Abstract
The nucleolus is the most prominent nuclear domain, where the core processes of ribosome biogenesis occur vigorously. All these processes are finely orchestrated by many nucleolar factors to build precisely ribosome particles. In animal cells, perturbations of ribosome biogenesis, mostly accompanied by structural disorders of the nucleolus, cause a kind of cellular stress to induce cell cycle arrest, senescence, or apoptosis, which is called nucleolar stress response. The best-characterized pathway of this stress response involves p53 and MDM2 as key players. p53 is a crucial transcription factor that functions in response to not only nucleolar stress but also other cellular stresses such as DNA damage stress. These cellular stresses release p53 from the inhibition by MDM2, an E3 ubiquitin ligase targeting p53, in various ways, which leads to p53-dependent activation of a set of genes. In plants, genetic impairments of ribosome biogenesis factors or ribosome components have been shown to cause characteristic phenotypes, including a narrow and pointed leaf shape, implying a common signaling pathway connecting ribosomal perturbations and certain aspects of growth and development. Unlike animals, however, plants have neither p53 nor MDM2 family proteins. Then the question arises whether plant cells have a nucleolar stress response pathway. In recent years, it has been reported that several members of the plant-specific transcription factor family NAC play critical roles in the pathways responsive to various cellular stresses. In this mini review, we outline the plant cellular stress response pathways involving NAC transcription factors with reference to the p53-MDM2-dependent pathways of animal cells, and discuss the possible involvement of a plant-unique, NAC-mediated pathway in the nucleolar stress response in plants.
Keywords: nucleolar stress response, nucleolus, NAC transcription factor, ribosome biogenesis, ribosomal protein, Pre-rRNA processing, cell proliferation, development
The core processes of ribosome biogenesis, such as ribosomal RNA (rRNA) transcription, pre-rRNA processing, and ribosome assembly, take place in the nucleolus, which is the most prominent nuclear domain. These processes are finely controlled by many nucleolar factors, to build precisely small and large ribosome particles. Perturbations of any of the steps of ribosome biogenesis in the nucleolus cause a type of stress called nucleolar stress or ribosomal stress, which stimulates a specific signaling pathway in animal cells. Recent studies have implicated a plant-unique pathway in the nucleolar stress response of plant cells. In this mini review, we will discuss how plant cells sense and respond to nucleolar stress, with reference to the p53-dpeendent nucleolar stress response pathway of animal cells.
Ribosome biogenesis in the nucleolus
The nucleolus is formed around the nuclear rDNA regions where tandem repeats of rRNA genes lie, and serves as a site for the main part of biogenesis of the ribosome, a huge ribonucleoprotein machinery that executes translation of the nucleotide sequence of mRNAs into the amino acid sequence of proteins. In the nucleolus, ribosome biogenesis starts with the transcription of pre-rRNAs from rRNA genes, followed by their processing and assembly with ribosomal proteins (RPs) into two ribosome subunits, the small subunit (SSU) and the large subunit (LSU).
Among the four species of rRNA, i.e., 5S, 5.8S, 18S, and 25–28S rRNAs, only 5S rRNA is transcribed separately, whereas the other three rRNAs are transcribed as a single precursor molecule. This precursor undergoes sequential processing events, such as base modification, cleavage, and trimming, which are tightly linked with the stepwise assembly of RPs onto pre-rRNAs. These events are guided by ribosome biogenesis factors (RBFs), which are enriched in the nucleolus, including small nucleolar RNAs (snoRNAs), snoRNA-associated proteins, and many other proteins. The processing pathway of pre-rRNAs and the repertoire of RBFs are partly different but largely common between yeast, animals, and plants (Henras et al., 2015; Weis et al., 2015). The nucleolar activities of pre-rRNA processing and RP assembly produce near-complete forms of the SSU and LSU. The SSU comprises 18S rRNA and 33 types of RPs, whereas the LSU comprises 5S, 5.8S, and 25–28S rRNAs and 46–48 types of RPs. The SSU and LSU RPs are highly conserved throughout eukaryotes, with only a few exceptions.
Nucleolar stress response pathway in animal cells
The elaborately regulated biogenesis of ribosomes is disturbed under unhealthy conditions for various reasons, such as nutrient starvation, hypoxia, heat shock, chemical suppression of ribosome biogenesis, genetic impairments of RBFs, and deficiencies of RPs (Mayer and Grummt, 2005; Boulon et al., 2010). In animal cells, such perturbations of ribosome biogenesis, regardless of origin, are mostly accompanied by structural disorders of the nucleolus, although they are not always obvious, and cause a particular type of stress called nucleolar stress or ribosomal stress, which stimulates specific signaling pathways leading to cell-cycle arrest, senescence, or apoptosis (Olausson et al., 2012; James et al., 2014). These stress signaling pathways are classified into two types, p53-dependent and -independent ones (Olausson et al., 2012; James et al., 2014; Russo and Russo, 2017).
