium localize abundantly in colon cancer epithelial cells (Abed et al., 2016). However, the potential effect of F. nucleatum on chemotherapy is not examined in human literature. Here, we tested whether and how F. nucleatum affected chemotherapy in CRC patients. We have found that the amount of F. nucleatum is increased in CRC patients with recurrence post-chemotherapy, compared with those with non-recurrence post-chemotherapy. We have demonstrated that F. nucleatum plays a critical role in mediating CRC chemoresistance in response to small drug chemotherapeutics via a selective target loss of miR-18a* and miR-4802, and activation of the autophagy pathway.
RESULTS
F. nucleatum Is Associated with Colorectal Cancer Recurrence and Patient Outcome
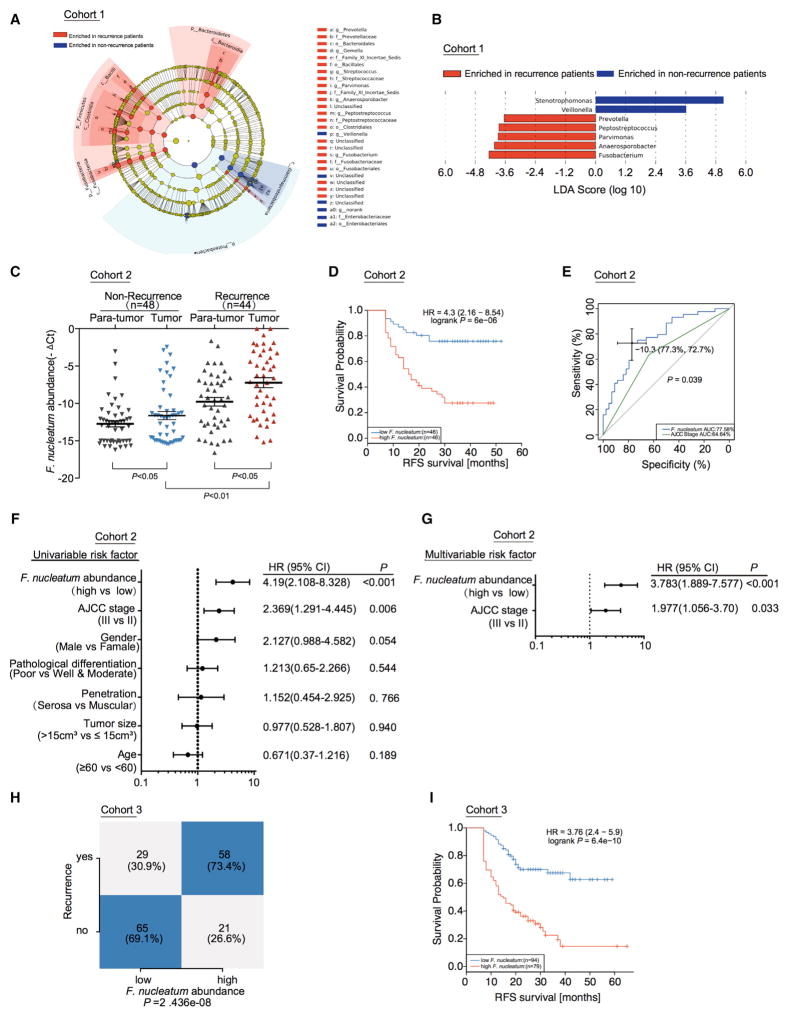
To examine the potential relationship between the gut microbiota alteration and CRC recurrence, we re-analyzed our previous data (Yu et al., 2015) and compared the pyrosequenced data by using a Roche 454 GS FLX in 16 CRC tissues from patients with recurrence and 15 CRC tissues without recurrence (Cohort 1, Figure 1A, Table S1). We used the LEfSe algorithm (Segata et al., 2011) to define the potential differential bacterium patterns between recurrent and non-recurrent CRC patients in Cohort 1. We found that Fusobacterium, Anaerosporobacter, Parvimonas, Peptostreptococcus, and Prevotella were enriched in recurrent CRC tissues as compared to non-recurrent CRC tissues (Figure 1B). Anaerosporobacter is rarely associated with human disease (Jeong et al., 2007). We further studied Fusobacterium nucleatum, Prevotella intermedia, Parvimonas micra, Peptostreptococcus anaerobius. Real-time PCR showed that F. nucleatum was the most enriched bacterium among the four bacteria in patients with recurrent CRC (Figure S1A), compared with patients without recurrence. This suggests that F. nucleatum may play a role in CRC recurrence. F. nucleatum is the most dominant phylotype in CRC (Kostic et al., 2012). We quantified the amount of F. nucleatum in 48 CRC tissues from patients without recurrence (group 1) and 44 CRC tissues from patients with recurrence (group 2) (Cohort 2, Table S2). In agreement with our data in Cohort 1 and previous reports (Castellarin et al., 2012; Kostic et al., 2012), the amount of F. nucleatum in CRC tissues was higher in recurrent patients than that in non-recurrent patients (Figure 1C). Furthermore, there was an enrichment of F. nucleatum in CRC tissues compared with adjacent normal tissues in both recurrence and non-recurrence groups (Figure 1C). CRC recurrence is attributed to chemoresistance. Thus, F. nucleatum correlates with CRC recurrence. The high amount of F. nucleatum may potentially promote CRC chemoresistance.
Figure 1. F. nucleatum Is Associated with Cancer Recurrence and Patient Outcome.
(A) A cladogram representation of data in CRC patients with recurrence (16) versus no recurrence (15) by 16S rDNA sequencing. Taxa enriched in patients with recurrence (Red) and without recurrence (Blue). The brightness of each dot is proportional to its effect size.
(B) Linear discriminant analysis (LDA) coupled with the effect size measurements identifies the significant abundance of data in A. Taxa enriched in recurrent (Red) and non-recurrent (Blue) patients are indicated with negative (Red) or positive (Blue) LDA scores, respectively. Only taxa greater than LDA threshold of 3.5 are shown.
(C) Statistical analysis of the amount of F. nucleatum in Cohort 2, nonparametric Mann–Whitney test.
(D) Recurrence-Free Survival (RFS) was compared between patients with low and high amount of F. nucleatum in Cohort 2, Log-rank test.
(E) Receiver operating characteristic (ROC) analysis was conducted based on the amount of F. nucleatum and AJCC in colorectal cancer.
(F) Univariate analysis was performed in Cohort 2. The bars correspond to 95% confidence intervals.
(G) Multivariate analysis was performed in Cohort 2. The bars correspond to 95% confidence intervals.
(H) Statistical analysis was conducted based on the amount of F. nucleatum and recurrence rate in Cohort 3 by the cut off value of F. nucleatum defined in Cohort 2, Chi-square test.
(I) RFS was compared between patients with low and high abundance of F. nucleatum in 173 patients with colorectal cancer (Cohort 3) by the cut off value of F. nucleatum defined in Cohort 2, Log-rank test.
See also Figure S1.
We next evaluated the relationship between the amount of F. nucleatum and different clinicopathological features in Cohort 2. The amount of F. nucleatum was positively associated with the American Joint Committee on Cancer (AJCC) stage and tumor size (Figure S1B). A high amount of F. nucleatum was strongly associated with shorter recurrence free survival (RFS) (Figure 1D). The five-year recurrence survival was substantially shorter in the F. nucleatum-high group than the F. nucleatum-low group. Receiver operating characteristic (ROC) curve analysis was conducted to predict the potential CRC recurrence using either AJCC stage or the amount of F. nucleatum (Figure 1E, Table S3). We observed that the area under curve (AUC) of F. nucleatum-based prediction was higher than that of the AJCC-stage based model (0.776 versus 0.646, p = 0.039). Youden Index was used to determine the optimal cutoff point and −10.3 [−deltaCT value] was selected based on the abundance of F. nucleatum that provided the best balance between the sensitivity and the specificity to predict CRC recurrence (Table S3). In addition, univariate (Figure 1F) and multivariate (Figure 1G) regression analyses of Cohort 2 demonstrated that the amount of F. nucleatum was an independent predictor of CRC aggressiveness with significant hazard ratios for predicting clinical outcome. Its predictive value was comparable to that of the AJCC stage. Thus, the data in Cohort 2 not only confirm our observation in Cohort 1 but also define the potential value of the amount of F. nucleatum in predicting CRC recurrence.
To further validate if F. nucleatum had a similar prediction value in cancer recurrence in a different and large patient population, we analyzed an additional cohort with 173 patients (Cohort 3, Table S4). The samples in Cohort 3 were classified into high and low-risk subsets according to the cut-off value (−10.3[−deltaCT value]) of F. nucleatum abundance derived from Cohort 2 (Table S3). We found that the recurrence rate in the high-risk group was significantly higher than the low-risk group (73.4% versus 30.9%, p = 2.436e-8) (Figure 1H). Again, the amount of F. nucleatum was higher in recurrent patients than non-recurrent patients in Cohort 3 (Figure S1C). We confirmed that the high amount of F. nucleatum was associated with shorter RFS (Figure 1I). Univariate (Figure S1D) and multivariate (Figure S1E) Cox regression analyses in Cohort 3 revealed that the amount of F. nucleatum was an independent predictor of CRC aggressiveness. Our data indicate that F. nucleatum is pathologically and clinically associated with cancer recurrence and patient outcome.
