Abstract.
Accurate two-dimensional to three-dimensional (2-D/3-D) registration of preoperative 3-D data and intraoperative 2-D x-ray images is a key enabler for image-guided therapy. Recent advances in 2-D/3-D registration formulate the problem as a learning-based approach and exploit the modeling power of convolutional neural networks (CNN) to significantly improve the accuracy and efficiency of 2-D/3-D registration. However, for surgery-related applications, collecting a large clinical dataset with accurate annotations for training can be very challenging or impractical. Therefore, deep learning-based 2-D/3-D registration methods are often trained with synthetically generated data, and a performance gap is often observed when testing the trained model on clinical data. We propose a pairwise domain adaptation (PDA) module to adapt the model trained on source domain (i.e., synthetic data) to target domain (i.e., clinical data) by learning domain invariant features with only a few paired real and synthetic data. The PDA module is designed to be flexible for different deep learning-based 2-D/3-D registration frameworks, and it can be plugged into any pretrained CNN model such as a simple Batch-Norm layer. The proposed PDA module has been quantitatively evaluated on two clinical applications using different frameworks of deep networks, demonstrating its significant advantages of generalizability and flexibility for 2-D/3-D medical image registration when a small number of paired real-synthetic data can be obtained.
Keywords: 2-D/3-D registration, image-guided procedures, convolutional neural networks, pairwise domain adaptation module
1. Introduction
Given the recent development in medical imaging technologies, image-guided procedures are becoming increasingly popular with reduced invasiveness and postprocedure complications.1 2-D/3-D registration, which aligns the preoperative 3-D data and the intraoperative 2-D data into the same coordinate system, is one of the key enabling technologies of image-guided procedures.2 The modalities of preoperative 3-D data include computed tomography (CT), magnetic resonance imaging, positron emission tomography of patients, and computer-aided design (CAD) models of medical devices, and the intraoperative 2-D data include ultrasound (US) and x-ray. By aligning the 2-D and 3-D data, accurate 2-D/3-D fusion can provide complimentary information for advanced image-guided radiation therapy, radiosurgery, endoscopy, and interventional radiology.3 Figure 1 demonstrates two examples of 2-D/3-D registration: (1) estimation of six degrees of freedom (DoF) pose of a transesophageal echocardiography (TEE) transducer in an x-ray image by registering its CAD model with the x-ray image, which is the key enabling technology for fusing live US and x-ray images for image-guided therapy; (b) registration of spine vertebra in CT and x-ray images, which has a wide range of applications in interventional imaging when spine is a visible object in the imaging field.
Fig. 1.
Examples of 2-D/3-D medical image registration applications, with 2-D x-ray images on the left and 2-D/3-D overlay results on the right. (a) TEE transducer registration: the 3-D CAD model of TEE is overlaid in yellow, and the blue and red cones are the TEE projection target using the ground truth pose and the estimated pose of the TEE transducer. TEE projection target is defined as the four corners of the TEE imaging cone at 60 mm depth. (b) Spine vertebra registration: the CT volume of the spine is overlaid in yellow on 2-D x-ray spine images.
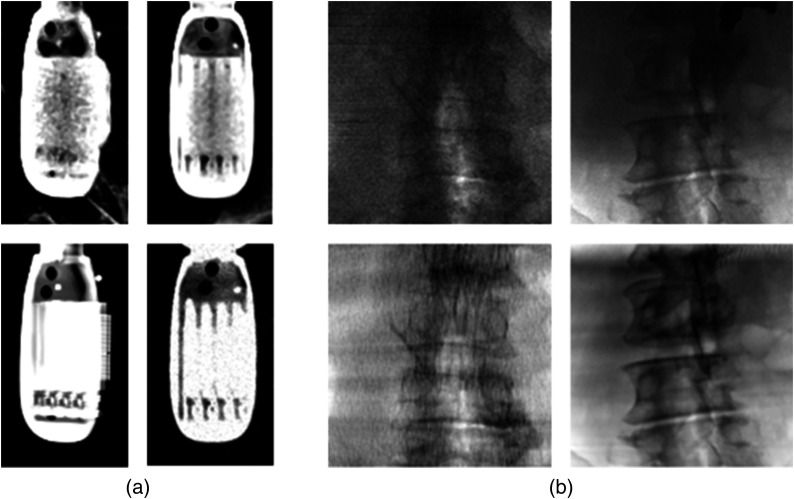
Optimization-based methods have been extensively studied for 2-D/3-D registration in the past decades. In these methods, a simulated x-ray image, referred to as digitally reconstructed radiograph (DRR), is derived from the 3-D x-ray attenuation map by simulating the attenuation of virtual x-rays. An optimizer is employed to maximize an intensity-based similarity measure between the DRR and x-ray images.4,5 Although optimization-based methods are accurate, their computational efficiency is limited since they usually need many iterations of DRR generation and similarity computation. In addition, pose initialization in a close neighborhood of the correct pose is often required due to the small capture range. Recent development in deep learning-based 2-D/3-D registration methods have shown promising improvement in both computational efficiency and capture range compared to conventional optimization-based methods.2,6–8 While deep learning offers large modeling capacity, sufficient training of such a deep model requires a large number of labeled data, which may be expensive or even impractical to collect from clinical procedures, especially for 2-D x-ray images that tend to lack information in depth. Therefore, the aforementioned data-driven approaches are often trained with synthetically generated data before they are tested on real clinical data. Specifically, synthetic data are generated by rendering DRR images from preoperative 3-D data (e.g., CAD models and CT) with random poses to simulate real x-ray images. Even though variations such as background and contrast medium are randomly added to make the appearance of the synthetic data more realistic, domain differences between real and synthetic data still exist. Compared with synthetically generated data, TEE probe in real x-ray images [Fig. 2(a)] are blurred with artifacts in the center of the probe, whereas spine vertebra in the real x-ray images [Fig. 2(b)] have distinct sharpness and noises. In addition, artifacts and other devices could be presented in real clinical x-ray images. These domain differences lead to the domain shifting problem that downgrades the performance of the trained deep models on real clinical data.
