Abstract
Chromosome dynamics during meiotic prophase I are associated with a series of major events such as chromosomal reorganization and condensation, pairing/synapsis and recombination of the homologs, and chromosome movements at the nuclear envelope (NE). The NE is the barrier separating the nucleus from the cytoplasm and thus plays a central role in NE-associated chromosomal movements during meiosis. Previous studies have shown in various species that NE-linked chromosome dynamics are actually driven by the cytoskeleton. The linker of nucleoskeleton and cytoskeleton (LINC) complexes are important constituents of the NE that facilitate in the transfer of cytoskeletal forces across the NE to individual chromosomes. The LINCs consist of the inner and outer NE proteins Sad1/UNC-84 (SUN), and Klarsicht/Anc-1/Syne (KASH) domain proteins. Meiosis-specific adaptations of the LINC components and unique modifications of the NE are required during chromosomal movements. Nonetheless, the actual role of the NE in chromosomic dynamic movements in plants remains elusive. This review summarizes the findings of recent studies on meiosis-specific constituents and modifications of the NE and corresponding nucleoplasmic/cytoplasmic adaptors being involved in NE-associated movement of meiotic chromosomes, as well as describes the potential molecular network of transferring cytoplasm-derived forces into meiotic chromosomes in model organisms. It helps to gain a better understanding of the NE-associated meiotic chromosomal movements in plants.
Keywords: nuclear envelope, chromosome dynamics, meiosis prophase I, SUN proteins, KASH proteins, meiotic modification, cytoplasmic adaptors, nucleoplasmic adaptors
Introduction
Meiosis has the following characteristics, one round of DNA replication and two rounds of chromosome separation (Roeder, 1997). Prophase I is the longest and most complex phase of meiosis, which is vital to ensure the faithful completion of meiosis. A series of chromosome dynamics-associated events such as chromosomal reorganization and condensation, establishment of meiotic-specific chromosome structure, homologous chromosome pairing, and dynamic chromosome movements is closely integrated and finely spatiotemporally controlled during meiotic prophase I (Padmore et al., 1991; Dawe et al., 1994; Hunter and Kleckner, 2001; Blat et al., 2002; Borner, 2006; Golubovskaya et al., 2006; Kleckner, 2006; Zickler, 2006; Tiang et al., 2012). During meiosis, telomeres attach to the nuclear envelope (NE), which in turn drives chromosome movement (Tiang et al., 2012). The NE is a highly conserved eukaryotic structure that protects DNA from enzymatic degradation (Stewart et al., 2007; Wilson and Dawson, 2011). Recent studies have shown that the NE fulfills distinct functions by regulating sets of the proteins that are embedded in the NE. Furthermore, the NE is a crucial determinant for reproduction and fertility; its particular components, the Klarsicht/ANC-1/Syne-1 homology (KASH) proteins and Sad-1/UNC-84 homology (SUN) proteins, play a key role in meiotic chromosome movements (Razafsky and Hodzic, 2009; Kracklauer et al., 2013; Subramanian and Hochwagen, 2014). Nonetheless, the precise role of the NE in chromosome dynamics remains elusive. Here, we review recent studies on meiosis-specific constituents and modifications involving the NE and related nucleoplasmic/cytoplasmic adaptors, as well as propose a molecular network of cytoplasm-derived forces that influence NE-linked meiotic chromosomal movements.
An overview of the NE structure
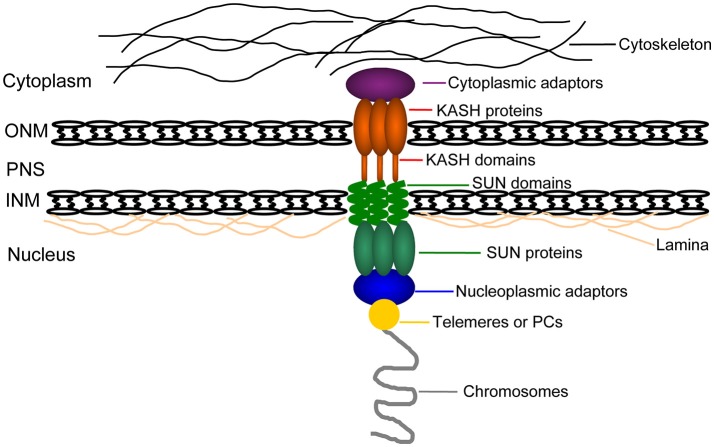
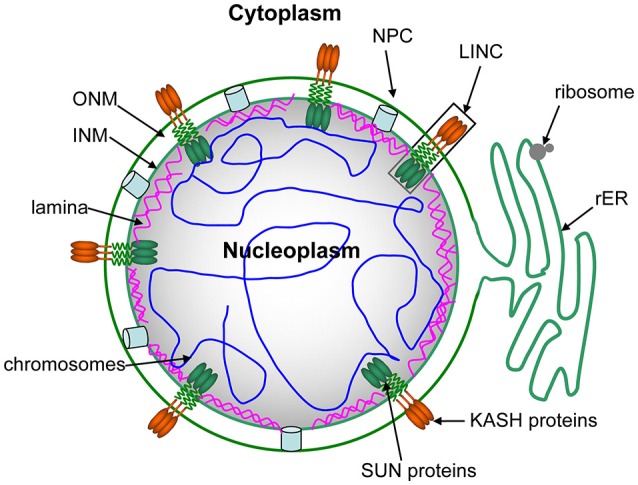
In eukaryotes, the nucleus is a characteristic feature of eukaryotic cells that is enclosed by the NE. Figure 1 shows the structure of the NE during interphase. The NE is a highly conserved eukaryotic double membrane that separates and protects the genetic material of cells (Stewart et al., 2007; Wilson and Dawson, 2011). The general structure of the NE consists of the inner nuclear membrane (INM), outer nuclear membrane (ONM), and the perinuclear space (PNS), which is about 50 nm in thickness and situated between the INM and ONM (Figure 1). The double nuclear membranes are connected by nuclear pore complexes (NPCs) and linkers of nucleoskeleton and cytoskeleton (LINC) complexes (Figure 1; Crisp et al., 2006). NPCs serve as the fusion site of the INM and ONM and form transport channels for macromolecules that move to and from the nucleus and cytoplasm. LINCs stabilize the structure of the NE, play important roles in cell division, and establish cellular polarity, fertilization, cellular migration, and differentiation by connecting the INM and ONM (Crisp et al., 2006; Rothballer et al., 2013; Sosa et al., 2013). However, despite these junctions, the ONM and INM are still divergent. The ONM is a specialized extension of the endoplasmic reticulum (ER), which is studded with ribosomes that facilitate protein synthesis (Park and Craig, 2010). The ONM also binds cytoskeletal components such as microtubules (MTs), as well as acts as a nucleation center of MTs during cell division (Han and Dawe, 2011; Masoud et al., 2013). A series of proteins in the INM interact with various nuclear constituents, including chromosomes and the nucleoskeleton, to ensure the link between the NE and the corresponding nuclear materials (Starr, 2009; Bickmore and van Steensel, 2013). The nuclear lamina as a protein network juxtaposed to the INM nucleoplasmic side. However, currently understanding of the nuclear lamina in plants is limited. An INM-linked dense meshwork was founded in plants by electron microscopy, that is similar to animal laminae (Ciska and de la Espina, 2014).
Figure 1.
The interphase structure of the NE. The NE consists of the inner nuclear membrane (INM), outer nuclear membrane (ONM) and the perinuclear space (PNS). The NE is embedded with nuclear pore complexes (NPCs), SUN proteins in the INM and KASH proteins in the ONM. LINC complexes are made of SUN proteins and KASH proteins, transferring cytoplasm-derived forces inti the chromosomes in the nucleoplasm. The ONM facing the cytoplasm is connected with the rough endoplasmic reticulum (rER). The nuclear lamina is a protein network that is situated close to the INM nucleoplasmic side. In plants, little is known about the nuclear lamina. However, electron microscopy has revealed there is an INM-associated dense meshwork, similar to the animal lamina.
Recent studies have shown that the NE is not only a physical nucleocytoplasmic barrier, but also a multifunctional platform (Fransz and de Jong, 2011; Gross and Bhattacharya, 2011). The NE thus allows specific proteins to be embedded in the ONM and INM, respectively, thereby establishing specific cytoplasm-facing and nucleoplasm-facing functions. A collection of specific integral membrane proteins in the NE include nuclear pore complexes (NPCs), SUN proteins (Razafsky and Hodzic, 2009; Starr and Fridolfsson, 2010) in the INM, and KASH proteins (Wilhelmsen et al., 2006; Rothballer and Kutay, 2013) in the ONM. SUN proteins and KASH proteins form LINC complexes (Crisp et al., 2006). Thus, animal NE proteins transport nucleocytoplasmic macromolecules, are involved in chromosomal dynamics, regulate transcription, and induce aging and nuclear migration (Gruenbaum et al., 2005; Andres and Gonzalez, 2009; Hetzer and Wente, 2009; Starr, 2009). Furthermore, certain NE components play a key role in chromosome pairing and synapsis of homologs during meiosis (Subramanian and Hochwagen, 2014). The LINC complex is an important NE component that has been implicated in the directed movement of meiotic chromosomes within the nucleus (Razafsky and Hodzic, 2009; Kracklauer et al., 2013).