F. nucleatum Promotes Cancer Autophagy Activation
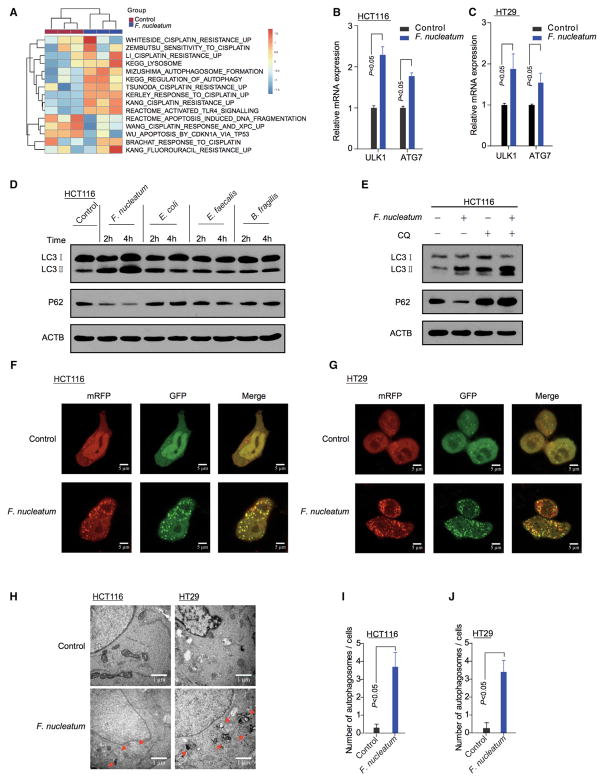
We hypothesized that F. nucleatum was biologically involved in the development of colon cancer chemoresistance. To test this hypothesis, we co-cultured colon cancer cells with F. nucleatum, performed a RNA-seq analysis, and compared the gene expression profiles between the colon cancer cells co-cultured with or without F. nucleatum. Co-culture with F. nucleatum downregulated 992 gene expressions and upregulated 1,466 gene expressions in HT29 cells (adjusted p value < 0.05, raw data accessible via GEO: GSE90944) (Table S5). Single sample gene set enrichment analysis (ssGSEA) revealed that the gene sets including MizushimaI_Autophagosome_Formation, KEGG_Lysosome, KEGG_regulation_of_Autophagy, and Hallmark_apoptosis were enriched in CRC cells co-cultured with F. nucleatum (Figure 2A). Given the role of the autophagy pathway in cellular survival (Song et al., 2009), our data suggest that F. nucleatum may cause autophagy pathway activation and potentially support cancer chemoresistance. In line with this, western blot analysis showed that F. nucleatum increased the expression of multiple autophagy signaling elements including pAMPK, pULK1, ULK1, and ATG7 in HCT116 cells and HT29 cells. These cells exhibited low LC3 protein cleavage level at the basal condition (Figures S2A and S2B). Real-time PCR confirmed that F. nucleatum affected the ULK1 and ATG7 mRNA levels (Figures 2B and 2C). These data suggest that F. nucleatum may drive autophagy activation in CRC cells. To examine this possibility, we performed autophagy functional assays in CRC cells co-cultured with F. nucleatum. Increased LC3-II and decreased p62 expression was detected in the F. nucleatum-co-cultured HCT116 cells (Figure 2D) and HT29 cells (Figure S2C) in a concentration-dependent manner (Figure S2D). These effects were not found in the CRC cells co-cultured with P. intermedia, P. micra, P. anaerobius, Escherichia coli, Enterococcus faecalis, or Bacteroides fragilis (Figure 2D, Figures S2C, S2E and S2F). Furthermore, we treated HCT116 cells (Figure 2E) and HT29 cells (Figure S2G) with Chloroquine (CQ), an autophagy lysosomal inhibitor. Addition of CQ blocked the autophagic flux in the F. nucleatum-cultured cells (Figure 2E). We established HCT116 cells and HT29 cells that stably expressed a tandem mRFP-EGFP-LC3 construct. We found that F. nucleatum induced the autophagic flux in HCT116 cells (Figure 2F) and HT29 cells (Figure 2G). In addition, autophagosomes were evaluated in HCT116 cells and HT29 cells co-cultured with or without F. nucleatum (Figure 2H). Transmission electron microscopy showed an increase in the formation of autophagic vesicles in the F. nucleatum-co-cultured HCT116 cells (Figure 2I) and HT29 cells (Figure 2J). Collectively, our data indicate that F. nucleatum activates the autophagy pathway in CRC cells.
Figure 2. F. nucleatum Promotes Cancer Autophagy Activation.
(A) ssGSEA analysis was conducted to show the relationship between the amount of F. nucleatum and autophagy-related pathways in CRC tissues.
(B, C) Real-Time PCR was performed in HCT116 cells (B) and HT29 (C) cells cultured with F. nucleatum, nonparametric Mann–Whitney test.
(D) Western blot was performed on autophagy element expression in HCT116 cells co-cultured with F. nucleatum, E. coli, E. faecalis or B. fragilis.
(E) Western blot was performed in HCT116 cells co-cultured with F. nucleatum in the presence of CQ.
(F and G) HCT116 cells (F) and HT29 cells (G) that stably expressed mRFP-EGFP-LC3 fusion protein were co-cultured with F. nucleatum. Confocal microscopic analysis is shown (2000 × magnification). Bar scale, 5 μm.
(H) Autophagosomes were observed by transmission electron microscopy (17500 × magnification) in HCT116 cells (left) and HT29 cells (right) cultured with F. nucleatum. Bar scale, 1 μm.
(I and J) Statistical analysis was performed to calculate the number of autophagosomes in HCT116 cells (I) and HT29 cells (J) shown by transmission electron microscopy, nonparametric Mann–Whitney test.
See also Figure S2.
F. nucleatum Induces Cancer Chemoresistance via the Autophagy Pathway
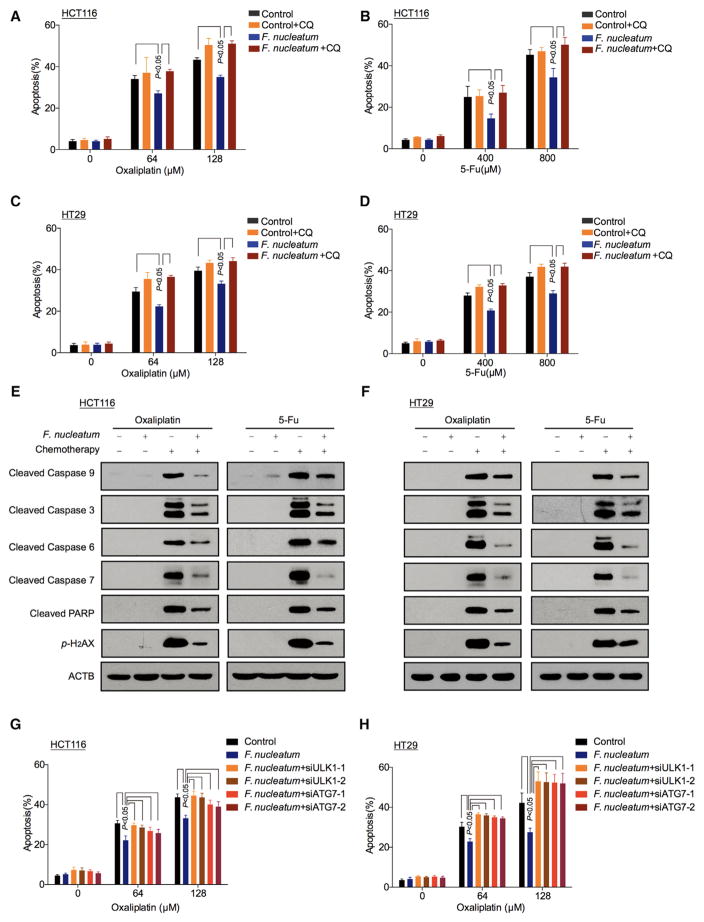
We next hypothesized that F. nucleatum induced cancer chemoresistance via the autophagy pathway. To initially determine whether F. nucleatum induces cancer chemoresistance, different multiplicity of infection (MOI) of F. nucleatum was used in the co-culture with CRC cells. We observed that F. nucleatum (MOI = 100) had no effect on HCT116 cell and HT29 cell proliferation (Figures S3A and S3B). We examined the potential role of this dose of F. nucleatum in CRC chemoresistance. As expected, Oxaliplatin (Figure 3A) and 5-FU (Figure 3B) induced HCT116 cell apoptosis. Co-culture with F. nucleatum, but not with P. intermedia, P. micra, P. anaerobius, and medium control (Figures S3C–S3F), reduced HCT116 cell apoptosis induced by these chemotherapeutic agents. Moreover, F. nucleatum had no protective effect on HCT116 cells and HT29 cells treated with Doxorubicin (Figures S3G and S3H). The data indicate that F. nucleatum inuduces CRC resistance to Oxaliplatin and 5-FU.
Figure 3. F. nucleatum Induces Chemoresistance in Colorectal Cancer Cells via Activation of the Autophagy Pathway.
(A–D) Apoptosis was detected by flow cytometry in HCT116 cells (A, B) and HT29 cells (C, D). The cells were co-cultured with F. nucleatum or treated with CQ, and different concentrations of Oxaliplatin (A and C) and 5-FU (B, D). nonparametric Mann–Whitney test.
(E and F) Cleaved caspases and p-H2AX expression were detected by western blot in HCT116 cells (E) and HT29 cells (F). The cells were co-cultured with F. nucleatum or treated with CQ, and different concentrations of Oxaliplatin and 5-FU.
(G and H) Apoptosis was detected by flow cytometry in HCT116 cells (G) and HT29 cells (H). The cells were transfected with ULK1 and ATG7 siRNAs, and subsequently co-cultured with F. nucleatum and different concentrations of Oxaliplatin, nonparametric Mann–Whitney test.
See also Figure S3.
SW480 cells are relatively sensitive to Oxaliplatin treatment (Moutinho et al., 2014). To quantitatively evaluate the potential effect of F. nucleatum on Oxaliplatin-induced SW480 cell chemoresistance, we generated the Oxaliplatin-resistant SW480 cells from the parential SW480 cells by continuous exposure to gradually increased concentrations of Oxaliplatin. We compared Oxaliplatin-induced cell apoptosis among the parental SW480 cells, the Oxaliplatin-resistant SW480 cells, and the F. nucleatum-co-cultured parental SW480 cells in the presence of different concentrations of Oxaliplatin (Figures S3I and S3J). We found that 109.9 μM Oxaliplatin resulted in 50%, 32.9%, and 35.5% cell apoptosis in the parental SW480 cells, the Oxaliplatin-resistant SW480 cells, and the F. nucleatum-co-cultured parental SW480 cells, respectively (Figure S3I). Given that the Oxaliplatin-resistant SW480 cells and the F.nucleatum-cultured parental SW480 cells were similarly resistant to Oxaliplatin treatment, the data suggest that F. nucleatum efficiently enables chemoresistance to the parental SW480 cells. In further support of this possibility, the EC50 of Oxaliplatin was 109.9 μM, 190.9 μM, and 177.2 μM for the parental SW480 cells, the Oxaliplatin-resistant SW480 cells, and the F.nucleatum-cultured parental SW480 cells, respectively (Figure S3J). In addition, we inoculated the parental SW480 cells into nude mice and treated the mice with different doses of Oxaliplatin with or without F. nucleatum. F. nucleatum alone had no effect on tumor growth (Figures S3K and S3L). As expected, tumor growth was significantly inhibited by both low and high doses of Oxaliplatin, and these effects were efficiently aborgated by F. nucleatum (Figures S3M and S3N). The data suggest that F. nucleatum endows potent chemoresistance to several CRC cells.
