Summary
During meiosis, the formation of crossovers (COs) generates genetic variation and provides physical links that are essential for accurate chromosome segregation. COs occur in the context of a proteinaceous chromosome axis. The transcriptomes and proteomes of anthers and meiocytes comprise several thousand genes and proteins, but because of the level of complexity relatively few have been functionally characterized. Our understanding of the physical and functional interactions between meiotic proteins is also limited. Here we use affinity proteomics to analyse the proteins that are associated with the meiotic chromosome axis protein, ASY1, in Brassica oleracea anthers and meiocytes. We show that during prophase I ASY1 and its interacting partner, ASY3, are extensively phosphorylated, and we precisely assign phosphorylation sites. We identify 589 proteins that co‐immunoprecipitate with ASY1. These correspond to 492 Arabidopsis orthologues, over 90% of which form a coherent protein–protein interaction (PPI) network containing known and candidate meiotic proteins, including proteins more usually associated with other cellular processes such as DNA replication and proteolysis. Mutant analysis confirms that affinity proteomics is a viable strategy for revealing previously unknown meiotic proteins, and we show how the PPI network can be used to prioritise candidates for analysis. Finally, we identify another axis‐associated protein with a role in meiotic recombination. Data are available via ProteomeXchange with identifier PXD006042.
Keywords: meiosis, chromosome axis, phosphorylation, LC‐MS/MS, protein–protein interaction, Brassica oleracea, Arabidopsis thaliana
Significance statement
The proteinaceous meiotic chromosome axis is critically important in coordinating meiotic recombination and progression yet due to the level of complexity our understanding of the physical and functional protein interactions involved in this process is limited. Here we immunotarget the Brassica ASY1 protein to analyse the protein network associated with the axis during prophase I, identifying phosphosites in ASY1 and its interacting partner ASY3, revealing a novel axis protein and providing other insights into plant meiosis.
Introduction
During meiosis, homologous recombination (HR) generates crossovers (COs) that provide genetic variation and promote accurate chromosome segregation at the first meiotic division. The HR pathway has been studied extensively in Saccharomyces cerevisiae, and is thought to be broadly similar in plants (Osman et al., 2011). HR occurs within the context of profound changes in chromosome organization (Kleckner, 2006). Following replication, sister chromatids are linked by the cohesin complex (Haering and Jessberger, 2012). At the leptotene stage of prophase I, the sister chromatids become organized into linear looped arrays that are conjoined at the loop bases by a proteinaceous axis running along their length. At zygotene, the pairs of homologous chromosomes begin to align and become tightly linked by the synaptonemal complex (SC). This is a highly conserved tripartite structure comprising the chromosome axes with transverse filament (TF) proteins bridging the region between the axes (Page and Hawley, 2004). In many organisms, including plants, mutations leading to defects in axis/SC proteins often have a profound effect on CO formation, whereas in turn recombination pathway mutants can disrupt chromosome morphogenesis (Couteau et al., 1999; Grelon et al., 2001; Armstrong et al., 2002; Li et al., 2004; Higgins et al., 2005; Ferdous et al., 2012).
CO formation is highly coordinated, such that chromosome pairs receive at least one, termed the ‘obligate’ CO (Jones and Franklin, 2006). CO designation is thought to occur early in prophase I, and reduces the probability that another CO will occur in an adjacent region, a phenomenon known as CO interference (reviewed in Berchowitz and Copenhaver, 2010). Precisely how these outcomes are achieved remains to be fully elucidated. Nevertheless, proteins associated with the chromosome axis and SC clearly play an important role (Zickler and Kleckner, 2016). The most extensively studied plant meiotic chromosome axis protein is ASY1 (PAIR2 in rice). Arabidopsis asy1 mutants fail to synapse and have severely reduced CO formation (Ross et al., 1997). In the absence of ASY1, the DMC1 recombinase fails to become stably established on the chromosomes, with the result that interhomologue recombination is severely compromised (Sanchez‐Moran et al., 2007). Although not required for axis formation per se, ASY1 association with the chromatin is concurrent with axis morphogenesis. Immunolocalization of male meiocytes indicates that it first appears as punctate foci in G2 before progressing to a more linear signal along the entire length of the chromosome axes by leptotene (Armstrong et al., 2002). ASY1 remains detectable throughout prophase I, but remodelling of the axis by the AAA+ ATPase, PCH2, during zygotene appears to progressively deplete it from the axis, such that its signal is more obviously associated with the chromatin loops, a process necessary for the normal extension of the SC and the patterned formation of COs (Lambing et al., 2015). Taken together, these studies indicate that ASY1 plays important roles in the coordination of axis/SC morphogenesis and recombination to produce the meiosis‐specific bias that favours interhomologue recombination and the maturation of CO‐designated recombination intermediates.
To date, insight into meiosis in plants has largely derived from mutant analysis of individual genes (Mercier et al., 2015), identified either through sequence conservation with other species or from mutant or suppressor genetic screens (De Muyt et al., 2009; Crismani et al., 2012; Girard et al., 2014). Global approaches to identify plant meiotic genes and proteins have also been adopted. The transcriptomes of developing anthers undergoing meiosis and of isolated meiocytes have been analysed using microarrays and RNAseq (Chen et al., 2010; Tang et al., 2010; Aya et al., 2011; Deveshwar et al., 2011; Libeau et al., 2011; Yang et al., 2011 and Dukowic‐Schulze et al., 2014). These studies reveal a highly complex picture, identifying in the order of 1000–2000 meiotically implicated genes. Moreover, the fact that the relationship between mRNA transcription and the cellular level of the corresponding proteins is nonlinear, the possibility of alternatively spliced meiotic protein variants (Kalsotra and Cooper, 2011; Schmid et al., 2013; Sprink and Hartung, 2014 and Wang et al., 2014), plus evidence from budding yeast and other species that post‐translational modifications of meiotic proteins play a key role in their function (for example, Rockmill and Roeder, 1991; Lin et al., 2010; Attner et al., 2013), increases the complexity still further. Proteomic studies present a similarly complex picture (Zhang et al., 2017). Analysis of the proteome and phosphoproteome of Oryza sativa (rice) anthers identified 4984 proteins and 3203 phosphoproteins associated with early anther development and meiosis (Ye et al., 2015), whereas a further study focusing on rice meiocytes identified 1316 proteins (Collado‐Romero et al., 2014).
Here we aimed to reduce complexity by using affinity‐based proteomics to immuno‐target the key meiotic axis protein, ASY1, to enrich for associated protein complexes using Brassica oleracea, which we have previously shown can be used to provide an enriched source of meiotic tissue for proteomic analysis (Sánchez‐Morán et al., 2005; Osman et al., 2009). We anticipated that this strategy might also begin to reveal the physical interactions that occur between proteins during prophase I of meiosis. We identify the BoASY1 co‐immunoprecipitating proteins and show that their Arabidopsis counterparts form a coherent protein–protein interaction (PPI) network that, importantly, can be used to prioritize candidates for verification of a meiotic role. We present ICU2, the DNA polymerase α subunit, as proof of principle of this approach. We also describe the discovery of another axis‐associated protein with an apparent role in meiotic recombination and identify multiple phosphorylation sites in BoASY1 and its interacting partner, BoASY3, providing further insights into meiosis in higher plants.
Results
Identification of BoASY1 co‐immunoprecipitating proteins
We carried out co‐immunoprecipitation (co‐IP) of ASY1 from B. oleracea to enrich for associated protein complexes (Figure 1a). Anthers (n = 200) were used either intact or their contents were extruded to further enrich for meiotic cells (hereafter, these samples are referred to as ‘anthers’ or ‘meiocytes’, respectively). Proteins were extracted under non‐denaturing conditions to preserve meiotic complexes. With the high level of sequence identity between BoASY1 and AtASY1 (83.6%) we could target BoASY1 using an anti‐AtASY1 antibody (Figure S1; Armstrong et al., 2002). Parallel control co‐IPs were carried out using non‐specific IgG. Proteins were analysed by in‐solution mass spectrometry (MS) and identified using a combined database comprising Brassica rapa sequences (Wang et al., 2011) and Brassica sequences obtained from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov) in 2010. Putative orthologues in Arabidopsis thaliana were identified using the best blastp score. Five anther and four meiocyte data sets were collected. ASY1 was identified in all data sets with 55 peptides in total and up to 75% sequence coverage (Figure S2a), confirming that targeting was successful. In addition, each data set contained several hundred BoASY1 sample‐specific proteins. These ranged from proteins present in all data sets and identified by a relatively large number of unique peptides to those that appeared only once with two unique peptides. To obtain an indication of protein reliability, the raw data from all experiments were searched together and peptide/protein label‐free quantification was carried out using the in‐house tool peakjuggler (unpubl. data; Andersen et al., 2017), to determine peak area. Six of the nine data sets were obtained using three technical replicates of each sample, allowing the statistical significance of proteins to be determined using limma analysis (Smyth Gordon, 2004). Proteins showing a fold‐change of ≥5 in sample relative to control and P < 0.01 in at least one data set were considered significant in label‐free quantification. In addition, to avoid inadvertently excluding any meiotically relevant proteins, we decided to retain all ASY1 sample‐specific proteins satisfying the minimum identification threshold of two peptides while excluding all proteins identified in any of the control samples. This also allowed us to consider data from the three remaining data sets that lacked replicates. Any proteins that were accepted purely on this qualitative basis were considered less reliable than the quantitatively significant group. Nevertheless, our decision to retain them appeared justified when several were subsequently confirmed as having a meiotic role (see below). Details of both sets of accepted proteins are presented in Table S1.
Figure 1.

Identification of BoASY1 co‐immunoprecipitating proteins.
(a) Summary of workflow for co‐immunoprecipitation (co‐IP) experiments. (b) Numbers of ASY1 sample‐specific proteins identified in Brassica meiotic tissues, with corresponding numbers of putative Arabidopsis orthologues in parentheses. (c) Molecular function of putative Arabidopsis orthologues indicated by gene ontology classification. (d) ASY1 co‐IP network, with nodes representing proteins and edges representing interactions. ASY1 (green) and cluster of cohesin, histone and replication‐related proteins (orange) are highlighted. (e) Detailed view of cluster. Proteins are named according to TAIR.