Fig. 2.
Comparison of real clinical data and synthetically generated data. (a) Real TEE transducer x-ray (left) versus synthetically generated DRRs (right). (b) Real spine vertebra x-ray (left) versus synthetically generated DRRs (right).
In our preliminary work,6 we exploit the ability to generate a corresponding synthetic data for each labeled real data with the exact same pose parameters and define a pairwise loss measuring the distance between features from paired real and synthetic data to represent the performance gap between the domains. In this paper, we further propose a pairwise domain adaptation (PDA) module to bridge the performance gap. The proposed PDA module is (1) powerful with additional network capacity to model complex domain variances, (2) flexible for different deep learning-based 2-D/3-D registration frameworks, (3) easy to be plugged into any pretrained convolutional neural networks (CNN) model, and (4) trainable with hierarchical pairwise losses using only a few real-synthetic pairs. The proposed PDA module is evaluated on two different deep learning frameworks with two different clinical problems: CNN-based residual regression for TEE transducer registration and deep reinforcement learning (DRL)-based policy learning for spine vertebra registration. The experiment results demonstrate PDA module’s advantage in generalization and superior performance for real clinical data. The proposed PDA module has the potential to benefit any medical imaging problems where paired real-synthetic data can be obtained.
The remainder of the paper is organized as follows. Section 2 provides a literature review of deep learning-based 2-D/3-D registration and deep domain adaptation methods. Section 3 presents the proposed PDA module with the two deep learning-based 2-D/3-D registration problems. Section 4 presents the experimental results, and Sec. 5 concludes the paper.
2. Related Works
2.1. Deep Learning-Based 2-D/3-D Medical Imaging Registration
Recently, deep learning-based methods have shown promising results in 2-D/3-D registration. A CNN-based regression approach was proposed to model the nonconvex mappings between registration parameters and image residual features.2 They further improve the capture range and computational efficiency by modeling the complex mapping problem with a hierarchical CNN regression architecture.6 The reported framework shows the state-of-the-art performance in both accuracy and speed for 2-D/3-D registration of rigid objects. A Markov decision process (MDP) formulation for image registration was introduced for 3-D/3-D registration,7 where an artificial agent is trained to perform registration by taking a series of actions in the MDP environment. The agent-based approach was furthermore extended to 2-D/3-D registration.8 It is widely recognized that successful training of deep learning models often requires a large number of labeled data, which for many applications (e.g., image-guided surgery) are difficult or impractical to collect. The above methods exploit a large number of synthetically generated data for training. However, the domain difference between synthetic and real data often causes a performance gap when applying the trained model on real data. In Ref. 6, we first introduced the PDA to improve the registration accuracy. Pairwise domain distance is employed to fine-tune the last convolutional layer of the pretrained CNN model. Compared with Ref. 6, the proposed PDA module in this paper is a flexible plug-and-play module that can be applied to general network structures. It also provides additional network complexity to better model domain variances. In the experiment section, we evaluate the proposed PDA module with two different 2-D/3-D registration frameworks6,7 and demonstrate that the proposed PDA module can significantly improve the performance of CNN models pretrained with synthetic data.
2.2. Deep Domain Adaptation
Domain adaptation methods have been invested to address the problem of domain shifting by establishing knowledge transfer from the source domain training data to the target domain testing data to extract domain invariant features. In the literature, deep domain adaptation works can be categorized into two directions: discrepancy-based and adversarial-based.9 The strategy of discrepancy-based method is to guide the model training toward the target domain by minimizing a defined domain distance. Deep domain confusion employs maximum mean discrepancy (MMD) as the domain loss for adaptation.10 MMD measures domain difference by calculating the norm of the difference between the means of the two domains. The deep adaptation network minimizes MMD with multiple kernels and expends the MK-MMD loss on multiple layers in CNN.11 Deep correlation alignment method simply matches the mean and covariance of the data distributions of the two domains.12 In contrast, adversarial-based methods aim to encourage domain confusion through an adversarial objective, i.e., a binary domain classifier.13,14 Since these methods generally model the domain distance over source and target data distributions in an unsupervised fashion, they still require a large number of data from both domains. In addition, these methods tend to compare deep features at high level layers of CNN [mostly fully connected (FC) layers]. Since FC layers are more task-specific, employing FC features will limit the flexibility of the adapted model.