Chromosome dynamics in meiosis
DNA is replicated once, but chromosomes are segregated twice during meiosis (Roeder, 1997). Meiotic divisions are subdivided into meiosis I and meiosis II. Homologous chromosomes are separated in meiosis I, and sister chromatids are segregated from each other in meiosis II. A series of coordinated processes are required during the two meiotic divisions. Prophase I, metaphase I, anaphase I, and telophase I occur in meiosis I. Prophase I as the longest and most complex phase and is further subdivided into five distinguished stages according to the degree of chromatin condensation. The stages in succession are leptotene, zygotene, pachytene, diplotene, and diakinesis (Baarends and Grootegoed, 2003; Wijnker and Schnittger, 2013).
Chromosome dynamics including reorganization and condensation of chromosomes, homologous chromosome pairing, chromosome movements, and establishment of meiosis-specific chromosome structure occur during prophase I of meiosis (Tiang et al., 2012). Homologous chromosome pairing (Dawe et al., 1994) is tightly associated with the process of meiotic recombination (Tiang et al., 2012). Meiosis involves unique chromosome dynamic processes such as pairing/ synapsis and recombination of homologs that occur during meiotic prophase I, as have been extensively characterized in model systems involving Saccharomyces cerevisiae, Schizosaccharomyces pombe, and C. elegans (Hiraoka and Dernburg, 2009; Koszul and Kleckner, 2009). These meiosis-specific events are closely integrated and finely controlled temporally and spatially (Padmore et al., 1991; Hunter and Kleckner, 2001; Blat et al., 2002; Borner, 2006; Kleckner, 2006; Zickler, 2006). Synapsis and recombination ensure the establishment of chiasmata that hold homologous chromosomes together, thereby facilitating correct segregation (Tiang et al., 2012).
Telomere movements at the NE during meiosis
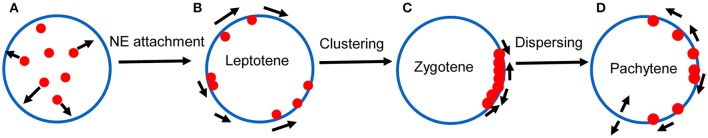
Telomeres are blocks of highly conserved repetitive DNA sequences at chromosome ends that protect chromosomes from nucleolytic degradation and fusion. The behavior of centromeres and telomeres largely controlls chromosomal dynamics of prophase I (Siderakis and Tarsounas, 2007). Previous studies have shown in various species that the cytoskeleton induces chromosomal movements using telomere-NE attachments (Bhalla and Dernburg, 2008; Koszul and Kleckner, 2009; Sheehan and Pawlowski, 2009; Woglar and Jantsch, 2014). During meiosis prophase I, telomere positions undergo dynamic changes, including telomeric attachment, clustering, dispersal, and redistribution across the nuclear periphery (Figure 2). During meiotic interphase, telomeres are distributed across the nucleolus instead of the NE. Prior to pairing, telomeres attach to the NE at the onset of leptotene stage. As leptotene proceeds, telomeres are attached to the NE and are stably linked to it. These tethered telomeres move within the INM and gather at a certain region, creating a characteristic flower-like structure, known as the bouquet of telomeres (Bass et al., 2000; Golubovskaya et al., 2002; Harper et al., 2004; Richards et al., 2012). Telomere clustering starts at the late leptotene stage, always overlaps with the zygotene stage, and usually persists until pachytene (Bass, 2003). The telomere bouquet always appears during the zygotene stage, after which telomeres are then scattered again. Despite telomere clustering may be observed at the early pachytene stage, if homologous chromosomes are completely paired at the end of pachytene, telomeres are dispersed evenly across the NE again while additional nuclear deformations and rotations occur.
Figure 2.
Telomere movement at the NE during meiotic prophase. The four different movement classes are indicated as (A–D). Red dots indicate the positions of the telomeres relative to the NE. The relative direction of telomeric movements is indicated by black arrows. (A) Telomeres scattering in the nucleus move to the NE at the onset of leptotene stage. (B) Telomeres are tethered to the NE and stably connected to it at the late leptotene stage. The telomere clustering starts in the late leptotene stage, always overlaps with the zygotene stage and usually persists until pachytene. (C) The tightest clustering of telomeres is usually observed at the zygotene stage. (D) At pachytene, telomeres are motile and scattered over the NE again, while additional nuclear deformations and rotations occur (black arrows). For further information please see the Scherthan (2007).
The characteristic telomere-guided chromosome movements are an evolutionarily highly conserved hallmark of meiotic prophase I (Scherthan et al., 1996; Koszul and Kleckner, 2009). The telomere “bouquet” stage has been observed in all organisms studied regardless of whether they have big (maize) or small (fission yeast) genomes (Scherthan, 2001), except C. elegans and Drosophila, which both employ non-canonical methods of homology searching (Mckee, 2004).
Functional significance of the telomere bouquet
Bouquet formation of telomeres feature chromosomal movements within the NE, which might facilitate homologous chromosome pairing and synapsis (Scherthan, 2001; Lee et al., 2012). Several lines of evidence show that one of the most likely functions of the bouquet is to warrant the efficient initiation of pairing and synapsis of between homologous chromosomes (Tabata, 1962; Carlton and Cande, 2002; Moens et al., 2011). Mutants with defects in bouquet generation always show defects in chromosome pairing, which suggests the possible role of the bouquet in chromosome pairing (Harper et al., 2004; Klutstein and Cooper, 2014). Several mutants, for example, plural abnormalities of meiosis 1 (pam I) (Golubovskaya et al., 2002), desynaptic 1 (dsy1) (Bass et al., 2003), and poor homologous synapsis1 (phs1) (Pawlowski et al., 2004) exhibit significant defects in homologous pairing in maize. Correspondingly, clusters of telomeres persist in pairing-defective spo11 mutants of Sordaria and S. cerevisiae (Trelles-Sticken et al., 1999). Therefore, it seems likely that the bouquet physically brings homologous chromosomes into close proximity at a certain region of the NE, supporting homologous chromosome pairing and synapsis, double-strand break (DSB) repair, and recombination (Scherthan et al., 1996; Bass et al., 2000), thereby preventing and dissolving heterologous associations of non-homologous chromosomes (Zickler and Kleckner, 1998; Moens et al., 2011). However, the actual function of the meiotic bouquet is still not entirely clear.
LINC complexes
It has been shown in several species that the cytoskeleton induces dynamic motility of chromosomes via telomere-NE attachments (Bhalla and Dernburg, 2008; Koszul and Kleckner, 2009; Sheehan and Pawlowski, 2009; Woglar and Jantsch, 2014). The NE is the barrier separating the nucleus from the cytoplasm that plays a central role in the NE-associated chromosomal movements. Significantly, NE-linked chromosome dynamics are actually driven by the cytoskeleton during meiotic progression (Trelles-Sticken et al., 1999; Conrad et al., 2008; Koszul et al., 2008; Lee et al., 2012). The implication here is that there must be mechanisms that transmit cytoskeletal forces across the NE to individual chromosomes. The special double-layer-membrane structure of the NE raises the question of how can various regions of chromosomes, telomeres in particular, be physically connected to the cytoskeleton during meiosis. Because the NE remains intact during the process of synapsis, there has to be a molecular machinery spanning both the INM and ONM and interacting with chromatin and other cytoskeletal components, respectively. The LINC complexes consist of SUN domain family proteins in the INM and KASH domain homology proteins in the ONM (Burke and Roux, 2009; Razafsky and Hodzic, 2009; Starr and Fridolfsson, 2010).
The LINC complexes span the INM and ONM and form the bridge between the nucleoskeleton and the cytoskeleton through the SUN-KASH domain interaction in the NE lumen (Razafsky and Hodzic, 2009; Starr and Fridolfsson, 2010; Kim et al., 2015). In this way, mechanical forces from the cytoskeleton are directly transduced to the NE and then into chromosomes. A chain of interactions from the cytoskeletal elements to the nucleoskeleton as follows, various components of the cytoskeleton interact with the cytoplasmic domains of KASH proteins, which in turn induces SUN proteins in the INM to interact with KASH proteins at their C-termini in the PNS and with specific nuclear contents at the N-termini in the nucleoplasm (Haque et al., 2006; Bone et al., 2014). The LINC complexes are responsible for the transfer of this force across the nuclear envelope and enable a direct communication and connection between nuclear and cytoplasmic content.