The canonical and most extensively studied pathway of animal nucleolar stress response is the p53-dependent pathway. The well-known antitumor transcription factor p53 and its destabilizer MDM2 play a pivotal role in this pathway. In normal conditions, the activity of p53 is suppressed to a low level under the control of MDM2, which acts as a component of E3 ubiquitin ligase and targets p53, leading to the ubiquitination and proteasomal degradation of p53. Upon nucleolar stress, RPs are released from the nucleolus into the nucleoplasm. Several of the RPs released, including RPL5, RPL11, RPL23, and RPS7, bind directly to MDM2 as effectors, thus preventing its action on p53 (Lohrum et al., 2003; Dai and Lu, 2004; Jin et al., 2004; Chen et al., 2007). As a result, p53 becomes stable and active in regulating the expression of genes that are involved in cell-cycle arrest, senescence, and apoptosis (Zhang and Lu, 2009; Deisenroth and Zhang, 2010; Golomb et al., 2014). Genes that are expressed at relatively high levels of p53 activity induce senescence or apoptosis, whereas genes that are expressed at low p53 levels induce cell-cycle arrest (Lai et al., 2007).
In recent years, increasing evidence has indicated that additional mechanisms not involving p53 participate in nucleolar stress response. For instance, RPs released from the nucleolus upon nucleolar stress, such as RPL11 and RPS14, were shown to repress the activity of the oncoprotein transcription factor c-Myc, which is crucial for the expression of many genes involved in cell growth and proliferation, through direct binding to the c-Myc protein and/or controlling microRNA-induced silencing complex (miRISC)-mediated turnover of c-Myc mRNA (Dai et al., 2007; Challagundla et al., 2011; Zhou et al., 2013). It was also reported that in response to nucleolar stress, released RPs can induce cell cycle arrest via regulation of the E2F transcription factor E2F-1 or CDK inhibitors p21Waf1/Cip1 and p27Kip1, independently of p53 (Iadevaia et al., 2010; Donati et al., 2011; Russo et al., 2013).
Effects of the perturbation of ribosome biogenesis in plants
In plants, especially in the model plant Arabidopsis, many ribosome-related mutants, which are impaired in an RBF- or RP-encoding gene, have been isolated and characterized. Most of these mutants exhibit a similar spectrum of phenotypes, including a narrow and pointed leaf shape and retardation of root growth (Byrne, 2009; Horiguchi et al., 2012; Tsukaya et al., 2013; Weis et al., 2015). A severe loss of the adaxial–abaxial polarity of leaves under the genetic background of asymmetric leaves 1 (as1) or as2 is also a notable phenotype that is shared by the ribosome-related mutants (Pinon et al., 2008; Horiguchi et al., 2011; Huang et al., 2014; Matsumura et al., 2016). These common phenotypic features of various ribosome-related mutants imply the existence of a common mechanism that regulates growth and development in response to various perturbations of ribosome biogenesis.
At the subcellular level, structural changes of the nucleolus have been reported for various RBF mutants (Table 1). In most cases, enlargement of the nucleolus was observed in association with excessive accumulation of intermediates of pre-rRNA processing. This nucleolar enlargement is sometimes linked with the development of a nucleolar cavity, i.e., nucleolar vacuolation (Ohbayashi et al., 2011). Moreover, in the Arabidopsis mutant that carries a disrupted nucleolin gene (AtNUC-L1), nucleolar disorganization and decondensation of the rDNA chromatin structure were shown to co-occur (Pontvianne et al., 2007). These findings indicate that perturbation of rRNA biogenesis generally induces structural disorders of the nucleolus in plant cells, as well as in animal cells (Nishimura et al., 2015).
Table 1.
List of Arabidopsis genes that have been shown to participate in the function and/or structural integrity of the nucleolus.