3. Methods
3.1. Problem Statement
3.1.1. 2-D/3-D registration problem definition
A common x-ray imaging system is shown in Fig. 3. Assuming that the beam divergence is corrected by the x-ray imaging system and the x-ray sensor has a logarithm static response, the generation of x-ray image can be defined as follows:
| (1) |
where is the intensity of the x-ray image, is the intensity of the x-ray image at point , is the ray from the x-ray source to point , parameterized by , and is the attenuation coefficient of the x-ray. Denoting the x-ray attenuation map of the object to be imaged as , and the 3-D transformation from the object coordinate system to the x-ray imaging coordinate system as , the attenuation coefficient at point in the x-ray imaging coordinate system is
| (2) |
Combining Eqs. (1) and (2), we have
| (3) |
In 2-D/3-D registration problems, is determined by the x-ray imaging system, is provided by the 3-D preoperative data (e.g., CT intensity), and the transformation is to be estimated from the input x-ray image . Note that given , , and , a synthetic x-ray image (often referred to as DRR) can be computed following Eq. (3) using the ray-casting algorithm.15
Fig. 3.
Projection geometry of 2-D/3-D registration.
A rigid-body 3-D transformation can be parameterized by a vector with three in-plane and three out-of-plane transformation parameters.2 In particular, in-plane transformation parameters include two translation parameters, and , and 1 rotation parameter, ; out-of-plane transformation parameters include one out-of-plane translation parameter, , and two out-of-plane rotation parameters, and . The effects of in-plane transformation parameters are approximately 2-D rigid-body transformations while the effects of out-of-plane translation and rotations are scaling and shape changes, respectively.
3.1.2. CNN regression-based 2-D/3-D registration for TEE transducer
TEE and x-ray fluoroscopy are the two major live imaging modalities for image-guided catheter-based interventions. TEE can provide detailed visualization for soft tissue anatomies while x-ray can capture medical devices. Accurate 2-D/3-D registration of TEE transducer from x-ray images can enable advanced image guidance, e.g., fused visualization and joint analysis of anatomy and devices. A CNN regression-based approach was proposed to estimate the transformation parameters .6 Figure 4(a) shows the structure of the CNN regressor. Given an x-ray image of the TEE transducer with ground truth pose parameter and initial estimation of pose parameters , a DRR is rendered at pose , and the CNN regressor is trained to model estimate the pose parameter residual by comparing the x-ray and DRR images. Region-of-interest (ROI) extraction is performed via probabilistic boosting tree (PBT) detectors.5 More specifically, a series of cascaded PBT classifiers are trained to classify the initial in-plane and out-of-plane translation () using Haar-like and rotated Haar-like features. Based on these four parameters, the ROI of the TEE transducer in the input x-ray image can be extracted. Following ROI extraction, normalization is performed to the x-ray and DRR patches. Intensity values of the patches are normalized from to . Background pixels (intensity value in and ) are ignored during normalization. This normalization process is to ensure that the input data to CNN model has consistent brightness and contrast. The x-ray and DRR patches are further resized to pixels. Image residual feature is calculated via subtraction and fed into CNN regressor. The CNN regressor aims to model the mappings between registration parameters and the image residual feature. The CNN regressor has 10 convolutional layers with kernels and incremental feature map numbers . Pooling layers with kernels are added after every two convolutional layers. Following the convolutional layers and pooling layers, there is one FC layer with 1024 neurons, and the last FC layer then outputs the registration parameters. More details of the framework can be found in the preliminary version of this paper.6 To generate synthetic training data, two images are rendered: a DRR image with a random starting pose , and a synthetic x-ray image with ground truth parameter . The image residual feature can be calculated as
| (4) |
Fig. 4.
Problem framework of (a) CNN regression-based 2-D/3-D registration for TEE transducer and (b) DRL-based 2-D/3-D registration for spine vertebra.
Then, the CNN regressor can be trained with the following loss function:
| (5) |
where is the number of training samples, is the CNN weights to be learned, is the output of the CNN regressor with input image residual feature , and denotes Euclidean distance.