The group taken forward for further analysis therefore comprised 589 Brassica proteins corresponding to 492 Arabidopsis gene loci. Note that all but one Brassica protein could be assigned a putative Arabidopsis orthologue; the discrepancy between the number of Brassica and Arabidopsis proteins is explained partly by the database containing sequences from several different Brassica species and partly by an ancient Brassicaceae lineage‐specific whole‐genome triplication event (Liu et al., 2014; Parkin et al., 2014), such that in some cases several Brassica proteins indicate the same Arabidopsis orthologue. As expected, there was some overlap between anther and meiocyte data sets, with 421 of the total 589 Brassica proteins (71.5%) identified in both tissue types (336 of 492 for Arabidopsis; Figure 1b); however, 125 Brassica proteins (21.2%) were identified only in meiocytes despite ASY1 being detected equally well in both tissues (117 proteins, 23.8%, for Arabidopsis).
Chromosome axis and SC‐associated proteins co‐immunoprecipitate with ASY1
Gene ontology (GO) classification of the 492 Arabidopsis orthologues using The Arabidopsis Information Resource website (TAIR, https://www.arabidopsis.org) indicated that the group of ASY1 co‐IP proteins covered a range of molecular functions (Figure 1c). Analysis of GO enrichment relative to the Arabidopsis genome was conducted using panther accessed through the GO consortium website. Further analysis was carried out using the 453 orthologues identified from meiocytes. In both cases, a large number of GO terms were found to be enriched, but notable amongst the Biological Process terms showing the highest fold enrichment were several relating to DNA processing and nucleus organisation (Table S2a), and ‘DNA‐dependent ATP‐ase activity’ was one of the most highly enriched terms for Molecular Function (Table S2b). Several large protein complexes and functional pathways or families were well represented in the ASY1 co‐IP data, so where appropriate we used a combination of the KEGG pathway database (http://www.kegg.jp/kegg/pathway.html) and examination of the relevant literature to group Arabidopsis orthologues accordingly (Table S2).
We identified 12 proteins with a prior confirmed meiotic role in Arabidopsis, including several axis and SC‐associated proteins (Table S2). From the cohesin complex we identified sub‐units SMC1, SMC3 and SCC3 and one of the five Arabidopsis SPO76 cohesin cofactor proteins, PDS5C (Chelysheva et al., 2005; Lam et al., 2005; Pradillo et al., 2015). The SC transverse filament protein, ZYP1a, and the condensin I subunit, CAP‐D2, were also detected (Higgins et al., 2005; Smith et al., 2014), as were the axis protein, ASY3, and PCH2, an AAA+ ATPase with a role in prophase I axis remodelling, both of which we characterized and published during the course of this study (Ferdous et al., 2012; Lambing et al., 2015). Given the functional relationship of the HR pathway and the developing axis and SC, it was encouraging that several meiotic recombination proteins immunoprecipitated with ASY1, notably PRD3, required for DNA double‐strand break (DSB) formation (De Muyt et al., 2009), and the recombinase DMC1 (Klimyuk and Jones, 1997; Doutriaux et al., 1998). Finally, we identified two peptides of the CDK1 homologue, CDKA;1, previously implicated as having a role in meiotic progression (Cromer et al., 2012). Most of the previously confirmed meiotic proteins were identified either from both tissue types or solely from meiocyte samples; however, CAP‐D2 was identified only from intact anthers.
Other proteins that have (or are predicted to have) a close association with chromatin were present in the ASY1 co‐IP data (Table S2), including proteins involved in DNA replication and repair, chromatin remodelling proteins, putative transcription factors and regulators, and histone proteins. There were several proteins implicated in the RNA‐dependent DNA methylation (RdDM) pathway, including AGO4 (Table S2). Argonaute proteins have been shown to have important pre‐meiotic and meiotic roles in a range of organisms, including several plant species (Nonomura et al., 2007; Olmedo‐Monfil et al., 2010; Singh et al., 2011; Oliver et al., 2014, 2016; Liu and Nonomura, 2016).
Twenty 26S proteasome and 11 ubiquitination‐related proteins were identified (Table S2), suggesting a close association between these proteins and the meiotic chromosome axis. In animals and yeast, the importance of the ubiquitin–proteasome system (UPS) in regulating key aspects of meiosis, such as recombination and meiotic progression, is well established (Bose et al., 2014), and is now also beginning to be elucidated in plants (Wang and Yang, 2006; Zhao et al., 2006; He et al., 2016).
ASY1 co‐IP proteins form a coherent protein–protein interaction network
As the ASY1 co‐IP proteins covered a wide range of protein types and GO terms, we carried out network analysis to determine whether they were predicted to interact based on existing data in the public domain. We used an open‐source database of known and predicted protein interactions: STRING (Szklarczyk et al., 2015). The STRING network was created using the 492 putative Arabidopsis orthologues of BoASY1 co‐IP proteins (Table S1). The network was visualized using cytoscape (Appendix S1; Figure 1d): 92.7% of proteins (456) formed a single network, with relatively few ‘orphans’ (36), suggesting that they had immunoprecipitated as a coherent group, providing further evidence that the co‐IP approach was successful. It is worth noting that six proteins in the ‘orphans’ group lacked annotation or were otherwise ‘unknown’, and a further five uncharacterized proteins were linked to the main network only by virtue of co‐expression or by being co‐mentioned in public text collections (Appendix S1; Table S2). Given that axis and SC proteins tend to be poorly conserved, these proteins were interesting candidates for further study (see below).
The ASY1 co‐IP network can be used to prioritize candidates for functional analysis
Candidate proteins were investigated for a meiotic role by cytological examination of chromosome spreads of male meiocytes from homozygous Arabidopsis T‐DNA insertion lines. Initially, we chose candidates based largely on confidence of identification (Table S1) and absence of a previously published role, but we also considered their potential function as inferred from conserved domains, etc. We found several with a strong meiotic mutant phenotype, including ASY3 and PCH2 (Ferdous et al., 2012; Lambing et al., 2015). In addition, from a sample of 10 candidates exhibiting only a modest or no reduction in fertility, four displayed a relatively minor mutant phenotype, where meiotic defects were clearly observed at the cytological level but were present in only a subset of meiocytes (<10%). Defects included chromosome fragmentation, unresolved interlocks, interbivalent connections, univalency and chromosome bridges at the division stages. Results from the four candidates are summarized in Figures S2 and S4, and Table S4. Interestingly, a phospho‐modified peptide was detected in the N terminus of the Brassica orthologue of one of the candidates (gi257685916; At5g59210), an structural maintenance of chromosomes (SMC) domain protein, and other phosphopeptides were identified in the N‐ and C‐terminal regions of Bra004279 (At1g68060, MAP70‐1) a microtubule‐associated protein (Figure S2b, c). Data from IntAct (Arabidopsis Interactome Mapping Consortium, 2011; Orchard et al., 2014), revealed a two‐hybrid array interaction between At5g59210 and At1g68060, thus supporting a direct physical interaction between the B. oleracea orthologues of these two proteins in our study.
As mentioned above, many of the ASY1 co‐IP proteins were identified with few unique peptides, appearing in only one or two data sets, and as such might be considered relatively low‐confidence candidates (Table S1). We therefore investigated whether we could use the ASY1 co‐IP STRING network to prioritize candidates for analysis, particularly as the process of identifying and screening homozygous mutants is labour and time intensive. In the network, ASY1 and PCH2 are located in a cluster that contains several histone‐related proteins, cohesin complex components and proteins associated with DNA replication, including RFC complex and MCM family proteins (Figure 1e; Table S2). A member of the MCM family, MCM8, is involved in DMC1‐independent DSB repair in Arabidopsis (Crismani et al., 2013), whereas the large subunit of the heteropentameric RFC complex, RFC1, is required for meiotic DSB repair (Liu et al., 2013) and interference‐sensitive COs (Wang et al., 2012b). Topoisomerase II is necessary for resolving heterochromatic DNA entanglements during female meiosis I in Drosophila melanogaster (Hughes and Hawley, 2014), for meiotic chromosome condensation and separation in mice (Li et al., 2013), and has a role in mediating CO interference in budding yeast (Zhang et al., 2014). Given the clear link between proteins in this cluster and aspects of meiotic DNA metabolism, we decided to investigate other cluster members and, indeed, mutant analysis of several of these, for example ICU2, did suggest a meiotic role.
ICU2 is required for normal progression through meiosis
ICU2 (the catalytic subunit of DNA polymerase α) is involved in mediating epigenetic states in Arabidopsis (Barrero et al., 2007; Liu et al., 2010 and Hyun et al., 2013), and has a potential role in HR (Liu et al., 2010). To determine whether ICU2 is involved in meiosis we analysed icu2‐1, homozygous for a non‐lethal missense allele of the gene (Barrero et al., 2007). icu2‐1 has a pleiotropic phenotype, including early flowering, leaf incurvature, homeotic transformations of some floral parts, reduced plant height and reduced fertility (Barrero et al., 2007). In our hands, the fertility of icu2‐1 was 45.4% (seed count per silique, n = 50). Analysis of male meiosis in icu2‐1 indicated that during most of prophase I the mutant was indistinguishable from wild‐type (WT) Arabidopsis (background En2, 2n = 10; Figure 2). Chromosomes appeared as thin threads in leptotene (Figure 2a, d), with homologues becoming fully paired and synapsed by pachytene (Figure 2b, e). ZYP1 immunolocalisation at this stage suggested synapsis was complete (Figure 2c, f). Chromosomes then began to desynapse and condense, and at metaphase I five aligned bivalents were observed in the WT (Figure 2g). Separation of the homologues at anaphase I (Figure 2h) followed by separation of sister chromatids at the second division then resulted in a tetrad of the four haploid products of meiosis (Figure 2i). In icu2‐1, however, 44.4% of first divisions appeared aberrant (n = 30); nuclei did not exhibit five normal bivalents at metaphase I (Figure 2j), and as homologues began to separate at anaphase I, fragmentation and abnormal chromosomal connections were observed (Figure 2k). This resulted in unbalanced nuclei with fragmented chromosomes at the tetrad stage (Figure 2l). The programmed formation of meiotic DSBs and subsequent recombination and synapsis is dependent on the activity of the SPO11 complex (Bergerat et al., 1997). To determine whether the fragmentation/connections observed in icu2‐1 resulted from unrepaired breaks incurred during pre‐meiotic replication or resulted from defective repair of SPO11‐induced DSBs during recombination, we generated a spo11‐1‐4 icu2‐1 double mutant. Chromosome spreads of spo11‐1‐4 icu2‐1 indicated that most meiocytes had a similar phenotype to the spo11‐1‐4 single mutant, with 10 achiasmate univalents rather than five bivalents at metaphase I (Figure 2m, n). Only 10.0% of nuclei exhibited fragmentation and/or unresolved connections in the double mutant (Figure 2o) compared with 48.3% of nuclei in the icu2‐1 single mutant (Figure 2p; χ2 (1) = 19.52, P < 0.0001, n = 60). Therefore the spo11‐1‐4 mutation can largely rescue the phenotype of the icu2‐1 single mutant, indicating that ICU2 has a role in meiotic recombination.