SUN domain proteins
Molecular characteristics of sun proteins
SUN proteins as important INM-integral components of LINC complexes that exhibit highly conserved structure and function (Starr, 2009). SUN proteins comprise an N-terminal region and a C-terminal region that are separated by one or more transmembrane domains (TMDs) (Tzur et al., 2006; Worman and Gundersen, 2006). The N-termini of SUN proteins are variable and directly or indirectly interact with lamins, which are the components of the nucleoskeleton (Lee et al., 2002; Crisp et al., 2006; Haque et al., 2006; Bone et al., 2014) and tether chromosomes to the nuclear periphery (Bupp et al., 2007; King et al., 2008; Morimoto et al., 2012; Link et al., 2014). The C-terminal region contains the well-conserved SUN domain, which extends into the PNS that interacts with KASH proteins. Most SUN proteins have coiled-coil domains (CCDs) at their N-termini, which facilitate in domain trimerization (Sosa et al., 2012; Zhou et al., 2012b).
Two divergent classes of SUN proteins have been identified by homology searching in plants: classical SUN proteins which contain SUN domains at the C-terminus (Murphy et al., 2010), and a second group of SUN proteins, with SUN domains in the center of the SUN protein, and thus designated as mid-SUN proteins (Murphy et al., 2010). The function of mid-SUN proteins is far less well-understood than the Cter-SUNs. Mid-SUN proteins differ from Cter-SUN proteins in both structure and localization. Mid-SUN proteins frequently contain three TMDs and plant mid-SUN proteins usually contain a conserved PM3-associated domain (PAD) (Murphy et al., 2010; Graumann et al., 2014). In addition, mid-SUN proteins are located in both the NE and the ER (Murphy et al., 2010; Graumann et al., 2014).
Members and functions of sun proteins
SUN domain proteins have been identified in various species (Table 1). Three Arabidopsis SUN proteins (AtSUN3, AtSUN4, and AtSUN5) and three maize SUN proteins (ZmSUN3, ZmSUN4, and ZmSUN5) belong to the mid-SUN group (Murphy et al., 2010; Murphy and Bass, 2012; Graumann et al., 2014). The presence of several SUN protein members in a single organism (often at least five in humans) and their ability to form multimers implicate these are involved in a wide range of important cellular functions. Reports have shown that SUN proteins are implicated in interactions with lamins, nuclear positioning, spindle architecture, apoptosis, centrosome linkage to the nucleus, and maintenance of even spacing between the INM and ONM (Table 1). In addition, SUN proteins are required in a number of systems to attach telomeres or pairing centers to the NE during meiosis (Chikashige et al., 2006; Ding et al., 2007; Penkner et al., 2007; Conrad et al., 2008; Koszul et al., 2008). For example, SUN1, SUN2, Sad1, and Mps3 tether chromosomes to the nuclear periphery by interacting with telomere-binding proteins (Bupp et al., 2007; King et al., 2008; Morimoto et al., 2012; Link et al., 2014). The SUN protein trimer can usually bind three KASH domains of KASH homology proteins in the PNS (Sosa et al., 2012; Zhou et al., 2012a). In maize, ZmSUN2 produces a unique belt-like structure at the NE that undergoes remarkable dynamic changes during meiosis (Murphy et al., 2014). Accordingly, AtSUN1 and AtSUN2 have been localized to meiotic prophase I-specific regions (Varas et al., 2015). In maize, ZmSUN3 as a mid-SUN protein, has been supposed to play an important role in meiotic divisions (Murphy and Bass, 2012). Of the five identified SUN proteins of mammals, SUN1 and SUN2 proteins have been demonstrated to be the only ones that are also expressed in meiotic cells, thereby indicating dual somatic and meiotic functions (Schmitt et al., 2007; Chi et al., 2009; Yu et al., 2011). To date, studies involving SUN1- and SUN1/SUN2-deficient mice have revealed that although SUN2 functions in part similarly to SUN1 in meiosis, SUN2 can not effectively compensate for the loss of SUN1 in meiosis (Schmitt et al., 2007; Chi et al., 2009; Lei et al., 2009). However, a single mutation for either SUN1 or SUN2 genes has no effect on reproduction or meiosis in A. thaliana (Varas et al., 2015). Several groups have then hypothesized that SUN1 and SUN2 assemble heteromultimeric complexes (Wang et al., 2006; Lu et al., 2008). Taking into account that in mice, SUN2 protein shares its localization with SUN1 protein and meiotic KASH5 protein, it is then speculated that during normal meiosis SUN1 and SUN2 form heterotrimers which interact with KASH5 protein to assemble meiotic LINCs. In the absence of SUN1, LINCs may only consist of SUN2 and KASH5, still attaching telomeres of chromosomes to the NE, yet in a less effective way than complete SUN1/SUN2-KASH5 complexes. And then this could explain the partial redundancy between SUN1 and SUN2 in mice. Further research is required to determine how these SUN family members coordinate in the near future.
Table 1.
Members and functions of the SUN protein family.
| Members | Functions | References |
|---|---|---|
| Mammals | ||
| SUN1 SUN2 | Movement and attachment of telomere in meiosis; nuclear anchorage and migration; integrity of the NE; recruit KASH proteins | Hodzic et al., 2004; Padmakumar et al., 2004; Crisp et al., 2006; Haque et al., 2006; Ding et al., 2007; Zhang et al., 2009; Morimoto et al., 2012 |
| SUN3 | Links the nucleus to posterior manchette during sperm head formation | Göb et al., 2010 |
| SPAG4 | Not at the NE, function unknown | Shao et al., 1999 |
| SPAG4L | Not at the NE; Links the acrosomic vesicle to the spermatid nucleus; involved in acrosome biogenesis | Frohnert et al., 2011 |
| Drosophila | ||
| Klaroid | Nuclear anchorage during Drosophila oogenesis.; nuclear migration | Patterson et al., 2004; Yu et al., 2006; Kracklauer et al., 2007 |
| SPAG4/Giacomo | Not at the NE; involved in centriolar-nuclear attachment during spermatogenesis | Malone et al., 2003 |
| C. elegans | ||
| UNC-84 | Nuclear positioning; nuclear anchorage and migration | Starr et al., 2001; Starr and Han, 2002 |
| SUN-1/matefin | Links the centrosome to nucleus; homologous chromosome pairing and synapsis in meiosis; apoptosis | Malone et al., 2003; Tzur et al., 2006; Penkner et al., 2009; Sato et al., 2009 |
| S. pombe | ||
| Sad1 | Spindle architecture; meiotic chromosome pairing and synapsis | Shimanuki et al., 1997; Miki et al., 2004; Chikashige et al., 2006; Ding et al., 2007 |
| S. cerevisiae | ||
| Mps3 | Linkage to the NE of SPB; SPB duplication; telomere attachment to and clustering within the NE | Jaspersen et al., 2006; Conrad et al., 2008; Wanat et al., 2008; Horigome et al., 2011 |
| Arabidopsis | ||
| AtSUN1 AtSUN2 | Recruit KASH proteins to the NE; nuclear elongation and movement; meiotic recombination and synapsis | Graumann et al., 2010; Oda and Fukuda, 2011; Zhou et al., 2012a, 2015a,b; Tamura et al., 2013; Varas et al., 2015 |
| AtSUN3, AtSUN4, AtSUN5 | Mid-SUN proteins; seed development and involved in nuclear morphology | Graumann, 2014; Zhou et al., 2015b |
| Maize | ||
| ZmSUN1 ZmSUN2 | Involved in meiotic telomere dynamics | Murphy et al., 2014 |
| ZmSUN3 ZmSUN4 ZmSUN5 | Mid-SUN proteins; ZmSUN3 plays a role in meiosis; ZmSUN4/ZmSUN5: unknown functions | Murphy et al., 2010; Murphy and Bass, 2012 |
| Dictyostelium | ||
| Sun-1 | Centrosome attachment; genome stability | Xiong et al., 2008 |
KASH domain proteins
Molecular characteristics of KASH proteins
Four criteria were employed to define KASH proteins (Starr, 2011). First, KASH proteins are positioned at the ONM. Second, the C-terminal KASH domain is essential for interaction between KASH and SUN proteins. Third, the KASH domains ensure their localization to the ONM (Crisp et al., 2006). Fourth, N-terminal domains of KASH proteins are not highly conserved and are linked to the cytoskeleton. The KASH domain usually includes a hydrophobic transmembrane domain and a sequence of 6–30 amino acids in the PNS. The perinuclear 6- to 30-amino acid domain of KASH proteins is usually highly conserved, for example, 13 of 20 residues are identical between C. elegans ANC-1 and human Syne/Nesprin-1/-2. The terminal region of the perinuclear sequence of the KASH domain consists of a highly conserved four-amino acid motif PPPX in most animals; however, specifically, the penultimate proline appears to be widely conserved across kingdoms, which is essential in mediating SUN-KASH interaction (Lenne et al., 2000; Razafsky and Hodzic, 2009; Starr and Fridolfsson, 2010; Sosa et al., 2012). Apart from the PPPX motif, the last C-terminal four amino acids of plant KASH proteins are usually XVPT (X represents V/A/L/P) (Zhou et al., 2012a; Zhou and Meier, 2013). Similar to SUN proteins, KASH domain proteins can also form multimers (Djinovic-Carugo et al., 2002; Mislow et al., 2002). The SUN-KASH complexe usually comprise SUN protein trimers and KASH protein trimers. SUN-KASH interactions occur when the KASH domain fits into a hydrophobic pocket that is assembled by three SUN proteins.