3.1.3. DRL-based 2-D/3-D registration for spine vertebra
In image-guided spine surgery, registration of 3-D preoperative CT data and 2-D intraoperative x-ray image can provide valuable assistance such as vertebra localization and device path planning. To address this problem, an MDP agent-based framework was proposed to train the artificial agent, which can iteratively choose the optimal action to recover 6DoF parameters of the target vertebra.7 Specifically, the process of 2-D/3-D registration is formulated as an MDP, where at every time point with a pose , an artificial intelligent agent modeled by a deep neural network observes the x-ray image and DRR rendered with the pose , and produces an action to modify the pose. At a time point , the state is defined by the current transformation . Rewards of actions can be calculated by
| (6) |
where is the ground truth transformation, defines the distance of ground truth transformation and current transformation, and is the new transformation after taking action . In this work, the action set has 12 candidate transformations with for each of the 6DoF parameters. More detailed formulations can be found in Ref. 7. As shown in Fig. 4(b), the CNN architecture has two branches for input x-ray image and DRR image separately. A minmax normalization is applied on the x-ray and DRR images to normalized their intensities to . The input x-ray and DRR images are resized to pixels. Each branch has four convolutional layers with kernels and increasing feature map size . After each convolutional layer, a pooling layer with kernels is added. The convolutional features are then concatenated and fed into four FC layers with decreasing number of neurons . The output of the last FC layer corresponds to the rewards of the 12 candidate actions. Similar to Eq. (5), the training loss is defined by
| (7) |
As shown in Fig. 4, the above two methods are different in the following aspects:
-
1.
The tasks of CNN models are different. In TEE registration, the CNN model outputs the 6DoF transformation parameters; in spine registration, the CNN model outputs 12 rewards for 12 candidate transformations.
-
2.
The CNN architectures are different. In TEE registration, the CNN model takes a single residual image as the input; in spine registration, there are two branches to process x-ray and DRR images separately.
-
3.
The learning methods are different. In TEE registration, the CNN model is trained with supervised learning; in spine registration, the CNN model is trained with reinforcement learning.
In the following sections, we will present the proposed PDA model that can significantly improve the 2-D/3-D registration performance for both applications with a few real data (around 100), despite the differences in CNN architecture, input-output model, and learning methods.
3.2. Pairwise Domain Loss
To handle domain shifting problem between synthetic and real data, typical deep domain adaptation methods focus on unsupervised distribution modeling and require a large number of target domain data in order to model the distribution reasonably well. Another intuitive solution is to fine-tune the CNN model pretrained with the synthetic data using labeled real data. However, with very limited number of labeled real data (tens to hundreds), performance improvement of naive fine-tuning could still be limited without exploiting other priors. In this paper, our aim is to design a domain adaptation method that is suitable for 2-D/3-D registration with very limited real data, by exploiting the fact that paired real and synthetic data can be generated. Specifically, for a real x-ray image with a known ground truth pose for the object to be registered, we can render a DRR image with the same 6DoF pose. As shown in Fig. 2, the image appearance difference (e.g., object appearance, artifacts, noises, and background) is the only difference between the generated real-synthetic image pairs that causes the performance gap. If we consider a CNN model to be a trained feature extractor followed by a regressor/classifier , our aim is to train a domain invariant feature extractor , which has consistent performance over paired real data and synthetic data , and has similar behavior as the pretrained feature extractor over synthetic data
| (8) |
In addition, for a well-trained regressor , the results from real data and synthetic data should be close to ground truth
| (9) |
Thus, to train the domain invariant feature extractor with real-synthetic pairs set , a pairwise domain loss can be defined by
| (10) |
Minimizing forces to extract domain invariant features. To ensure that the adapted feature extractor retains a consistent performance for the original task, we add the pretraining loss over synthetic data as a regularization term
| (11) |
where is a parameter to balance the level of domain adaptation and the performance on the original CNN task. Unlike many deep domain adaptation methods that adapt higher level task-specific FC layers, we focus on convolutional features that (1) can better model appearance difference between domains in image registration problems (Fig. 2) and (2) can be applied across different tasks (e.g., estimation of different pose parameters) using the same training data.
3.3. Pairwise Domain Adaptation Module
We propose a PDA module that can be plugged into CNN models and adds extra network capacity for the purpose of domain adaptation without modifying the weights of the pretrained model. In this way, the pretrained and will remain unchanged and focus on the original CNN tasks, whereas the PDA module will focus on extracting domain invariant features for domain adaptation. The design of the PDA module is demonstrated in Fig. 5(a). The PDA module consists of convolutional layers to transform features extracted from pretrained into domain invariant features that generalize better on real clinical data.
Fig. 5.
(a) Illustration of PDA module plugged into a pretrained CNN model. (b) Illustration of PDA module with basic loss. (c) Illustration of PDA module with multilayer loss ( module).
In the PDA module, we replace the pretraining loss in Eq. (11) with a synthetic feature distance , which is the Euclidean distance between original synthetic feature and transformed synthetic feature [Fig. 5(b)]
| (12) |
where denotes the synthetic dataset and denotes the size of . Together the loss function becomes
| (13) |
This loss term encourages the PDA module to find a balance between two goals: (1) the real feature is transferred to be as close as possible to the corresponding synthetic feature and (2) the synthetic feature largely remains unchanged. Since is independent from the task-specific FC layers, the PDA module can be applied across different tasks using the same training data.