| Gene symbol | AGI code | Protein product | Protein localization | Mutant allele | Mutation type | Mutant effects on | Mutant suppression by sriw1 | References | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Nucleolar structure | Nucleolar functions | Growth and development | ||||||||
| RID2 | At5g57280 | Methyltransferase | Nucleolus | rid2-1 | Base substitution (missense) | Enlargement accompanied by vacuolation | Impairment of pre-rRNA processing | Temperature-dependent defects in leaf blade development and root growth in seedlings, and in callus formation, adventitious root formation and shoot regeneration in tissue culture, and severe loss of leaf abaxial–adaxial polarity in as1 and as2 | Yes | Konishi and Sugiyama, 2003; Ohbayashi et al., 2011, 2017; Shinohara et al., 2014; Matsumura et al., 2016 |
| rid2-2, rid2-3 | T-DNA insertion | — | — | Female gametophyte lethality, maternal sterility, or maternal embryonic lethality | — | Ohbayashi et al., 2011 | ||||
| RID3 | At3g49180 | WD40 repeat protein | Nucleus | rid3 | Base substitution (missense) | Enlargement | Impairment of pre-rRNA processing | Temperature-dependent defects in leaf blade development and root growth in seedlings, and in shoot regeneration in tissue culture | Yes | Tamaki et al., 2009; Shinohara et al., 2014; Ohbayashi et al., 2017 |
| RH10 | At5g60990 | DEAD-box RNA helicase | Nucleolus | rh10-1 | Base substitution (missense) | Enlargement | Impairment of pre-rRNA processing | Temperature-dependent defects in leaf blade development and severe loss of leaf abaxial–adaxial polarity in as1 and as2 | Yes | Matsumura et al., 2016; Ohbayashi et al., 2017 |
| ATNUC-L1 | At1g48920 | Nucleolin | Nucleolus | Atnuc-L1-1 | T-DNA insertion | Disorganization (rDNA heterochromatin decondensation) | Impairment of pre-rRNA processing (blockage of cleavage at least at the P site) and increased transcription of specific rRNA variants | Pleiotropic defects in growth and development, including abnormal leaf morphogenesis, and severe loss of leaf abaxial–adaxial polarity in as1 and as2 | — | Pontvianne et al., 2007, 2010; Matsumura et al., 2016 |
| parl1-1 | 2 bp insertion | — | — | Abnormal vein patterning, pointed narrow leaf, retarded root growth, and reduced fertility | — | Petricka and Nelson, 2007 | ||||
| parl1-2 (Atnuc-L1-2) | T-DNA insertion | Disorganization (rDNA heterochromatin decondensation) | Impairment of pre-rRNA processing and increased transcription of specific rRNA variants | Abnormal vein patterning, pointed narrow leaf, retarded root growth, reduced fertility, and severe loss of leaf abaxial–adaxial polarity in as1 and as2 | — | Petricka and Nelson, 2007; Pontvianne et al., 2010; Matsumura et al., 2016 | ||||
| DOMINO1 | At5g62440 | Small plant-specific protein | Nucleus | domino1 | T-DNA insertion | Enlargement | Inactivation of ribosome biogenesis | Retarded embryogenesis | — | Lahmy et al., 2004 |
| NOF1 | At1g17690 | DUF1253-containing protein | Nucleolus | nof1-1 | T-DNA insertion (in the promoter region) | Enlargement | Increase in rRNA transcription | Embryonic lethality (irregular pattern and/or additional cell division, and lack of cell adhesion) | — | Harscoët et al., 2010 |
| nof1-2 | T-DNA insertion | — | — | Female gametophyte lethality | — | Harscoët et al., 2010 | ||||
| APUM23 | At1g72320 | Pumilio protein | Nucleolus | apum23-1, apum23-2 | T-DNA insertion | Enlargement | Impairment of processing/degradation of pre-rRNAs | Pleiotropic defects in growth and development, including abnormal leaf morphogenesis | — | Abbasi et al., 2010 |
| apum23-3 | Base substitution (missense) | — | Impairment of pre-rRNA processing | Pleiotropic defects in growth and development, including abnormal leaf morphogenesis and severe loss of leaf abaxial–adaxial polarity in as1 and as2 | — | Huang et al., 2014 | ||||
| TOZ | At5g16750 | WD40 repeat protein | Nucleolus | toz | Ds insertion | Enlargement | Impairment of pre-rRNA processing | Embryonic lethality (aberrant cell division) | — | Griffith et al., 2007 |
| ATLa1 | At4g32720 | La motif protein | Nucleoplasm and nucleolar cavity (?) | atla1-1, atla1-2 | T-DNA insertion | Enlargement | Interference with ribosome biogenesis (?) | Embryonic lethality (arrest at the early globular stage) | — | Fleurdépine et al., 2007 |
| ATREN1 | At1g77570 | Heat-shock transcription factor | Nucleolus | atren1 | T-DNA insertion | Enlargement | Interference with ribosome biogenesis (?) | Defects in male gametophyte development | — | Renák et al., 2014 |
Taking together the morphological and cytological characteristics of ribosome-related mutants, we can speculate that perturbations of ribosome biogenesis and the resultant structural disorders of the nucleolus affect several aspects of plant growth and development via a specific signaling pathway. If this is true, then what is the pathway? By analogy with animal cells, it seems reasonable to assume the presence of a nucleolar stress response pathway at work in plant cells. However, as plants have no homologs of p53 or MDM2 (Huart and Hupp, 2013), the plant pathway should not involve a p53- and MDM2-dependent mechanism and might be distinct from the nucleolar stress response pathway of animal cells.