To address whether F. nucleatum modulates CRC chemoresistance via the autophagy pathway, we co-cultured HCT116 cells and HT29 cells with F. nucleatum and treated these cells with Oxaliplatin and 5-FU in the presence of CQ. We found that the F. nucleatum-induced chemoresistant effect was abolished by CQ treatment in HCT116 cells (Figures 3A and 3B) and HT29 cells (Figures 3C and 3D). Moreover, western blotting showed that Oxaliplatin and 5-FU induced the cleavage of caspase 9, caspase 3, caspase 6, caspase 7, PARP, and p-H2AX, and these effects were blocked by F. nucleatum co-culture in HCT116 cells (Figure 3E) and HT29 cells (Figure 3F). Thus, F. nucleatum may prevent CRC cells from chemotherapy-induced apoptosis via the autophagy pathway.
To examine whether the autophagy elements such as ULK1 and ATG7 may participate in F. nucleatum-induced chemoresistance in CRC cells, we analyzed LC3 cleavage status in ULK1 and ATG7 siRNA-transfected CRC cells cultured with F. nucleatum. As expected, ULK1 and ATG7 siRNAs decreased the two gene expression in CRC cells (Figures S3O and S3P). F. nucleatum co-culture increased the cleavage of LC3, ULK1, and ATG7 expression in wild-type ULK1 and ATG7 expressing CRC cells. ULK1 and ATG7 siRNAs blocked F. nucleatum-induced LC3 cleavage (Figures S3Q and S3R). The data indicate that F. nucleatum may induce autophagy activation via increasing ULK1 and ATG7 expression. Next, we treated CRC cells with chemotherapy agents. We found that F. nucleatum decreased CRC cell apoptosis in response to Oxaliplatin (Figures 3G and 3H) and 5-FU treatment (Figures S3S and S3T). This effect was abolished in ULK1 or ATG7-siRNA-transfected cells (Figures 3G and 3H; Figures S3S and S3T). Thus, the data strongly suggest that F. nucleatum may promote CRC chemoresistance by activating the autophagy pathway, and the autophagy elements ULK1 and ATG7 participate in the F. nucleatum-mediated chemoresistance in CRC cells.
F. nucleatum Activates Cancer Autophagy via a Selective Loss of miR-18a* and miR-4802
To explore the mechanism by which F. nucleatum induced upregulation of pULK1, ULK1, and ATG7 at both the mRNA and protein levels, we constructed the recombination luciferase reporter plasmids, pGL3-ULK1p and pGL3-ATG7p, containing the promoter region of ULK1 or ATG7. Luciferase assay showed that F. nucleatum co-culture had no effect on transcriptional activity of pGL3-ULK1p and pGL3-ATG7p in CRC cells (Figures S4A and S4B). This suggests that F. nucleatum-increased ULK1 or ATG7 mRNA is not dependent on the transcriptional activation of ULK1 or ATG7 promoter.
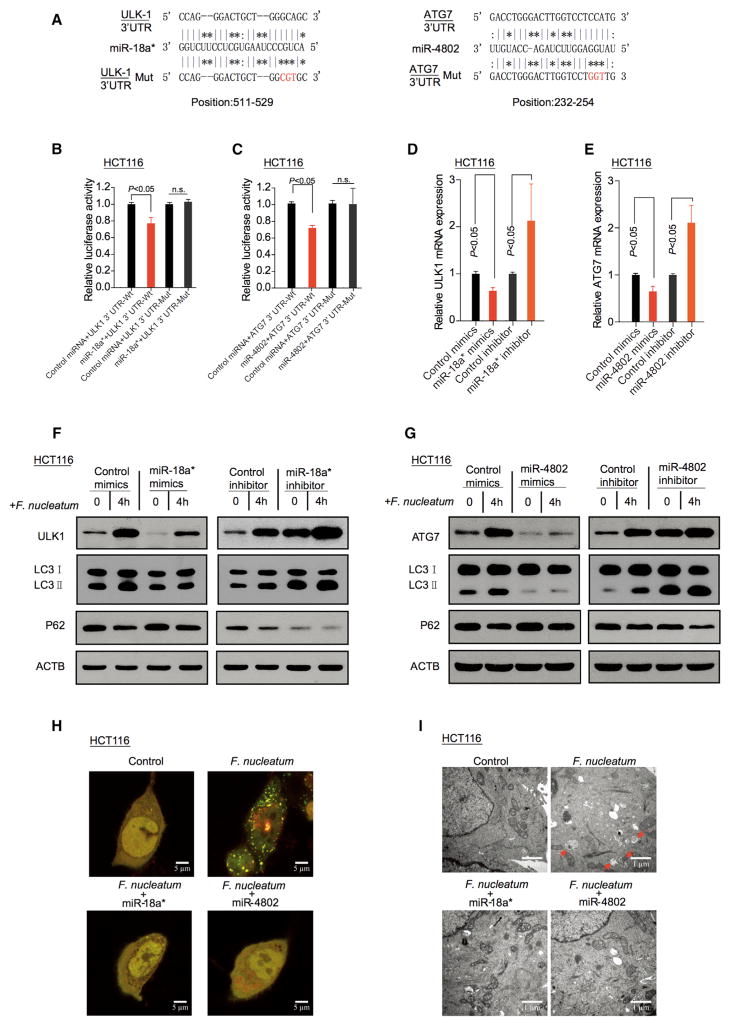
MiRNAs often regulate gene expression by binding to the RISC complex and directing sequence-specific cleavage of target mRNA or repressing the target mRNA translation (Bartel, 2009; Krek et al., 2005). We hypothesized that dysregulated miRNAs may contribute to F. nucleatum-increased ULK1 and ATG7 expression. To test this hypothesis, we performed a global miRNA expression profiling of CRC tissues with a high amount of F. nucleatum from six recurrent patients, and of CRC tissues with a low amount of F. nucleatum from six non-recurrent patients (Cohort 1, Figure S4C, left). Sixty-eight miRNAs were significantly downregulated in the CRC tissues with a high amount of F. nucleatum as compared to that with a low amount of F. nucleatum (Figure S4C, right; Table S6). Next, we used the FindTar3 (http://bio.sz.tsinghua.edu.cn/) and miRDB databases (http://mirdb.org/miRDB/) to identify potential miRNAs, which may regulate ULK1 and ATG7. After overlapping these potential ULK1- and ATG7-regulatory miRNAs with the identified 68 downregulated miRNAs, we found four and three miRNAs, which may regulate ULK1 and ATG7, respectively (Figure S4C, right). We validated these seven miRNAs with real-time PCR. We found that miR-4802 and miR-18a* were the most significantly down-regulated miRNAs in response to F. nucleatum intervention in HCT116 cells (Figure S4D) and HT29 cells (Figure S4E). Target prediction programs and sorting algorithm suggested potential specific targets for miR-18a* and miR-4802 in the seed regions within the 3′UTR regions of ULK1 and ATG7 genes, respectively (Figure 4A). Luciferase reporter assays demonstrated that miR-18a* and miR-4802 suppressed the luciferase activity in HCT116 cells (Figures 4B–4C) and HT29 cells (Figures S4F and S4G) transfected with wild-type ULK1 and ATG7 reporter plasmids, but not with the mutant reporter ones. Real-time PCR showed that miR-18a* and miR-4802 decreased ULK1 and ATG7 mRNA levels, and the mRNA expression levels of ULK1 and ATG7 were rescued by miR-18a* and miR-4802 inhibitors in HCT116 cells (Figures 4D and 4E) and HT29 cells (Figures S4H and S4I). In addition, western blotting revealed that overexpression of miR-18a* or miR-4802 suppressed F. nucleatum-induced conversion of LC3-I to LC3-II and simultaneously increased p62 protein expression in HCT116 cells (Figures 4F and 4G) and HT29 cells (Figures S4J and S4K). Accumulation of autophagosomes was reduced in miR-18a* and miR-4802 overexpressing HCT116 cells (Figures 4H and 4I) and HT29 cells (Figures S4L and S4M) co-cultured with F. nucleatum, compared to controls. Thus, the data indicate that F. nucleatum activates cancer autophagy via a selective loss of miR-18a* and miR-4802.
Figure 4. F. nucleatum Activates Cancer Autophagy via Downregulation of miR-18a* and miR-4802.
(A) The predicted binding sequences for miR-18a* (left) and miR-4802 (right) within the human ULK1 and ATG7 3′UTR, respectively. Seed sequences are highlighted.
(B) Luciferase activity was measured in HCT116 cells transfected with miR-18a* mimics or control miRNA. The luciferase reporters expressing wild-type or mutant human ULK1 3′UTRs were used. The luciferase activity was normalized based on the control miRNA transfection. n.s., not significant.
(C) Luciferase activity was measured in HCT116 cells transfected with miR-4802 mimics or control miRNA. The luciferase reporters expressing wild-type or mutant human ATG7 3′UTRs were used.
(D) Real-time PCR was performed in HCT116 cells to detect the expression of ULK1 gene after transfected with miR-18a* mimics or inhibitor, nonparametric Mann–Whitney test.
(E) Real-time PCR was performed in HCT116 cells to detect the expression of ATG7 gene after transfected with miR-4802 mimics or inhibitor, nonparametric Mann–Whitney test.
(F and G) HCT116 cells were transfected with mimics or inhibitor of miR-18a* (F) and miR-4802 (G), respectively. After culturing with F. nucleatum, autophagy and target proteins were detected by western blot in HCT116 cells.