The kernel size and feature map numbers of the convolutional layers in the PDA module are set to be identical with the last convolutional layer in the pretrained CNN model to keep feature map dimension consistent after adaptation. In this paper, we set kernel size to be and feature map size to be 128. In order to train the PDA module, we initialize the convolution kernels as identity matrices. More specifically, for a convolutional layer in PDA module with weights , we set
| (14) |
and set the rest of the weights to be 0. The purpose of identity initialization is to ensure that at the beginning of the training, the PDA module preserves the meaningful input synthetic features. In this way, the training mainly focuses on reducing the domain distance and is easier to converge.
In Fig. 5(c), we further enhance the PDA module by introducing a multilayer loss where pairwise feature distance and synthetic feature distance are calculated for each layer in the PDA module
| (15) |
| (16) |
where and denote the adapted and original feature extractors at layer . The PDA module loss can be defined as
| (17) |
where is the number of convolutional layers in the PDA module, and in all experiments. By introducing multilayer loss, domain distance of the lower layers in the PDA module can be more flexibly modeled. In addition, the weights are updated with gradients calculated in each layer that can reduce the effect of vanishing gradients and leads to a better supervision for the training of the PDA module, similar to resNet.8 In the experiments, we denote the PDA module with multilayer loss as module. The proposed PDA module has the following merits:
-
1.
The direct measurement of domain distance on paired data allows training of the PDA module using a small number of real data, by focusing on the key image appearance differences between domains excluding other factors such as poses. In contrast, previous domain adaptation methods typically employ statistical domain distance measurement, which requires a large number of data from both domains.
-
2.
The pairwise loss allows the domain adaptation to be performed on convolutional layers, which are more correlated to image appearance (i.e., synthetic versus real data). In contrast, previous domain adaptation methods typically only adapt FC layers because the statistical distance measurement is not reliable on high dimensional feature maps.
-
3.
The proposed PDA module is a flexible plug-and-play module that can be applied to general network structures. Since the PDA module is added and trained after the main network is trained, it does not affect the network design and training of the main network.
4. Experiments and Discussion
4.1. Experimental Setup
We evaluated the proposed PDA and modules on two clinical datasets for TEE and spine registration. To pretrain the CNN models, we followed the training procedure in the previous papers6,8 where one million synthetic data were generated with random poses and backgrounds. Stochastic gradient descent16 was employed to update weights with task loss for 100,000 iterations with the batch size of 50. The details of the two datasets are as follows:
-
1.
The TEE dataset consists of 1663 x-ray images. A 3-D CAD model of TEE transducer was used to generate DRR images and synthetic training data. To demonstrate the performance of the PDA module, in this paper, we only focus on the global level CNN regressors for out-of-plane parameters , which are most difficult to estimate from a single 2-D x-ray image. Therefore, the starting pose was set to , and the capture range was and . The performance was evaluated via root mean square error (RMSE) of and .
-
2.
The spine dataset consists of 420 x-ray images with 42 corresponding C-arm CT volumes. We sampled the x-ray images with the primary C-arm angle in , and trained the agent with a capture range of 20 mm for translation parameters error () and 10 deg for rotation parameters error (). The performance was evaluated via the target registration error (TRE), computed as the RMSE of the location of seven landmarks on a chosen spine vertebra, and the error rate was measured using the criteria as TRE > 10 mm, following Ref. 8. The ground truth was provided by the calibration of the C-arm system.
4.2. Performance Analysis
We first conducted property analysis of the PDA module using the TEE data. Figure 6 shows the task loss , feature distance , and synthetic feature distance on testing data during training. Two randomly selected sequences with around 100 image frames were used for domain adaptation. The feature distance on testing data reduces during training, demonstrating that using the direct supervision provided by the pairwise domain loss, the PDA module can be effectively trained with a small number of data to reduce domain distance on unseen testing data without noticeable overfitting. The synthetic feature distance starts from 0 due to the identity initialization of PDA module and stays at a small value during training, showing that the PDA module can preserve the feature of synthetic data while adapting the feature of real data toward that of the corresponding synthetic data. The training curves also demonstrate that the feature distance and task loss are strongly correlated (i.e., they are reduced in parallel), which indicates that the proposed pairwise domain distance is an effective domain distance measurement, and that minimizing it can effectively improve model generalization on real data.
Fig. 6.
Training feature distance and synthetic feature distance , testing feature distance and testing loss of the proposed approach on TEE dataset over iterations.