Implication of ANAC082 as a mediator of the nucleolar stress response in plants
Very recently, the important evidence of the nucleolar stress response pathway in plants came from molecular genetic studies of Arabidopsis mutants, root initiation defective 2 (rid2) and suppressor of rid two 1 (sriw1). rid2 is a temperature-sensitive mutant that is impaired in pre-rRNA processing because of a missense mutation in the gene encoding a putative RNA methyltransferase and is characterized phenotypically by severe defects in cell proliferation and a striking enlargement and vacuolation of the nucleolus at high temperatures (Ohbayashi et al., 2011). sriw1 is a rid2 suppressor mutant that was isolated from a mutagenized population of rid2 (Ohbayashi et al., 2017). The sriw1 mutation markedly alleviated the cell proliferation defects of rid2 without relieving the impaired pre-rRNA processing. It was further shown that the sriw1 mutation had the abilities to restore growth and development not only in rid2 but also generally in various ribosome-related mutants, including both RBF mutants and RP mutants, and to confer weak resistance to chemicals that interfere with ribosome biogenesis or ribosomal function. sriw1 was identified as a loss-of-function mutation of the gene encoding ANAC082, which belongs to the plant-specific transcription factor family NAM/ATAF/CUC (NAC). The expression level of ANAC082 was temperature-dependently increased in temperature-sensitive RBF mutants. These findings collectively implicate ANAC082 as a key mediator that works downstream of perturbations of ribosome biogenesis and nucleolar disorders, thus leading to growth defects and developmental alterations in plants; i.e., plant cells are considered to respond to nucleolar stress via a plant-unique signaling pathway mediated by ANAC082.
Possible mechanisms of sensing nucleolar stress in plants
To understand the nucleolar stress response in plants, how plant cells sense perturbed ribosome biogenesis and nucleolar disorders is one of the most critical questions. Among the fragmentary pieces of information available currently regarding the plant pathway of the nucleolar stress response, a clue to one of its early steps can be found in the elevated expression of ANAC082 in RBF mutants (Ohbayashi et al., 2017). Transcriptional regulation and/or post-transcriptional regulation may contribute to this upregulation of ANAC082 expression. With respect to post-transcriptional regulation, the existence of a conserved upstream open reading frame (uORF) in the upstream region of the main ORF in the ANAC082 mRNA is of considerable note (Takahashi et al., 2012). This uORF was demonstrated to have an amino-acid-sequence-dependent, negative effect on the expression of the downstream main ORF (Ebina et al., 2015). Most of the regulatory uORFs studied to date cause ribosome stalling at their termination codon, which impedes the access of ribosomes to the main ORF and often induces nonsense-mediated mRNA decay (Gao and Geballe, 1996; Law et al., 2001; Gaba et al., 2005; Uchiyama-Kadokura et al., 2014). Such uORF-dependent control can be affected by ribosomal defects, as was reported for the expression of uORF-containing genes that encode auxin signaling factors in the Arabidopsis RP mutant rpl24b (Nishimura et al., 2004, 2005; Zhou et al., 2010). Taking these findings into consideration, the uORF of ANAC082 might be a candidate site of nucleolar stress sensing. A possible underlying mechanism is that, under nucleolar stress, because of a shortage of functional ribosomes, imbalance of ribosomal subunits, or some other abnormal situation of ribosomes, the constraint of ANAC082 expression by ribosome stalling at the uORF is loosened, resulting in the activation of ANAC082.
It is also possible that the activity of ANAC082 is regulated via a protein–protein interaction upon nucleolar stress, given the knowledge that NAC transcription factors generally form a homodimer or heterodimer through interaction at the N-terminal NAC domain (Ernst et al., 2004). ANAC082 has a potential of binding to several species of NAC transcription factors, including NAC1, CUP-SHAPED COTYLEDON 2 (CUC2), ANAC103, and VASCULAR-RELATED NAC-DOMAIN (VND) proteins (Yamaguchi et al., 2015). Moreover, ANAC082 is part of the list of transcription factors that can bind to the WWE domain-containing, non-NAC proteins RADICAL-INDUCED CELL DEATH 1 (RCD1), and SIMILLAR TO RCD-ONE 1 (SRO1) (Jaspers et al., 2009). If some of these proteins act as a partner of ANAC082, it is also possible that the partner, instead of ANAC082, is responsible for the process of nucleolar stress sensing.
