Abstract
Flax (Linum usitatissimum L.) is an important cash crop, and its agronomic traits directly affect yield and quality. Molecular studies on flax remain inadequate because relatively few flax genes have been associated with agronomic traits or have been identified as having potential applications. To identify markers and candidate genes that can potentially be used for genetic improvement of crucial agronomic traits, we examined 224 specimens of core flax germplasm; specifically, phenotypic data for key traits, including plant height, technical length, number of branches, number of fruits, and 1000-grain weight were investigated under three environmental conditions before specific-locus amplified fragment sequencing (SLAF-seq) was employed to perform a genome-wide association study (GWAS) for these five agronomic traits. Subsequently, the results were used to screen single nucleotide polymorphism (SNP) loci and candidate genes that exhibited a significant correlation with the important agronomic traits. Our analyses identified a total of 42 SNP loci that showed significant correlations with the five important agronomic flax traits. Next, candidate genes were screened in the 10 kb zone of each of the 42 SNP loci. These SNP loci were then analyzed by a more stringent screening via co-identification using both a general linear model (GLM) and a mixed linear model (MLM) as well as co-occurrences in at least two of the three environments, whereby 15 final candidate genes were obtained. Based on these results, we determined that UGT and PL are candidate genes for plant height, GRAS and XTH are candidate genes for the number of branches, Contig1437 and LU0019C12 are candidate genes for the number of fruits, and PHO1 is a candidate gene for the 1000-seed weight. We propose that the identified SNP loci and corresponding candidate genes might serve as a biological basis for improving crucial agronomic flax traits.
Keywords: flax (Linum usitatissimum L.), agronomic traits, GWAS, SLAF-seq, candidate genes
Introduction
Flax (Linum usitatissimum L.) is one of the oldest plants cultivated for fiber and edible oil and remains an important cash crop worldwide. Breeding selection for fiber flax or linseed flax has resulted in two plant types, which differ considerably in agronomic performance (Diederichsen and Ulrich, 2009). Compared with linseed flax cultivars, fiber flax plants are typically taller, with fewer branches and fruits and lower seed production (Booth et al., 2004). Therefore, agronomic traits directly affect the seed yield of linseed flax and the bast fiber quality of fiber flax. In recent years, traditional breeding methods have been employed to introduce genetic changes that improve the agronomic traits of flax. However, agronomic features are complex, quantitative traits that are controlled by multiple genes. Consequently, traditional breeding approaches do not satisfy the demand for improving flax traits. Thus, far a number of genetic studies on agronomic traits of flax have been reported. For example, amplified fragment length polymorphism (AFLP) and simple-sequence repeat (SSR) markers were used to perform QTL analysis for four flax traits, revealing several yield-related QTLs (Gehringer et al., 2006). In addition, 464 SSR markers were employed to perform QTL analysis for nine traits in a natural population composed of 390 flax germplasm resources that were planted in eight environments (Soto-Cerda et al., 2014); in that study, the authors identified 12 markers that were closely linked to six traits. A genome-wide scan was performed using 407 core germplasm resources and 448 SSR markers before association mapping was conducted to elucidate the non-neutral genomic regions potentially underlying divergent selection between fiber and linseed cultivars, and the candidate genes involved in the biosynthesis of the cell wall, lignin, and fatty acids were analyzed (Soto-Cerda et al., 2013). Furthermore, Deng et al. (2014) used 61 pairs of SSR primers, 91 pairs of expressed sequence tag (EST)-SSR primers, and 102 pairs of genomic-SSR primers to perform association analysis for yield-related traits in 182 core germplasm resources of flax; the authors identified 57 high-quality allelic variations, including 31 showing yield-enhancing effects and 26 showing the opposite. Nevertheless, these association studies were all based on common molecular markers (e.g., SSR, EST-SSR) that might not be sufficient in light of the rapid development of new sequencing technologies. Furthermore, the investigations cited above were not enough to elucidate the genes related to the complicated agronomic traits of flax.
In recent years, genome-wide association studies (GWAS) based on next-generation sequencing (NGS) technology have become the new approach for improving crop traits. GWAS is suitable for phenotypic data under multiple environments, thereby reducing environment-induced errors and enhancing results accuracy (Hall et al., 2010). Once the genotype data of a population are available, GWAS can be performed to examine multiple traits (Atwell et al., 2010). As such, the approach provides a basis for elucidating the genetic structures of the complex traits of a crop. The resulting association alleles can be used for marker-assisted molecular breeding and are crucial for innovating germplasm resources and improving cultivars. So far, GWAS related to important agronomic traits have been reported in several crops, including maize (Xue et al., 2013; Farfan et al., 2015), rice (Huang et al., 2010; Han et al., 2016), and soybean (Sonah et al., 2015; Zhang et al., 2015). However, an NGS-based GWAS analysis examining agronomic traits in flax (L. usitatissimum) has not been reported.
Previous GWAS have mostly been based on SNP array technology, which can detect known SNP loci but not new loci (Vilkki et al., 2013; Zhang et al., 2016). In light of this limitation, a high-throughput sequencing-based technology known as specific locus-amplified fragment sequencing (SLAF-seq) was developed (Sun et al., 2013). In comparison with other technologies, SLAF-seq has the following advantages: (i) generation of high-density SNP loci numbering in the millions after one sequencing reaction, (ii) capability of detecting novel SNP loci in unknown mutation-harboring loci compared with SNP arrays, (iii) suitability for any species regardless of the presence of a reference genome, and (iv) a higher rate of identified SNP loci that become genuine association markers. As a consequence, the technology has been applied to many crops including rice, soybean, sesame, cucumber, Brassica napus, etc. (Zhang et al., 2013; Xu et al., 2014; Geng et al., 2016; Han et al., 2016; Li et al., 2016).
Here, we examined 224 core germplasm resources of flax grown under different environmental conditions for the main agronomic traits of plant height, technical length, number of branches, number of fruits, and 1000-grain weight. Subsequently, SLAF-seq was employed to perform GWAS and recover potential alleles controlling these traits. To our knowledge, this is the first SLAF-seq-based GWAS with the goal to identify SNP loci and candidate genes linked to important agronomic flax traits. The results provide a basis for molecular marker (related to main agronomic traits)-assisted breeding and improvement of the main agronomic traits in flax.
Materials and methods
Experimental materials and survey of traits
The core germplasm resources of 224 flax accessions were collected from institutions in China and other countries (Table S1). They were sown at the Harbin Experimental Base of Heilongjiang Academy of Agricultural Sciences (45°65′N, 126°68′E), Harbin, China, in April of 2015 and 2016 (2015HRB, 2016HRB) as well as at the Lanxi Experimental Base (6°27′N, 126°28′E), Lanxi, China, in April of 2016 (2016LX). The average annual rainfall from seeding to harvest was 390.1 mm in 2015HRB and 453 mm in 2016HRB, and the average annual temperature was 5.26° and 4.99°C. The average annual rainfall from seeding to harvest was 504.4 mm in 2016LX, and the average annual temperature was 3.11°C. The experiment at each location used a randomized completed block design with three replicates. Each cultivar was planted in triplicate in 2-m lines, with a 20-cm inter-line gap. The field management was the same as the local field management. At the maturing stage, ten plants were selected from each replicate for phenotyping. The five agronomic traits (plant height, technical length, branch number, fruit number, 1,000-grain weight) were investigated. Plant height was measured as the distance between the cotyledon scar and the top of the first-degree branch. Technical length was measured as the distance between the cotyledon scar of the flax plant and the base of the first-degree branch below the inflorescence. The branch number was the number of first-degree branches on the top of the main stem. The fruit number was the number of all fruits that had seeds on the top of the main stem. The 1,000-grain weight was the absolute weight of 1,000 seeds (water content 9%) that were mature, full and clean.
Extraction of genomic DNA
Genomic DNA was isolated from fresh leaves harvested from 20-day-old seedlings of flax. The Tiangen plant total genomic DNA extraction kit (Tiangen Biotech Co. Ltd., Beijing, China) was used for the genomic DNA extraction. A NanoDrop 2000 (Thermo Scientific, Massachusetts) was used to determine the DNA concentration and quality to ensure that DNA samples met the requirements of the sequencing reaction (concentration ≥ 18 ng/μL; volume ≥ 30 μL).