(H) HCT116 cells that stably expressed mRFP-EGFP-LC3 fusion protein were transfected with miR-18a* and miR-4802 mimics. After culturing with F. nucleatum, autophagosomes were observed under confocal microscope (2000 × magnification) in HCT116 cells. Bar scale, 5 μm.
(I) Autophagosomes were observed by transmission electron microscopy (17500 × magnification) in HCT116 cells transfected with miR-18a* (left) and miR-4802 (right) mimics, and then co-cultured with F. nucleatum. Bar scale, 1 μm.
See also Figure S4.
MiR-18a* and miR-4802 Regulate F. nucleatum-Mediated Chemoresistance
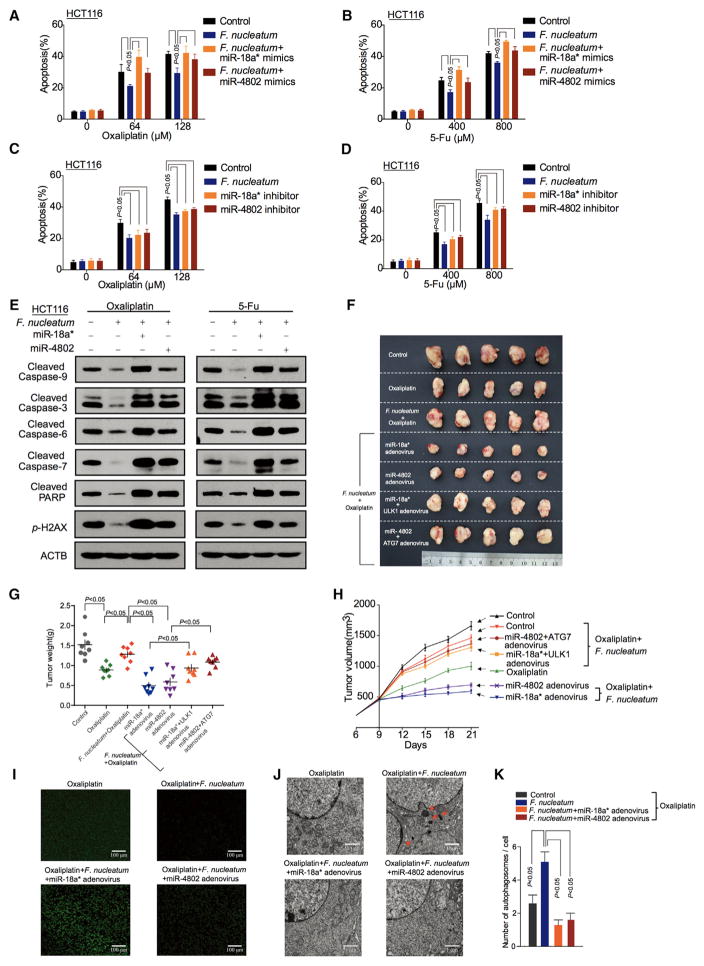
The selective loss of miR-18a* and miR-4802 expression in F. nucleatum-cultured cell lines led us to hypothesize that miR-18a* and miR-4802 may regulate F. nucleatum-mediated chemoresistance. To test this hypothesis, miR-18a* and miR-4802 mimics or inhibitors were transfected in CRC cells cultured with F. nucleatum. These microRNA mimics and inhibitors had no effect on CRC proliferation (Figure S5A). However, miR-18a* and miR-4802 mimics increased Oxaliplatin- and 5-FU-induced apoptosis in HCT116 cells (Figures 5A and 5B) and HT29 cells (Figures S5B and S5C) cultured with F. nucleatum, compared with controls. Consistent with these data, a loss-of-function study revealed that anti-miR-18a* and anti-miR-4802 increased chemoresistance in HCT116 cells (Figures 5C and 5D) and HT29 cells (Figures S5D and S5E) cultured with F. nucleatum. Western blotting showed that the inhibitory effects of F. nucleatum on the chemotherapy-induced caspase and PARP cleavage and p-H2AX were abolished by miR-18a* and miR-4802 mimics transfection in HCT116 cells (Figure 5E) and HT29 cells (Figure S5F).
Figure 5. miR-18a* and miR-4802 Regulate F. nucleatum-Mediated Chemoresistance.
(A–D) Apoptosis was detected by flow cytometry in HCT116 cells. HCT116 cells were transfected with mimics (A, B) or inhibitors (C, D) of miR-18a* and miR-4802, co-cultured with F. nucleatum, and treated with different concentrations of Oxaliplatin (A, C) and 5-FU (B, D). nonparametric Mann–Whitney test.
(E) Western blot was performed in HCT116 cells. HCT116 cells were transfected with mimics or inhibitors of miR-18a* and miR-4802, co-cultured with F. nucleatum, and treated with different concentrations of Oxaliplatin (left) and 5-FU (right).
(F) Representative data of tumors in mice under different conditions. Figure 5(F) and Figure S5G shared experimental controls.
(G and H) Statistical analysis of tumor weights (G) and volumes (H) in different groups, n = 8/group nonparametric Mann–Whitney test.
(I) TUNEL assays were performed to detect tumor cell apoptosis in xenograft tumor tissues. The mice received different treatments.
(J) Transmission electron microscopy was performed to show the autophagosomes in xenograft tumor tissues. The mice received different treatments (17500 × magnification). Bar scale, 1 μm.
(K) Statistical analysis of autophagosomes. Autophagosomes were detected by transmission electron microscopy in xenograft tumor tissues, nonparametric Mann–Whitney test.
See also Figure S5–6.
In the CRC xenograft mouse models, HCT116 cells and HT29 cells were inoculated into nude mice, followed by treatment with miRNAs, chemotherapeutic agents, F. nucleatum-co-culture, and other manipulations. There was no difference in tumor weight (Figures S5G and S5H) and tumor growth (Figure S5I) among different control groups. Interestingly, tumor growth was significantly decreased by Oxaliplatin (Figures S5J–S5L) and 5-FU treatment (Figures S5M–S5O), and these decreases were blocked by F. nucleatum treatment in vivo. Thus, F. nucleatum participates in CRC chemoresistance in response to Oxaliplatin and 5-FU therapy. Furthermore, F. nucleatum-induced CRC chemoresistance was rescued by CQ, an autophagy pathway inhibitor (Figures S5J–S5O). In addition, F. nucleatum-induced CRC chemoresistance was reversed by miR-18* and miR-4802 adenovirus transduction in tumor bearing mouse models treated with Oxaliplatin (Figures 5F–5H) or 5-FU (Figures S6A–S6C). Overexpression of ULK1 and ATG7 blocked miR-18a* and miR-4802-mediated reversion of F. nucleatum-stimulated chemoresistance in tumor tissues (Figures 5F–5H, Figures S6A–S6C). Furthermore, western blotting confirmed that miR-18a* and miR-4802 rescued the inhibitory effects of F. nucleatum on the apoptotic gene expression (Figure S6D), tumor apoptosis (Figure 5I, Figure S6E), and auto-phagosome formation (Figures 5J and 5K, Figures S6F and S6G) in response to chemotherapy. These data support that miR-18a* and miR-4802 regulate F. nucleatum-mediated chemoresistance by blocking F. nucleatum-induced autophagy activation.
TLR4 and MYD88 Pathway Is Involved in F. nucleatum-Mediated Chemoresistance
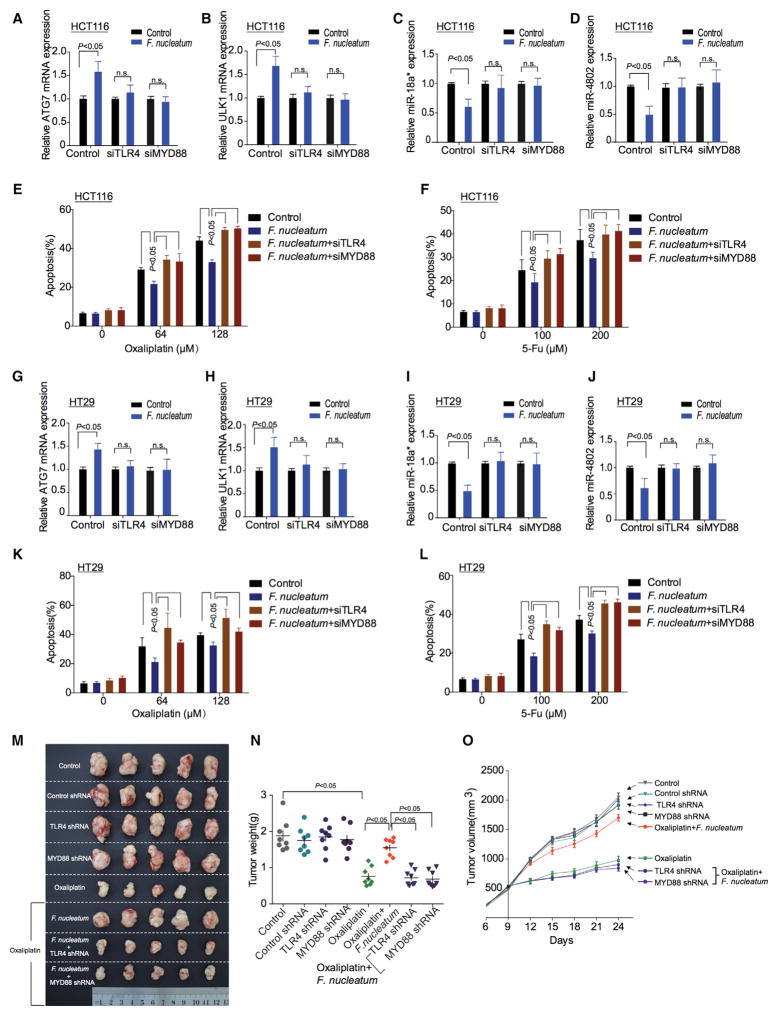
The TLR4 and MYD88 innate immune signaling pathway is activated in response to F. nucleatum intervention (Abreu and Peek, 2014; Liu et al., 2007). In line with this, we found that the levels of TLR4 and MYD88 transcripts (Figures S7A and S7B) and proteins (Figures S7C and S7D) were enhanced in response to F. nucleatum treatment in HCT116 cells and HT29 CRC cells. To examine whether the TLR4 and MYD88 pathway participated in F. nucleatum-induced autophagy activation, we transfected CRC cells with TLR4 and MYD88 siRNA and co-cultured the cells with F. nucleatum. Western blotting showed that F. nucleatum-mediated autophagy activation was reduced in TLR4 or MYD88 siRNA-transfected CRC cells (Figures S7C and S7D). Knockdown of TLR4 or MYD88 expression blocked F. nucleatum-induced ULK1 and ATG7 upregulation, F. nucleatum-induced miRNA-18a* and miRNA-4802 loss, and F. nucleatum-induced chemoresistance in HCT116 cells (Figures 6A–6F) and HT29 cells (Figures 6G–6L). Furthermore, F. nucleatum-induced CRC chemoresistance was rescued by knocking down TLR4 or MYD88 in the CRC xenograft mouse model, as shown by reduced tumor weight (Figures 6M and 6N, Figures S7E and S7F) and tumor volume (Figure 6O, Figure S7G). The data indicate that F. nucleatum-induced loss of miRNA-18a* and miRNA-4802, F. nucleatum-activated autophagy pathway, and F. nucleatum–mediated chemoreistance are dependent on the TLR4 and MYD88 signaling pathway.