Multiple NACs in plant cellular stress responses corresponding to multiple roles of p53 in animal cellular stress responses
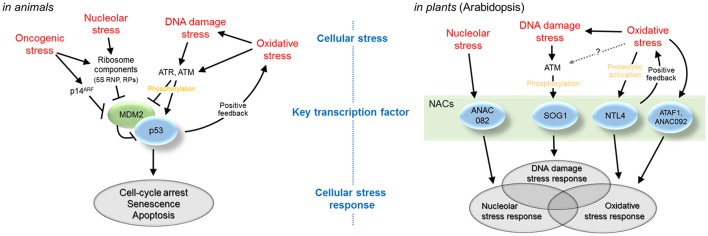
In animal cells, p53 participates as a nodal regulator not only in the nucleolar stress response pathway but also in pathways that respond to several other cellular stresses, such as oncogenic stress, DNA damage stress, and oxidative stress (Serrano et al., 1997; Horn and Vousden, 2007; Hu et al., 2012). Oncogenic stress, which is caused by the inappropriate expression of oncogenes or proto-oncogenes, is transduced to the activation of p53 mainly through the transcriptional upregulation and protein stabilization of the tumor suppressor p14ARF and the consequent inhibition of MDM2 activity by p14ARF (Sherr and Weber, 2000; Gallagher et al., 2006; Chen et al., 2010). Another route connecting oncogenic stress to p53 activation depends on the binding of the 5S ribonucleoprotein complex (5S RNP), consisting of 5S rRNA, RPL5, and RPL11, to MDM2 (Nishimura et al., 2015). In the pathway of DNA damage stress response, sensing DNA double-strand breaks, persisting single-stranded DNA, or stalled replication forks, the PI3K-related protein kinase ATM or ATR is activated and phosphorylates specific sites of p53 and MDM2, which results in the activation and stabilization of p53 (Shiloh, 2001; Maréchal and Zou, 2013). Oxidative stress caused by reactive oxygen species (ROS) induces the activation of p53 via the DNA damage stress response pathway, which is triggered by oxidation damage of DNA and via direct activation of ATM by oxidation (Barzilai and Yamamoto, 2004; Guo et al., 2010). Importantly, the ROS-induced p53 signaling acts toward generating ROS through the downstream effector p66shc, which comprises a positive feedback loop (Migliaccio et al., 1999; Nemoto and Finkel, 2002).
Among the cellular stresses with responses that rely on p53 in animal cells, DNA damage stress and oxidative stress also occur in plant cells. Therefore, as is the case for the nucleolar stress response, the question arises regarding how plant cells respond to these stresses in the absence of p53 homologs. In Arabidopsis, it was demonstrated by molecular genetic studies that the DNA damage stress response is governed by SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1)/ANAC008, a transcription factor of the NAC family (Preuss and Britt, 2003; Yoshiyama et al., 2009). Upon DNA damage stress, the SOG1 protein is hyperphosphorylated in an ATM-dependent manner and thereby activated to regulate gene expression, which resembles the regulatory mechanism of p53 by ATM and ATR in animal cells (Yoshiyama et al., 2013). Based on these findings, SOG1 is often discussed as a functional counterpart of p53, although SOG1, and p53 have no sequence similarity (Yoshiyama, 2015). Oxidative stress induces DNA damage and elicits the DNA damage stress response in plant cells, as well as in animal cells. Moreover, in plants, oxidative stress promotes leaf senescence independently of DNA damage. Several NAC transcription factors, such as ARABIDOPSIS THALIANA ACTIVATION FACTOR 1 (ATAF1)/ANAC002 and ANAC092, have been reported as mediators of this process (Balazadeh et al., 2010; Garapati et al., 2015). Of these, the most notable one is the membrane-bound NAC protein NAC WITH TRANSMEMBRANE MOTIF 1-LIKE 4 (NTL4)/ANAC053, as this transcription factor is activated through oxidative proteolysis by ROS and promotes ROS production, thus representing a positive feedback loop, similar to that of the p53-dependent oxidative stress response pathway in animal cells (Lee et al., 2012, 2014).
In summary, in the context of cellular stress responses, animal cells utilize p53 to regulate responses to nucleolar stress, DNA damage stress, and oxidative stress, while plant cells employ multiple NAC transcription factors for these roles of p53 (Figure 1). Cellular stresses such as nucleolar stress, DNA damage stress, and oxidative stress are both extrinsic and intrinsic to basic cellular activities of ribosome biogenesis, genome replication, and energy metabolism; thus, they are unavoidable in all organisms. To sense and cope with cellular stresses, animals have evolved signaling systems that share a limited number of transcription factors, including p53, as central regulators. In contrast, plants have evolved stress signaling systems that use different transcription factors for different stresses. This strategy seems related to the diversification of transcription factors of particular groups in plants. In this respect, the NAC family has received increasing attention. NAC is one of the largest families of plant transcription factors and includes more than 100 members (Zhu et al., 2012). Many of the NAC-family members have been previously implicated in responses to abiotic and biotic stresses triggered by external causes, such as drought, salt, cold, and pathogen infection (Nakashima et al., 2012; Puranik et al., 2012; Nuruzzaman et al., 2013). Recently discovered NAC-dependent pathways of intrinsic cellular stress responses have added more emphasis to the variety of the NAC transcription factors in plant stress responses.
Figure 1.
NAC-dependent pathways of plant cellular stress responses and p53-dependent pathways of animal cellular stress responses. Animal cells utilize p53 to regulate responses to nucleolar stress, DNA damage stress, and oxidative stress, while plant cells employ multiple NAC transcription factors for these roles of p53.