Modification of the genomic DNA
First, the restriction enzyme HaeIII was used to digest the genomic DNA in a 50 μL aqueous solution containing 500 ng of genomic DNA, 41 μL of NEB Buffer (10 ×), and 0.12 μL of HaeIII (1 U/μL). This solution was incubated at 37°C for 15 h. The resulting DNA was column-purified using a QIAGEN kit and solubilized in 50 μL of EB (0.01 mol/L). The sticky ends of the digested DNA fragments were filled in, and their 5′ ends were then phosphorylated using a 100 μL solution containing 30 μL of purified DNA (50 ng/μL), 10 μL of T4 DNA Ligase Buffer (containing 10 mmol/L final concentration ATP), 4 μL dNTP Mix (10 mmol/L), 5 μL of T4 DNA polymerase (5 U/μL), 1 μL of Klenow fragment (5 U/μL), and 5 μL of T4 polynucleotide kinase (10 U/μL). This solution was incubated at 20°C for 30 min in a thermocycler. The DNA was column-purified using a QIAGEN kit and solubilized in 33 μL of EB (0.01 mol/L). Next, a base was added to the 3′ ends of the 5′ phosphorylated DNA fragments, allowing them to connect to the Solexa adaptor, which has a T base at its 5′ end, using the following conditions: 32 μL of purified DNA (50 ng/μL), 5 μL of Klenow Buffer (10 ×), 10 μL of dATP (1 mmol/L), and 3 μL of Klenow Exo (5 U/μL). The solution was placed in a 37°C water bath for 30 min. The DNA was column-purified using a QIAGEN kit and solubilized in 10 μL of EB (0.01 mol/L). Finally, the Solexa adapter was attached to the DNA fragments to allow them to hybridize in the flow cells in the sequencing reactions. The reaction conditions were as follows: the solution containing 10 μL of purified DNA (50 ng/μL), 25 μL of DNA Ligase Buffer (2 ×), 10 μL of Adapter (5 pmol/μL), and 5 μL of DNA Ligase (5 U/μL) was incubated at 20°C for 15 min in the polymerase chain reaction (PCR) system. The DNA was column-purified using a QIAGEN kit and solubilized in 30 μL of EB (0.01 mol/L).
PCR amplification and sequencing
Based on the restriction analysis of PAProC (http://www.paproc.de/) (Nussbaum et al., 2001), DNA fragments of 500–580 bp were gel purified and PCR amplified using a forward primer (5′-AATGATACGGCGACCACCGA-3′) and a reverse primer (5′-CAAGCAGAAGACGGCATACG-3′). PCR amplification was performed in a 40 μL aqueous solution containing 8 μL of purified DNA (50 ng/μL), 1.5 μL of forward primer (50 pmol/μL), 1.5 μL of reverse primer (50 pmol/μL), 9 μL of dNTP mix (10 mmol/L) and 20 μL of Phusion DNA polymerase (2 U/μL). The amplification procedure was as follows: pre-denaturation at 98°C for 30 s, followed by 18 amplification cycles of denaturation at 98°C for 10 s, annealing at 65°C for 30 s, and extension at 72°C for 30 s, before a final extension at 72°C for 5 min. After the reaction, the DNA was column-purified using a QIAGEN kit and solubilized in 30 μL of EB (0.01 mol/L). Purified DNA samples were quantified using the Qubit system before bridge amplification was performed on the surface of the flow cells to generate DNA clusters. The PCR products were re-purified and then prepared for paired-end sequencing on an Illumina HiSeq 2500 sequencing platform (Illumina, San Diego, CA, USA).
Data processing and data submission information
Raw sequencing reads were separated using barcode sequences: Illumina SLAF libraries were barcoded with standard Illumina multiplex adaptors and pooled for sequencing in sets of three samples to generate an average of 6-fold sequence coverage per sample (Purcell et al., 2007; Healey et al., 2014). Low-quality reads (QC score < 20) were removed before SOAP 2.20 (Sun et al., 2013) was employed to align the resulting reads with the reference genome of Linum usitatissimum v1.0 (https://phytozome.jgi.doe.gov/pz/portal.html#!search?show=KEYWORD) (Wang et al., 2012). A read was considered valid if both ends mapped onto the genome and could be used to define SLAF markers. Based on the results of alignment and correction, groups with a mean sequencing depth of 4 were recruited to define SLAF markers. Next, the number of SLAF markers per 100 K genome was recorded to obtain the distribution of SLAF markers in the scaffolds. Finally, SNP loci were detected in the collection of the 224 specimens using pre-defined SLAF markers, whereby the number of SNP loci per 100 K genome was documented. Raw Illumina sequences were deposited in the National Center for Biotechnology Information (NCBI) and can be accessed in the database (https://www.ncbi.nlm.nih.gov/) under accession SRP116365 or SRS2474942 for leaf.
Population structure analysis and significant SNP discovery
The population structure analysis used 146,959 SNPs to infer the genetic background of an accession that belongs to a cluster under a given number of populations (K). The number of genetic clusters was predefined as K = 1–5 for all accessions and was calculated using Admixture software (Hardy and Vekemans, 2002; Alexander et al., 2009).
LD (linkage disequilibrium) between pairs of SNPs was estimated by using squared allele frequency correlations (r2) in Tassel version 3.0 (Bradbury et al., 2007). Each significant SNP was evaluated for the extent of local LD. The region was defined as extending to where LD between nearby SNPs and the lead SNP decayed to r2 > 0.8, MAF > 0.05. Only SNPs with an MAF more than 0.05 and <10% missing data were used. The SNP nomenclature used in this study is based on the number of scaffolds that contained an SNP plus the position of the polymorphism in the scaffold.
Genome-wide association analyses
The efficient model was performed with both GLM and MLM using Tassel software. The population structure matrix generated from Admixture was used as the Q matrix for the GLM model. P-values of P ≤ 1.268 × 10−5 (P = 0.01/n; n = total markers used, which is roughly a Bonferroni correction, corresponding to -log10 (P) = 5, red line) and P ≤ 1.268 × 10−6 (P = 0.1/n; n = total markers used, which is roughly a Bonferroni correction, corresponding to -log10 (P) = 6, blue line) were defined as the genome-wide control threshold and suggestive threshold, respectively. The genes within 10 Kb of a significant SNP's flanking region were reported as candidate genes.
Results
Descriptive statistics of agronomic traits
Under three different environmental conditions, plant height had a minimum value of 42.20 cm, a maximum value of 125.40 cm, and a maximum coefficient of variation of 18.09%; technical length had a minimum value of 27.60 cm, a maximum value of 103.20 cm, and a maximum coefficient of variation of 22.76%; number of branches had a minimum value of 2, a maximum value of 12, and a maximum coefficient of variation of 55.57%; number of fruits had a minimum value of 2, a maximum value of 39, and a maximum coefficient of variation of 53.47%; and 1,000-grain weight had a minimum value of 3.18 g, a maximum value of 9.21 g, and a maximum coefficient of variation of 16.27%. The results therefore indicated that the test germplasm resources of flax contained extraordinary genetic variation (Table 1).
Table 1.
Results of five important agronomic traits derived from the flax germplasm resources.
| Environments | Traits | Minimum value | Maximum value | Range | Mean value | Standard deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|---|---|
| 2015HRB | Plant height (cm) | 42.20 | 109.50 | 67.30 | 81.94 | 14.82 | 18.09 |
| Technical length (cm) | 27.60 | 94.80 | 67.20 | 64.38 | 14.27 | 22.19 | |
| Number of branches (unit) | 2.00 | 12.00 | 10.00 | 4.50 | 1.08 | 24.00 | |
| Number of fruits (unit) | 4.00 | 29.00 | 25.00 | 9.00 | 3.73 | 41.44 | |
| 1,000-grain weight (g) | 3.94 | 8.91 | 4.97 | 5.00 | 0.77 | 15.32 | |
| 2016HRB | Plant height (cm) | 49.50 | 120.10 | 70.60 | 89.77 | 15.65 | 17.43 |
| Technical length (cm) | 35.20 | 103.20 | 68.00 | 70.52 | 16.05 | 22.76 | |
| Number of branches (unit) | 2.00 | 12.00 | 10.00 | 4.00 | 1.13 | 28.25 | |
| Number of fruits (unit) | 2.00 | 30.00 | 29.00 | 6.16 | 3.12 | 50.64 | |
| 1,000-grain weight (g) | 3.18 | 8.92 | 5.74 | 4.96 | 0.79 | 15.92 | |
| 2016LX | Plant height (cm) | 54.20 | 125.40 | 71.2 | 91.45 | 14.46 | 15.81 |
| Technical length (cm) | 30.30 | 100.40 | 70.1 | 65.81 | 14.92 | 22.67 | |
| Number of branches (unit) | 2.00 | 10.00 | 8.00 | 5.47 | 3.04 | 55.57 | |
| Number of fruits (unit) | 4.00 | 39.00 | 35.00 | 10.06 | 5.38 | 53.47 | |
| 1,000-grain weight (g) | 3.82 | 9.21 | 5.39 | 5.10 | 0.83 | 16.27 |
Sequencing results
SNP detection was performed in the collection of 224 germplasm resources of flax based on the predefined 346,639 SLAF tags, which generated a total of 584,987 SNP loci (MAF ≥ 0.05). Considering both the SLAF and SNP data, we defined the SLAF markers associated with SNPs as polymorphic SLAF markers to thereby examine the SLAF polymorphisms. Our analysis yielded a total of 146,959 polymorphic SLAF markers with a mean depth of 7.2. After quality control, there were 34,932 SNP loci used for subsequent GWAS analyses (Table S2).