Members and functions of KASH proteins
To date, KASH domain proteins have been identified in various species (Table 2). These KASH proteins are involved in different processes, such as nuclear migration, linkage to the nucleus, attaching nuclei to actin filaments and so on (Table 2).
Table 2.
Members and functions of the KASH protein family.
| Members | Functions | References |
|---|---|---|
| Mammals | ||
| Syne-1 (Nesprin-1) Syne-2 (Nesprin-2) | Attach nuclei to actin filaments; nuclear migration and nucleokinesis | Apel et al., 2000; Zhang et al., 2007, 2009 |
| Nesprin-3 | A versatile connector between the nucleus and the cytoskeleton | Ketema and Sonnenberg, 2011 |
| Nesprin-4 | Binding kinesin; cell polarization | Roux et al., 2009 |
| KASH 5 | Dynein-driven telomere dynamics in meiosis | Morimoto et al., 2012 |
| Drosophila | ||
| Klarsicht | Anchoring microtubules to the NE; nuclear migration and centrosome attachment | Mosleybishop et al., 1999; Patterson et al., 2004; Elhananytamir et al., 2012 |
| MSP-300 | Nuclear anchorage | Yu et al., 2006 |
| C. elegans | ||
| KDP-1 | Cell- cycle progression | Mcgee et al., 2009 |
| ANC-1 | Nuclear anchorage | Starr and Han, 2002 |
| UNC-83 | Nuclear migration | Starr et al., 2001; Meyerzon et al., 2009 |
| ZYG-12 | Links centrosomes to nuclei; meiotic chromosome paring and synapsis | Malone et al., 2003; Sato et al., 2009; Zhou et al., 2009 |
| S. pombe | ||
| Kms1 | Meiotic dynein-driven chromosome movement and pairing | Miki et al., 2004; Chikashige et al., 2006 |
| Kms2 | Meiotic and mitotic chromosome movements | Miki et al., 2004; Chikashige et al., 2006; King et al., 2008 |
| S. cerevisiae | ||
| Csm4 | Meiotic actin-driven chromosome movements and pairing | Conrad et al., 2008; Koszul et al., 2008 |
| Dictyostelium | ||
| Interaptin | Function unknown | Rivero et al., 1998 |
| Arabidopsis | ||
| WIP1-3 | Anchors WIT1-2 to the NE; anchoring RanGAP to the NE | Yu et al., 2011; Zhou et al., 2012b, 2015b |
| SINE1 | Actin-dependent nuclear positioning | Zhou et al., 2014 |
| SINE2 | Contributes to innate immunity against an oomycete pathogen | Zhou et al., 2014 |
| AtTIK | Function unknown | Graumann et al., 2014 |
The less similarity between KASH domains is very weak, suggesting that many KASH proteins have yet to be discovered. For example, C. elegans ZYG-12 and S. cerevisiae Csm4 poorly aligns with other KASH domains, but these fit the criteria for KASH proteins (Starr and Fischer, 2005; Conrad et al., 2008; Koszul et al., 2008). Tryptophan–proline–proline (WPP)-interacting proteins (WIP)1-3 and SUN-interacting NE 1-2 proteins (SINE 1-2) are plant-specific KASH proteins that share a low degree of similarity with metazoan KASH proteins (Graumann et al., 2010; Oda and Fukuda, 2011; Zhou et al., 2012a, 2014; Zhou and Meier, 2013). These proteins reside in the ONM via SUN-KASH interactions, fulfilling the criteria for KASH proteins mentioned. AtTIK is a novel Arabidopsis KASH domain protein that has been identified using a split-ubiquitin-based membrane yeast two-hybrid screen (Graumann et al., 2014).
Among these reported KASH proteins, only mammalian KASH5, C. elegans ZYG-12, and yeast Kms1, Kms2, and Csm4 have been confirmed to be involved in meiosis (Table 2).
Meiosis-specific adaptations involving the NE
Although the NE has a highly conserved basic structure in eukaryotes, it also undergoes meiosis-specific adjustment to facilitate chromosome dynamics. The nuclear lamina is a protein network that is juxtaposed to the INM nucleoplasmic side. It is mainly composed of lamin proteins. In animals, the nuclear lamina undergoes significant modifications in lamin B1 (B-type lamin) and lamin C2 (A-type lamin isoform) during meiosis, and lamin C2 is exclusively expressed in meiotic cells. This implicates that NE is modified to adapt to the requirements of meiosis (Furukawa et al., 1994; Alsheimer and Benavente, 1996). Current understanding of the functions of the nuclear lamina is limited in plants. It has been postulated that the nuclear matrix component protein (NMCP) family members are likely the best appropriate candidates for plant lamins (Ciska and de la Espina, 2014). Fluorescence resonance energy transfer experiments have shown that the N-termini of AtSUN1 and AtSUN2 co-localize with CRWN1, which is a member of the NMCP family in Arabidopsis (Graumann, 2014). However, its physical co-localization does not demonstrate that AtSUN1 and AtSUN2 directly or indirectly interact with CRWN1. Investigations on meiosis-specific adjustments with respect to components and functions of the nuclear lamina in plants are limited.
LINC complexes are important components of the NE that also undergo remarkable adaptations to the requirements of meiosis. SUN proteins and KASH proteins are encoded by various genes that are differentially expressed in various cell types and tissues (Roux et al., 2009; Göb et al., 2010, 2011; Frohnert et al., 2011; Kracklauer et al., 2013; Duong et al., 2014). LINC complexes generally exhibit features that involve specific cellular processes. Meiotic chromosomal movements within the NE are driven by cytoskeletal forces that span the double NE and are transferred to the chromosomes via specific LINC complexes (Kracklauer et al., 2013; Yamamoto, 2014). Unique reconstruction of the NE structure and formation of meiosis-specific LINC complexes are required during telomere attachment, movements, clustering, and reposition (Hiraoka and Dernburg, 2009). The meiosis-specific LINC complexes are modulated with respect to their constituent proteins and interaction partners (Table 3). LINC complexes are species-specific. In mice, meiosis-specific LINC complexes are composed of SUN1 and/or SUN2, and KASH5, which promote chromosome pairing and synapsis (Ding et al., 2007; Schmitt et al., 2007; Morimoto et al., 2012). The SUN protein Sad1 directly interacts with a KASH protein Kms1, assembling a functional meiotic LINC complex in S. pombe (Miki et al., 2004). The KASH domain protein ZYG-12 as a SUN1-interacting meiotic LINC component in C. elegans (Malone et al., 2003). The Zea mays SUN protein, ZmSUN3 is necessary for homologous chromosome synapsis, recombination, and chromosome segregation (Murphy et al., 2010; Murphy and Bass, 2012). However, the real meiotic KASH partner of ZmSUN3 remains elusive. In Arabidopsis, AtSUN1 and AtSUN2 are both associated with meiosis (Duroc et al., 2014; Varas et al., 2015). At the same time, the Arabidopsis genome encodes four KASH proteins, three WIP proteins (AtWIP1, AtWIP2, and AtWIP3) and one AtTIK protein, which all interact with AtSUN1 (Zhou et al., 2012a; Graumann et al., 2014). However, their definitive meiosis-specific functions remain unclear.
Table 3.