In addition, we compared the proposed transfer learning method with fine-tuning of the CNN model using task loss on real data. Fine-tuning is to retrain the pretrained CNN model with a small learning rate. In comparison, the proposed transfer learning method is to fix the pretrained CNN model and insert the PDA module after the convolutional layers. Other domain adaptation methods reported in the literature using statistics-based or adversarial-based domain distance require a large number of data and cannot be applied to our problems with limited data. Table 1 shows the performance comparison of module and fine-tuning method using an increasing number of training data. As shown in Table 1, the RMSE of module reduces rapidly when the data number is still relatively small and using around 100 data is sufficient to train the module effectively. In comparison, the RMSE of fine-tuning method reduces slowly when the data number is less than 400 and only becomes comparable with that of when the number of data is increased to 800. This shows that fine-tuning using real data requires a relatively large number of data (i.e., ) to achieve its optimal performance, whereas the proposed module can effectively transfer the model to the target domain using a much smaller number of data (i.e., ).
Table 1.
Performance (RMSE) comparison of module and fine-tune with different number of training data.
| Number of sequence | 1 | 2 | 4 | 6 | 8 | 10 | 12 |
|---|---|---|---|---|---|---|---|
| Number of real x-ray images | 64 | 97 | 194 | 296 | 412 | 647 | 806 |
| M2. fine-tuning using task loss | N/A | 6.87 | 6.28 | 5.84 | 5.53 | 4.86 | 4.00 |
| M5. module | 6.80 | 4.59 | 4.45 | 4.30 | 4.16 | 4.04 | 3.96 |
4.3. Evaluation of the Proposed Methods
In this section, we tested two baseline methods and three proposed methods: M1: baseline CNN trained purely on synthetic data without domain adaptation; M2: fine-tuning of the CNN model on real-data using the task loss;17 M3: fine-tuning of the CNN model on real-data using the pairwise domain loss [Eq. (11)]; M4: PDA module [Fig. 5(b)]; M5: module [Fig. 5(c)]. For PDA and modules, three convolutional layers were used. In addition, from performance analysis in the previous section, it is shown that domain adaptation using 100 to 150 real data can already achieve close-to-optimal performance. Thus, for the TEE dataset, we randomly sampled two sequences with in total x-ray images for domain adaptation. For the spine dataset, we randomly selected 140 x-ray images for domain adaptation. Since the real x-ray data are limited and unevenly distributed in both datasets, to better evaluate the proposed methods and compare with the existing methods, we employ the rest of the real data for testing. We update the weights for 40,000 iterations to guarantee that the training is converged and select the model at the end of the training to test the performance. The test was repeated three times to crossvalidate the proposed methods.
The results for TEE and spine are summarized in Tables 2 and 3, respectively. First, we compared the performance with and without the proposed pairwise domain loss. Comparing M2 and M3 shows that by using the pairwise domain loss, the RMSE in TEE registration was reduced by 18.63% (i.e., from 6.87 deg to 5.59 deg), and the error rate and mean TRE in spine registration were reduced by 11.39% (i.e., from 15.71% to 13.92%) and 1.76% (i.e., from 6.80 to 6.68 mm), respectively. This demonstrates the effectiveness of the pairwise domain loss on domain adaptation using a small number of paired data from both domains.
Table 2.
Quantitative results of the proposed PDA module on the problem of CNN regression-based 2-D/3-D registration for TEE transducer.
| Method | RMSE (deg) |
|---|---|
| M1. baseline CNN w.o. adaptation | 7.60 |
| M2. fine-tuning using task loss | 6.87 |
| M3. fine-tuning using pairwise domain loss | 5.59 |
| M4. PDA module | 4.79 |
| M5. module | 4.59 |
Note: The best performance is highlighted in bold.
Table 3.
Quantitative results of the proposed PDA module on the problem of DRL-based 2-D/3-D registration for spine vertebra.
| Method | Error rate () | Mean TRE (mm) |
|---|---|---|
| M1. baseline CNN w.o. adaptation | 16.07% | 7.45 |
| M2. fine-tuning using task loss | 15.71% | 6.80 |
| M3. fine-tuning using pairwise domain loss | 13.92% | 6.68 |
| M4. PDA module | 12.26% | 5.93 |
| M5. module | 11.20% | 5.65 |
Note: The best performance is highlighted in bold.
The comparison between M3 and M4 shows that the PDA module further improves the domain adaptation performance. In particular, the RMSE for TEE registration was reduced by 14.31% (i.e., from 5.59 deg to 4.79 deg), and the error rate and mean TRE for spine registration was reduced by 11.93% (i.e., from 13.92% to 12.26%) and 11.23% (i.e., from 6.68 to 5.93 mm), respectively. This is due to the extra modeling power provided by the PDA module solely for domain adaptation purpose. In addition, the result of M5 shows that the module further improves domain adaptation performance with a multilayer loss in Eq. (17), which shows that the multilayer loss can better model domain distance in the underlying layers and leads to a better supervision for the domain adaptation training. When comparing M5 with M2, the RMSE for TEE registration was reduced by 33.19% (i.e., from 6.87 deg to 4.59 deg), and the error rate and mean TRE in spine registration was reduced by 28.71% (i.e., from 15.71% to 11.20%) and 16.91% (i.e., from 6.80 to 5.65 mm). This demonstrates the proposed has significant improvement over the baseline method fine-tuning using task loss. In summary, the proposed pairwise domain loss and module are shown to be effective to improve generalization of the deep learning-based 2-D/3-D registration methods on real clinical data.