Analysis of population structure
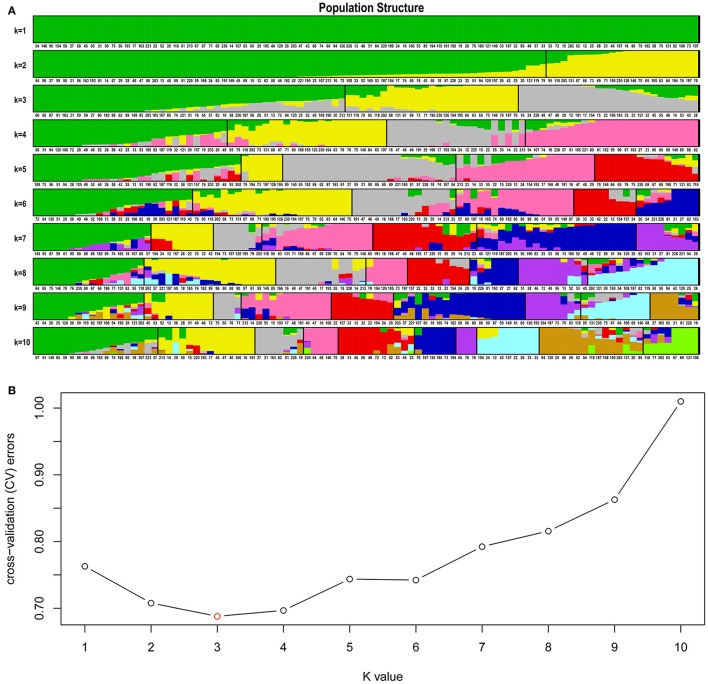
The Admixture software was used to analyze the population clustering and structure of the 224 germplasm resources (Figures 1A,B). Specifically, clustering was first performed assuming that the number of clusters (K) was between 1 and 10. Then, the results were cross-validated to determine that the optimal K-value was 3 (according to the valley of the error rates of cross-validation). In other words, our results implied that the collection most likely originated from three ancestors. Given that population stratification might affect the accuracy of association analysis, we generated QQ plots of individual traits (Supplementary Figures 1–5). The results indicated that the observation values (ordinate) generally matched with the corresponding expected values (abscissa), suggesting that the association analysis did not produce any false negativity due to population stratification. Hence, the GWAS results were reliable.
Figure 1.
Cluster analysis and population structure of 224 germplasm resources of flax. (A) Cluster map of the population structure (each color represents a cluster, and each row represents the result of a given number of clusters (K, K = 1–10); (B) Population structure plot [ordinate represents cross-validation error rate (CV-value); abscissa represents number of clusters (K)].
Genome-wide association analysis
SNP loci displaying significant correlation with plant height
The GLM and MLM models of TASSEL were employed to perform GWAS, which revealed that nine SNP loci were significantly associated with plant height (P < 1.26E-06). The relevant Manhattan plots and QQ plots of the two models and three environments are shown in Supplementary Figure 1. The GLM generated nine SNP loci in the three environments, including six in 2015HRB, one in 2016HRB, and two in 2016LX. However, there was not a single SNP that occurred in more than one environment. In comparison, the MLM only generated two SNP loci (scaffold344_309662 and scaffold51_1349321) in 2015HRB, both of which were also identified by the GLM in 2015HRB. The genes closest to the two SNP loci include UGT (UDP-glycosyltransferase) and PL (Pectate lyase). Moreover, the genes closest to the other seven SNP loci were CBP (Calcineurin B-like protein), PI-PLC X (PI-PLC X domain-containing protein), SPP (Squamosa promoter-binding-like protein), PPR (PPR repeat family), PSP (Pectate lyase superfamily protein), UF (Ubiquitin family), and CS (Cellulose synthase) (Table 2).
Table 2.
Associated single nucleotide polymorphisms (SNPs) and the nearest genes for plant height traits of flax.
| Model | Environment | SNP position (bp) | scaffold | Location | P-value | Nearest gene | Distance to SNP (kb) |
|---|---|---|---|---|---|---|---|
| GLM | 2015HRB | scaffold112_114241 | scaffold112 | 114241 | 7.43E-07 | Calcineurin B-like protein (CBP) | upstream 0.27 |
| scaffold1491_318496 | scaffold1491 | 318496 | 1.91E-07 | PI-PLC X domain-containing protein (PI-PLC X) | interior | ||
| scaffold31_1800846 | scaffold31 | 1800846 | 6.65E-07 | Squamosa promoter-binding-like protein (SPP) | downstream 0.642 | ||
| scaffold344_309662 | scaffold344 | 309662 | 1.11E-07 | UDP-glycosyltransferase 1 (UDP) | downstream 4.558 | ||
| scaffold51_1349321 | scaffold51 | 1349321 | 8.08E-07 | Pectate lyase (PL) | downstream 8.566 | ||
| scaffold59_572553 | scaffold59 | 572553 | 1.06E-07 | PPR repeat family (PPR) | upstream 0.52 | ||
| 2016HRB | scaffold156_641874 | scaffold156 | 641874 | 7.27E-07 | Pectate lyase superfamily protein (PSP) | downstream 0.51 | |
| 2016LX | scaffold147_367986 | scaffold147 | 367986 | 1.10E-07 | Ubiquitin family (UF) | upstream 8.3 | |
| scaffold859_123972 | scaffold859 | 123972 | 4.22E-07 | cellulose synthase (CS) | downstream 0.56 | ||
| MLM | 2015HRB | scaffold344_309662 | scaffold344 | 309662 | 1.11E-07 | UDP-glycosyltransferase (UDP) | downstream 4.558 |
| scaffold51_1349321 | scaffold51 | 1349321 | 8.08E-07 | Pectate lyase (PL) | downstream 8.566 |
SNP loci displaying significant correlation with technical length
Three SNP loci, identified by only the GLM in two environments, were found to be significantly associated with technical length (P < 1.26E-06) (Supplementary Figure 2). Among these, scaffold297_275113 and scaffold361_14957 were identified in 2015 HRB, whereas scaffold273_68457 was identified in 2016 HRB. Genes closest to the three SNP loci included HP (Hypothetical protein), VTP (Vesicle transport protein), and MIF (Macrophage migration inhibitory factor) (Table 3).
Table 3.
Associated single nucleotide polymorphisms (SNPs) and the nearest genes for the technical length trait of flax.
| Model | Environment | SNP position (bp) | scaffold | Location | P-value | Nearest gene | Distance to SNP (kb) |
|---|---|---|---|---|---|---|---|
| GLM | 2015HRB | scaffold297_275113 | scaffold297 | 275113 | 3.96E-07 | hypothetical protein(HP) | downstream 1.83 |
| scaffold361_14957 | scaffold361 | 14957 | 9.44E-07 | Vesicle transport protein(VTP) | upstream 1.19 | ||
| 2016HRB | scaffold273_68457 | scaffold273 | 68457 | 1.18E-06 | Macrophage migration inhibitory factor(MIF) | interior |
SNP loci displaying significant correlation with number of branches
Twenty-one SNP loci exhibited a significant association with the number of branches (P < 1.26E-06). The relevant Manhattan plots and QQ plots of the two models and three environments are shown in Supplementary Figure 3. GLM identified nine SNP loci in 2015HRB, two SNP loci in 2016HRB, and nine SNP loci in 2016LX. Among these, eight SNP loci were identified in both 2015HRB and 2016LX (scaffold116_30201, scaffold156_1203677, scaffold1863_545, scaffold353_773806, scaffold42_494571, scaffold464_754364, scaffold635_43971, and scaffold977_784147). MLM identified six SNP loci in 2015HRB, two SNP loci in 2016HRB, and seven SNP loci in 2016LX. Among these, six SNP loci were identified in both 2015HRB and 2016LX (scaffold116_30201, scaffold156_1203677, scaffold1863_545, scaffold353_773806, scaffold464_754364, and scaffold977_784147). There were eight SNP loci co-identified by both the GLM and MLM (scaffold116_30201, scaffold156_1203677, scaffold1863_545, scaffold353_773806, scaffold464_754364, scaffold977_784147, scaffold359_282990, and scaffold359/289139). In addition, six SNP loci displayed associations in both models and were identified in two environments (2015HRB and 2016LX) (scaffold116_30201, scaffold156_1203677, scaffold1863_545, scaffold353_773806, scaffold464_754364, and scaffold977_784147). The genes closest to the six SNP loci were GRAS (GRAS domain family), GST (Glutathione S-transferase), PORR (Plant organelle RNA recognition domain), PIP5K (Phosphatidylinositol-4-phosphate 5-Kinase), XTH (Xyloglucan endotransglucosylase/hydrolase), and DDR (DNA-damage-repair) (Table 4).