Future perspectives
In conclusion, recent studies collectively suggest that plant cells use a specific pathway for nucleolar stress response involving ANAC082, a member of the plant-unique transcription factor family NAC. Considering the possible role of uORF in the regulation of ANAC082 expression, the molecular mechanism of this pathway may be quite different from that of the p53-dependent pathway of nucleolar stress response in animal cells; nevertheless, ANAC082 might be regarded as a counterpart of p53 because of their common role as critical transcriptional regulators of the nucleolar stress response. During plant evolution, NAC transcription factors have been highly diversified, and different NAC factors have been assigned to different pathways of stress responses in plants, several of which correspond to the different roles of p53 in the stress responses of animal cells. In this sense, ANAC082 might also be considered a player of the NAC team that is responsible for the tasks carried out by p53 in animal cells.
The ANAC082-dependent pathway underlying the plant nucleolar stress response is a recent concept, and its molecular details remain totally unknown. A particularly important issue is the elucidation of how nucleolar stress is sensed in plants to activate the ANAC082 pathway. Further studies focusing on this problem may lead to the understanding of nucleolar surveillance in plant cells and open new horizons in plant nucleolar biology.
Author contributions
IO and MS conceived this review and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abbasi N., Kim H. B., Park N. I., Kim H. S., Kim Y. K., Park Y. I., et al. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64, 960–976. 10.1111/j.1365-313X.2010.04393.x [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Wu A., Mueller-Roeber B. (2010). Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 5, 733–735. 10.4161/psb.5.6.11694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai A., Yamamoto K. (2004). DNA damage response to oxidative stress. DNA Repair 3, 1109–1115. 10.1016/j.dnarep.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. E. (2009). A role for the ribosome in development. Trends Plant Sci. 14, 512–519. 10.1016/j.tplants.2009.06.009 [DOI] [PubMed] [Google Scholar]
- Challagundla K. B., Sun X. X., Zhang X., DeVine T., Zhang Q., Sears R. C., et al. (2011). Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol. Cell. Biol. 31, 4007–4021. 10.1128/MCB.05810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Shan J., Zhu W. G., Qin J., Gu W. (2010). Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 464, 624–627. 10.1038/nature08820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., et al. (2007). Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037. 10.1038/sj.onc.1210327 [DOI] [PubMed] [Google Scholar]
- Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007). Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 26, 3332–3345. 10.1038/sj.emboj.7601776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. S., Lu H. (2004). Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482. 10.1074/jbc.M403722200 [DOI] [PubMed] [Google Scholar]
- Deisenroth C., Zhang Y. (2010). Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 29, 4253–4260. 10.1038/onc.2010.189 [DOI] [PubMed] [Google Scholar]
- Donati G., Brighenti E., Vici M., Mazzini G., Treré D., Montanaro L., et al. (2011). Selective inhibition of rRNA transcription downregulates E2F-1: a new p53-independent mechanism linking cell growth to cell proliferation. J. Cell Sci. 124, 3017–3028. 10.1242/jcs.086074 [DOI] [PubMed] [Google Scholar]
- Ebina I., Takemoto-Tsutsumi M., Watanabe S., Koyama H., Endo Y., Kimata K., et al. (2015). Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 43, 1562–1576. 10.1093/nar/gkv018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H. A., Olsen A. N., Skriver K., Larsen S., Lo Leggio L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. 10.1038/sj.embor.7400093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurdépine S., Deragon J. M., Devic M., Guilleminot J., Bousquet-Antonelli C. (2007). A bona fide La protein is required for embryogenesis in Arabidopsis thaliana. Nucleic Acids Res. 35, 3306–3321. 10.1093/nar/gkm200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M. S. (2005). Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20, 449–460. 10.1016/j.molcel.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Gallagher S. J., Kefford R. F., Rizos H. (2006). The ARF tumor suppressor. Int. J. Biochem. Cell Biol. 38, 1637–1641. 10.1016/j.biocel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Gao J., Geballe A. P. (1996). Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell Biol. 16, 603–608. 10.1128/MCB.16.2.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapati P., Xue G. P., Munné-Bosch S., Balazadeh S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139. 10.1104/pp.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L., Volarevic S., Oren M. (2014). p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 588, 2571–2579. 10.1016/j.febslet.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Griffith M. E., Mayer U., Capron A., Ngo Q. A., Surendrarao A., McClinton R., et al. (2007). The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell 19, 2246–2263. 10.1105/tpc.106.042697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010). ATM activation by oxidative stress. Science 330, 517–521. 10.1126/science.1192912 [DOI] [PubMed] [Google Scholar]