Figure 2.

Cytological analysis of icu2‐1 showing male meiotic chromosome spreads stained with 4′,6‐diamidino‐2‐phenylindole (DAPI).
(a–c, g–i) Wild type (WT). (d–f, j–l) icu2‐1. (c, f) Immunolocalization of ZYP1 (green), DAPI (blue). (m–p) First division in single and double mutants of SPO11‐1 and ICU2. (m) spo11‐1‐4 and (n) spo11‐1‐4 icu2‐1 nucleus with 10 univalents. (o) spo11‐1‐4 icu2‐1 and (p) icu2‐1 nucleus with unresolved chromosomal connections (yellow arrows) and fragmentation (orange arrows). Scale bars: 10 μm.
Analysis of ICU2 therefore provides ‘proof of principle’ of using the co‐IP networks to prioritize particular proteins or key interactions for verification and analysis, and appears to justify our choice of acceptance level in the co‐IP analysis as the protein was identified in the lower confidence qualitative group with only three peptides (see above and Table S1).
We also analysed a homozygous mutant of MCM2, albeit in less detail than icu2‐1, and observed mild meiotic defects and a 10% reduction in fertility (Figure S4; Table S4).
Identification of an axis‐associated protein
As axis and SC proteins tend to be poorly conserved at the primary sequence level, we were interested to note that the ASY1 co‐IP data included several uncharacterized proteins lacking known functional domains (Table S2). As mentioned above, one such protein was subsequently characterized as ASY3 (Ferdous et al., 2012). Another protein, encoded by At2g33793, was found to share 23.9% identity and 40.1% similarity with the C‐terminal predicted coiled‐coil region of AtASY3 (Figure S6a). At2g33793 formed links with several other STRING network proteins on the basis of co‐expression, including TOPII, MCM5, CAP‐D2 and PRD3 (Appendix S1). During this project, At2g33793 was independently identified by Mathilde Grelon's group (INRA, France), and was subsequently referred to as ASY4.
Preliminary characterization of a weak mutant allele of ASY4 (Figure S6b, c) indicated normal vegetative growth and silique length, but a slight reduction in fertility based on seed count per silique (mean of 57.50 compared with 60.54 in the WT, P < 0.001, n = 50). Analysis of meiocytes from asy4 confirmed a meiotic role (Figure 3). During prophase I, asy4 appeared similar to the WT, and by pachytene homologues appeared paired and synapsed based on ZYP1 immunolocalization (Figure 3a–f), although we cannot rule out the possibility of short stretches of chromosomes remaining unsynapsed in some nuclei. Most asy4 nuclei completed meiosis apparently normally; however, at metaphase I, unlike the situation in the WT where five aligned bivalents were invariably observed, a small proportion of asy4 nuclei contained univalents (2.3%, n = 130; Figure 3g, h). Abnormal inter‐bivalent connections were also apparent (Figure 3i). As homologues separated at anaphase I, chromosome bridges were observed in 15.6% (n = 32) of nuclei (Figure 3k, l; χ2 (1) for aberrant nuclei at the first division = 6.28, P = 0.012, n = 162).
Figure 3.

Cytological analysis of asy4 showing male meiotic chromosome spreads.
(a–f) During early meiotic stages asy4 appears similar to the wild type (WT), with chromosomes becoming paired and synapsed by pachytene. (g) WT metaphase I. (h–i) asy4 metaphase I with (h) univalents and (i) inter‐bivalent connections. (j) WT anaphase I. (k–l) asy4 anaphase I, with (k) chromosome bridges and (l) stray chromosome or large fragment and chromosome bridge. DNA is stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). In (c) and (f) the immunolocalization of ZYP1 (green) marks the synaptonemal complex. Arrows indicate relevant features. Scale bars: 10 μm.
We then explored the interaction between ASY4 and the other axis proteins, ASY1 and ASY3. Previously we showed that ASY1 and ASY3 directly interact via the C‐terminal predicted coiled‐coil region of ASY3 (Ferdous et al., 2012). Given the high sequence similarity between this region of ASY3 and ASY4, we investigated whether ASY4 could also directly interact with ASY1 or, indeed, with ASY3. Yeast two‐hybrid analysis found no direct interaction between ASY1 and ASY4 (Figure 4a). In contrast, in an analysis of full‐length cDNAs from ASY3 and ASY4, yeast growth was enabled even under high‐stringency selection, demonstrating a direct physical interaction between their encoded proteins. Furthermore, as in the interaction between ASY3 and ASY1 (Ferdous et al., 2012), the predicted coiled‐coil region of ASY3 (residues 623–793) was sufficient for interaction with ASY4 (Figure 4b). These results confirm that ASY4 is axis‐associated with a potential role in meiotic recombination.
Figure 4.

Yeast 2‐hybrid analysis of ASY4. Plasmid constructs were co‐transformed into yeast cells and plated on SD–Leu/–Trp (–LT), SD–Leu/–Trp/–His (–LTH) and SD–Leu/–Trp/–His/–Ade (–LTHA).
(a) ASY4 and ASY1: absence of growth on –LTH and –LTHA, but growth on the control medium, –LT, suggested that there was no direct interaction between ASY4 and ASY1. (b) ASY4 and ASY3: growth on –LTH and –LTHA confirmed that the predicted coiled coil‐containing region of ASY3 (amino‐acid residues 623–793) is sufficient for interaction with ASY4.
BoASY1 and BoASY3 are phosphorylated at multiple sites
In other organisms the phosphorylation of chromosome axis proteins is important in regulating their activity during meiosis (Rogers et al., 2002; Brar et al., 2006; Carballo et al., 2008; Katis et al., 2010; Fukuda et al., 2012; Penedos et al., 2015; Sakuno and Watanabe, 2015). We were therefore interested in whether MS analysis would enable us to identify phospho‐modified residues in BoASY1 and its interacting partner BoASY3. This proved to be the case. For BoASY1, phospho‐modified forms of 13 different peptides were identified with a total of 18 distinct phospho‐modified Serine (S) or Threonine (T) sites (Table 1). For two of the peptides the precise position of the phosphate group was unclear, but in the majority of cases the position of the phosphorylated residue could be unambiguously determined. Several peptides had doubly phosphorylated forms. Of the 18 phospho‐modified residues, four corresponded to S/TQ motifs, the preferred sites of phosphorylation for the ATM/ATR family of DNA damage response serine/threonine kinases. Notably, all of the phospho‐modified S/TQ sites were located within two S/TQ cluster domains (SCDs), defined as a region where three or more S/TQ motifs occur within a span of up to 100 residues (Traven and Heierhorst, 2005; Figure 5). SCDs are known targets of ATM/ATR. SCD1 is located near the centre of the protein, between HORMA and SWIRM domains, and comprises four S/TQ sites (S267, T272, T294 and S300), with phospho‐modification detected at T294 and S300. AtASY1 also has a SCD in this region, although it differs slightly from the BoASY1 SCD, containing only three S/TQ motifs (S267, T269 and T295) and lacking a site corresponding to S300 in BoASY1 (Figure 5). BoASY1 T294 is conserved, however, corresponding to T295 in AtASY1. Both BoASY1 and AtASY1 also contain a second SCD consisting of three S/TQ motifs close to the C terminus. Two of the S/TQ motifs in this SCD (S569 and S572) lie in close proximity on the same BoASY1 peptide (Figure 5; Table 1), and we identified phospho‐modification at S568, S569 and S572.
Table 1.