Table 4.
Associated single nucleotide polymorphisms (SNPs) and the nearest genes for the number of branches trait of flax.
| Model | Environment | SNP position (bp) | scaffold | Location | P-value | Nearest gene | Distance to SNP (kb) |
|---|---|---|---|---|---|---|---|
| GLM | 201HRB | scaffold116_30201 | scaffold116 | 30201 | 3.86E-11 | GRAS domain family (GRAS) | upstream 9.57 |
| scaffold156_1203677 | scaffold156 | 1203677 | 2.29E-11 | Glutathione S-transferase (GST) | downstream 0.52 | ||
| scaffold1863_545 | scaffold1863 | 545 | 8.39E-11 | Plant organelle RNA recognition domain (PORR) | upstream 6.46 | ||
| scaffold212_601171 | scaffold212 | 601171 | 1.63E-07 | Cytochrome P450 (P450) | upstream 4.36 | ||
| scaffold353_773806 | scaffold353 | 773806 | 7.04E-11 | Phosphatidylinositol-4-phosphate 5-Kinase (PIP5K) | downstream 6.62 | ||
| scaffold42_494571 | scaffold42 | 494571 | 1.79E-07 | Glycerophosphodiester phosphodiesterase (GP) | interior | ||
| scaffold464_754364 | scaffold464 | 754364 | 7.77E-07 | xyloglucan endotransglucosylase/hydrolase (XTH) | interior | ||
| scaffold635_43971 | scaffold635 | 43971 | 1.08E-06 | Ricinus communis acid phosphatase (RCAP) | interior | ||
| scaffold977_784147 | scaffold977 | 784147 | 2.69E-10 | DNA-damage-repair (DDR) | downstream 1.65 | ||
| 2016HRB | scaffold212_216830 | scaffold212 | 216830 | 6.81E-07 | Transferase family (TF) | upstream 6.80 | |
| scaffold359_282990 | scaffold359 | 282990 | 7.47E-12 | Aldehyde dehydrogenase (AD) | interior | ||
| 2016LX | scaffold116_30201 | scaffold116 | 30201 | 3.86E-11 | GRAS domain family (GRAS) | upstream 9.57 | |
| scaffold156_1203677 | scaffold156 | 1203677 | 2.29E-11 | Glutathione S-transferase (GST) | downstream 0.52 | ||
| scaffold1863_545 | scaffold1863 | 545 | 8.39E-11 | Plant organelle RNA recognition domain (PORR) | upstream 6.46 | ||
| scaffold353_773806 | scaffold353 | 773806 | 7.04E-11 | Phosphatidylinositol-4-phosphate 5-Kinase (PIP5K) | downstream 6.62 | ||
| scaffold42_494571 | scaffold42 | 494571 | 1.79E-07 | Glycerophosphodiester phosphodiesterase (GP) | interior | ||
| scaffold464_754364 | scaffold464 | 754364 | 7.77E-07 | xyloglucan endotransglucosylase/hydrolase (XTH) | interior | ||
| scaffold635_43971 | scaffold635 | 43971 | 1.08E-06 | Ricinus communis acid phosphatase (RCAP) | interior | ||
| scaffold977_784147 | scaffold977 | 784147 | 2.69E-10 | DNA-damage-repair (DDR) | downstream 1.65 | ||
| scaffold359_289139 | scaffold359 | 289139 | 2.30E-08 | Protein of unknown function | upstream 1.25 | ||
| MLM | 2015HRB | scaffold116_30201 | scaffold116 | 30201 | 3.86E-11 | GRAS domain family (GRAS) | upstream 9.57 |
| scaffold156_1203677 | scaffold156 | 1203677 | 2.29E-11 | Glutathione S-transferase (GST) | downstream 0.52 | ||
| scaffold1863_545 | scaffold1863 | 545 | 8.39E-11 | Plant organelle RNA recognition domain (PORR) | upstream 6.46 | ||
| scaffold353_773806 | scaffold353 | 773806 | 7.04E-11 | Phosphatidylinositol-4-phosphate 5-Kinase (PIP5K) | downstream 6.62 | ||
| scaffold464_754364 | scaffold464 | 754364 | 7.77E-07 | xyloglucan endotransglucosylase/hydrolase (XTH) | interior | ||
| scaffold977_784147 | scaffold977 | 784147 | 2.69E-10 | DNA-damage-repair (DDR) | downstream 1.65 | ||
| 2016HRB | scaffold977_469888 | scaffold977 | 469888 | 3.79E-07 | Lus10031183.BGIv1.0 | upstream 1.2 | |
| scaffold359_282990 | scaffold359 | 282990 | 7.47E-12 | Lus10013155.BGIv1.0 | interior | ||
| 2016LX | scaffold116_30201 | scaffold116 | 30201 | 3.86E-11 | GRAS domain family (GRAS) | upstream 9.57 | |
| scaffold156_1203677 | scaffold156 | 1203677 | 2.29E-11 | Glutathione S-transferase (GST) | downstream 0.52 | ||
| scaffold1863_545 | scaffold1863 | 545 | 8.39E-11 | Plant organelle RNA recognition domain (PORR) | upstream 6.46 | ||
| scaffold353_773806 | scaffold353 | 773806 | 7.04E-11 | Phosphatidylinositol-4-phosphate 5-Kinase (PIP5K) | downstream 6.62 | ||
| scaffold464_754364 | scaffold464 | 754364 | 7.77E-07 | xyloglucan endotransglucosylase/hydrolase (XTH) | interior | ||
| scaffold977_784147 | scaffold977 | 784147 | 2.69E-10 | DNA-damage-repair (DDR) | downstream 1.65 | ||
| scaffold359_289139 | scaffold359 | 289139 | 2.30E-08 | Lus10013156.BGIv1.0 | upstream 1.25 |
SNP loci displaying significant correlation with number of fruits
Nine SNP loci exhibited significant associations with the number of fruits (P < 1.26E-06). The corresponding Manhattan plots and QQ plots of the two models and three environments are shown in Supplementary Figure 4. The GLM identified three SNP loci in 2015HRB, two SNP loci in 2016HRB, and four SNP loci in 2016LX. Among these, scaffold137_111000 and scaffold225_427119 were identified in both 2015HRB and 2016LX. In addition, scaffold156 and scaffold413 were both identified in 2016HRB and 2016LX, but each had different association loci in the two environments. MLM identified association SNP loci in only two environments, including three SNP loci in 2015HRB and four SNP loci in 2016LX. Among these, scaffold137_111000 and scaffold225_427119 were identified in both 2015HRB and 2016LX. An overview of the results showed that five SNP loci were identified by both models (scaffold137_111000, scaffold225_427119, scaffold687_123666, scaffold156_1203677, and scaffold413_388319). The genes closest to the five SNP loci were TATP (Transmembrane amino acid transporter protein), Contig1437 (Linum usitatissimum clone Contig1437 microsatellite sequence), LU0019C12 (Linum usitatissimum clone LU0019C12 mRNA sequence), FH (Fumarate hydratase), and RP (Ribosomal protein) (Table 5).
Table 5.