Figure 6. TLR4 and MYD88 Pathway Is Involved in F. nucleatum-Mediated Chemoresistance.
(A–D) Real-time PCR was performed to detect ATG7 (A), ULK1 (B), miR-18a* (C), and miR-4802 (D) expression in HCT116 cells. HCT116 cells were co-cultured with F. nucleatum and transfected with TLR4 and MYD88 siRNAs, respectively.
(E and F) Apoptosis was detected by flow cytometry in HCT116 cells. The cells were co-cultured with F. nucleatum after TLR4 and MYD88 siRNAs transfection, and subsequently treated with different concentrations of Oxaliplatin (E) or 5-FU (F), nonparametric Mann–Whitney test.
(G–J) Real-time PCR was performed to detect expression of ATG7 (G), ULK1 (H), miR-18a* (I), and miR-4802 (J) expression in HT29 cells. HT29 cells were co-cultured with F. nucleatum and transfected with TLR4 and MYD88 siRNAs, respectively.
(K and L) Apoptosis was detected by flow cytometry in HT29 cells. The cells were co-cultured with F. nucleatum after TLR4 and MYD88 siRNAs transfection, and subsequently treated with different concentrations of Oxaliplatin (K) or 5-FU (L), nonparametric Mann–Whitney test.
(M) Representative data of tumors in nude mice bearing HCT116 cells in different groups. Figure 6M and Figure S6A shared experimental controls.
(N and O) Statistical analysis of mouse tumor weights (N) and volumes (O) in different groups, n = 8/group, nonparametric Mann–Whitney test.
See also Figure S7.
The Levels of F. nucleatum, MicroRNAs, and Autophagy Components Correlate and Are Clinically Relevant in CRC Patients
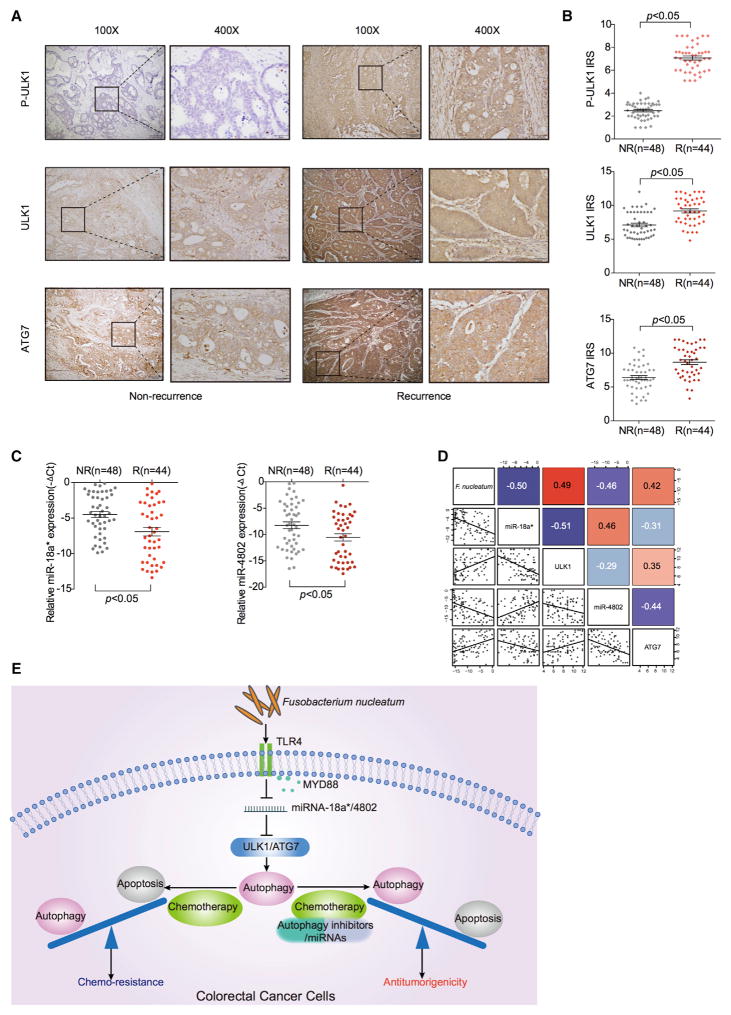
To investigate clinical significance of F. nucleatum, microRNAs (miR-18a* and miR-4802), and autophagy components (ULK1 and ATG7) in CRC patients, we studied CRC tissues and normal colorectal tissues adjacent to cancer lesions in Cohort 2. We quantified F. nucleatum and the expression of miR-18a* and miR-4802 with real-time PCR and detected the protein expression of pULK1, ULK1, and ATG7 by immunohistochemistry in CRC tissues and normal tissues. We found that the high amount of F. nucleatum, the high expression of p-ULK1, ULK1, and ATG7, and the low levels of miR18a* and miR-4802 expression were more likely detected in patients with recurrence, compared with patients without recurrence (Figures 7A–7C). The amount of F. nucleatum in CRC tissues negatively correlated with the levels of miR-18a* and miR-4802 and positively correlated with ULK1 and ATG7 expression (Figure 7D). The data demonstrate that the amount of F. nucleatum and the expression levels of miR-18a* and miR-4802 positively and negatively correlate with autophagy pathway activation status, respectively. Altogether, we reason that F. nucleatum may act on CRC via TLR4 and MYD88, cause a selective loss of miR-18a* and miR-4802 expression, subsequently result in autophagy activation, and consequently promote chemoresistance in patients with colorectal cancer (Figure 7E).
Figure 7. The Levels of F.nucleatum, miR-18a*, miR-4802, and Autophagy Components Correlate and Are Relevant in CRC Patients.
(A) Representative immunohistochemistry of p-ULK1 (upper), ULK1 (middle), and ATG7 (lower) proteins in CRC tissues from patients without recurrence and with recurrence (Cohort 2). NR, non-recurrence; R, recurrence.
(B) Statistical analysis of immunohistochemical immunoreactive score of Remmele and Stegner (IRS) scores of pULK1 (upper), ULK1 (middle), and ATG7 (lower) proteins in Cohort 2. NR, non-recurrence; R, recurrence.
(C) Statistical analysis of miR-18a* (left) and miR-4802 (right) expression by real-time PCR in Cohort 2. NR, non-recurrence; R, recurrence.
(D) Correlations among F. nucleatum, miR-18a*, miR-4802, ULK1, and ATG7 levels in human colorectal cancer tissues (Cohort 2).
(E) Schematic diagram of the relationship among F. nucleatum, autophagy and chemoresistance.
DISCUSSION
Capecitabine (and 5-FU) in combination with platinum-based chemotherapy has been widely used to treat different types of cancer including CRC (Kelland, 2007). Although CRC patients’ initial responses to surgical debulking and chemotherapy is often effective, relapse with drug-resistant cancer usually occurs and patients succumb to disease (Bertotti and Sassi, 2015; Jemal et al., 2009). Unfortunately, CRC patients are generally not responsive to novel immune checkpoint therapy (Zou et al., 2016). Conventional chemotherapy remains the first line therapy for patients with CRC. Thus, understanding the mechanisms of chemoresistance in CRC is essential to optimizing current therapeutic strategies.
Cancer genetic and epigenetic alterations in CRC chemotherapeutic response have been extensively reported (Bardelli and Siena, 2010; Dallas et al., 2009; Esteller, 2008; Linardou et al., 2008; Van Geelen et al., 2004; Weichert et al., 2008). Recent mouse studies have shown that the gut microbiota may modulate local immune responses and in turn affect chemotherapy (Iida et al., 2013; Viaud et al., 2013) and immunotherapy (Sivan et al., 2015; Vétizou et al., 2015). Human studies demonstrate that the adaptive immune system can also regulate chemosensitivity of human ovarian cancer (Wang et al., 2016). However, the potential role of the gut microbiota in CRC chemoresistance is poorly understood. Through a combination of genomic, bioinformatic, biological, in vivo models and clinical studies, we have demonstrated that autophagy-related pathways are enriched and activated in CRC patients with a high amount of F. nucleatum, and that F. nucleatum promotes CRC chemoresistance.
Metagenomic and transcriptomic analyses have revealed that intestinal microbes, especially F. nucleatum, are involved in CRC development (Castellarin et al., 2012; Kostic et al., 2012). F. nucleatum attaches to the host epithelial E-cadherin and promotes colorectal carcinogenesis via the fusobacterial adhesin FadA(Rubinstein et al., 2013). The interaction between a host polysaccharide, Gal-GalNAc with fusobacterial lectin (Fap2) facilitates F. nucleatum enrichment in CRC tissues (Abed et al., 2016). However, it is unknown whether and how F. nucleatum may mediate chemoresistance in CRC. We report that F. nucleatum induces LC3-II expression, autophagic flux, and autophagosome synthesis in CRC cells. Accordingly, F. nucleatum stimulates expression of the autophagy-related proteins, pULK1, ULK1, and ATG7 in CRC, and biochemical and genetic autophagy inhibition enhances the sensitivity of F. nucleatum-treated CRC cells to 5-FU and Oxaliplatin. Thus, we conclude that autophagy contributes to F. nucleatum-mediated CRC resistance to Oxaliplatin and 5-FU regimens. Our data may explain why CRC patients with a high amount of F. nucleatum experience poor outcomes (Mima et al., 2016).