Phosphorylation sites identified in BoASY1 and BoASY3
| Protein | Site | Phosphopeptide(s) | Tissue | ptmRS: Best site probabilities |
|---|---|---|---|---|
| BoASY1 | S17 | EAEITEQD(S)LLLTR | A | S9(Phospho), 100.00 |
| S253 and S260 | STGPN(S)VHDEQP(S)DSDSEISQTK | M | S6 (Phosho), 97.33; S13 (Phosho), 99.82 | |
| S260 | STGPNSVHDEQP(S)DSDSEISQTK | A and M | S13(Phospho), 99.99 | |
| S260 and S262 | STGPNSVHDEQP(S)D(S)DSEISQTK | S13(Phospho), 100.00; S15(Phospho), 100.00 | ||
| S262 | STGPNSVHDEQPSD(S)DSEISQTK | S15(Phospho), 99.85 | ||
| S262 and S264 | STGPNSVHDEQPSD(S)D(S)EISQTK (with S264) | S15(Phospho), 82.82; S17(Phospho), 90.01 | ||
| T294 | ETQFLVAAVEKQEDDDGEVDEDN(T)QDPVESQQQLER | A and M | T24(Phospho), 100.00 | |
| QEDDDGEVDEDN(T)QDPVESQQQLER | T13(Phospho), 100.00 | |||
| S300 | QEDDDGEVDEDNTQDPVE(S)QQQLER | A and M | S19(Phospho), 100.00 | |
| QEDDDGEVDEDN(T)QDPVE(S)QQQLER (with T294) | T13(Phospho), 100.00; S19(Phospho), 100.00 | |||
| S442 or S443 | MVQEGYVED(S)SNRR or MVQEGYVEDS(S)NRR | A | S10(Phospho), 50.00; S11(Phospho), 50.00 | |
| T493 | TNGQDAKL(T)PDVSTR | A and M | T9(Phospho), 100.00 | |
| L(T)PDVSTR | T2(Phospho), 100.00 | |||
| S504 | GGIH(S)IGSDLTR | S5(Phospho), 98.97 | ||
| S504 and S507 | GGIH(S)IG(S)DLTR | A and M | S5(Phospho), 100.00; S8(Phospho), 99.88 | |
| S507 | GGIHSIG(S)DLTR | S8(Phospho), 100.00 | ||
| S526 | SAMHQNGSVL(S)EQTISK | M | S11(Phospho), 99.98 | |
| T536 | ANN(T)PMSSNAQPVASR | A and M | T4(Phospho), 100.00 | |
| S539 | ANNTPM(S)SNAQPVASR | A and M | S7 (Phospho), 99.39 | |
| S547 or S550 | ANNTPMSSNAQPVA(S)RESFAVK or | M | S15(Phospho), 50.00; S18(Phospho), 50.00 | |
| ANNTPMSSNAQPVASRE(S)FAVK | ||||
| S568 and S569 | ICTDAGTD(S)(S)QASQDRR | A and M | S9 (Phospho), 99.89; S10 (Phospho), 91.24 | |
| S569 | ICTDAGTDS(S)QASQDR | A and M | S10(Phospho), 96.10 | |
| S572 | ICTDAGTDSSQA(S)QDRR | A and M | S13(Phospho), 98.72 | |
| BoASY3 | S15 | SFGSNFHPS(S)QPR | M | S10(Phospho), 94.53 |
| S156 | GNEMDK(S)PER | A and M | S7(Phospho), 100.00 | |
| S205 | A(S)PEYNEDVNSETPEVVK | M | S2(Phospho), 99.67 | |
| T231 or S232 | LNQDK(T)SNDDPLTK or LNQDKT(S)NDDPLTK | M | T6(Phospho), 50.00; S7(Phospho), 50.00 | |
| S251 or S253 | HHSDTIETD(S)E(S)PEVATR | M | S10(Phospho), 49.72; S12(Phospho), 49.72 | |
| S432 and S441 | EK(S)VEPENDFQ(S)PTFGYK | A | S3(Phospho), 100.00; S12(Phospho), 92.49 |
Phospho‐modified residues are indicated by parentheses. Some peptides were confirmed as doubly phosphorylated. For a few peptides a phospho‐modification could be confirmed, but the precise location within the peptide could not be determined. All sites were identified by ptmRS and manually verified. A (anther) and M (meiocyte) indicate the tissue(s) from which the phosphopeptides were identified.
Figure 5.

Phosphorylation sites identified in BoASY1. The full‐length sequence of BoASY1 is shown aligned with AtASY1 and its budding yeast orthologue ScHop1 (clustal omega). BoASY1 phospho‐modified residues are highlighted in blue (note that two phospho‐sites could not be precisely determined, as indicated above the sequence). ScHop1 phospho‐sites (Carballo et al., 2008) are highlighted in green. S/TQ cluster domains in BoASY1 and AtASY1 are indicated by red lines above the sequence. Predicted HORMA (residues 13‐222) and SWIRM (residues 389‐438) domains are highlighted in yellow in AtASY.
The remaining phospho‐modified sites in BoASY1 also tend to occur in clusters. Of particular note are S260, S262 and S264, located immediately upstream of SCD1 (Figure 5; Table 1). Phosphorylation at these sites was complex, with both singly and various doubly phosphorylated peptides observed. Interestingly, the multiple acidic residues surrounding the phosphoserines matches the hallmark motif of casein kinase 2 (CK2; Pinna, 2002). Two more loose clusters, each consisting of three phospho‐modified sites within a stretch of up to 15 residues, occur between the SWIRM domain and SCD2. One site in each cluster (T493 and T536) is at a consensus minor CDK1 motif (S/TP) (Figure 5; Table S1).
In BoASY3 we identified phospho‐modified forms of six different peptides (Table 1). Most carried a single modification, but one was doubly phosphorylated at S432 and S441. The positions of most sites could be unequivocally determined, but in two cases was ambiguous (T231 or S232; S251 or S253). Four of the sites are at consensus CDK1 motifs: S205, S253 and S441 are at minor motifs and S156 is at a full motif (S/TPXK/R; Figure 6). Only one site, S15, is at an S/TQ motif.
Figure 6.
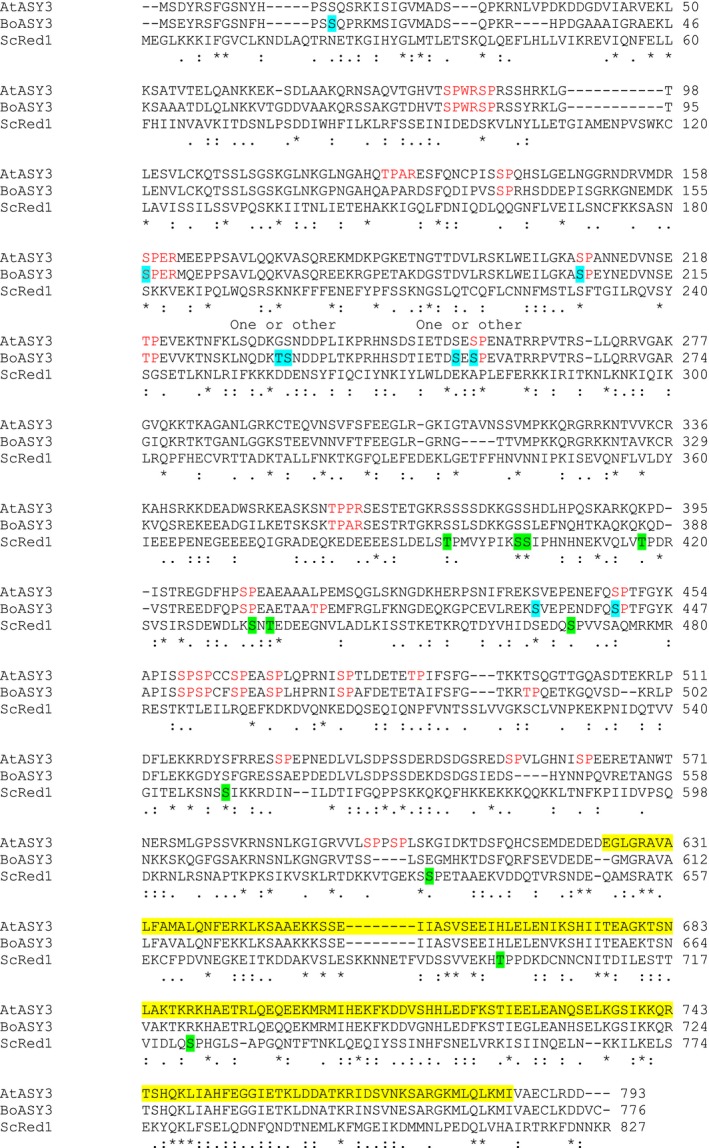
Phosphorylation sites identified in BoASY3. The full‐length sequence of BoASY3 is shown aligned with AtASY3 and ScRed1, the likely functional homologue of ASY3 in yeast (clustal omega). BoASY3 phospho‐modified residues are highlighted in blue. (Note that two BoASY3 phospho‐sites could not be precisely determined, as indicated above the sequence). ScRed1 putative cdc28 sites or cdc28‐independent, but experimentally verified, meiosis‐dependent phospho‐sites (Lai et al., 2011) are highlighted in green. Red text indicates minimal (S/T‐P) or full (S/T‐P‐X‐K/R) consensus CDK1 motifs in BoASY3 and AtASY3. A predicted coiled coil region is highlighted in yellow in AtASY3.
Discussion
In plants, as in other sexually reproducing organisms, the frequency and distribution of COs during meiosis is governed by the functional inter‐relationship between the recombination machinery and the proteinaceous structures that organize the chromosomes during prophase I of meiosis. Thus far our understanding of plant meiosis largely derives from the analysis of around 90 plant meiotic genes, primarily identified through mutant analysis. Although effective, this approach is hampered by several factors. For instance, many of the proteins that are crucial for meiosis are likely to be involved in essential processes in somatic cells. Some genes are duplicated, functionally redundant or, when mutated, produce only subtle phenotypes with little impact on fertility (at least under standard glasshouse growth conditions). Here we have demonstrated that affinity proteomics can be used as an additional approach to identify proteins that play a role in meiosis, and that by targeting a specific component of the meiotic machinery it is possible to begin to define the protein–protein interaction network in which the protein participates.
Proteins that co‐immunoprecipitate with ASY1 can be organized into a coherent interaction network
Previously, global analyses of the proteomes of developing rice anthers during meiosis and isolated rice meiocytes have identified several thousand proteins, highlighting the complexity of the meiotic proteome (Collado‐Romero et al., 2014; Ye et al., 2015). Nevertheless, it is likely that the picture is incomplete as Ye et al. (2015) identified peptides corresponding to just 10 of at least 28 characterized rice meiotic proteins (Luo et al., 2014), and homologues of only 14 and seven Arabidopsis and budding yeast meiotic proteins, respectively.
Adopting a strategy based on affinity purification of meiotic complexes has provided a viable alternative approach, in that focusing on proteins associated with a key meiotic protein, in this case ASY1, substantially reduces complexity and facilitates functional analysis. This approach has enabled us to define a PPI network of 492 nodes that incorporates ASY1. That a substantial proportion of the network proteins are likely to have a meiotic role has been validated using a combination of prior functional knowledge (for example, the presence of CAP‐D2, ZYP1 and PRD3; Higgins et al., 2005; De Muyt et al., 2009; Smith et al., 2014) and functional analysis (this study; Ferdous et al., 2012; Lambing et al., 2015). It is apparent from this analysis that although some network proteins such as ASY3, PCH2, ICU2 and PRD3 have major meiotic roles, this is not the case for a significant proportion. Mutant analysis of a small sample suggests that many of the network proteins may have only a minor effect on meiosis; however, this is based on a preliminary analysis, and hence we cannot rule out that the mild mutant phenotypes are the result of functional redundancy or the fact that the analyses were conducted only under standard growth conditions. It is important to note that given their subtle mutant phenotypes, a meiotic role for these proteins is unlikely to have been detected using previous approaches, such as fertility screening.