Constituents of meiotic-specific LINC complexes in various organisms.
| S. pombe | S. cerevisiae | C. elegans | Mice | Arabidopsis | Maize | |
|---|---|---|---|---|---|---|
| SUN domain proteins | Sad1 | Mps3 | Metafin/SUN-1 | SUN1, SUN2 | AtSUN1, AtSUN2 | ZmSUN1, ZmSUN2, ZmSUN3 |
| KASH domain proteins | Kms1, Kms2 | Csm4 | ZYG-12 | KASH5 | AtWIP1-3 | U |
U, Unidentified.
Kinase-associated meiosis-specific modifications of the NE
The meiosis-specific functions of the ubiquitously expressed SUN proteins indicate that SUN proteins undergo post-translational modifications to mediate their meiotic functions. The Polo-like family of Ser/Thr kinase (PLK) of C. elegans co-localizes with PCs during meiosis, bringing about aggregation of SUN-1/ZYG-12 within the NE, thereby mediating dynein-driven chromosomal motions (Harper et al., 2011; Labella et al., 2011; Wynne et al., 2012; Rog and Dernburg, 2015). Phosphorylation modifications of the SUN1 nucleoplasmic domain through checkpoint protein kinase (CHK) family members CHK-2 and PLK-2 influence SUN1 motions within the INM during meiosis in C. elegans (Penkner et al., 2009; Sato et al., 2009; Labella et al., 2011). A recent study has shown that CHK-2 is a master regulator of meiosis in C. elegans, which first phosphorylates PC-binding zinc finger proteins HIM-8 and ZIMs, which in turn recruits PLK-2 to PCs (Kim et al., 2015).
Cyclin-dependent kinases (CDKs) are another group of highly conserved serine/threonine protein kinases that have been detected in various species from yeast to humans and play key roles in regulating the cell cycle and the cell division. During mammalian meiotic prophase I, CDK2 plays a critical role in meiosis-associated telomeric dynamics and meiosis-specific modifications of the NE components (Ashley et al., 2001; Berthet et al., 2003; Ortega et al., 2003; Viera et al., 2009, 2015). In mice, CDK2 mediates the accurate dynamic distribution of SUN1 protein via phosphorylation of SUN1 protein. SUN1 persists at the NE as a cap from the leptotene to pachytene phases in the absence of CDK2 in mice. CDK2 also affects the assembly of the meiosis-specific nuclear lamina. In the absence of CDK2, the distribution of lamin C2, a meiosis-specific isoform of lamin A and LAP2 (lamin-associated protein) are severely impaired, with a complete lack of LAP2 (Viera et al., 2015). However, the possible pathways to determine altered distribution of lamin C2 in meiosis are unknown.
Meiosis-specific adaptors between telomeres/PCs and LINC
Telomeres link chromosomes to the NE through the LINCs. From yeast to humans, telomeres (or pairing centers in the worm) are always anchored to the NE by specific adaptors in the meiosis prophase I. The linkers connecting telomeres to LINCs are mainly composed telomere-binding proteins. In S. pombe, the linker between LINCs and telomeres is mediated by telomeric proteins Taz-1 and Rap-1, and the meiosis-specific proteins, Bouquet1-4 (Bqt1-4) (Chikashige et al., 2006, 2009). In C. elegans, chromosomes are connected to the NE through chromosome-specific pairing centers (PCs), instead of telomeres. Accordingly, LINCs tether chromosomes to the NE through PC-specific proteins, ZIM-1, ZIM-2, ZIM-3, and HIM-8 (Phillips et al., 2005; Phillips and Dernburg, 2006; Penkner et al., 2007; Sato et al., 2009; Baudrimont et al., 2010). During meiosis in S. cerevisiae, Ndj1 as a meiosis-specific adaptor connects LINCs to telomeres (Conrad et al., 2007, 2008). In mammals, telomere repeat-binding bouquet formation protein 1/2 (TERB1/2) and membrane-anchored junction protein (MAJIN) form a complex, TERB1/2-MAJIN, which serves as a meiosis-specific link between telomeres and LINCs (Daniel et al., 2014; Shibuya et al., 2014, 2015). In addition, meiotic LINCs of mammals are able to interact with meiosis-specific laminae. It is unknown whether meiosis-specific lamina proteins have an effect on telomere connection with LINCs. Currently, how telomeres are modified to mediate telomeric attachment to the NE during meiosis in plants remains unclear.
Meiosis-specific adaptors between the cytoskeleton and the LINC
Anchoring linkers bridging LINCs and the cytoskeleton are responsible for transferring cytoskeletal forces to the NE, which then mediates meiotic chromosome movements along the NE during prophase I stages that comprise cytoskeleton or associated motor proteins (Koszul and Kleckner, 2009; Kracklauer et al., 2013). The LINC-complex is bound to the actin cytoskeleton via the atypical KASH protein Csm4 and actin in S. cerevisiae (Conrad et al., 2007, 2008). The LINC-complex is connected to microtubules (MTs) in the cytoplasm through Kms1 (KASH protein) and dynein light chain-family protein Dlc1 in S. pombe (Miki et al., 2002), KASH5, and dynein in mammals (Morimoto et al., 2012; Rothballer and Kutay, 2013), ZYG-12 KASH protein and dynein motors in C. elegans (Sato et al., 2009; Wynne et al., 2012), KASH proteins AtWIP-1, AtWIP-2 and a kinesin1-like protein AtPSS1 in Arabidopsis (Duroc et al., 2014; Wang et al., 2014).
An integrated mechanical system transferring cytoplasm forces into meiotic chromosomes
The mechanisms responsible for dynamic chromosome movements have been partially deciphered in model organisms (Figure 3). The LINC complex couples the microtubule network and chromosomes. Nucleoplasmic adaptors tether telomeres or PCs (in C. elegans) to LINCs. Cytoplasmic adaptors connect cytoskeleton or cytoskeleton-associated proteins to LINC. The network between the cytoskeleton and chromosomes is telomeres/PCs-nucleoplasmic adaptors-NE-cytoplasmic adaptors-cytoskeleton. The molecular link system by which these forces are implemented differs in constituents in various organisms, telomeres-Taz1/Rap1/Bqt(1-4)-Sad1-Kms1/2-dynein (Dlc1)-MTs in S. pombe; PCs-ZIM(1-3)/HIM8-SUN1-ZYG12-Dynein-MTs in C. elegans; telomeres-Ndj1-Mps3-Csm4-actin-actin cable in S. cerevisiae; telomeres-TERB1/2/MAJIN-SUN1/SUN2-KASH5-dynein-MTs in mice; and telomeres-?-AtSUN1/AtSUN2-AtWIP1/2-kinesin (AtPSS1)-MTs in Arabidopsis. Whether and how NMCP family proteins and modification of SUN proteins are involved in the above molecular link system in plants remain unclear.
Figure 3.
A schematic representation of the link transferring cytoplasm forces into meiotic chromosomes. Telomeres or PCs (gold circle) connect to the NE through nucleoplasmic adaptors (schematized with a blue oval) and the nucleoplasmic domains (in green ovals) of SUN-domain proteins spanning the INM (in green; shown as a trimer). KASH domain proteins span the ONM (in red; shown as a trimer). Then SUN domains (in green helix) can interact with KASH domains (in red stub) in the PNS. Cytoplasmic adaptors (in purple) connect the cytoplasmic domains (in red ovals) of KASH proteins to the cytoskeleton (in black lines). The nucleoplasmic domains of SUN proteins can also interact with lamins (in orange). Cytoskeleton, cytoplasmic adaptors, SUN-KASH protein bridges, nucleoplasmic adaptors and telomeres/PCS form the central link that spans the nuclear envelope, transferring cytoplasm-derived forced into chromosomes. NE, nuclear envelope; INM, inner nuclear membrane; ONM, outer nuclear membrane; PNS, the perinuclear space.
Conclusions and future perspectives
Telomere-led chromosomal dynamics within the NE and mediated by LINCs are pivotal for meiosis and thus fertility. The NE as a regulatory platform is finely modified with respect to its constituents in meiosis. Meiosis-specific adaptations of the LINC components, cytoplasmic linkers, and nucleoplasmic linkers contribute to these movements. Our current knowledge of the LINC network can serve as a starting point for future studies in plants. KASH proteins are not well conserved and thus warrant identification of additional novel family members. There are still a number of issues concerning the meiotic adaptions of the NE that need to be addressed. How are ubiquitously expressed NE components regulated during meiosis? Are plant NMCP family proteins involved in telomeric attachment to the NE, similar to the lamina proteins? Are there more adaptor molecules participating in the LINC network?
Authors contributions
XY and XZ wrote the manuscript. RY, KL, HG, and JL contributed to the preparation of this manuscript. FL, YW, and GW organized and reviewed the manuscript. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Natural Science Foundation of Hubei Province (grant numbers 2013CFB423 and 2014CFB320), the National Natural Science Foundation of China (grant numbers 31400243 and 31201152), the National Grand Project of Science and Technology (2018ZX08012001 and 2018ZX0801104B), Breeding special grants of seven major crops (2017YFD0102000), and the Major Research Project of CAAS Science and Technology Innovation Program supported this study.