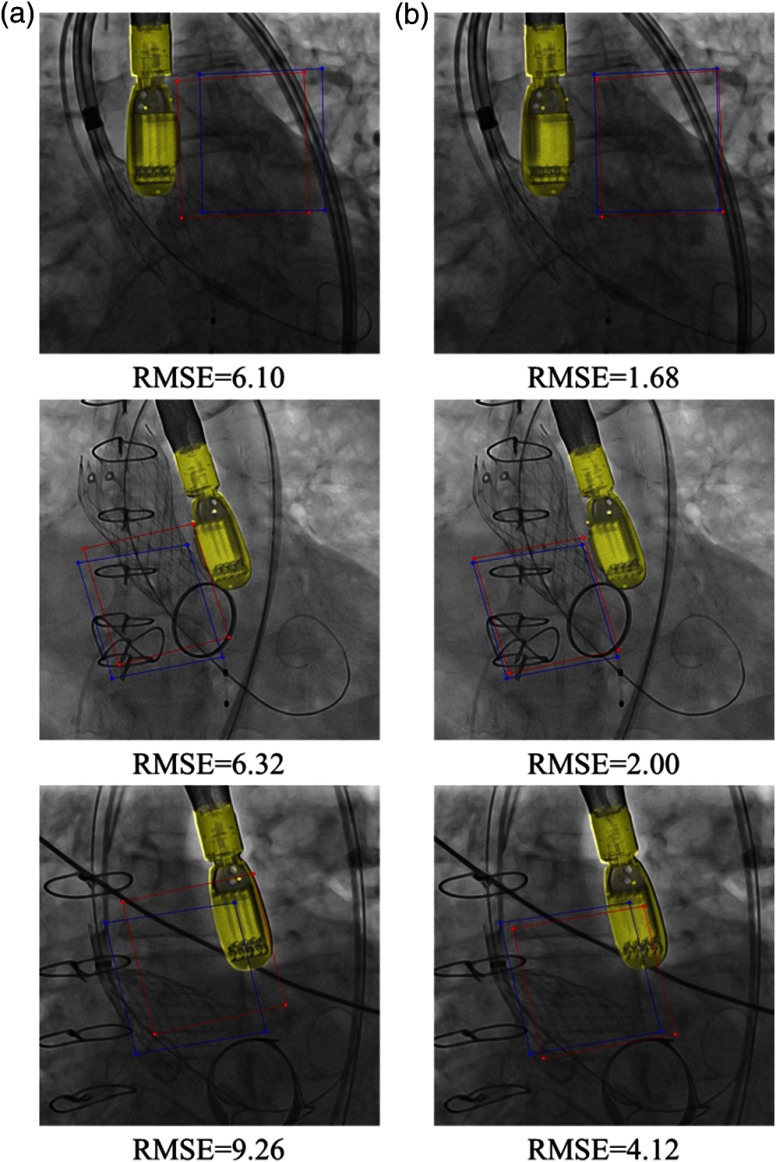
Samples of qualitative results of module on TEE and spine data are shown in Figs. 7 and 8, respectively. Figure 7 shows that the accuracy of TEE transducer pose estimation is significantly improved after applying module. Figure 8 shows that without the module, the agent could register the spine vertebra in 3-D CT with a wrong vertebra in the x-ray image, due to the appearance difference in synthetic training data and the real testing x-ray image. In comparison, with the module, the agent successfully registers the spine vertebra.
Fig. 7.
Example results of TEE registration with (a) original CNN model and with (b) module.
Fig. 8.
Example results of spine vertebra registration with (a) original CNN model and (b) with module. The blue and red crosses are the target and estimated vertebra center, respectively.
5. Conclusion
In this paper, we presented a PDA module to tackle the domain shifting problem for CNN-based 2-D/3-D registration. A pairwise domain loss was proposed to effectively model domain difference between synthetic generated pretraining data and real clinical data. In addition, a PDA module was proposed to learn domain invariant features using only a few paired real and synthetic data. The proposed PDA module was evaluated on two different 2-D/3-D registration problems, demonstrating its advantages in generalization and flexibility for clinical applications. The proposed PDA module can be plugged into any pretrained CNN models and has the potential to benefit any medical imaging problem where a small number of paired real-synthetic data can be obtained.
Biographies
Jiannan Zheng received his BSc degree in electrical engineering from Xi’an Jiaotong University, China, in 2010, and his MSc degree in mechanical engineering from Concordia University, Canada, in 2012. He is currently pursuing his PhD with the University of British Columbia, Canada. His current research interest includes medical imaging, deep learning, and computer vision. The work presented in this paper was done during his internship at Siemens Healthineers in Princeton, New Jersey, USA.