Associated single nucleotide polymorphisms (SNPs) and the nearest genes for the number of fruits trait of flax.
| Model | Environment | SNP position (bp) | scaffold | Location | P-value | Nearest gene | Distance to SNP (kb) |
|---|---|---|---|---|---|---|---|
| GLM | 2015HRB | scaffold137_111000 | scaffold137 | 111000 | 6.28E-08 | Transmembrane amino acid transporter protein (TATP) | upstream 0.65 |
| scaffold225_427119 | scaffold225 | 427119 | 1.91E-07 | Linum usitatissimum clone Contig1437 microsatellite sequence | downstream 1.53 | ||
| scaffold687_121617 | scaffold687 | 121617 | 7.22E-07 | Linum usitatissimum clone LU0019C12 mRNA sequence | upstream 0.36 | ||
| 2016HRB | scaffold156_761294 | scaffold156 | 761294 | 2.76E-07 | Lus10040627.BGIv1.0 | downstream 0.54 | |
| scaffold413_1116527 | scaffold413 | 1116527 | 2.97E-07 | Fumarate hydratase (FH) | interior | ||
| 2016LX | scaffold137_111000 | scaffold137 | 111000 | 6.28E-08 | Transmembrane amino acid transporter protein (TATP) | upstream 0.65 | |
| scaffold225_427119 | scaffold225 | 427119 | 1.91E-07 | Linum usitatissimum clone Contig1437 microsatellite sequence | downstream 1.53 | ||
| scaffold156_1203677 | scaffold156 | 1203677 | 1.14E-12 | Ribosomal protein (RP) | upstream 1.14 | ||
| scaffold413_388319 | scaffold413 | 388319 | 1.03E-06 | Chitinase class I (CCI) | upstream 1.02 | ||
| MLM | 2015HRB | scaffold137_111000 | scaffold137 | 111000 | 6.28E-08 | Transmembrane amino acid transporter protein (TATP) | upstream 0.65 |
| scaffold225_427119 | scaffold225 | 427119 | 1.91E-07 | Linum usitatissimum clone Contig1437 microsatellite sequence | downstream 1.53 | ||
| scaffold687_123666 | scaffold687 | 123666 | 3.22E-07 | Linum usitatissimum clone LU0019C12 mRNA sequence | interior | ||
| 2016LX | scaffold137_111000 | scaffold137 | 111000 | 6.28E-08 | Transmembrane amino acid transporter protein (TATP) | upstream 0.65 | |
| scaffold225_427119 | scaffold225 | 427119 | 1.91E-07 | Linum usitatissimum clone Contig1437 microsatellite sequence | downstream 1.53 | ||
| scaffold156_1203677 | scaffold156 | 1203677 | 1.14E-12 | Lus10040728.BGIv1.0 | upstream 1.14 | ||
| scaffold413_388319 | scaffold413 | 388319 | 1.03E-06 | Lus10028377.BGIv1.0 | upstream 1.02 |
SNP loci showing significant correlation with 1,000-grain weight
Twenty-three SNP loci exhibited significant associations with 1000-grain weight (P < 1.26E-06). The corresponding Manhattan plots and QQ plots of the two models and three environments are shown in Supplementary Figure 5. The GLM identified ten SNP loci in 2015HRB, five SNP loci in 2016HRB, and eight SNP loci in 2016LX. Among these, four loci, namely scaffold112_184204, scaffold1143_190268, scaffold1317_154716, and scaffold1519_272169, were repeatedly identified in the three environments; scaffold123_1191347 was repeatedly identified in both 2015HRB and 2016HRB. The MLM identified eight SNP loci in 2015 HRB, seven SNP loci in 2016HRB, and four SNP loci in 2016LX. Among these, four loci, namely scaffold112_184204, scaffold1155_171787, scaffold132_713877, and scaffold1519_272169, were identified in all three environments; eight SNP loci were identified by both models (scaffold112_184204, scaffold123_1191347, scaffold1317_154716, scaffold1519_272169, scaffold1155_171787, scaffold132_713877, scaffold1491_58878, and scaffold15_1207948). The genes closest to the eight SNP loci were HP (Hypothetical protein), NAD-DEF (NAD dependent epimerase/dehydratase family), TS (Terpene synthase), TPI (Trypsin and protease inhibitor), STK (Serine/threonine protein kinase), CAP (CDP-alcohol phosphatidyltransferase), PHO1 (SPX and EXS domain-containing protein), and ARP (Autophagy-related protein) (Table 6).
Table 6.
Associated single nucleotide polymorphisms (SNPs) and the nearest genes for the 1,000-grain weight trait of flax.
| Model | Environment | SNP position (bp) | scaffold | Location | P-value | Nearest gene | Distance to SNP (kb) |
|---|---|---|---|---|---|---|---|
| GLM | 2015HRB | scaffold101_354340 | scaffold101 | 354340 | 3.68E-09 | Uncharacterized protein (UP) | interior |
| scaffold112_184204 | scaffold112 | 184204 | 4.55E-09 | hypothetical protein (HP) | interior | ||
| scaffold1143_190268 | scaffold1143 | 190268 | 2.83E-07 | serine/threonine-protein kinase (STK) | downstream 0.04 | ||
| scaffold1155_171787 | scaffold1155 | 171787 | 1.23E-08 | NAD dependent epimerase/dehydratase family (NAD-DEF) | interior | ||
| scaffold123_1191347 | scaffold123 | 1191347 | 5.48E-08 | Probable terpene synthase (TS) | interior | ||
| scaffold1317_154716 | scaffold1317 | 154716 | 7.62E-10 | Trypsin and protease inhibitor (TPI) | upstream 0.04 | ||
| scaffold132_713877 | scaffold132 | 713877 | 1.52E-11 | serine/threonine-protein kinase MPS1-like (STK-MPS1) | interior | ||
| scaffold1491_58878 | scaffold1491 | 58878 | 3.67E-10 | CDP-alcohol phosphatidyltransferase (CAP) | upstream 3.01 | ||
| scaffold15_1207948 | scaffold15 | 1207948 | 3.65E-08 | SPX and EXS domain-containing protein (PHO1) | interior | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-10 | Autophagy-related protein (ARP) | interior | ||
| 2016HRB | scaffold112_184204 | scaffold112 | 184204 | 5.32E-09 | Lus10018116.BGIv1.0 | interior | |
| scaffold1143_190268 | scaffold1143 | 190268 | 1.61E-07 | Serine/threonine protein kinase (STK) | downstream 0.04 | ||
| scaffold123_1191347 | scaffold123 | 1191347 | 3.51E-08 | Terpene synthase (TS) | interior | ||
| scaffold1317_154716 | scaffold1317 | 154716 | 8.00E-09 | Trypsin and protease inhibitor (TPI) | upstream 0.04 | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-09 | Autophagy-related protein (ARP) | interior | ||
| 2016LX | scaffold101_354340 | scaffold101 | 354340 | 3.68E-09 | Uncharacterized protein | interior | |
| scaffold112_184204 | scaffold112 | 184204 | 4.55E-09 | hypothetical protein (HP) | interior | ||
| scaffold1143_190268 | scaffold1143 | 190268 | 2.83E-07 | serine/threonine-protein kinase (STK) | downstream 0.04 | ||
| scaffold1155_171787 | scaffold1155 | 171787 | 1.23E-08 | NAD dependent epimerase/dehydratase family (NAD-DEF) | interior | ||
| scaffold1317_154716 | scaffold1317 | 154716 | 7.62E-10 | Trypsin and protease inhibitor (TPI) | upstream 0.04 | ||
| scaffold132_713877 | scaffold132 | 713877 | 1.52E-11 | serine/threonine-protein kinase MPS1-like (STK-MPS1) | interior | ||
| scaffold1491_58878 | scaffold1491 | 58878 | 3.67E-10 | CDP-alcohol phosphatidyltransferase (CAP) | upstream 3.01 | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-10 | Autophagy-related protein (ARP) | interior | ||
| MLM | 2015HRB | scaffold112_184204 | scaffold112 | 184204 | 4.55E-09 | hypothetical protein (HP) | interior |
| scaffold1155_171787 | scaffold1155 | 171787 | 1.23E-08 | NAD dependent epimerase/dehydratase family (NAD-DEF) | interior | ||
| scaffold123_1191347 | scaffold123 | 1191347 | 5.48E-08 | Probable terpene synthase (TS) | interior | ||
| scaffold1317_154716 | scaffold1317 | 154716 | 7.62E-10 | Trypsin and protease inhibitor (TPI) | upstream 0.04 | ||
| scaffold132_713877 | scaffold132 | 713877 | 1.52E-11 | serine/threonine-protein kinase MPS1-like (STK-MPS1) | interior | ||
| scaffold1491_58878 | scaffold1491 | 58878 | 3.67E-10 | CDP-alcohol phosphatidyltransferase (CAP) | upstream 3.01 | ||
| scaffold15_1207948 | scaffold15 | 1207948 | 3.65E-08 | SPX and EXS domain-containing protein 1 (PHO1) | interior | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-10 | Autophagy-related protein (ARP) | interior | ||
| 2016HRB | scaffold112_184204 | scaffold112 | 184204 | 5.32E-09 | Lus10018116.BGIv1.0 | interior | |
| scaffold123_1191347 | scaffold123 | 1191347 | 3.51E-08 | Lus10042202.BGIv1.0 | interior | ||
| scaffold1317_154716 | scaffold1317 | 154716 | 8.00E-09 | Lus10007888.BGIv1.0 | upstream 0.04 | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-09 | Lus10007527.BGIv1.0 | interior | ||
| scaffold132_713877 | scaffold132 | 713877 | 1.52E-11 | serine/threonine-protein kinase MPS1-like(STK-MPS1) | interior | ||
| scaffold1491_58878 | scaffold1491 | 58878 | 3.67E-10 | CDP-alcohol phosphatidyltransferase (CAP) | upstream 3.01 | ||
| scaffold15_1207948 | scaffold15 | 1207948 | 3.65E-08 | SPX and EXS domain-containing protein 1 (PHO1) | interior | ||
| 2016LX | scaffold112_184204 | scaffold112 | 184204 | 4.55E-09 | hypothetical protein (HP) | interior | |
| scaffold1155_171787 | scaffold1155 | 171787 | 1.23E-08 | NAD dependent epimerase/dehydratase family (NAD-DEF) | interior | ||
| scaffold132_713877 | scaffold132 | 713877 | 1.52E-11 | serine/threonine-protein kinase MPS1-like (STK-MPS1) | interior | ||
| scaffold1519_272169 | scaffold1519 | 272169 | 2.52E-10 | Autophagy-related protein (ARP) | interior |
Candidate gene prediction
In this study, a total of 42 SNP loci were found to display significant association with five important agronomic traits (P < 1.26E-06). The Manhattan plots of the SNP loci (Supplementary Figures 1–5) as well as Tables 2–6 revealed that relatively more SNP loci were found to be linked to number of branches and 1000-grain weight (over twenty for each trait). In comparison, only nine SNP loci showed significant association with plant height or number of fruits, as did only three SNP loci with technical length. Next, candidate genes were screened in the 10 kb zone of each of the 15 SNP loci. The resulting candidate genes were then screened further using co-identification in both the GLM and MLM as well as co-occurrences in at least two of the three environments, whereby 15 final candidate genes were obtained (Table S3).