References
- Alsheimer M., Benavente R. (1996). Change of karyoskeleton during mammalian spermatogenesis: expression pattern of nuclear lamin C2 and its regulation. Exp. Cell Res. 228, 181–188. 10.1006/excr.1996.0315 [DOI] [PubMed] [Google Scholar]
- Andrés V., González J. M. (2009). Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 187, 945–957. 10.1083/jcb.200904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel E. D., Lewis R. M., Grady R. M., Sanes J. R. (2000). Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986–31995. 10.1074/jbc.M004775200 [DOI] [PubMed] [Google Scholar]
- Ashley T., Walpita D., De Rooij D. G. (2001). Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 114, 685–693. [DOI] [PubMed] [Google Scholar]
- Baarends W. M., Grootegoed J. A. (2003). Chromatin dynamics in the male meiotic prophase. Cytogenet. Genome Res. 103, 225–234. 10.1159/000076808 [DOI] [PubMed] [Google Scholar]
- Bass H. W. (2003). Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell. Mol. Life Sci. 60, 2319–2324. 10.1007/s00018-003-3312-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H. W., Bordoli S. J., Foss E. M. (2003). The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L) cause distinct telomere-misplacement phenotypes during meiotic prophase. J. Exp. Bot. 54, 39–46. 10.1093/jxb/erg032 [DOI] [PubMed] [Google Scholar]
- Bass H. W., Riera-Lizarazu O., Ananiev E. V., Bordoli S. J., Rines H. W., Phillips R. L., et al. (2000). Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113(Pt 6), 1033–1042. [DOI] [PubMed] [Google Scholar]
- Baudrimont A., Penkner A., Woglar A., Machacek T., Wegrostek C., Gloggnitzer J., et al. (2010). Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 6:e1001219. 10.1371/journal.pgen.1001219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis L. (2003). Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785. 10.1016/j.cub.2003.09.024 [DOI] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F. (2008). Prelude to a division. Cell Dev. Biol. 24, 397–424. 10.1146/annurev.cellbio.23.090506.123245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore W. A., van Steensel B. (2013). Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284. 10.1016/j.cell.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R. U., Hunter N., Kleckner N. (2002). Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802. 10.1016/S0092-8674(02)01167-4 [DOI] [PubMed] [Google Scholar]
- Bone C. R., Tapley E. C., Gorjánácz M., Starr D. A. (2014). The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol. Biol. Cell 25, 2853–2865. 10.1091/mbc.E14-05-0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner G. V. (2006). Balancing the checks: surveillance of chromosomal exchange during meiosis. Biochem. Soc. Trans. 34, 554–556. 10.1042/BST0340554 [DOI] [PubMed] [Google Scholar]
- Bupp J. M., Martin A. E., Stensrud E. S., Jaspersen S. L. (2007). Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179, 845–854. 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Roux K. J. (2009). Nuclei take a position: managing nuclear location. Dev. Cell. 17, 587–597. 10.1016/j.devcel.2009.10.018 [DOI] [PubMed] [Google Scholar]
- Carlton P. M., Cande W. Z. (2002). Telomeres act autonomously in maize to organize the meiotic bouquet from a semipolarized chromosome orientation. J. Cell Biol. 157, 231–242. 10.1083/jcb.200110126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y. H., Cheng L. I., Myers T., Ward J. M., Williams E., Su Q., et al. (2009). Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development 136, 965–973. 10.1242/dev.029868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y., Tsutsumi C., Yamane M., Okamasa K., Haraguchi T., Hiraoka Y. (2006). Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125, 59–69. 10.1016/j.cell.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Chikashige Y., Yamane M., Okamasa K., Tsutsumi C., Kojidani T., Sato M., et al. (2009). Membrane proteins Bqt3 and−4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 187, 413–427. 10.1083/jcb.200902122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M., de la Espina M. D. S. (2014). The intriguing plant nuclear lamina. Front. Plant Sci. 5:166. 10.3389/fpls.2014.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Chao G., Shinohara M., Kosaka H., Shinohara A., et al. (2008). Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133, 1175–1187. 10.1016/j.cell.2008.04.047 [DOI] [PubMed] [Google Scholar]
- Conrad M. N., Lee C. Y., Wilkerson J. L., Dresser M. E. (2007). MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 8863–8868. 10.1073/pnas.0606165104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J. B., Shanahan C., Burke B., et al. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172, 41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K., Tränkner D., Wojtasz L., Shibuya H., Watanabe Y., Alsheimer M., et al. (2014). Mouse CCDC79 (TERB1) is a meiosis-specific telomere associated protein. BMC Cell Biol. 15:17. 10.1186/1471-2121-15-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe R. K., Sedat J. W., Agard D. A., Cande W. Z. (1994). Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76, 901–912. 10.1016/0092-8674(94)90364-6 [DOI] [PubMed] [Google Scholar]
- Ding X., Xu R., Yu J., Xu T., Zhuang Y., Han M. (2007). SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872. 10.1016/j.devcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K., Gautel M., Ylänne J., Young P. (2002). The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 513, 119–123. 10.1016/S0014-5793(01)03304-X [DOI] [PubMed] [Google Scholar]
- Duong N. T., Morris G. E., Lam L. T., Zhang Q., Sewry C. A., Shanahan C. M., et al. (2014). Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS ONE. 9:e94380. 10.1371/journal.pone.0094380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y., Lemhemdi A., Larchevêque C., Hurel A., Cuacos M., Cromer L., et al. (2014). The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genet. 10:e1004674. 10.1371/journal.pgen.1004674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhananytamir H., Yu Y. V., Shnayder M., Jain A., Welte M., Volk T. (2012). Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 198, 833–846. 10.1083/jcb.201204102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., de Jong H. (2011). From nucleosome to chromosome: a dynamic organization of genetic information. Plant J. 66, 4–17. 10.1111/j.1365-313X.2011.04526.x [DOI] [PubMed] [Google Scholar]
- Frohnert C., Schweizer S., Hoyer-Fender S. (2011). SPAG4L/SPAG4L-2 are testis-specific SUN domain proteins restricted to the apical nuclear envelope of round spermatids facing the acrosome. Mol. Hum. Reprod. 17, 207–218. 10.1093/molehr/gaq099 [DOI] [PubMed] [Google Scholar]
- Furukawa K., Inagaki H., Hotta Y. (1994). Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp. Cell Res. 212, 426–430. 10.1006/excr.1994.1164 [DOI] [PubMed] [Google Scholar]
- Göb E., Meyernatus E., Benavente R., Alsheimer M. (2011). Expression of individual mammalian Sun1 isoforms depends on the cell type. Commun. Integr. Biol. 4, 440–442. 10.4161/cib.15369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göb E., Schmitt J., Benavente R., Alsheimer M. (2010). Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE 5:e12072. 10.1371/journal.pone.0012072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I. N., Hamant O., Timofejeva L., Wang C. J., Braun D., Meeley R., et al. (2006). Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119, 3306–3315. 10.1242/jcs.03054 [DOI] [PubMed] [Google Scholar]
- Golubovskaya I. N., Harper L. C., Pawlowski W. P., Schichnes D., Cande W. Z. (2002). The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162, 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K. (2014). Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS ONE 9:e93406. 10.1371/journal.pone.0093406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K., Runions J., Evans D. E. (2010). Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 61, 134–144. 10.1111/j.1365-313X.2009.04038.x [DOI] [PubMed] [Google Scholar]
- Graumann K., Vanrobays E., Tutois S., Probst A. V., Evans D. E., Tatout C. (2014). Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J. Exp. Bot. 65, 6499–6512. 10.1093/jxb/eru368 [DOI] [PubMed] [Google Scholar]
- Gross J., Bhattacharya D. (2011). Endosymbiont or host: who drove mitochondrial and plastid evolution? Biol. Direct. 6:12. 10.1186/1745-6150-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6, 21–31. 10.1038/nrm1550 [DOI] [PubMed] [Google Scholar]
- Han Z., Dawe R. K. (2011). Mechanisms of plant spindle formation. Chromosome Res. 19, 335–344. 10.1007/s10577-011-9190-y [DOI] [PubMed] [Google Scholar]
- Haque F., Lloyd D. J., Smallwood D. T., Dent C. L., Shanahan C. M., Fry A. M., et al. (2006). SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26, 3738–3751. 10.1128/MCB.26.10.3738-3751.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L., Golubovskaya I., Cande W. Z. (2004). A bouquet of chromosomes. J. Cell Sci. 117, 4025–4032. 10.1242/jcs.01363 [DOI] [PubMed] [Google Scholar]
- Harper N. C., Rillo R., Jover-Gil S., Assaf Z. J., Bhalla N., Dernburg A. F. (2011). Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 21, 934–947. 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W., Wente S. R. (2009). Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17, 606–616. 10.1016/j.devcel.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A. F. (2009). The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605. 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Hodzic D. M., Yeater D. B., Bengtsson L., Otto H., Stahl P. D. (2004). Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279, 25805–25812. 10.1074/jbc.M313157200 [DOI] [PubMed] [Google Scholar]