A further important point is that the presence of a protein within the PPI network does not necessarily imply a direct molecular interaction with ASY1 or indeed any other component. Additional analyses are required to determine such interactions. For example, Y2H analysis confirmed a direct interaction between ASY1 and ASY3 (Ferdous et al., 2012), whereas this is not the case for ASY1 and ZYP1. Indeed, installation of ZYP1 to form the SC is dependent on PCH2‐mediated depletion of ASY1 from the axis (Lambing et al., 2015).
Using the network led us to prioritize ICU2, the catalytic subunit of Arabidopsis DNA polymerase α, for analysis, and we confirmed that it has a role in meiotic recombination. RFC1 was proposed to be involved in DNA lagging‐strand synthesis during double Holliday junction formation (Wang et al., 2012a), and DNA leading‐strand synthesis was found to be important for the formation of interference‐sensitive COs in Arabidopsis (Huang et al., 2015). The precise role of ICU2 will require further study.
Cytological analysis suggested a potentially interesting mutant phenotype for At5g59210, the protein product of which is predicted to contain extensive coiled‐coil regions. Further work will be required to fully characterize the role of this protein, but the identification of a phospho‐site near the N‐terminus of its Brassica orthologue is interesting, particularly as IntAct indicates an interaction with MAP70‐1, a plant‐specific microtubule‐associated protein, the Brassica orthologue of which was also found to be phosphorylated. We have not yet investigated MAP70‐1 for a meiotic role. Any analysis would need to address the fact that it is part of a multigene family, sharing a high degree of identity with three other proteins (Korolev et al., 2005), and that the Brassica orthologues of all four proteins were present in the ASY1 co‐IP data (Tables S1 and S2). Consistent with the identification of microtubule‐associated proteins, several β‐tubulin and α‐tubulin proteins were also present in the co‐IP data.
Of the 492 Arabidopsis loci submitted for STRING analysis, 11 were unknown/uncharacterized and six could not be incorporated into the PPI network. These proteins were thought to be good candidates for investigation for a meiotic role, which led to the identification and preliminary characterization of ASY4. Although we were able to analyse only a weak mutant allele of ASY4, its cytological phenotype of impaired CO formation, together with its high degree of similarity to the C terminal of ASY3, and the confirmation of a direct Y2H interaction between the two proteins, strongly supports an axis‐associated role for the protein. No direct Y2H interaction with ASY1 was detected, suggesting that ASY4 may have been co‐immunoprecipitated by an indirect interaction with ASY1 via ASY3, thus illustrating a further advantage of using an affinity proteomics approach in that by targeting meiotic complexes, secondary and even higher order protein interactors might be identified. An indirect interaction with ASY1 might explain why we identified relatively few unique peptides of ASY4 compared with ASY1 and ASY3 (3, 55 and 39, respectively), although this could also have been influenced by its smaller size (ASY4 has a predicted molecular weight of 24.69 kDa compared with 67.21 kDa for ASY1 and 88.00 kDa for ASY3).
Protein phosphorylation
Mass spectrometry (MS) revealed that in vivo BoASY1 is extensively phosphorylated at prophase I of meiosis. Amongst the 18 identified sites there were four S/TQ motifs distributed between two SCDs. SCD1 is located near the centre of the protein, between the HORMA and SWIRM domains, and SCD2 is near the C terminus. Sequence alignment suggests a comparable arrangement in AtASY1, although there appears to be an additional S/TQ motif in SCD1 of BoASY1 (Figure 5). Although further studies will be required to determine whether phosphorylation of the S/TQ residues in SCD1 and SCD2 is of functional significance, comparison with Hop1 in budding yeast (Figure 5) suggests that this is possible, at least for some of the S/TQ sites in SCD1 (Carballo et al., 2008). Hop1 contains eight S/TQ motifs; three of them form an SCD located just downstream of the HORMA domain, and all three are phosphorylated in vivo during meiosis. Phosphorylation at T318 has the greatest effect in promoting Hop1‐dependent interhomologue recombination (Carballo et al., 2008), and phosphorylation at S298 promotes stable interaction of HOP1 and Mek1 effector kinase on the chromosomes following initial phospho‐T318 mediated Mek1 recruitment (Penedos et al., 2015). Similar to Hop1, SCD1 is found near the centre of the protein between the HORMA and SWIRM domains in BoASY1 and AtASY1. A full‐length alignment of BoASY1 with Hop1 (Figure 5) suggests that residue T294 in SCD1 corresponds in position to Hop1 T318. As is the case for T318 in Hop1, a flanking S/TQ motif (S300) is also phosphorylated. Although it is reasonable to speculate that the S/TQ sites within SCD1 and the Hop1 SCD may be functionally comparable, this is clearly not the case for SCD2, which is absent from the budding yeast protein. SCD2 is conserved in the rice HORMA domain protein, OsPAIR2, however, and has also been reported to undergo phosphorylation (Ye et al., 2015). This occurred at S579, which corresponds to S572 in BoASY1. A second OsPAIR2 phospho‐site was detected nearby at S569, a non‐S/TQ site. Nevertheless, the significance of phosphorylation in the ASY1 C terminal remains obscure, as it appears that this SCD is present in the orthologues of some plant species, but not in others.
The significance of the tendency for the non‐S/TQ phosphosites in BoASY1 to also occur in clusters remains to be determined. The cluster S260, S262 and S264 just upstream of SCD1 (Figure 5; Table 1) is particularly interesting because the multiple acidic residues surrounding the phospho‐serines matches the hallmark motif of casein kinase II (Pinna, 2002). CK2 motifs were recently identified amongst irradiation and ATM/ATR‐dependent upregulated phosphorylation sites in Arabidopsis, although it remains to be seen whether these sites are actually targeted by CK2 in an ATM/ATR‐dependent manner (Roitinger et al., 2015). Phosphorylation at minor CDK1 sites within the two clusters situated between the SWIRM domain and SCD2 may also be of significance, particularly as CDKA;1 was amongst the proteins we identified, albeit with only two peptides.
The most striking feature of the BoASY3 phospho‐sites is that all seven are located in the N‐terminal region of the protein (Figure 6). This is consistent with an earlier functional analysis of ASY3 that showed that the C‐terminal coiled‐coil region of the protein (residues 623–793) is involved in its interaction with ASY1 in Arabidopsis (Ferdous et al., 2012), and may therefore be inaccessible for signalling. Four of the sites were at consensus CDK1 motifs: one at a full motif and three at minor motifs. A single phospho‐site, at position S81, was identified in the rice ASY3 orthologue, PAIR3, and was also at a minor CDK1 motif (Ye et al., 2015). Red1, the budding yeast orthologue of BoASY3, contains seven putative target sites of Cdc28 (CDK1) and at least four Cdc28‐independent phosphorylation sites (Lai et al., 2011); however, in a full‐length alignment of BoASY3 and Red1, only S432 and S441 lie in close proximity to a Red1 phosphosite (S469; Figure 6). Of these, S441 is a minimal S/TP motif, like Red1 S469. Functional analysis of phosphorylation in Red1 suggested that it was non‐essential for its functions in meiosis (Lai et al., 2011), so it will be interesting to investigate any potential role for ASY3 and ASY1 phosphorylation in future studies.
Besides CDKA;1, several other kinases and phosphatases were identified in the ASY1 co‐IP data, including the protein phosphatase 2A subunits PP2AA2 and PP2A‐3 (Tables S1 and S2). PP2A has been implicated in a number of meiotic roles in animals and yeast (e.g. Lu et al., 2002; Kitajima et al., 2006; Riedel et al., 2006; Nolt et al., 2011; Tang et al., 2016), and is regulated by the UPS during mouse oocyte maturation (Yu et al., 2015). It remains to be established whether it, or any of the kinases identified in our study, has a role in plant meiosis, however.
Proteins associated with other cellular processes
The ASY1 co‐IP data contained multiple components of several large complexes and functional pathways, such as the 26S proteasome, the ubiquitination system and the spliceosome (Table S2). Because of their participation in a wide range of cellular functions, one could argue that these proteins were recovered simply as a result of non‐specific protein interactions. It is therefore important to emphasize that they were identified either as significant in label‐free quantification or as ASY1 sample‐specific (absent from all control data sets), suggesting that at least some of the complex/pathway components were isolated on the basis of a specific interaction with the ASY1 meiotic complex. Indeed, an examination of the literature provides several indications of a close association between these particular protein complexes/pathways and meiotic chromatin. For example, HEI10, a mammalian RING domain protein with E3 ubiquitin‐ligase activity (Ward et al., 2007), and related proteins in budding yeast (Zip3), Sordaria macrospora, Arabidopsis and rice (HEI10) are required for CO formation, and have been shown to localize to discrete foci along meiotic chromosomes (Agarwal and Roeder, 2000; Chelysheva et al., 2012; Wang et al., 2012a; De Muyt et al., 2014; Qiao et al., 2014). Furthermore, studies indicate that Zip3 in yeast and RNF212 (its mammalian orthologue) and HEI10 in mouse mediate the recruitment of proteasomes to chromosome axes to regulate axis morphogenesis, homologue pairing, synapsis and meiotic recombination (Ahuja et al., 2017; Rao et al., 2017). Examination of the Caenorhabditis elegans germline provides further evidence that proteasome recruitment to the chromosome axes is an evolutionarily conserved feature of meiosis (Ahuja et al., 2017).