Discussion
Genome-wide association study (GWAS) is based on molecular markers, SNPs, that are present throughout a genome and facilitate direct association analysis for complex traits. It is considered an effective method to determine molecular markers that influence crucial traits (Gu et al., 2011; Liu et al., 2013). As a consequence, SLAF-seq-based GWAS has been launched in several crops, such as maize, rice, and soybean. Zhao et al. (2015) used this approach to examine 330 soybean cultivars to identify genes related to resistance against sclerotinia stem rot: the dominant locus Oswm13-1 was identified and four resistance candidate genes were acquired. Likewise, the method was used to analyze 440 soybean germplasm resources of various origins to identify genes related to resistance against soybean cyst nematode (SCN, Heterodera glycines Ichinohe) (Han et al., 2015); the authors identified 19 SNP loci significantly associated with SCN resistance. Su et al. (2016) used SLAF-seq to identify 81,675 SNP loci in cotton, performing GWAS for 355 cotton germplasm resources to identify 11 SNP loci associated with five earliness-related traits. Moreover, Yang et al. (2016) used this approach to analyze 419 core germplasm resources of rice to identify the novel gene LAC6 as associated with amylose content and show LOC_Os06g11340 to be a likely candidate gene for LAC6. These results indicated that SLAF-seq-based GWAS is a well-developed technology to identify high-quality alleles. In comparison with rice, soybean, maize, and cotton, flax has not received adequate research efforts using GWAS to identify high-quality alleles (no GWAS have yet been reported for flax). In this study, we employed NGS sequencing technology coupled with SLAF-seq to perform GWAS to identify SNP loci associated with important traits and determined their candidate genes.
Population stratification and genetic relationship are two key factors affecting the accuracy of population structure. We used Admixture software to analyze the population structure of flax. The results of population structure analyses showed that flax accessions were clearly divided into three groups—oil using, fiber using and oil-fiber using groups—at K = 3. The result indicated a strong divergence between different flax groups. Correspondingly, if the influence of population structure is not considered, then the stratification effect may be misinterpreted as genetic events, leading to pseudo-negativity in the association analysis. Hence, QQ plots of the five important traits under different environmental conditions were generated to validate the accuracy of the population correction. The results of the QQ plots showed that, overall, the observed values matched the expected values except for a few outliers at the ends. In other words, the correction of the population structure produced reliable results; thus, the association analysis did not produce any false associations because of population stratification.
GLM and MLM are the most commonly used algorithmic models in GWAS. The advantage of GLM is that it is more comprehensive and can obtain more SNPs associated with the traits, but its accuracy in identifying SNP loci is worse than MLM (Huang et al., 2010; Yang et al., 2010; Zhang et al., 2010; Liu et al., 2016). MLM can improve the accuracy of the analysis but can also miss some important SNP loci do to the strict screening conditions. Multiple algorithmic models should be used to conduct GWAS data analysis in actual application (Dhanapal and Crisosto, 2013; Hecht et al., 2013; Zhang et al., 2017). However, we found that the observed p-value from GLM greatly deviated from the expected p-value, while the p-value from the MLM model was close to the expected p-value (Supplementary Figures 1–5). The results indicated that the false positives were well controlled in the MLM model in our study.
The two association models (i.e., GLM and MLM) and the phenotypic data derived from three environments (i.e., 2015HRB, 2016HRB, and 2016LX) were used to perform GWAS for five important flax traits, generating a total of 107 loci (42 individual SNP loci) that displayed significant association with the five important flax traits (P < 1.26E-06). Afterwards, a more stringent screening was performed, in which the 42 SNP loci were subjected to analyses of co-identification by both the GLM and MLM as well as co-occurrence in 2015HRB, 2016HRB, and 2016LX. Ultimately, we identified two SNP loci associated with plant height, six SNP loci associated with the number of branches, five SNP loci associated with the number of fruits, and eight SNP loci associated with the 1,000-grain weight. Given that the aforementioned SNP loci displayed repeated occurrences (in both models and/or in at least two environments), they potentially have pivotal influences on the relevant agronomic flax traits. As such, they can be recruited as candidate genetic markers impacting these five important flax traits. The remaining SNP loci only had single occurrences of association (in only one model and in only one environment); therefore, their reliability must be investigated further.
Next, candidate genes were screened in the 10 kb zone of each of the SNP loci, which generated 15 potential candidate genes. Among these, there were two candidate genes for plant height, UGT and PL. It was reported that the overexpression of UGT84B1 and UGT74E2 in Arabidopsis thaliana (A. thaliana)causes phenotypes with shorter stature and more shoot branches. UGT84B1 overexpressors also have wrinkled leaves and reduced root gravitropism (Jin et al., 2013). In addition, UGT74D1 has been shown to modulate the metabolic pathway of auxin (IAA) in A thaliana to influence its development (Tanaka et al., 2014). Transgenic rice lines ectopically over-expressing the cZOGT1 and cZOGT2 genes exhibit short shoot phenotypes, delay of leaf senescence, and a decrease in crown root number. These results suggest that cZOGT activity has a physiological impact on growth and development of rice (Kudo et al., 2012). As such, UGT, in a similar fashion to its homologs in other plants, is likely involved in developmental regulation in flax, thereby affecting plant height. In addition, studies in rice and A. thaliana have shown that PL is intricately associated with plant development (Palusa et al., 2007; Leng et al., 2017) and that PL promotes plant growth and development via adjusting the cell division rate and cell wall relaxation (Sun and Nocker, 2010; Sun et al., 2010). These reports corroborated our finding that PL was a candidate gene for plant height in flax. Association analysis of technical length only identified one gene, MIF. Previous studies revealed that MIF is related to human disease and immunity (Roberts et al., 2017; Shin et al., 2017); if the gene is overexpressed, it may lead to the expansion and proliferation of cancer cells. However, the gene has not been examined in plants; thus, further studies are needed regarding whether MIF indeed affects the technical length of flax. Association analysis of the number of branches identified four candidate genes. Among these, GRAS is a transcription factor unique to plants and plays pivotal roles in development and signal transduction. LS and MOC1 are both members of the GRAS protein family. Lack of expression of LS prevents the formation of the axillary meristem and in turn decreases the number of axillary buds (Schmitz and Theres, 1999; Greb et al., 2003). Lack of expression of MOC1 results in the almost complete loss of tillering in rice plants because the gene is responsible for regulating that biological process via promoting the cell cycle (Li et al., 2003; Sun et al., 2010). These findings therefore indicate that MOC1 is involved in branch formation, directly affects the trait of the number of branches, and is a crucial candidate gene for this trait in flax. In plants, a major function of GST is to detoxify exogenous toxins and harmful endogenous metabolites. Specifically, mercapto groups of GST can be catalyzed to bind to a variety of endogenous electrophilic compounds and lipophilic substrates (Dixon et al., 2002; Moons, 2003). However, the involvement of GST in the branching or development of plants has not been reported; thus, further studies are needed to verify its association with the number of branches in flax. In A. thaliana, XTH9 is expressed in the shoot apical meristem of flower buds and flower stalks and is related to the elongation of these tissues; its loss of expression results in a phenotype of short internodal cell length (Hyodo et al., 2003). Moreover, the overexpression of its Brassica campestris homolog, BcXTH1, in A. thaliana leads to the elongation of flower stalks and an increase in plant height (Shin et al., 2006). Hence, the findings in A. thaliana studies suggest that XTH possibly plays an important role in dictating the number of branches in flax and corroborate our results that XTH is a candidate gene for this trait. Of the three genes showing association with the number of fruits, TATP has not been reported in plants. In addition, Contig1437 and LU0019C12 were both cloned from flax, but there are no studies examining their functions. Therefore, they remain to be validated by functional studies. Of the five candidate genes possibly associated with 1000-grain weight, SPX proteins contain a C-terminal EXS domain, which is a part of the PHO1 family. In A. thaliana and rice, PHO1 participates in transfer and signal transduction of phosphate from roots to the aboveground parts (Hamburger et al., 2002; Svistoonoff et al., 2007; Secco et al., 2010). Because the uptake of phosphorus can clearly improve seed yield in crops, these previous studies on PHO1 are consistent with our findings that it is an important candidate gene for the 1,000-grain weight. However, HP, TS, CAP, and STK have not been previously associated with seed yield.