- Horigome C., Okada T., Shimazu K., Gasser S. M., Mizuta K. (2011). Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. EMBO J. 30, 3799–3811. 10.1038/emboj.2011.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70. 10.1016/S0092-8674(01)00430-5 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Martin A. E., Glazko G., Giddings T. H., Jr., Morgan G., Mushegian A., et al. (2006). The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174, 665–675. 10.1083/jcb.200601062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema M., Sonnenberg A. (2011). Nesprin-3: a versatile connector between the nucleus and the cytoskeleton. Biochem. Soc. Trans. 39, 1719–1724. 10.1042/BST20110669 [DOI] [PubMed] [Google Scholar]
- Kim D. I., Birendra K. C., Roux K. J. (2015). Making the LINC: SUN and KASH protein interactions. Biol. Chem. 396, 295–310. 10.1515/hsz-2014-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. C., Drivas T. G., Blobel G. (2008). A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 134, 427–438. 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. (2006). Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194. 10.1007/s00412-006-0055-7 [DOI] [PubMed] [Google Scholar]
- Klutstein M., Cooper J. P. (2014). The chromosomal courtship dance-homolog pairing in early meiosis. Curr. Opin. Cell Biol. 26, 123–131. 10.1016/j.ceb.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kleckner N. (2009). Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 19, 716–724. 10.1016/j.tcb.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K. P., Prentiss M., Kleckner N., Kameoka S. (2008). Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133, 1188–1201. 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer M. P., Banks S. M., Xie X., Wu Y., Fischer J. A. (2007). Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1, 75–85. 10.4161/fly.4254 [DOI] [PubMed] [Google Scholar]
- Kracklauer M. P., Link J., Alsheimer M. (2013). LINCing the nuclear envelope to gametogenesis. Curr. Top. Dev. Biol. 102, 127–157. 10.1016/B978-0-12-416024-8.00005-2 [DOI] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., Zetka M. (2011). Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell 21, 948–958. 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Conrad M. N., Dresser M. E. (2012). Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 8:e1002730. 10.1371/journal.pgen.1002730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Starr D., Cohen M., Liu J., Han M., Wilson K. L., et al. (2002). Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell 13, 892–901. 10.1091/mbc.01-06-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K., Zhang X., Ding X., Guo X., Chen M., Zhu B., et al. (2009). SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 10207–10212. 10.1073/pnas.0812037106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne P. F., Raae A. J., Altmann S. M., Saraste M., Hörber J. K. (2000). States and transitions during forced unfolding of a single spectrin repeat. FEBS Lett. 476, 124–128. 10.1016/S0014-5793(00)01704-X [DOI] [PubMed] [Google Scholar]
- Link J., Leubner M., Schmitt J., Göb E., Benavente R., Jeang K. T., et al. (2014). Analysis of meiosis in SUN1 deficient mice reveals a distinct role of SUN2 in mammalian meiotic LINC complex formation and function. PLoS Genet. 10:e1004099. 10.1371/journal.pgen.1004099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Gotzmann J., Sironi L., Jaeger V. M., Schneider M., Luke Y., et al. (2008). Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim. Biophys. Acta 1783, 2415–2426. 10.1016/j.bbamcr.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Malone C. J., Misner L., Le Bot N., Tsai M. C., Campbell J. M., Ahringer J., et al. (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Masoud K., Herzog E., Chaboute M. E., Schmit A. C. (2013). Microtubule nucleation and establishment of the mitotic spindle in vascular plant cells. Plant J. 75, 245–257. 10.1111/tpj.12179 [DOI] [PubMed] [Google Scholar]
- Mcgee M. D., Stagljar I., Starr D. A. (2009). KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J. Cell Sci. 122, 2895–2905. 10.1242/jcs.051607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckee B. D. (2004). Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677, 165–180. 10.1016/j.bbaexp.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Meyerzon M., Fridolfsson H. N., Ly N., Mcnally F. J., Starr D. A. (2009). UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 136, 2725–2733. 10.1242/dev.038596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F., Kurabayashi A., Tange Y., Okazaki K., Shimanuki M., Niwa O. (2004). Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics 270, 449–461. 10.1007/s00438-003-0938-8 [DOI] [PubMed] [Google Scholar]
- Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. (2002). The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13, 930–946. 10.1091/mbc.01-11-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislow J. M., Kim M. S., Davis D. B., Mcnally E. M. (2002). Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 115, 61–70. [DOI] [PubMed] [Google Scholar]
- Moens P. B., Bernelotmoens C., Spyropoulos B. (2011). Chromosome core attachment to the meiotic nuclear envelope regulates in Chloeaktis (Orthoptera). Genome 32, 601–610. 10.1139/g89-488 [DOI] [Google Scholar]
- Morimoto A., Shibuya H., Zhu X., Kim J., Ishiguro K., Han M., et al. (2012). A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198, 165–172. 10.1083/jcb.201204085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosleybishop K. L., Li Q., Patterson L., Fischer J. A. (1999). Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr.Biol. 9, 1211–1220. 10.1016/S0960-9822(99)80501-6 [DOI] [PubMed] [Google Scholar]
- Murphy S. P., Bass H. W. (2012). The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. J. Cell Sci. 125, 3681–3690. 10.1242/jcs.108290 [DOI] [PubMed] [Google Scholar]
- Murphy S. P., Gumber H. K., Mao Y., Bass H. W. (2014). A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.). Front. Plant Sci. 5:314. 10.3389/fpls.2014.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. P., Simmons C. R., Bass H. W. (2010). Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol. 10:269. 10.1186/1471-2229-10-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2011). Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 66, 629–641. 10.1111/j.1365-313X.2011.04523.x [DOI] [PubMed] [Google Scholar]
- Ortega S. P. I., Odajima J., Martin A., Dubus P., Sotillo R., Barbero J. L., et al. (2003). Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 33, 25–31. 10.1038/ng1232 [DOI] [PubMed] [Google Scholar]
- Padmakumar V. C., Abraham S. S., Noegel A. A., Tunggal B., Karakesisoglou I., Korenbaum E. (2004). Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 295, 330–339. 10.1016/j.yexcr.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. (1991). Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66, 1239–1256. 10.1016/0092-8674(91)90046-2 [DOI] [PubMed] [Google Scholar]
- Park S. H., Craig B. (2010). Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11, 515–521. 10.1038/embor.2010.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K., Molofsky A. B., Robinson C., Acosta S., Cater C., Fischer J. A. (2004). The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15, 600–610. 10.1091/mbc.E03-06-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W. P., Golubovskaya I. N., Timofejeva L., Meeley R. B., Sheridan W. F., Cande W. Z. (2004). Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92. 10.1126/science.1091110 [DOI] [PubMed] [Google Scholar]
- Penkner A., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., et al. (2009). Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139, 920–933. 10.1016/j.cell.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., et al. (2007). The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12, 873–885. 10.1016/j.devcel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Dernburg A. F. (2006). A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11, 817–829. 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., Meneely P. M., et al. (2005). HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051–1063. 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D., Hodzic D. (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186, 461–472. 10.1083/jcb.200906068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D. M., Greer E., Martin A. C., Moore G., Shaw P. J., Howard M. (2012). Quantitative dynamics of telomere bouquet formation. PLoS Comput. Biol. 8, 1776–1776. 10.1371/journal.pcbi.1002812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Kuspa A., Brokamp R., Matzner M., Noegel A. A. (1998). Interaptin, an actin-binding protein of the alpha-actinin superfamily in Dictyostelium discoideum, is developmentally and cAMP-regulated and associates with intracellular membrane compartments. J. Cell Biol. 142, 735–750. 10.1083/jcb.142.3.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S. (1997). Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Rog O., Dernburg A. (2015). Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 10, 1639–1645. 10.1016/j.celrep.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A., Kutay U. (2013). The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122, 415–429. 10.1007/s00412-013-0417-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A., Schwartz T. U., Kutay U. (2013). LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function. Nucleus 4, 29–36. 10.4161/nucl.23387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Crisp M. L., Liu Q., Kim D., Kozlov S., Stewart C. L., et al. (2009). Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. U.S.A. 106, 2194–2199. 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C. M., Rillo R., Carlton P. M., Wynne D. J., et al. (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919. 10.1016/j.cell.2009.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. (2001). A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2, 621–627. 10.1038/35085086 [DOI] [PubMed] [Google Scholar]