Around 400 splicing‐related proteins have been predicted or confirmed in Arabidopsis (Wang and Brendel, 2004; Koncz et al., 2012). We identified 25 spliceosome‐related proteins (Table S2), including PRL1 from the spliceosome‐activating NineTeen Complex (NTC) core and three NTC‐associated proteins, as defined by Monaghan et al., (2009). There is increasing evidence of a role for the NTC in the coordination of DNA damage responses (reviewed in Koncz et al., 2012). Interestingly, Ye et al. (2015) found that the RNA splicing pathway was extensively phosphorylated in rice anthers at around the time of meiosis (as indeed were the DNA synthesis and RdDM pathways, which are also well represented in our data). Although any association of spliceosome factors with meiotic chromosomes or a DNA repair role in meiosis remains to be established, our analysis may be a pointer in this direction.
Technical considerations
Although we anticipated that it might be feasible to use anthers to identify proteins that co‐IP with ASY1, we were concerned that meiocytes represent only a small proportion of the tissue, potentially compromising our ability to identify less abundant meiotic proteins. Our data reveal that although several of the proteins with a prior confirmed role in meiosis could be identified from anthers, most were identified exclusively from the meiocyte‐enriched samples. This was also the case for ICU2 and ASY4. Other experiments carried out in our lab suggest that certain meiotic proteins, notably ZYP1, are more easily recovered from intact anthers than extruded meiocytes, however, and hence there may be technical reasons favouring their detection from this tissue. In this context, it is interesting that during native meiotic protein extraction in budding yeast, the ZYP1 orthologue Zip1 appears to be far less stable than ASY1 and ASY3 homologues Hop1 and Red1 (Lin et al., 2010). Thus, although it is clear that the additional effort in preparing meiocyte‐enriched material was justified, it seems that analysis of both tissue types provides the most comprehensive picture.
The decision to retain the lower confidence group of all ASY1 sample‐specific proteins satisfying the minimum identification threshold of two peptides appeared justified for two reasons. First, several of them were found to have a meiotic role, and second, over 90% of the identified proteins formed a single PPI network; however, we cannot rule out that in some cases low‐confidence proteins may have appeared to be sample specific by chance. There are several biological reasons why genuine interactors might appear with low confidence. Low‐abundance proteins and proteins that form only transient interactions with the ASY1 complex may be under‐represented in samples (extreme examples being protein kinases and phosphatases). Indirect interactors or proteins that form only weak interactions with the complex may also be more difficult to detect. Further sample enrichment to target a substage of prophase I may begin to address some of these limitations, as would targeting other proteins in the ASY1 interaction network in order to confirm and extend the network, enabling a comprehensive picture of meiotic interactions to emerge. This, together with ongoing technical improvements in sample preparation methods and MS analysis, should help to increase our ability to identify genuine protein interactors that currently lie at the borderline of detection.
Experimental procedures
Plant material, nucleic acid extraction and mutation site mapping
Arabidopsis thaliana ecotypes Columbia (0) and Enkheim‐2 (En‐2) and B. oleracea var. alboglabra A12DHd were used for WT analysis. Arabidopsis seed stocks were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Plants were grown, Arabidopsis material was harvested and nucleic acid extractions were carried out as previously described (Higgins et al., 2004). T‐DNA insertion sites of mutant lines were confirmed by PCR and, in the case of asy4, by sequencing. The missense mutation of icu2‐1 was confirmed by sequencing and tetra‐primer ARMS‐PCR (Ye et al., 2001). Primer details are listed in Appendix S2.
Co‐immunoprecipitation analysis
Brassica meiotic tissue was collected as previously described (Sánchez‐Morán et al., 2005). Co‐IP analysis was based on a previously described method, with minor modifications (Osman et al., 2013). Full details of the procedure are available in Appendix S2.
Bioinformatic analysis
Brassica proteins were used to identify putative A. thaliana orthologues using best blastp 2.6.0 score (with an acceptance threshold of an E‐value of 1e −5) against TAIR 10 protein sequences (https://www.arabidopsis.org). GO categorization of A. thaliana orthologues was carried out using the TAIR website (https://www.arabidopsis.org/tools/bulk/go/index.jsp). GO enrichment analysis was carried out using panther accessed through the GO consortium website (http://geneontology.org). The KEGG pathway database was used to predict functional pathways for Arabidopsis orthologues (http://www.kegg.jp/kegg/pathway.html; Kanehisa et al., 2016). PPI networks of Arabidopsis orthologues were generated using STRING 10.5 (http://string-db.org; Szklarczyk et al., 2015), using default settings. The resulting network and protein description files were used to produce the networks in cytoscape 3.5.1 (http://www.cytoscape.org). Sequence alignments were carried out using clustal omega (Sievers et al., 2011) or emboss needle (Rice et al., 2000), accessed through the EMBL‐EBI website (https://www.ebi.ac.uk).
Antibody production
The AtZYP1B C‐terminal antibody was produced using a previously described procedure (Ferdous et al., 2012) with primers ZYP1B‐C‐F and ZYP1B‐C‐R (Appendix S2). Polyclonal antiserum against the recombinant protein was raised in rabbit (Orygen Antibodies Ltd.; http://www.orygen.co.uk).
Cytological procedures
Cytological procedures were carried out as previously described (Higgins et al., 2004). Antibodies were used as follows: anti‐AtASY1 (rat, 1/1000 dilution) and anti‐AtZYP1B‐C (rabbit, 1/500 dilution). DNA was stained with 1 μg ml−1 4′,6‐diamidino‐2‐phenylindole (DAPI) in Vectashield.
Yeast 2‐hybrid analysis
Yeast 2‐hybrid analysis was carried out as previously described (Ferdous et al., 2012). Details of primers used for plasmid construction are presented in Appendix S2.
Statistical procedures
Fertility in WT and mutant plants was compared using single‐factor anova. Chi‐square (χ2) tests were carried out using graphpad prism 7 (https://graphpad.com) using Yate's correction.
Accession numbers
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2016), with the identifier PXD006042. The following lines were used for mutant analysis: At5g46070, SALK_016366; At3g52140, SALK_046271; At5g42220, SALK_151742; At5g59210, GABI_094G05; mcm2 (At1g44900), SALK_023429; icu2‐1 (At5g67100), N329; spo11‐1‐4 (At3g13170), WiscDsLox461‐464J19 (Roberts 2010); asy4 (At2g33793), SAIL_886_D04.
Conflicts of interest
The authors declare no known conflicts of interest.
Supporting information
Figure S1. Targeting BoASY1 using an anti‐AtASY1 antibody.
Figure S2. Protein sequence coverage.
Figure S3. Mutant analysis of three meiotic candidates.
Figure S4. Mutant analysis of At5g59210.
Figure S5. Mutant analysis of meiotic candidate MCM2.
Figure S6. Alignment of ASY4 (At2g33793) with ASY3 and mapping of T‐DNA insertion SAIL_886_D04 in asy4.
Table S1. ASY1 sample‐specific Brassica proteins with their putative Arabidopsis orthologues.
Table S2. Gene ontology enrichment analysis of ASY1 sample‐specific proteins.
Table S3. Functional grouping of ASY1 sample‐specific proteins.
Table S4. Summary of analysis of meiotic candidates.
Appendix S1. PPI network of ASY1 sample‐specific proteins in Cytoscape format.
Appendix S2. Supporting experimental procedures.
Acknowledgements
Horticultural/technical support was provided by Karen Staples and Steve Price, University of Birmingham. We wish to thank Professors Graham King (Southern Cross University, Australia), Isobel Parkin (University of Saskatchewan, Canada) and Xiaowu Wang (Chinese Academy of Agricultural Sciences, China) for their help with Brassica sequences. We are grateful to Dr Eugenio Sanchez‐Moran and Professor George Bassel (University of Birmingham) for helpful discussions. Research leading to these results has received funding from the European Community's Seventh Framework Programme FP7/2007‐2013 under grant agreement number KBBE‐2009‐222883, and from the Biotechnology and Biological Sciences Research Council (BBSRC) under grant number ERA‐Caps‐13 BB/M004902/1.