Conclusion
In this study, we employed SLAF-seq to perform GWAS for five important agronomic traits in 224 germplasm resources of flax. Using two models (i.e., GLM and MLM) for flax grown in three environments (i.e., 2015HRB, 2016HRB, and 2016LX), we identified a total of 42 SNP loci displaying a significant association (P < 1.26E-06), including 15 SNP loci having co-identification either by both models or by co-occurrence in two or more environments. Next, candidate genes were screened in the 10 kb zone of each of the 15 SNP loci to identify 15 candidate genes possibly related to the five important agronomic traits. Our subsequent analyses determined that UGT and PL are candidate genes for plant height, GRAS and XTH are candidate genes for the number of branches, Contig1437 and LU0019C12 are candidate genes for the number of fruits, and PHO1 is a candidate gene for 1,000-grain weight. These SNP loci and candidate genes may serve as a biological basis for improving these important traits of flax.
Author contributions
ZD, ZY, LZ, and QT carried out most of the experimental work, and this study was conceived by JSu. Collections of flax germplasm resources were performed by DZ and XY. DX and JSun designed the research and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the fund for the 59th Batch of Certificate of China Postdoctoral Science Foundation (2016M591302), China Agriculture Research System (CARS-19-E01), Agriculture Scientific and Technological Innovation Project of Chinese Academy of Agricultural Sciences (ASTIP-IBFC01), and Introduction of Doctor's Personnel Scientific Research and Development Fund (201507-43). We also thank the National Bast Fiber Crops Germplasm Improvement Center of Flax Branch Center for kindly supplying the experimental platform.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02232/full#supplementary-material
References
- Alexander D. H., Novembre J., Lange K. (2009). Fast model-based estimation of ancestry in unrelated individual. Genome Res. 19, 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjálmsson B. J., Willems G., Horton M., Li Y., et al. (2010). Genome-wide association study of 107 phenotypes in arabidopsis thaliana inbred lines. Nature 465, 627–631. 10.1038/nature08800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth I., Harwood R. J., Wyatt J. L., Grishanov S. (2004). A comparative study of the characteristics of fibre-flax (Linum usitatissimum). Ind. Crops Prod. 20, 89–95. 10.1016/j.indcrop.2003.12.014 [DOI] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., Buckler E. S. (2007). Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Deng X., Qiu C. S., Chen X. B., Long S. H., Guo Y., Hao D. M., et al. (2014). Multiple analysis of relationship of agronomic traits and yield formation in flax(linum usitatissimum L.). Southwest China J. Agric. Sci. 27, 535–540. 10.16213/j.cnki.scjas.2014.02.038 [DOI] [Google Scholar]
- Dhanapal A. P., Crisosto C. H. (2013). Association genetics of chilling injury susceptibility in peach (Prunus persica (L.) Batsch) across multiple years. 3 Biotech, 3, 481–490. 10.1007/s13205-012-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichsen A., Ulrich A. (2009). Variability in stem fibre content and its association with other characteristics in 1177 flax (Linum usitatissimum L.) genebank accessions. Ind. Crops Prod. 30, 33–39. 10.1016/j.indcrop.2009.01.002 [DOI] [Google Scholar]
- Dixon D. P., Lapthorn A., Edwards R. (2002). Plant glutathione transferases. Genome Biol. 3:reviews3004-1. 10.1186/gb-2002-3-3-reviews3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan I. D., Gn D. L. F., Murray S. C., Isakeit T., Huang P. C., Warburton M., et al. (2015). Genome wide association study for drought, aflatoxin resistance, and important agronomic traits of maize hybrids in the sub-tropics. PLoS ONE 10:e0117737. 10.1371/journal.pone.0117737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehringer A., Friedt W., Lühs W., Snowdon R. J. (2006). Genetic mapping of agronomic traits in false flax (Camelina sativa subsp. sativa). Genome 49, 1555–1563. 10.1139/g06-117 [DOI] [PubMed] [Google Scholar]
- Geng X., Jiang C., Yang J., Wang L., Wu X., Wei W. (2016). Rapid identification of candidate genes for seed weight using the slaf-seq method in brassica napus. PLoS ONE 11:e0147580. 10.1371/journal.pone.0147580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., et al. (2003). Molecular analysis of the lateral suppressor gene in arabidopsis reveals a conserved control mechanism for axillary meristem formation. Gene Dev. 17, 1175–1187. 10.1101/gad.260703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Feng C., Ma L., Song C., Wang Y., Da Y., et al. (2011). Genome-wide association study of body weight in chicken f2 resource population. PLoS ONE 6:e21872. 10.1371/journal.pone.0021872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D., Tegström C., Ingvarsson P. K. (2010). Using association mapping to dissect the genetic basis of complex traits in plants. Brief. Funct. Genomics 9, 157. 10.1093/bfgp/elp048 [DOI] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., Macdonald-Comber P. J., Somerville C., Poirier Y. (2002). Identification and characterization of the arabidopsis pho1 gene involved in phosphate loading to the xylem. Plant Cell 14, 889–902. 10.1105/tpc.000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhao X., Cao G., Wang Y., Li Y., Liu D., et al. (2015). Genetic characteristics of soybean resistance to hg type 0 and hg type 1.2.3.5.7 of the cyst nematode analyzed by genome-wide association mapping. BMC Genomics 16:598. 10.1186/s12864-015-1800-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhang B., Zhao H., Ayaad M., Xing Y. (2016). Genome-wide association studies reveal that diverse heading date genes respond to short and long day lengths between indica and japonica rice. Front Plant Sci. 7:1270. 10.3389/fpls.2016.01270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy O. J., Vekemans X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Resour. 2, 618–620. 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- Healey A., Furtado A., Cooper T., Henry R. J. (2014). Protocol: a simple method for extracting next-generation sequencing quality genomic dna from recalcitrant plant species. Plant Methods 10, 1–8. 10.1186/1746-4811-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B. C., Campbell N. R., Holecek D. E., Narum S. R. (2013). Genome-wide association reveals genetic basis for the propensity to migrate in wild populations of rainbow and steelhead trout. Mol. Ecol. 22, 3061–3076. 10.1111/mec.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42:961. 10.1038/ng.695 [DOI] [PubMed] [Google Scholar]
- Hyodo H., Yamakawa S., Takeda Y., Tsuduki M., Yokota A., Nishitani K., et al. (2003). Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, xth9, in inflorescence apices is related to cell elongation in arabidopsis thaliana. Plant Mol. Biol. 52, 473–482. 10.1023/A:1023904217641 [DOI] [PubMed] [Google Scholar]
- Jin S. H., Ma X. M., Han P., Wang B., Sun Y. G., Zhang G. Z., et al. (2013). UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS ONE 8:e61705. 10.1371/journal.pone.0061705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T., Makita N., Kojima M., Tokunaga H., Sakakibara H. (2012). Cytokinin activity of cis-zeatin and phenotypic alterations induced by over-expression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 160, 112 10.1104/pp.112.196733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y., Yang Y., Ren D., Huang L., Dai L., Wang Y., et al. (2017). A rice pectate lyase-like gene is required for plant growth and leaf senescence. Plant Physiol. 174, 1151–1166. 10.1104/pp.16.01625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qian Q., Fu Z., Wang Y., Xiong G., Zeng D., et al. (2003). Control of tillering in rice. Nature 422, 618. 10.1038/nature01518 [DOI] [PubMed] [Google Scholar]