Shun Miao received his BS degree from Zhejiang University, China, in 2009, his MS degree from North Carolina State University in 2010, and PhD from the University of British Columbia, Canada, in 2016. He is a senior research scientist at Siemens Healthineers, Princeton, New Jersey, USA. His research interests include biomedical image processing and analysis, and their applications in image-guided interventions.
Z. Jane Wang received her BS degree from Tsinghua University, China, in 1996, and her MS and PhD degrees from the University of Connecticut in 2000 and 2002, respectively, all in electrical engineering. She was a research associate with the Electrical and Computer Engineering Department, University of Maryland, College Park. Since 2004, she has been with the Department Electrical and Computer Engineering, the University of British Columbia, Canada, where she is currently a full professor. Her research interests include statistical signal processing, machine learning, and medical imaging. She has been an associate editor of the IEEE Signal Processing Magazine, the IEEE Transactions on Signal Processing, and the IEEE Transactions on Circles and Systems II.
Rui Liao is a senior key expert/principal scientist at Siemens Healthineers, Princeton, USA. She obtained her master’s degree from Nanyang Technological University, Singapore, in 2000, and her PhD from Duke University, USA, in 2004, both in electrical engineering. Her research interests are in the broad area of biomedical imaging, statistical signal processing, image-guided interventions, multimodality image fusion, and augmented reality. She is a senior IEEE member and has been an associate editor of the IEEE Signal Processing Magazine and on the board of IEEE Multi-Media Signal Processing Technical Committee.
Disclosures
The authors declare that they have no conflict of interest. The article does not contain any studies with human participants or animals performed by any of the authors. This article does not contain patient data.
Disclaimer
This feature is based on research and is not commercially available. Due to regulatory reasons its future availability cannot be guaranteed.
References
- 1.Comaniciu D., et al. , “Shaping the future through innovations: From medical imaging to precision medicine,” Med. Image Anal. 33, 19–26 (2016). 10.1016/j.media.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 2.Miao S., Wang Z. J., Liao R., “A CNN regression approach for real-time 2D/3D registration,” IEEE Trans. Med. Imaging 35(5), 1352–1363 (2016). 10.1109/TMI.2016.2521800 [DOI] [PubMed] [Google Scholar]
- 3.Markelj P., et al. , “A review of 3D/2D registration methods for image-guided interventions,” Med. Image Anal. 16(3), 642–661 (2012). 10.1016/j.media.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 4.Gao G., et al. , “Registration of 3D trans-esophageal echocardiography to x-ray fluoroscopy using image-based probe tracking,” Med. Image Anal. 16(1), 38–49 (2012). 10.1016/j.media.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 5.Sun S., et al. , “Towards automated ultrasound transesophageal echocardiography and x-ray fluoroscopy fusion using an image-based co-registration method,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Springer, pp. 395–403 (2016). [Google Scholar]
- 6.Zheng J., Miao S., Liao R., “Learning CNNS with pairwise domain adaption for real-time 6dof ultrasound transducer detection and tracking from x-ray images,” in Int. Conf. on Medical Image Computing and Computer-Assisted Intervention, Springer, pp. 646–654 (2017). [Google Scholar]
- 7.Liao R., et al. , “An artificial agent for robust image registration,” in Association for the Advancement of Artificial Intelligence (AAAI), pp. 4168–4175 (2017). [Google Scholar]
- 8.Miao S., et al. , “Dilated FCN for multi-agent 2D/3D medical image registration,” (2017).
- 9.Csurka G., “Domain adaptation for visual applications: a comprehensive survey,” arXiv:1702.05374 (2017).
- 10.Tzeng E., et al. , “Deep domain confusion: maximizing for domain invariance,” arXiv:1412.3474 (2014).
- 11.Long M., et al. , “Learning transferable features with deep adaptation networks,” in Int. Conf. on Machine Learning, pp. 97–105 (2015). [Google Scholar]
- 12.Sun B., Saenko K., “Deep coral: correlation alignment for deep domain adaptation,” in Computer Vision–ECCV 2016 Workshops, pp. 443–450, Springer; (2016). [Google Scholar]
- 13.Ganin Y., et al. , “Domain-adversarial training of neural networks,” J. Mach. Learn. Res. 17(59), 1–35 (2016). 10.1007/978-3-319-58347-1_10 [DOI] [Google Scholar]
- 14.Tzeng E., et al. , “Adversarial discriminative domain adaptation,” in NIPS Workshop on Adversarial Training (WAT), (2017). [Google Scholar]
- 15.Kruger J., Westermann R., “Acceleration techniques for GPU-based volume rendering,” in Proc. of the 14th IEEE Visualization 2003 (VIS ’03), Vol. 38, IEEE Computer Society; (2003). 10.1109/VISUAL.2003.1250384 [DOI] [Google Scholar]
- 16.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, pp. 1097–1105 (2012). [Google Scholar]
- 17.Yosinski J., et al. , “How transferable are features in deep neural networks?” in Advances in Neural Information Processing Systems, pp. 3320–3328 (2014). [Google Scholar]