References
- Agarwal, S. and Roeder, G.S. (2000) Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell, 102, 245–255. [DOI] [PubMed] [Google Scholar]
- Ahuja, J.S. , Sandhu, R. , Mainpal, R. , Lawson, C. , Henley, H. , Hunt, P.A. , Yanowitz, J.L. and Börner, G.V. (2017) Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science, 355, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, P.R. , Tirian, L. , Vunjak, M. and Brennecke, J. (2017) A heterochromatin‐dependent transcription machinery drives piRNA expression. Nature, 549, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium . (2011) Evidence for network evolution in an Arabidopsis interactome map. Science, 333, 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, S.J. , Caryl, A.P. , Jones, G.H. and Franklin, F.C.H. (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis‐associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645. [DOI] [PubMed] [Google Scholar]
- Attner, M.A. , Miller, M.P. , Ee, L.S. , Elkin, S.K. and Amon, A. (2013) Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl Acad. Sci. 110, 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya, K. , Suzuki, G. , Suwabe, K. et al. (2011) Comprehensive network analysis of anther‐expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS ONE, 6, e26162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero, J.M. , González‐Bayón, R. , del Pozo, J.C. , Ponce, M.R. and Micol, J.L. (2007) INCURVATA2 encodes the catalytic subunit of DNA polymerase α and interacts with genes involved in chromatin‐mediated cellular memory in Arabidopsis thaliana . Plant Cell, 19, 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz, L.E. and Copenhaver, G.P. (2010) Genetic interference: don't stand so close to me. Curr. Genom. 11, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat, A. , de Massy, B. , Gadelle, D. , Varoutas, P.‐C. , Nicolas, A. and Forterre, P. (1997) An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature, 386, 414–417. [DOI] [PubMed] [Google Scholar]
- Bose, R. , Manku, G. , Culty, M. and Wing, S.S . (2014) Ubiquitin–proteasome system in spermatogenesis In Posttranslational Protein Modifications in the Reproductive System (Sutovsky P., ed.). New York, NY: Springer New York, pp. 181–213. [DOI] [PubMed] [Google Scholar]
- Brar, G.A. , Kiburz, B.M. , Zhang, Y. , Kim, J.‐E. , White, F. and Amon, A. (2006) Rec8 phosphorylation and recombination promote the step‐wise loss of cohesins in meiosis. Nature, 441, 532–536. [DOI] [PubMed] [Google Scholar]
- Carballo, J.A. , Johnson, A.L. , Sedgwick, S.G. and Cha, R.S. (2008) Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell, 132, 758–770. [DOI] [PubMed] [Google Scholar]
- Chelysheva, L. , Diallo, S. , Vezon, D. et al. (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118, 4621. [DOI] [PubMed] [Google Scholar]
- Chelysheva, L. , Vezon, D. , Chambon, A. , Gendrot, G. , Pereira, L. , Lemhemdi, A. , Vrielynck, N. , Le Guin, S. , Novatchkova, M. and Grelon, M. (2012) The arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Farmer, A.D. , Langley, R.J. , Mudge, J. , Crow, J.A. , May, G.D. , Huntley, J. , Smith, A.G. and Retzel, E.F. (2010) Meiosis‐specific gene discovery in plants: RNA‐Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol. 10, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado‐Romero, M. , Alós, E. and Prieto, P. (2014) Unravelling the proteomic profile of rice meiocytes during early meiosis. Front. Plant Sci. 5, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F. , Belzile, F. , Horlow, C. , Grandjean, O. , Vezon, D. and Doutriaux, M.‐P. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of arabidopsis. Plant Cell, 11, 1623–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani, W. , Girard, C. , Froger, N. , Pradillo, M. , Santos, J.L. , Chelysheva, L. , Copenhaver, G.P. , Horlow, C. and Mercier, R. (2012) FANCM limits meiotic crossovers. Science, 336, 1588. [DOI] [PubMed] [Google Scholar]
- Crismani, W. , Portemer, V. , Froger, N. , Chelysheva, L. , Horlow, C. , Vrielynck, N. and Mercier, R. (2013) MCM8 is required for a pathway of meiotic double‐strand break repair independent of DMC1 in Arabidopsis thaliana . PLoS Genet. 9, e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer, L. , Heyman, J. , Touati, S. et al. (2012) OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet. 8, e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt, A. , Pereira, L. , Vezon, D. et al. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana . PLoS Genet. 5, e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt, A. , Zhang, L. , Piolot, T. , Kleckner, N. , Espagne, E. and Zickler, D. (2014) E3 ligase Hei10: a multifaceted structure‐based signaling molecule with roles within and beyond meiosis. Genes Dev. 28, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveshwar, P. , Bovill, W.D. , Sharma, R. , Able, J.A. and Kapoor, S. (2011) Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 11, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux, M.P. , Couteau, F. , Bergounioux, C. and White, C. (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana . Mol. Gen. Genet. 257, 283–291. [DOI] [PubMed] [Google Scholar]
- Dukowic‐Schulze, S. , Sundararajan, A. , Mudge, J. et al. (2014) The transcriptome landscape of early maize meiosis. BMC Plant Biol. 14, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous, M. , Higgins, J.D. , Osman, K. et al. (2012) Inter‐homolog crossing‐over and synapsis in arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8, e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, T. , Pratto, F. , Schimenti, J.C. , Turner, J.M.A. , Camerini‐Otero, R.D. and Höög, C. (2012) Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLoS Genet. 8, e1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, C. , Crismani, W. , Froger, N. , Mazel, J. , Lemhemdi, A. , Horlow, C. and Mercier, R. (2014) FANCM‐associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucl. Acids Res. 42, 9087–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon, M. , Vezon, D. , Gendrot, G. and Pelletier, G. (2001) AtSPO11‐1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering, C.H. and Jessberger, R. (2012) Cohesin in determining chromosome architecture. Exp. Cell Res. 318, 1386–1393. [DOI] [PubMed] [Google Scholar]
- He, Y. , Wang, C. , Higgins, J.D. , Yu, J. , Zong, J. , Lu, P. , Zhang, D. and Liang, W. (2016) MEIOTIC F‐BOX is essential for male meiotic DNA double‐strand break repair in rice. Plant Cell, 28, 1879–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J.D. , Armstrong, S.J. , Franklin, F.C.H. and Jones, G.H. (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J.D. , Sanchez‐Moran, E. , Armstrong, S.J. , Jones, G.H. and Franklin, F.C.H. (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Cheng, Z. , Wang, C. , Hong, Y. , Su, H. , Wang, J. , Copenhaver, G.P. , Ma, H. and Wang, Y. (2015) Formation of interference‐sensitive meiotic cross‐overs requires sufficient DNA leading‐strand elongation. Proc. Natl Acad. Sci. 112, 12534–12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, S.E. and Hawley, R.S. (2014) Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet. 10, e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, Y. , Yun, H. , Park, K. , Ohr, H. , Lee, O. , Kim, D.‐H. , Sung, S. and Choi, Y. (2013) The catalytic subunit of arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development, 140, 156. [DOI] [PubMed] [Google Scholar]
- Jones, G.H. and Franklin, F.C.H. (2006) Meiotic crossing‐over: obligation and interference. Cell, 126, 246–248. [DOI] [PubMed] [Google Scholar]
- Kalsotra, A. and Cooper, T.A. (2011) Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 12, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Sato, Y. , Kawashima, M. , Furumichi, M. and Tanabe, M. (2016) KEGG as a reference resource for gene and protein annotation. Nucl. Acids Res. 44, D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis, V.L. , Lipp, J.J. , Imre, R. , Bogdanova, A. , Okaz, E. , Habermann, B. , Mechtler, K. , Nasmyth, K. and Zachariae, W. (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7‐Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell 18, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, T.S. , Sakuno, T. , Ishiguro, K.‐I. , Iemura, S.‐I. , Natsume, T. , Kawashima, S.A. and Watanabe, Y. (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature, 441, 46–52. [DOI] [PubMed] [Google Scholar]
- Kleckner, N. (2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma, 115, 175. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I. and Jones, J.D.G. (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon‐induced allelic variation and meiosis‐associated expression. Plant J. 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Koncz, C. , deJong, F. , Villacorta, N. , Szakonyi, D. and Koncz, Z. (2012) The spliceosome‐activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev, A.V. , Chan, J. , Naldrett, M.J. , Doonan, J.H. and Lloyd, C.W. (2005) Identification of a novel family of 70 kDa microtubule‐associated proteins in Arabidopsis cells. Plant J. 42, 547–555. [DOI] [PubMed] [Google Scholar]
- Lai, Y.J. , Lin, F.M. , Chuang, M.J. , Shen, H.J. and Wang, T.F. (2011) Genetic requirements and meiotic function of phosphorylation of the yeast axial element protein Red1. Mol. Cell. Biol. 31, 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, W.S. , Yang, X. and Makaroff, C.A. (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 118, 3037. [DOI] [PubMed] [Google Scholar]
- Lambing, C. , Osman, K. , Nuntasoontorn, K. et al. (2015) Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11, e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Chen, C. , Markmann‐Mulisch, U. , Timofejeva, L. , Schmelzer, E. , Ma, H. and Reiss, B. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl Acad. Sci. USA, 101, 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.M. , Yu, C. , Wang, Z.W. , Zhang, Y.L. , Liu, X.M. , Zhou, D. , Sun, Q.Y. and Fan, H.Y. (2013) DNA topoisomerase II is dispensable for oocyte meiotic resumption but is essential for meiotic chromosome condensation and separation in mice. Biol. Reproduct. 89(118), 111. [DOI] [PubMed] [Google Scholar]
- Libeau, P. , Durandet, M. , Granier, F. , Marquis, C. , Berthomé, R. , Renou, J.P. , Taconnat‐Soubirou, L. and Horlow, C. (2011) Gene expression profiling of Arabidopsis meiocytes. Plant Biol. 13, 784–793. [DOI] [PubMed] [Google Scholar]
- Lin, F.M. , Lai, Y.J. , Shen, H.J. , Cheng, Y.H. and Wang, T.F. (2010) Yeast axial‐element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 29, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. and Nonomura, K.I. (2016) A wide reprogramming of histone H3 modifications during male meiosis I in rice is dependent on the Argonaute protein MEL1. J. Cell Sci. 129, 3553. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Ren, X. , Yin, H. , Wang, Y. , Xia, R. , Wang, Y. and Gong, Z. (2010) Mutation in the catalytic subunit of DNA polymerase α influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 61, 36–45. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Deng, Y. , Li, G. and Zhao, J. (2013) Replication factor C1 (RFC1) is required for double‐strand break repair during meiotic homologous recombination in Arabidopsis. Plant J. 73, 154–165. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Liu, Y. , Yang, X. et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Comm. 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q. , Dunn, R.L. , Angeles, R. and Smith, G.D. (2002) Regulation of spindle formation by active mitogen‐activated protein kinase and protein phosphatase 2A during mouse oocyte meiosis. Biol. Reproduct. 66, 29–37. [DOI] [PubMed] [Google Scholar]