- Scherthan H. (2007). Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 64, 117–124. 10.1007/s00018-006-6463-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Weich S., Schwegler H., Heyting C., Härle M., Cremer T. (1996). Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134, 1109–1125. 10.1083/jcb.134.5.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J., Benavente R., Hodzic D., Höög C., Stewart C. L., Alsheimer M. (2007). Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. U.S.A. 104, 7426–7431. 10.1073/pnas.0609198104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X., Tarnasky H. A., Lee J. P., Oko R., van der Hoorn F. A. (1999). Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 211, 109–123. 10.1006/dbio.1999.9297 [DOI] [PubMed] [Google Scholar]
- Sheehan M. J., Pawlowski W. P. (2009). Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc. Natl. Acad. Sci. U.S.A. 106, 20989–20994. 10.1073/pnas.0906498106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H., Hernández-Hernández A., Morimoto A., Negishi L., Höög C., Watanabe Y. (2015). MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163, 1252–1266. 10.1016/j.cell.2015.10.030 [DOI] [PubMed] [Google Scholar]
- Shibuya H., Ishiguro K., Watanabe Y. (2014). The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 16, 145–156. 10.1038/ncb2896 [DOI] [PubMed] [Google Scholar]
- Shimanuki M., Miki F., Ding D. Q., Chikashige Y., Hiraoka Y., Horio T., et al. (1997). A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mole Gen. Genet. 254, 238–249. 10.1007/s004380050412 [DOI] [PubMed] [Google Scholar]
- Siderakis M., Tarsounas M. (2007). Telomere regulation and function during meiosis. Chromosome Res. 15, 667–679. 10.1007/s10577-007-1149-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B. A., Kutay U., Schwartz T. U. (2013). Structural insights into LINC complexes. Curr. Opin. Struc. Biol. 23, 285–291. 10.1016/j.sbi.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B., Rothballer A., Kutay U., Schwartz T. (2012). LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047. 10.1016/j.cell.2012.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A. (2009). A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122, 577–586. 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A. (2011). KASH and SUN proteins. Curr. Biol. 21, R414–R415. 10.1016/j.cub.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Fischer J. A. (2005). KASH'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays 27, 1136–1146. 10.1002/bies.20312 [DOI] [PubMed] [Google Scholar]
- Starr D. A., Fridolfsson H. N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26, 421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Han M. (2002). Role of ANC-1 in tethering nuclei to the actin cytoskeleton. J. Biol. Chem. 298, 406–409. 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- Starr D. A., Hermann G. J., Malone C. J., Fixsen W., Priess J. R., Horvitz H. R., et al. (2001). unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128, 5039–5050. [DOI] [PubMed] [Google Scholar]
- Stewart C. L., Roux K. J., Burke B. (2007). Blurring the boundary: the nuclear envelope extends its reach. Science 318, 1408–1412. 10.1126/science.1142034 [DOI] [PubMed] [Google Scholar]
- Subramanian V. V., Hochwagen A. (2014). The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 6:a016675. 10.1101/cshperspect.a016675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata M. (1962). Chromosome pairing in intercrosses between stocks of interchanges involving the same two chromosomes in maize. Cytologia 27, 410–417. 10.1508/cytologia.27.410 [DOI] [Google Scholar]
- Tamura K., Iwabuchi K., Fukao Y., Kondo M., Okamoto K., Ueda H., et al. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23, 1776–1781. 10.1016/j.cub.2013.07.035 [DOI] [PubMed] [Google Scholar]
- Tiang C. L., He Y., Pawlowski W. P. (2012). Chromosome organization and dynamics during interphase, mitosis, and meiosis in plants. Plant Physiol. 158, 26–34. 10.1104/pp.111.187161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidl J., Scherthan H. (1999). Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112(Pt 5), 651–658. [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Wilson K. L., Gruenbaum Y. (2006). SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 7, 782–788. 10.1038/nrm2003 [DOI] [PubMed] [Google Scholar]
- Varas J., Graumann K., Osman K., Pradillo M., Evans D. E., Santos J. L., et al. (2015). Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 81, 329–346. 10.1111/tpj.12730 [DOI] [PubMed] [Google Scholar]
- Viera A., Alsheimer M., Gómez R., Berenguer I., Ortega S., Symonds C. E., et al. (2015). CDK2 regulates nuclear envelope protein dynamics and telomere attachment in mouse meiotic prophase. J. Cell Sci. 128, 88–99. 10.1242/jcs.154922 [DOI] [PubMed] [Google Scholar]
- Viera A., Rufas J. I., Barbero J. L., Ortega S., Suja J. A. (2009). CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 122, 2149–2159. 10.1242/jcs.046706 [DOI] [PubMed] [Google Scholar]
- Wanat J. J., Kim K. P., Koszul R., Zanders S., Weiner B., Kleckner N., et al. (2008). Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 4:e1000188. 10.1371/journal.pgen.1000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu R., Wang J., Wang P., Shen Y., Liu G. (2014). The Arabidopsis kinesin gene AtKin - 1 plays a role in the nuclear division process during megagametogenesis. Plant Cell Rep. 33, 819–828. 10.1007/s00299-014-1594-7 [DOI] [PubMed] [Google Scholar]
- Wang Q., Du X., Cai Z., Greene M. I. (2006). Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol. 25, 554–562. 10.1089/dna.2006.25.554 [DOI] [PubMed] [Google Scholar]
- Wijnker E., Schnittger A. (2013). Control of the meiotic cell division program in plants. Plant Reprod. 26, 143–158. 10.1007/s00497-013-0223-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K., Ketema M., Truong H., Sonnenberg A. (2006). KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 119, 5021–5029. 10.1242/jcs.03295 [DOI] [PubMed] [Google Scholar]
- Wilson K. L., Dawson S. C. (2011). Evolution: functional evolution of nuclear structure. J. Cell Biol. 195, 171–181. 10.1083/jcb.201103171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., Jantsch V. (2014). Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma 123, 15–24. 10.1007/s00412-013-0436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H. J., Gundersen G. G. (2006). Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 16, 67–69. 10.1016/j.tcb.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Wynne D. J., Rog O., Carlton P. M., Dernburg A. F. (2012). Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J. Cell Biol. 196, 47–64. 10.1083/jcb.201106022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Rivero F., Euteneuer U., Mondal S., Mana-Capelli S., Larochelle D., et al. (2008). Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic 9, 708–724. 10.1111/j.1600-0854.2008.00721.x [DOI] [PubMed] [Google Scholar]
- Yamamoto A. (2014). Gathering up meiotic telomeres: a novel function of the microtubule-organizing center. Cell Mol. Life Sci. 71, 2119–2134. 10.1007/s00018-013-1548-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Lei K., Zhou M., Craft C. M., Xu G., Xu T., et al. (2011). KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum. Mol. Genet. 20, 1061–1073. 10.1093/hmg/ddq549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Starr D. A., Wu X., Parkhurst S. M., Zhuang Y., Xu T., et al. (2006). The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 289, 336–345. 10.1016/j.ydbio.2005.10.027 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., et al. (2009). SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187. 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., et al. (2007). Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development 134, 901–908. 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- Zhou K., Rolls M. M., Hall D. H., Malone C. J., Hanna-Rose W. (2009). A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186, 229–241. 10.1083/jcb.200902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Meier I. (2013). How plants LINC the SUN to KASH. Nucleus 4, 206–215. 10.4161/nucl.24088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Graumann K., Meier I. (2015a). The plant nuclear envelope as a multifunctional platform LINCed by SUN and KASH. J. Exp. Bot. 66, 1649–1659. 10.1093/jxb/erv082 [DOI] [PubMed] [Google Scholar]
- Zhou X., Graumann K., Evans D. E., Meier I. (2012a). Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 196, 203–211. 10.1083/jcb.201108098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Graumann K., Wirthmueller L., Jones J. D., Meier I. (2014). Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J. Cell Biol. 205, 677–692. 10.1083/jcb.201401138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Graves N. R., Meier I. (2015b). SUN anchors pollen WIP–WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube targeting and fertility. J. Exp. Bot. 66, 7299–7307. 10.1093/jxb/erv425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Du X., Cai Z., Song X., Zhang H., Mizuno T., et al. (2012b). Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J. Biol. Chem. 287, 5317–5326. 10.1074/jbc.M111.304543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. (2006). From early homologue recognition to synaptonemal complex formation. Chromosoma 115, 158–174. 10.1007/s00412-006-0048-6 [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1998). The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32, 619–697. 10.1146/annurev.genet.32.1.619 [DOI] [PubMed] [Google Scholar]