- Li Y., Zeng X. F., Zhao Y. C., Li J. R., Zhao D. G. (2016). Identification of a new rice low-tiller mutant and association analyses based on the slaf-seq method. Plant Mol. Biol. Rep. 35, 1–11. 10.1007/s11105-016-1002-2 [DOI] [Google Scholar]
- Liu R., Sun Y., Zhao G., Wang F., Wu D., Zheng M., et al. (2013). Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in beijing-you chickens. PLoS ONE 8:e61172. 10.1371/journal.pone.0061172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang M., Fan B., Buckler E. S., Zhang Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12:e1005767. 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A. (2003). Osgstu3, and osgtu4, encoding tau class glutathione s -transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots, 1. FEBS Lett. 553, 427–432. 10.1016/S0014-5793(03)01077-9 [DOI] [PubMed] [Google Scholar]
- Nussbaum A. K., Kuttler C., Hadeler K. P., Rammensee H. G., Schild H. (2001). Paproc: a prediction algorithm for proteasomal cleavages available on the www. Immunogenetics 53, 87–94. 10.1007/s002510100300 [DOI] [PubMed] [Google Scholar]
- Palusa S. G., Golovkin M., Shin S. B., Richardson D. N., Reddy A. S. (2007). Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in arabidopsis. New Phytol. 174, 537–550. 10.1111/j.1469-8137.2007.02033.x [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S., Leng L., Soroka C. J., Boyer J. L., Bucala R., Assis D. N. (2017). FRI-393-Macrophage migration inhibitory factor (MIF) modulates T-cell proliferation and hepatic inflammation in a model of autoimmune liver disease. J. Hepatol. 66, S363–S364. 10.1016/S0168-8278(17)31067-X [DOI] [Google Scholar]
- Schmitz G., Theres K. (1999). Genetic control of branching in Arabidopsis and tomato. Curr. Opin. Plant Biol. 2, 51–55. 10.1016/S1369-5266(99)80010-7 [DOI] [PubMed] [Google Scholar]
- Secco D., Baumann A., Poirier Y. (2010). Characterization of the rice pho1 gene family reveals a key role for ospho1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 152, 1693–1704. 10.1104/pp.109.149872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M. S., Kang Y., Leng L., Bucala R., Kang I. (2017). Macrophage migration inhibitory factor serves as an upstream regulator of NLRP3 expression and subsequent IL-1beta production in human monocytes in response to lupus U1-snRNP immune complex. J. Immunol. 198, 210. [Google Scholar]
- Shin Y. K., Yum H., Kim E. S., Cho H., Gothandam K. M., Hyun J., et al. (2006). Bcxth1, a brassica campestris homologue of arabidopsis xth9, is associated with cell expansion. Planta 224, 32–41. 10.1007/s00425-005-0189-5 [DOI] [PubMed] [Google Scholar]
- Sonah H., O'Donoughue L., Cober E., Rajcan I., Belzile F. (2015). Identification of loci governing eight agronomic traits using a gbs-gwas approach and validation by qtl mapping in soya bean. Plant Biotechnol. J. 13, 211–221. 10.1111/pbi.12249 [DOI] [PubMed] [Google Scholar]
- Soto-Cerda B. J., Diederichsen A., Ragupathy R., Cloutier S. (2013). Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol. 13:78. 10.1186/1471-2229-13-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Cerda B. J., Duguid S., Booker H., Rowland G., Diederichsen A., Cloutier S. (2014). Genomic regions underlying agronomic traits in linseed (Linum usitatissimum L.) as revealed by association mapping. J. Integr. Plant Biol. 56, 75–87. 10.1111/jipb.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Pang C., Wei H., Li L., Liang B., Wang C., et al. (2016). Identification of favorable snp alleles and candidate genes for traits related to early maturity via gwas in upland cotton. BMC Genomics 17:687. 10.1186/s12864-016-2875-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. L., Zhang W. P., Xiong G. S., Yan M. X., Qian Q., Li J. Y., et al. (2010). Identification and functional analysis of the moc1 interacting protein 1. J. Genet. Genomics 37, 69–77. 10.1016/S1673-8527(09)60026-6 [DOI] [PubMed] [Google Scholar]
- Sun L. X., Nocker S. V. (2010). Analysis of promoter activity of members of the pectate lyase-like (pll) gene family in cell separation in arabidopsis. BMC Plant Biol. 10, 152. 10.1186/1471-2229-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. W., Liu D. Y., Zhang X. F., Li W. B., Liu H., Hong W. G., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700. 10.1371/journal.pone.0058700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S., Creff A., Reymond M., Sigoillot-Claude C., Ricaud L., Blanchet A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792. 10.1038/ng2041 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Hayashi K., Natsume M., Kamiya Y., Sakakibara H., Kawaide H., et al. (2014). Ugt74d1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in arabidopsis. Plant Cell Physiol. 55, 218–228. 10.1093/pcp/pct173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilkki J., Iso-Touru T., Schulman N. F., Dolezal M. A., Bagnato A., Soller M., et al. (2013). Revisiting QTL Affecting Clinical Mastitis by High-Density GWAS and Resequencing in the Finnish Ayrshire Dairy Cattle, in International Plant and Animal Genome Conference Xxi. San Diego, CA. [Google Scholar]
- Wang Z., Hobson N., Galindo L., Zhu S., Shi D., McDill J., et al. (2012). The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 72, 461–473. 10.1111/j.1365-313X.2012.05093.x [DOI] [PubMed] [Google Scholar]
- Xu X., Xu R., Zhu B., Yu T., Qu W., Lu L., et al. (2014). A high-density genetic map of cucumber derived from specific length amplified fragment sequencing (slaf-seq). Front. Plant Sci. 5:768. 10.3389/fpls.2014.00768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Warburton M. L., Sawkins M., Zhang X., Setter T., Xu Y., et al. (2013). Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor. App. Genet. 126, 2587–2596. 10.1007/s00122-013-2158-x [DOI] [PubMed] [Google Scholar]
- Yang X., Nong B., Xia X., Zhang Z., Zeng Y., Liu K., et al. (2016). Rapid identification of a new gene influencing low amylose content in rice landraces (Oryza sativa l.) using genome-wide association study with specific-locus amplified fragment sequencing. Genome 60, 465–472. 10.1139/gen-2016-0104 [DOI] [PubMed] [Google Scholar]
- Yang X., Yan J., Shah T., Warburton M. L., Li Q., Li L., et al. (2010). Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 121, 417–431. 10.1007/s00122-010-1320-y [DOI] [PubMed] [Google Scholar]
- Zhang H., Fan X., Zhang Y., Jiang J., Liu C. (2017). Identification of favorable SNP alleles and candidate genes for seedlessness in Vitis vinifera L. using genome-wide association mapping. Euphytica 213:136 10.1007/s10681-017-1919-z [DOI] [Google Scholar]
- Zhang J., Song Q., Cregan P. B., Nelson R. L., Wang X., Wu J., et al. (2015). Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (glycine max) germplasm. BMC Genomics 16:217. 10.1186/s12864-015-1441-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Wu X., Ma R., Jiang Q., Wang Y. (2016). Identifying liver cancer-related enhancer snps by integrating gwas and histone modification chip-seq data. BioMed Res. Int. 2016, 1–6. 10.1155/2016/2395341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang L., Xin H., Li D., Ma C., Xia D., et al. (2013). Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (slaf) sequencing. BMC Plant Biol. 13:141. 10.1186/1471-2229-13-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C. Q., Todhunter R. J., Tiwari H. K., Gore M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat Genetics 42:355. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Han Y., Li Y., Liu D., Sun M., Zhao Y., et al. (2015). Loci and candidate gene identification for resistance to sclerotinia sclerotiorum in soybean (glycine max L. merr.) via association and linkage maps. Plant J. Cell Mol. Biol. 82, 245–255. 10.1111/tpj.12810 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.