- Harscoët E., Dubreucq B., Plauqui J.-C., Lepiniec L. (2010). NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS ONE 9:e12829 10.1371/journal.pone.0012829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A. K., Plisson-Chastang C., O'Donohue M. F., Chakraborty A., Gleizes P. E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242. 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Mollá-Morales A., Pérez-Pérez J. M., Kojima K., Robles P., Ponce M. R., et al. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736. 10.1111/j.1365-313X.2010.04457.x [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Van Lijsebettens M., Candela H., Micol J. L., Tsukaya H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 191–192, 24–34. 10.1016/j.plantsci.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Horn H. F., Vousden K. H. (2007). Coping with stress: multiple ways to activate p53. Oncogene 26, 1306–1316. 10.1038/sj.onc.1210263 [DOI] [PubMed] [Google Scholar]
- Hu W., Feng Z., Levine A. J. (2012). The regulation of multiple p53 stress responses is mediated through MDM2. Genes Cancer 3, 199–208. 10.1177/1947601912454734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Kerstetter R. A., Irish V. F. (2014). APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 65, 1181–1191. 10.1093/jxb/ert478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huart A.-S., Hupp T. R. (2013). Evolution of conformational disorder & diversity of the p53 interactome. Biodiscovery 8:5 10.7750/BioDiscovery.2013.8.5 [DOI] [Google Scholar]
- Iadevaia V., Caldarola S., Biondini L., Gismondi A., Karlsson S., Dianzani I., et al. (2010). PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene 29, 5490–5499. 10.1038/onc.2010.279 [DOI] [PubMed] [Google Scholar]
- James A., Wang Y., Raje H., Rosby R., DiMario P. (2014). Nucleolar stress with and without p53. Nucleus 5, 402–426. 10.4161/nucl.32235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P., Blomster T., Brosché M., Salojävi J., Ahlfors R., Vainonen J. P., et al. (2009). Unequally redundant RCD1 and SRO1 mediate stress and development responses and interact with transcription factors. Plant J. 60, 268–279. 10.1111/j.1365-313X.2009.03951.x [DOI] [PubMed] [Google Scholar]
- Jin A., Itahana K., O'Keefe K., Zhang Y. (2004). Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680. 10.1128/MCB.24.17.7669-7680.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Sugiyama M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130, 5637–5647. 10.1242/dev.00794 [DOI] [PubMed] [Google Scholar]
- Lahmy S., Guilleminot J., Cheng C. M., Bechtold N., Albert S., Pelletier G., et al. (2004). DOMINO1, a member of a small plant-specific gene family, encodes a protein essential for nuclear and nucleolar functions. Plant J. 39, 809–820. 10.1111/j.1365-313X.2004.02166.x [DOI] [PubMed] [Google Scholar]
- Lai P. B. S., Chi T. Y., Chen G. G. (2007). Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vivo. Apoptosis 12, 387–393. 10.1007/s10495-006-0571-1 [DOI] [PubMed] [Google Scholar]
- Law G. L., Raney A., Heusner C., Morris D. R. (2001). Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276, 38036–38043. 10.1074/jbc.M105944200 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee H. J., Huh S. U., Paek K. H., Ha J. H., Park C. M. (2014). The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 227, 76–83. 10.1016/j.plantsci.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Lee S., Seo P. J., Lee H. J., Park C. M. (2012). A NAC transcription factor promotes reactive oxygen species production during drought-induced leaf senescence. Plant J. 70, 831–844. 10.1111/j.1365-313X.2012.04932.x [DOI] [PubMed] [Google Scholar]
- Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003). Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587. 10.1016/S1535-6108(03)00134-X [DOI] [PubMed] [Google Scholar]
- Maréchal A., Zou L. (2013). DNA damage sensing by the ATM and ATR kinases. Cold Sprind Harb. Perspect. Biol. 5:a012716. 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., Ohbayashi I., Takahashi H., Kojima S., Ishibashi N., Keta S., et al. (2016). A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 5, 942–954. 10.1242/bio.019109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C., Grummt I. (2005). Cellular stress and nucleolar function. Cell Cycle 4, 1036–1038. 10.4161/cc.4.8.1925 [DOI] [PubMed] [Google Scholar]
- Migliaccio E., Giorgio M., Mele S., Pelicci G., Reboldi P., Pandlfi P. P., et al. (1999). The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402, 309–313. 10.1038/46311 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress response. Biochem. Biophys. Acta 1819, 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nemoto S., Finkel T. (2002). Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295, 2450–2452. 10.1126/science.1069004 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kumazawa T., Kuroda T., Katagiri N., Tsuchiya M., Goto N., et al. (2015). Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 10, 1310–1323. 10.1016/j.celrep.2015.01.055 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Wada T., Okada K. (2004). A key factor of translation, ribosomal protein L24, is involved in gynoecium development in Arabidopsis. Biochem. Soc. Trans. 32, 611–613. 10.1042/BST0320611 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Wada T., Yamamoto K. T., Okada K. (2005). The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17, 2940–2953. 10.1105/tpc.105.036533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M., Sharoni A. M., Kikuchi S. (2013). Role of NAC transcription factors in the regulation of biotic and abiotic stress response in plants. Front. Microbiol. 4:248 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi I., Konishi M., Ebine K., Sugiyama M. (2011). Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 67, 49–60. 10.1111/j.1365-313X.2011.04574.x [DOI] [PubMed] [Google Scholar]