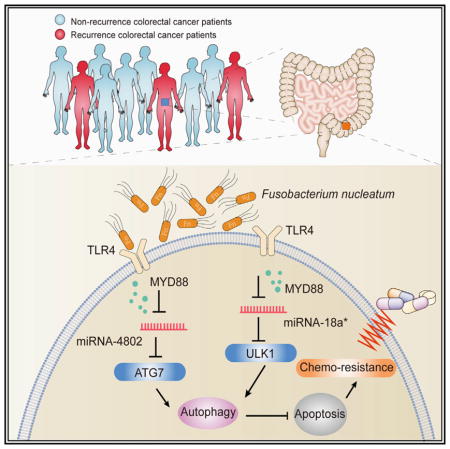
We have dissected the mechanisms by which F. nucleatum mediates the ULK1 and ATG7 pathway activation. F. nucleatum does not affect the transcription of ULK1 and ATG7. It has been reported that adherent-invasive E. coli (AIEC) may regulate human intestinal epithelial cell autophagy via modulating the expression of miR-30C and miR-130A (Nguyen et al., 2014). Our bioinformatic and functional studies have elucidated that miR-18a* and miR-4802 target ULK1 and ATG7, respectively, and are selectively lost due to F. nucleatum co-culture, and can biologically modulate CRC chemoresistance in vitro and in vivo. Thus, F. nucleatum medicates chemoresistance via selectively targeting specific miRNAs and autophagy elements. Given that the TLR4 and MYD88 innate immune signaling pathway is essential for F. nucleatum infection (Abreu and Peek, 2014; Liu et al., 2007), we have demonstrated that F.nucleatum-induced genomic loss of miR-18a* and miR-4802 depends on the TLR4 and MYD88 signaling pathway. Therefore, F. nucleatum orchestrates the TLR4-MYD88, miR18a* and miR4802, and ULK1/ATG7 autophagy network to biologically control CRC chemoresistance (Figure 7E).
In addition to its biological importance, our work may be relevant in clinical management of CRC patients. As the amount of F. nucleatum is associated with the risk of CRC recurrence, the measurement of F. nucleatum post-surgery may be an effective approach to predict patient outcome. Furthermore, our data raise an important clinical question: are conventional chemotherapeutic regimens including Capecitabine plus Oxaliplatin suitable for CRC patients with a high amount of F. nucleatum? Alternatively, we suggest that CRC patients with a high amount of F. nucleatum may be treated with conventional chemotherapy in combination with anti-F. nucleatum treatment and/or an autophagy inhibitor. Thus, it is important to detect F. nucleatum and its associated pathway and differentially manage patients with different levels of F. nucleatum.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENTS or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ACTB-HRP | Sigma-Aldrich | Cat#A1978 |
| Anti-LC3B | Sigma-Aldrich | Cat#L7543 |
| Anti-P62 | Cell Signaling Technology | Cat#8025 |
| Anti-Cleaved Caspase9 | Cell Signaling Technology | Cat#9505 |
| Anti-Cleaved Caspase3 | Cell Signaling Technology | Cat#9664 |
| Anti-Cleaved Caspase6 | Cell Signaling Technology | Cat#9761 |
| Anti-Cleaved Caspase7 | Cell Signaling Technology | Cat#8438 |
| Anti-Cleaved PARP | Cell Signaling Technology | Cat#5625 |
| Anti-p-H2AX(Ser139) | Cell Signaling Technology | Cat#9718 |
| Anti-p-AMPK(α)(Thr172) | Cell Signaling Technology | Cat#2535 |
| Anti-Beclin1 | Cell Signaling Technology | Cat#3495 |
| Anti-ATG5 | Cell Signaling Technology | Cat#12994 |
| Anti-ATG12 | Cell Signaling Technology | Cat#4180 |
| Anti-ATG16L1 | Cell Signaling Technology | Cat#8089 |
| Anti-ATG3 | Cell Signaling Technology | Cat#3415 |
| Anti-MYD88 | Cell Signaling Technology | Cat#3699 |
| Anti-ULK1 | Abcam | Cat#ab65056 |
| Anti-p-ULK1(Ser556) | Abcam | Cat#ab203207 |
| Anti-ATG7 | Abcam | Cat#ab52472 |
| Anti-TLR4 | Abcam | Cat#ab13556 |
| Biological Samples | ||
| Fresh colorectal cancer tissues | Renji Hospital affiliated with Shanghai Jiaotong University School of Medicine | N/A |
| Formalin-fixed paraffin-embedded colorectal cancer tissues | Renji Hospital affiliated with Shanghai Jiaotong University School of Medicine | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Oxaliplatin | Selleck Chemicals | Cat#S1224 |
| 5-FU | Selleck Chemicals | Cat#S1209 |
| Doxorubicin | Selleck Chemicals | Cat#S1208 |
| Chloroquine | Sigma-Aldrich | Cat#C6628 |
| Critical Commercial Assays | ||
| FITC Annexin V Apoptosis Detection Kit I | BD Biosciences | Cat#556547 |
| Fixable Viability Stain 510 | BD Biosciences | Cat#564406 |
| QIAamp DNA FFPE Tissue Kit | QIAGEN | Cat#56404 |
| miRNease FFPE Kit | QIAGEN | Cat#217504 |
| In Situ Cell Death Detection Kit, POD | Roche | Ca#11684817910 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GSE90944 |
| Experimental Models: Cell Lines | ||
| FHC | ATCC | CRL-1831 |
| HCT116 | ATCC | CCL-247 |
| HT29 | ATCC | HTB-38 |
| SW480 | ATCC | CCL-228 |
| SW1116 | ATCC | CCL-233 |
| Caco2 | ATCC | HTB-37 |
| LoVo | ATCC | CCL-229 |
| RKO | ATCC | CRL-2577 |
| DLD1 | ATCC | CCL-221 |
| Experimental Models: Organisms/Strains | ||
| Fusobacterium Nucleatum strain 25586 | ATCC | Cat#59899827 |
| Bacteroides fragilis strain 43860 | ATCC | Cat#5038385 |
| Enterococcus faecalis strain 47077 | ATCC | Cat#5091158 |
| Peptostreptococcus anaerobius strain 27337 | ATCC | Cat#63229810 |
| Parvimonas micra strain 33270 | ATCC | Cat#62282361 |
| Prevotella intermedia strain 49046 | ATCC | Cat#63032740 |
| Escherichia coli strain DH5α | TIANGEN | Cat#CB101 |
| BALB/c nude mice | Experimental Animal Centre of SIBS | N/A |
| Sequences of miRNA mimics, miRNA inhibitors, and mRNA siRNAs, see Table S7. | Genepharma | N/A |
| DNA primer sequences, See Table S7 | Sangon Biotech | N/A |
| miRNA and U6 Primers, See Table S7 | GeneCopoeia | Included in Table S7 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| FindTar3 | School of Life Science, Tsinghua University | http://bio.sz.tsinghua.edu.cn/ |
| miRDB | Department of Radiation Oncology, Washington University School of Medicine | http://mirdb.org/miRDB/ |
| FlowJo | FlowJo LLC | https://www.flowjo.com/ |
| ZEN 2011 Light Edition | ZEISS | https://www.zeiss.com/microscopy/ |
| R | R Development Core Team | https://www.r-project.org/ |
| TopHat2 | Kim et al., 2013 | http://tophat.cbcb.umd.edu |
| DeSeq2 | Love et al., 2014 | https://www.bioconductor.org/ |
| HTSeq | Anders et al., 2015 | http://www-huber.embl.de/HTSeq/ |
| GSVA | Hänzelmann et al., 2013 | https://www.bioconductor.org/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
As Lead Contact, Weiping Zou is responsible for all reagent and resource requests. Please contact Weiping Zou at at wzou@med.umich.edu with requests and inquiries.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
For the xenograft experiments, four-week-old male BALB/c nude mice were housed in laminar flow cabinets under specific pathogen-free conditions with food and water provided ad libitum. Colorectal cancer cells (1 × 107 SW480-male cells or 5 × 106 HCT116-male cells) were injected subcutaneously into the right axilla of each mouse to establish the CRC xenograft model. Six days after subcutaneous inoculation, mice were randomly divided into different groups for different sets of experiments. The relevant viral vectors and F. nucleatum were given by multipoint intratumoral injection, twice per week for three weeks. Chemotherapeutic agents and CQ were administered by intraperitoneal injection, twice per week for three weeks.
To explore the role of F. nucleatum in chemoresistance in Oxaliplatin-sensitive tumor in vivo, we used SW480-male cells (1×107) in the xenograft experiments. There were six groups: i) Saline (Control); ii) F. nucleatum bacteria solution; iii) low-dose Oxaliplatin (5 mg/kg); iv) low-dose Oxaliplatin (5 mg/kg) and F. nucleatum; v) high-dose Oxaliplatin (10 mg/kg); vi) high-dose Oxaliplatin(10 mg/kg) and F. nucleatum. Mice received intratumoral injection with F. nucleatum, and intraperitoneal injection with Oxaliplatin twice a week for three weeks.
In the following experiments, we used HCT116-male cells (5 × 106) to establish the xenograft models, and CQ (30 mg/kg), Oxaliplatin (7.5 mg/kg), and 5-FU (50 mg/kg) to treat the mice.
To explore the role of F. nucleatum, miRNAs, and autophagy in CRC chemoresistance in vivo, we used HCT116-male cells (5 × 106) in the xenograft experiments. There were eight groups: i) saline (Control); ii) F. nucleatum bacteria solution; iii) miRNA vector virus; iv) miR-18a* virus; v) miR-4802 virus; vi) control virus; vii) ATG7 overexpression virus; and viii) ULK1 overexpression virus.