- Ohbayashi I., Lin C. Y., Shinohara N., Matsumura Y., Machida Y., Horiguchi G., et al. (2017). Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29, 2644–2660. 10.1105/tpc.17.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olausson K. H., Nistér M., Lindström M. S. (2012). p53-dependent and -independent nucleolar stress response. Cells 1, 774–798. 10.3390/cells1040774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J. J., Nelson T. M. (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144, 173–186. 10.1104/pp.106.093575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V., Etchells J. P., Rossignol P., Collier S. A., Arroyo J. M., Martienssen R. A., et al. (2008). Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324. 10.1242/dev.016469 [DOI] [PubMed] [Google Scholar]
- Pontvianne F., Abou-Ellail M., Douet J., Comella P., Matia I., Chandrasekhara C., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6:e1001225. 10.1371/journal.pgen.1001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F. J., Echeverria M., et al. (2007). Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 18, 369–379. 10.1091/mbc.E06-08-0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S. B., Britt A. B. (2003). A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 164, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress response. Trends Plant Sci. 17, 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Renák D., Gibalová A., Solcová K., Honys D. (2014). A new link between stress response and nucleolar function during pollen development in Arabidopsis mediated by AtREN1 protein. Plant Cell Environ. 37, 670–683. 10.1111/pce.12186 [DOI] [PubMed] [Google Scholar]
- Russo A., Esposito D., Catillo M., Pietropaolo C., Crescenzi E., Russo G. (2013). Human rpL3 induces G1/S arrest or apoptosis by modulating p21waf1/cip1 levels in a p53-independent manner. Cell Cycle 12, 76–87. 10.4161/cc.22963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A., Russo G. (2017). Ribosomal proteins control or bypass p53 during nucleolar stress. Int. J. Mol. Sci. 18:140. 10.3390/ijms18010140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602. 10.1016/S0092-8674(00)81902-9 [DOI] [PubMed] [Google Scholar]
- Sherr C. J., Weber J. D. (2000). The ARF/p53 pathway. Curr. Opin. Genet. Dev. 10, 94–99. 10.1016/S0959-437X(99)00038-6 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. (2001). ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11, 71–77. 10.1016/S0959-437X(00)00159-3 [DOI] [PubMed] [Google Scholar]
- Shinohara N., Ohbayashi I., Sugiyama M. (2014). Involvement of rRNA biosynthesis in the regulation of CUC1 gene expressionand pre-meristematic cell mound formation during shoot regeneration. Front. Plant Sci. 5:159 10.3389/fpls.2014.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Takahashi A., Naito S., Onouchi H. (2012). BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinfomatics 28, 2231–2241. 10.1093/bioinformatics/bts303 [DOI] [PubMed] [Google Scholar]
- Tamaki H., Konishi M., Daimon Y., Aida M., Tasaka M., Sugiyama M. (2009). Identification of novel meristem factors involved in shoot regeneration through the analysis of temperature-sensitive mutants of Arabidopsis. Plant J. 57, 1027–1039. 10.1111/j.1365-313X.2008.03750.x [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Byrne M. E., Horiguchi G., Sugiyama M., Van Lijsebettens M., Lenhard M. (2013). How do “housekeeping” genes control organogenesis?–unexpected new findings on the role of housekeeping genes in cell and organ differentiation. J. Plant Res. 126, 3–15. 10.1007/s10265-012-0518-2 [DOI] [PubMed] [Google Scholar]
- Uchiyama-Kadokura N., Murakami K., Takemoto M., Koyanagi N., Murota K., Naito S., et al. (2014). Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol. 55, 1556–1567. 10.1093/pcp/pcu086 [DOI] [PubMed] [Google Scholar]
- Weis B. L., Kovacevic J., Missbach S., Schleiff E. (2015). Plant-specific feature of ribosome biogenesis. Trends Plant Sci. 20, 729–740. 10.1016/j.tplants.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Sumeda I., Nagahage P., Ohtani M., Ishikawa T., Uchimiya H., et al. (2015). Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotech. 32, 119–123. 10.5511/plantbiotechnology.15.0208a [DOI] [Google Scholar]
- Yoshiyama K., Conklin P. A., Huefner N. D., Britt A. B. (2009). Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 106, 12843–12848. 10.1073/pnas.0810304106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K. O. (2015). SOG1: a master regulator of the DNA damage response in plants. Genes Genet. Syst. 90, 209–216. 10.1266/ggs.15-00011 [DOI] [PubMed] [Google Scholar]
- Yoshiyama K. O., Kobayashi J., Ogita N., Ueda M., Kimura S., Maki H., et al. (2013). ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 14, 817–822. 10.1038/embor.2013.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H. (2009). Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377. 10.1016/j.ccr.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Roy B., von Arnim A. G. (2010). Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 10:193. 10.1186/1471-2229-10-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hao Q., Liao J. M., Liao P., Lu H. (2013). Ribosomal protein S14 negatively regulates c-Myc activity. J. Biol. Chem. 288, 21793–21801. 10.1074/jbc.M112.445122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Nevo E., Sun D., Peng J. (2012). Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 66, 1833–1848. 10.1111/j.1558-5646.2011.01553.x [DOI] [PubMed] [Google Scholar]