To explore the role of autophagy in F. nucleatum-mediated chemoresistance in vivo, we designed seven groups: i) Control group; ii) F. nucleatum group; iii) CQ group; iv) Oxaliplatin (or 5-FU) group; v) Oxaliplatin (or 5-FU) and F. nucleatum group; vi) Oxaliplatin (or 5-FU) and CQ group; vii) Oxaliplatin (or 5-FU), F. nucleatum and CQ group.
To explore the role of specific miRNAs and autophagy elements in F. nucleatum-mediated chemoresistance in vivo, we designed seven groups: i) Control group; ii) Oxaliplatin (or 5-FU) group; iii) Oxaliplatin (or 5-FU) and F. nucleatum group; iv) Oxaliplatin (or 5-FU), F. nucleatum and miR-18a* virus group; v) Oxaliplatin (or 5-FU), F. nucleatum and miR-4802 virus group; vi) Oxaliplatin (or 5-FU), F. nucleatum, miR-18a* virus and ULK1 overexpression virus group; vii) Oxaliplatin (or 5-FU), F. nucleatum, miR-4802 virus and ATG7 overexpression virus group.
To explore the role of the TLR4 signaling pathway in F. nucleatum-mediated chemoresistance in vivo, we designed eight groups: i) Control group; ii) control shRNA group; iii) TLR4 shRNA virus group; iv) MYD88 shRNA virus group; v) Oxaliplatin (or 5-FU) group; vi) Oxaliplatin (or 5-FU) and F. nucleatum group; vii) Oxaliplatin (or 5-FU), F. nucleatum, and TLR4 shRNA virus group; viii) Oxaliplatin (or 5-FU), F. nucleatum, and MYD88 shRNA virus group.
The length and width of the tumors (in millimeters) were measured every three days with calipers. Tumor volume was calculated using the formula (A × B2)/2, where A and B are the long and short dimensions, respectively. After three weeks, all mice were sacrificed and subcutaneous tumors were collected and weighed. The tumor volume and weight are presented as means ± SEM (n = 8). Mouse experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The study procedures were approved by the Institutional Animal Care and Use Committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University.
Bacterial Strains and Growth Conditions
Fusobacterium nucleatum strain ATCC 25586 (Castellarin et al., 2012), Bacteroides fragilis strain 43860, Enterococcus faecalis strain 47077, Peptostreptococcus anaerobius strain 27337, Parvimonas micra strain 33270, and Prevotella intermedia strain 49046 were purchased from American Type Culture Collection (ATCC, Manassas, VA). F. nucleatum and P. micra were cultured overnight at 37°C under anaerobic conditions (DG250, Don Whitley Scientific, West Yorkshire, UK) in brain heart infusion (BHI) broth supplemented with hemin, K2HPO4, Vitamin K1, and L-Cysteine(Rhee et al., 2009). P. anaerobius and P. intermedia were cultured in Wilkins-Chalgren anaerobe broth (CM0643, Thermo Fisher Scientific, West Palm Beach, FL) at 37°C under anaerobic conditions. B.fragilis was cultured overnight at 37°C under anaerobic conditions in ATCC 1490 Modified chopped meat medium. E. faecalis and the commensal Escherichia coli strain DH5α (Tiangen, China) were cultured in Luria-Bertani (LB) medium overnight at 37°C in shake cultivation at 220 rpm/min.
Cell Lines
Human colorectal cancer cell lines RKO-NA, SW1116-male, DLD1-male, SW480-male, Caco2-male, LOVO-male, HT29-female, and HCT116-male (ATCC) were cultured in RPMI-1640 medium (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere. Human colonic epithelial cell FHC (from 13 weeks gestation) was cultured in DMEM/F12 medium (GIBCO, Carlsbad, CA) supplemented with 25mM HEPES, 10 ng/ml cholera toxin, 0.005mg/ml insulin, 0.005 mg/ml transferrin, 100 ng/ml hydrocortisone and 10% FBS at 37°C in a humidified 5% CO2 atmosphere. To establish the Oxaliplatin-resistant SW480 cells, we cultured SW480 cells with gradually increased Oxaliplatin for two months. The EC50 of Oxaliplatin was 190.9 μM and 109.9 μM for the established Oxaliplatin-resistant SW480 cells and the parental SW480 cells, respectively.
Patients and Clinical Specimens
We studied 3 cohorts of patients with colorectal cancer from Renji Hospital affiliated to Shanghai Jiaotong University School of Medicine between 2012 and 2015. Cohort 1 and cohort 2 were from the Western Campus of Renji Hospital. Cohort 3 was from the Eastern Campus of Renji Hospital. There were 31 fresh tissues in Cohort 1 and 92 and 173 formalin-fixed paraffin-embedded tissues (FFPE) in Cohorts 2 and 3, respectively. We performed 16 s RNA sequencing studies in Cohort 1 to define which bacterium is predominant (and/or different) in the recurrent colorectal cancer tissues compared to the non-recurrent colorectal cancer tissues. We used Cohort 2 as a discovery clinical sample set to determine which levels of F. nucleatum are linked to chemoresistance-associated cancer recurrence. We used Cohort 3 as a validation dataset to evaluate whether the cut-off value generated from Cohort 2 could be applied and validated in an independent cohort with known clinical information such as recurrent or no-recurrent cancer. Thus, Cohorts 1, 2, and 3 serve as the models for our research exploration, discovery, and validation, respectively. All the patients information could be found in Tables S1–S2 and S4.
Patients were pathologically and clinically diagnosed with colorectal cancer. After surgical debulking, patients had undergone XELOX regimen therapy (Oxaliplatin 130 mg/m2 IV over two hours on the first day; Capecitabine 850~1000 mg/m2, twice daily p.o. for 14 days; repeated every three weeks). Informed consent was obtained from the patients before sample collection in accordance with institutional guidelines. Recurrence was monitored by imaging examination systems (Chest X-ray and CT), gastrointestinal endoscopy with biopsy, and telephone follow-up. The Ethics Committees in the Renji Hospital approved the study protocols. Written informed consents were obtained from all participants in this study. All the research was carried out in accordance with the provisions of the Helsinki Declaration of 1975.
METHOD DETAILS
High-Throughput Sequencing
For RNA sequencing, each sample was cleaned up on an RNeasy Mini Column (QIAGEN, Hilden, Germany), treated with DNase, and analyzed for quality on an Agilent 2100 Bioanalyzer. Samples were run on an Illumina HiSeq 3000 for 2 × 150-bp paired-end sequencing. The RNA-seq data analysis was performed according to the TopHat- HTSeq-DeSeq2 frame (Anders et al., 2013). Briefly, reads were mapped to the human genome (hg19) using TopHat v2.0.11(Kim et al., 2013) (http://tophat.cbcb.umd.edu) with the default options with a TopHat transcript index built from Ensembl_GRCh37. Count files of the aligned sequencing reads were generated by the htseq-count script from the Python package HTSeq with union mode, using the GTF annotation file(Anders et al., 2015). The read counts from each sequenced sample were combined into a count file, which was subsequently used for the differential expression analysis. Differential analyses were performed to the count files using DESeq2 packages, following standard normalization procedures(Love et al., 2014). Genes with less than 5 total counts in both conditions were removed from further analysis. The RNA sequence data have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE90944.
RNA Extraction and Real-Time PCR
Total RNA was extracted from the CRC lines using Trizol reagent (Invitrogen, Carlsbad, CA), and 1 μg of total RNA was reverse transcribed using the PrimeScript RT Reagent Kit (Perfect Real Time; Takara, Japan) to detect relative mRNAs. For the miRNAs, 1 μg of total RNA was reverse transcribed using All-in-One MiRNA Q-PCR Detection Kit (GeneCopoeia, Rockville, MD) according to manufacturer’s instructions in a total reaction volume of 25 μl. Quantitative real-time PCR was performed in triplicates on an Applied Biosystem 7900 quantitative PCR system (Applied Biosystems, Foster City, CA) as previously described (Sun et al., 2015). The Ct values obtained from different samples were compared using the 2−ΔΔCt method. β-actin and U6 served as internal reference genes, respectively.
Detection of F. nucleatum
The primer sequences of the reference gene, prostaglandin transporter (PGT) and the method for F. nucleatum detection were described previously (Castellarin et al., 2012; Chen et al., 2013). gDNA was extracted from fresh colorectal cancer tissue with the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) and from FFPE with QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany). gDNA from each specimen was subjected to qPCR to determine the amounts of F. nucleatum by detecting the 16S genes. Each reaction contained 40 ng of gDNA and was assayed in triplicate in 10 μL reactions containing 1 × Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, West Palm Beach, FL),0.4 μM each primer and was placed in a 96-well optical PCR plate. Amplification and detection of DNA was performed with the ABI StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA) under the following reaction conditions: 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and at 60°C for 1 min. The cycle threshold (Ct) values for F. nucleatum were normalized to the amount of human biopsy gDNA in each reaction by using PGT as a reference gene (Castellarin et al., 2012).
Western Blot and Chemical Reagents
Western blot was performed using standard techniques as described previously (Xiong et al., 2012). Cell extracts were collected and quantified using BCA Protein Assay Kit (Thermo Fisher Scientific, West Palm Beach, FL). 40 μg of protein was electrophoresed through 12% SDS polyacrylamide gels and were then transferred to PVDF membranes (Bio-Rad, Hercules, CA). The membranes were blocked in 5% fat-free milk for one hour and then incubated with primary antibodies at 4°C overnight. Secondary antibodies were labeled with HRP (KangChen, China) and the signals were detected using ECL Kit (Pierce Biotech, Rockford, IL). Subsequently, the images were analyzed by ImageJ 1.43 software. A β-actin antibody was used as a control for whole-cell lysates. The information on all antibodies is listed in RESOURCES TABLE.
Cell Proliferation Assay, Apoptosis Detection, and TUNEL Assay
Cell proliferation was assessed by Cell Counting Kit-8 (Dojindo, Japan) assay. Cells were seeded at 2000 cells/well into 96-well plates with 100 μL culture medium. The 10 μL of CCK-8 solution was added to the cells at specific time points and cells were incubated for 2 hr at 37°C. The reaction product was quantified according to the manufacturer’s instructions.
Apoptosis was examined by flow cytometric analysis. An Annexin V FITC/PI double stain assay (BD Biosciences, San Jose, CA) was performed following the manufacturer’s protocol. When the cells were treated with Doxorubicin, we performed FVS510 and Annexin V-FITC double staining to avoid the fluorescent signal of the drug. Tumor cell apoptosis in the xenograft tumor tissues was detected by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) technology using the in Situ Cell Death Detection Kit (Roche Molecular Biochemicals, Mannheim, Germany). The negative control was incubated with label solution (without terminal transferase) instead of the TUNEL reaction mixture.
Oligonucleotide Transfection
siRNAs, miRNA mimics, and inhibitors were purchased from Genepharma (Shanghai, China) (Table S7). Oligonucleotide transfection was performed using the DharmaFECT 1 siRNA transfection reagent (Invitrogen, Carlsbad, CA), while nonspecific siRNA or miRNA was used as negative controls.
Luciferase Assay
HCT116 cells or HT29 cells were co-transfected with 80 ng of the luciferase reporter plasmid, 10 ng of the pRL-TK-Renilla-luciferase plasmid (Promega, Madison, WI), and the indicated RNAs (final concentration of 20 nmol/L). Twenty-four hours after transfection, firefly and Renilla luciferase activities were quantified using the Dual-Luciferase Assay Kit (Promega, Madison, WI). Each transfection was performed in triplicates and repeated twice.
Electron Microscopy
Cells were treated as indicated and fixed with 2.5% glutaraldehyde containing 0.1 mol/L sodium cacodylate. Samples were fixed using 1% osmium tetroxide, followed by dehydration with an increasing concentration gradient of ethanol and propylene oxide. Samples were then embedded, cut into 50-nm sections, and stained with 3% uranyl acetate and lead citrate. Images were acquired using a JEM-1200 electron microscope (JEOL, Tokyo, Japan).
Confocal Microscope
HCT116 cells and HT29 cells were plated in 6-well plates and allowed to reach 50%–70% confluence at the time of transfection. MRFP-GFP-LC3 adenoviral vectors were purchased from HanBio Technology (Shanghai, China). The principle of the assay is based on different pH stability of green and red fluorescent proteins. The fluorescent signal of EGFP could be quenched under the acidic condition (pH below 5) inside the lysosome, and the mRFP fluorescent signal has no significant change under the acidic condition. In green and red-merged images, autophagosomes are shown as yellow puncta (i.e., RFP+GFP+), while autolysosomes are shown as red puncta (i.e., RFP+GFP−). Autophagic flux is increased when both yellow and red puncta are increased in cells, while autophagic flux is blocked when only yellow puncta are increased without red puncta alteration, or when both yellow and red puncta are decreased in cells (Zhou et al., 2012).
Adenoviral infection was performed according to the manufacturer’s instructions. CRC cells were incubated in growth medium with the adenoviruses for 2 hr at 37°C, and were grown in medium containing F. nucleatum and miRNAs at the indicated concentrations for 0–24 h at 37°C. LC3 puncta were examined with Zeiss LSM710 confocal microscope (Carl Zeiss) fitted with a 63 × oil immersion objective.
Adenovirus and Plasmids Construction
The control, control shRNA, control miRNA, miR-18a*, miR-4802, TLR4 shRNA, MYD88 shRNA adenovirus, as well as ULK1 and ATG7 overexpression adenovirus, and all plasmids were constructed by Shanghai Obio Techonology Company, Shanghai, China.
QUANTIFICATION AND STATISTICAL ANALYSES
Statistical Analysis
Statistical analyses were carried out using the program R (www.r-project.org). Data from at least three independent experiments performed in triplicates are presented as the means ± SE. Error bars in the scatterplots and the bar graphs represent SE. Data were examined to determine whether they were normally distributed with the One-Sample Kolmogorov-Smirnov test. If the data were normally distributed, comparisons of measurement data between two groups were performed using independent sample t test and the comparisons among three or more groups were first performed by one-way ANOVA test. If the results showed significant difference, when the data were skewed distribution, comparisons were performed by nonparametric test. Measurement data between two groups were performed using nonparametric Mann-Whitney test. To generate the ROC curves, patients were classified as recurrence time either longer or shorter than the median recurrence free survival, excluding patients who were alive for durations less than the median recurrence free survival at last follow-up. Single-sample gene set enrichment analysis (ssGSEA) was used to assess gene set activation scores in gene expression profiling data. ssGSEA calculates a sample level gene set score by comparing the distribution of gene expression ranks inside and outside the gene set. The ssGSEA score was calculated by Gene Set Variation Analysis (GSVA) R package (Hänzelmann et al., 2013). Statistical tests were two-tailed and a p value of less than 0.05 was considered statistically significant.
Data and Software Availability
Data from this study have been deposited in the Gene Expression Omnibus (GEO) databases under the following accession: GSE90944.
Supplementary Material
Highlights.
Specific gut microbes track with post-chemotherapy recurrence of colorectal cancer
F. nucleatum orchestrates the Toll-like receptor, microRNAs, and autophagy network to control cancer chemoresistance
Measuring and targeting F. nucleatum may be useful for patient prognosis and management
Acknowledgments
This project was supported in part by grants from the National Natural Science Foundation of China (81421001, 81320108024, 81530072, 81522008 31371273, 31371420, 81572303, and 81001070), the National Key Technology R&D Program (2014BAI09B05), the Program for Professor of Special Appointment (Eastern Scholar No. 201268 and 2015 Youth Eastern Scholar NO.QD2015003) at Shanghai Institutions of Higher Learning, the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant (no. 20152512, 20161309), the Chenxing Project of Shanghai Jiao-Tong University (H. Chen and J. Hong), and the National Cancer Institute (CA211016, W.Z). We thank Dr. Ming Zhong for collecting colocretal cancer tissues and patient information for this work. We thank Dr.Tingting Yan for graphic abstract conception.
Footnotes
Supplemental Information includes seven figures, seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2017.07.008.
AUTHOR CONTRIBUTIONS
Conceptualization, W.Z., J.H., J.-Y.F.; Methodology, I.K., N.N., J.H., H.C.; Investigation, T.Y., F.G., Y.Y., T.S., D.M., J.H., Y.Q., D.S.; Writing-Original Draft, J.H., H.C., F.G.; Statistical analyses, H.C., F.G.; Writing-Reviewing & Editing, I.K., N.N., Y.C., W.Z., J.-Y.F.; Funding Acquisition, J.-Y.F., J.H., H.C.,Y.C., W.Z.; Supervision, J.-Y.F., W.Z., J.H., and H.C.
References
- Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu MT, Peek RM., Jr Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534–1546. e1533. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertotti A, Sassi F. Molecular Pathways: Sensitivity and Resistance to Anti-EGFR Antibodies. Clin Cancer Res. 2015;21:3377–3383. doi: 10.1158/1078-0432.CCR-14-0848. [DOI] [PubMed] [Google Scholar]
- Cartwright TH. Treatment decisions after diagnosis of metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:155–166. doi: 10.1016/j.clcc.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, Yang L, Liu ZJ, Yuan YZ, Liu F, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- Dahan L, Sadok A, Formento JL, Seitz JF, Kovacic H. Modulation of cellular redox state underlies antagonism between oxaliplatin and cetuximab in human colorectal cancer cell lines. Br J Pharmacol. 2009;158:610–620. doi: 10.1111/j.1476-5381.2009.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, 2nd, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jeong H, Lim YW, Yi H, Sekiguchi Y, Kamagata Y, Chun J. Anaerosporobacter mobilis gen. nov., sp nov., isolated from forest soil. Int J Syst Evol Microbiol. 2007;57:1784–1787. doi: 10.1099/ijs.0.63283-0. [DOI] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- Liu H, Redline RW, Han YW. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J Immunol. 2007;179:2501–2508. doi: 10.4049/jimmunol.179.4.2501. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho C, Martinez-Cardús A, Santos C, Navarro-Pérez V, Martínez-Balibrea E, Musulen E, Carmona FJ, Sartore-Bianchi A, Cassingena A, Siena S, et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst. 2014;106:djt322–djt322. doi: 10.1093/jnci/djt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146:508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Rhee KJ, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LA, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG, Xavier RJ. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167(4):1125–1136. e1128. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L, Sun K, Shen F, Wu M, Wei L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- Sun TT, Tang JY, Du W, Zhao HJ, Zhao G, Yang SL, Chen HY, Hong J, Fang JY. Bidirectional regulation between TMEFF2 and STAT3 may contribute to Helicobacter pylori-associated gastric carcinogenesis. Int J Cancer. 2015;136:1053–1064. doi: 10.1002/ijc.29061. [DOI] [PubMed] [Google Scholar]
- Van Geelen CM, de Vries EG, de Jong S. Lessons from TRAIL-resistance mechanisms in colorectal cancer cells: paving the road to patient-tailored therapy. Drug Resist Updat. 2004;7:345–358. doi: 10.1016/j.drup.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walko CM, Lindley C. Capecitabine: a review. Clin Ther. 2005;27:23–44. doi: 10.1016/j.clinthera.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Wang W, Kryczek I, Dostál L, Lin H, Tan L, Zhao L, Lu F, Wei S, Maj T, Peng D, et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell. 2016;165:1092–1105. doi: 10.1016/j.cell.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Röske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- Xiong H, Hong J, Du W, Lin YW, Ren LL, Wang YC, Su WY, Wang JL, Cui Y, Wang ZH, Fang JY. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem. 2012;287:5819–5832. doi: 10.1074/jbc.M111.295964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YN, Yu TC, Zhao HJ, Sun TT, Chen HM, Chen HY, An HF, Weng YR, Yu J, Li M, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6:32013–32026. doi: 10.18632/oncotarget.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y, Chen Y, Deng X, Xia B, Lin J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy. 2012;8:1215–1226. doi: 10.4161/auto.20284. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Galluzzi L, Viaud S, Vétizou M, Daillère R, Merad M, Kroemer G. Cancer and the gut microbiota: an unexpected link. Sci Transl Med. 2015;7:271ps1. doi: 10.1126/scitranslmed.3010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.