- Luo, Q. , Li, Y. , Shen, Y. and Cheng, Z. (2014) Ten years of gene discovery for meiotic event control in rice. J. Genet. Genom. 41, 125–137. [DOI] [PubMed] [Google Scholar]
- Mercier, R. , Mézard, C. , Jenczewski, E. , Macaisne, N. and Grelon, M. (2015) The molecular biology of meiosis in plants. Ann. Rev. Plant Biol. 66, 297–327. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. , Xu, F. , Gao, M. , Zhao, Q. , Palma, K. , Long, C. , Chen, S. , Zhang, Y. and Li, X. (2009) Two Prp19‐like U‐box proteins in the MOS4‐associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolt, J.K. , Rice, L.M. , Gallo‐Ebert, C. , Bisher, M.E. and Nickels, J.T. (2011) PP2ACdc55 is required for multiple events during meiosis I. Cell Cycle, 10, 1420–1434. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.I. , Morohoshi, A. , Nakano, M. , Eiguchi, M. , Miyao, A. , Hirochika, H. and Kurata, N. (2007) A germ cell‐specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell, 19, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, C. , Santos, J. and Pradillo, M. (2014) On the role of some ARGONAUTE proteins in meiosis and DNA repair in Arabidopsis thaliana . Front. Plant Sci. 5, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, C. , Santos, J.L. and Pradillo, M. (2016) Accurate chromosome segregation at first meiotic division requires AGO4, a protein involved in RNA‐dependent DNA methylation in Arabidopsis thaliana . Genetics, 204, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo‐Monfil, V. , Duran‐Figueroa, N. , Arteaga‐Vazquez, M. , Demesa‐Arevalo, E. , Autran, D. , Grimanelli, D. , Slotkin, R.K. , Martienssen, R.A. and Vielle‐Calzada, J.‐P. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature, 464, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard, S. , Ammari, M. , Aranda, B. et al. (2014) The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucl. Acids Res. 42, D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, K. , Sanchez‐Moran, E. , Mann, S.C. , Jones, G.H. and Franklin, F.C.H. (2009) Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 28, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, K. , Higgins, J.D. , Sanchez‐Moran, E. , Armstrong, S.J. and Franklin, F.C.H. (2011) Pathways to meiotic recombination in Arabidopsis thaliana . New Phytol. 190, 523–544. [DOI] [PubMed] [Google Scholar]
- Osman, K. , Roitinger, E. , Yang, J. , Armstrong, S. , Mechtler, K. and Franklin, F.C.H . (2013) Analysis of meiotic protein complexes from arabidopsis and brassica using affinity‐based proteomics In Plant Meiosis: Methods and Protocols (Pawlowski W.P., Grelon M. and Armstrong S., eds). Totowa, NJ: Humana Press, pp. 215–226. [DOI] [PubMed] [Google Scholar]
- Page, S.L. and Hawley, R.S. (2004) The genetics and molecular biology of the synaptonemal complex. Ann. Rev. Cell Devel. Biol. 20, 525–558. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A.P. , Koh, C. , Tang, H. et al. (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea . Genome Biol. 15, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedos, A. , Johnson, A.L. , Strong, E. , Goldman, A.S. , Carballo, J.A. and Cha, R.S. (2015) Essential and checkpoint functions of budding yeast ATM and ATR during meiotic prophase are facilitated by differential phosphorylation of a meiotic adaptor protein, Hop1. PLoS ONE, 10, e0134297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna, L.A. (2002) Protein kinase CK2: a challenge to canons. J. Cell Sci. 115, 3873. [DOI] [PubMed] [Google Scholar]
- Pradillo, M. , Knoll, A. , Oliver, C. , Varas, J. , Corredor, E. , Puchta, H. and Santos, J.L. (2015) Involvement of the cohesin cofactor PDS5 (SPO76) during meiosis and DNA repair in Arabidopsis thaliana . Front. Plant Sci. 6, 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, H. , Prasada Rao, H.B.D. , Yang, Y. et al. (2014) Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat. Genet. 46, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, H.B.D.P. , Qiao, H. , Bhatt, S.K. et al. (2017) A SUMO‐ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science, 355, 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, P. , Longden, I. and Bleasby, A. (2000) EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Riedel, C.G. , Katis, V.L. , Katou, Y. et al. (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature, 441, 53–61. [DOI] [PubMed] [Google Scholar]
- Roberts, N.Y . (2010) Investigating the control of pairing and crossover formation in meiosis of Arabidopsis thaliana. Ph.D. Thesis. University of Birmingham, UK. http://etheses.bham.ac.uk/1009
- Rockmill, B. and Roeder, G.S. (1991) A meiosis‐specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 5, 2392–2404. [DOI] [PubMed] [Google Scholar]
- Rogers, E. , Bishop, J.D. , Waddle, J.A. , Schumacher, J.M. and Lin, R. (2002) The aurora kinase AIR‐2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitinger, E. , Hofer, M. , Köcher, T. , Pichler, P. , Novatchkova, M. , Yang, J. , Schlögelhofer, P. and Mechtler, K. (2015) Quantitative phosphoproteomics of the ataxia telangiectasia‐mutated (ATM) and ataxia telangiectasia‐mutated and rad3‐related (ATR) dependent DNA damage response in Arabidopsis thaliana . Mol. Cell Proteomics, 14, 556–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K.J. , Fransz, P. , Armstrong, S.J. , Vizir, I. , Mulligan, B. , Franklin, F.C. and Jones, G.H . (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T‐DNA‐transformed lines. Chromosome Res. 5, 551–559. [DOI] [PubMed] [Google Scholar]
- Sakuno, T. and Watanabe, Y. (2015) Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev. Cell, 32, 220–230. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Moran, E. , Santos, J.‐L. , Jones, G.H. and Franklin, F.C.H. (2007) ASY1 mediates AtDMC1‐dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Morán, E. , Mercier, R. , Higgins, J.D. , Armstrong, S.J. , Jones, G.H. and Franklin, F.C.H. (2005) A strategy to investigate the plant meiotic proteome. Cytogenet. Genome Res. 109, 181–189. [DOI] [PubMed] [Google Scholar]
- Schmid, R. , Grellscheid, S.N. , Ehrmann, I. et al. (2013) The splicing landscape is globally reprogrammed during male meiosis. Nucl. Acids Res. 41, 10170–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mole. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M. , Goel, S. , Meeley, R.B. , Dantec, C. , Parrinello, H. , Michaud, C. , Leblanc, O. and Grimanelli, D. (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell, 23, 443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.J. , Osman, K. and Franklin, F.C.H. (2014) The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J. 80, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth Gordon, K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 1–25. [DOI] [PubMed] [Google Scholar]
- Sprink, T. and Hartung, F. (2014) The splicing fate of plant SPO11 genes. Front. Plant Sci. 5, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D. , Franceschini, A. , Wyder, S. et al. (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucl. Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Zhang, Z.‐Y. , Zhang, W.‐J. , Zhao, X.‐M. , Li, X. , Zhang, D. , Liu, Q.‐Q. and Tang, W.‐H. (2010) Global gene profiling of laser‐captured pollen mother cells indicates molecular pathways and gene subfamilies involved in rice meiosis. Plant Physiol. 154, 1855–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, A. , Shi, P. , Song, A. , Zou, D. , Zhou, Y. , Gu, P. , Huang, Z. , Wang, Q. , Lin, Z. and Gao, X. (2016) PP2A regulates kinetochore‐microtubule attachment during meiosis I in oocyte. Cell Cycle, 15, 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven, A. and Heierhorst, J. (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA‐damage‐response proteins. BioEssays, 27, 397–407. [DOI] [PubMed] [Google Scholar]
- Vizcaíno, J.A. , Csordas, A. , del‐Toro, N. , Dianes, J.A. , Griss, J. , Lavidas, I. , Mayer, G. , Perez‐Riverol, Y. , Reisinger, F. and Ternent, T . (2016) 2016 update of the PRIDE database and its related tools. Nucl. Acids Res. 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B.‐B. and Brendel, V. (2004) The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre‐mRNA splicing. Genome Biol. 5, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Yang, M. (2006) The Arabidopsis SKP1‐LIKE1 (ASK1) protein acts predominately from leptotene to pachytene and represses homologous recombination in male meiosis. Planta, 223, 613–617. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wang, H. , Wang, J. et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Wang, M. , Tang, D. , Shen, Y. , Miao, C. , Hu, Q. , Lu, T. and Cheng, Z. (2012a) The role of rice HEI10 in the Formation of meiotic crossovers. PLoS Genet. 8, e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, Z. , Huang, J. , Shi, Q. , Hong, Y. , Copenhaver, G.P. , Gong, Z. and Ma, H. (2012b) The DNA replication factor RFC1 is required for interference‐sensitive meiotic crossovers in Arabidopsis thaliana . PLoS Genet. 8, e1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , You, C. , Chang, F. , Wang, Y. , Wang, L. , Qi, J. and Ma, H. (2014) Alternative splicing during Arabidopsis flower development results in constitutive and stage‐regulated isoforms. Front. Genet. 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J.O. , Reinholdt, L.G. , Motley, W.W. et al. (2007) Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Lu, P. , Wang, Y. and Ma, H. (2011) The transcriptome landscape of Arabidopsis male meiocytes from high‐throughput sequencing: the complexity and evolution of the meiotic process. Plant J. 65, 503–516. [DOI] [PubMed] [Google Scholar]
- Ye, S. , Dhillon, S. , Ke, X. , Collins, A.R. and Day, I.N.M. (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 29 e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Zhang, Z. , Long, H. , Zhang, Z. , Hong, Y. , Zhang, X. , You, C. , Liang, W. , Ma, H. and Lu, P. (2015) Proteomic and phosphoproteomic analyses reveal extensive phosphorylation of regulatory proteins in developing rice anthers. Plant J. 84, 527–544. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Ji, S.‐Y. , Sha, Q.‐Q. , Sun, Q.‐Y. and Fan, H.‐Y. (2015) CRL4–DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation. Nat. Commun. 6, 8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Wang, S. , Yin, S. , Hong, S. , Kim, K.P. and Kleckner, N. (2014) Topoisomerase II mediates meiotic crossover interference. Nature, 511, 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Hu, M. , Feng, X. , Gong, A. , Cheng, L. and Yuan, H . (2017) Proteomes and phosphoproteomes of anther and pollen: availability and progress. Proteomics, 17, 1770151 https://doi.org/10.1002/pmic.201600458 [DOI] [PubMed] [Google Scholar]
- Zhao, D. , Yang, X. , Quan, L. , Timofejeva, L. , Rigel, N.W. , Ma, H. and Makaroff, C.A. (2006) ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol. Biol. 62, 99–110. [DOI] [PubMed] [Google Scholar]
- Zickler, D. and Kleckner, N. (2016) A few of our favorite things: pairing, the bouquet, crossover interference and evolution of meiosis. Sem. Cell Dev. Biol. 54, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Targeting BoASY1 using an anti‐AtASY1 antibody.
Figure S2. Protein sequence coverage.
Figure S3. Mutant analysis of three meiotic candidates.
Figure S4. Mutant analysis of At5g59210.
Figure S5. Mutant analysis of meiotic candidate MCM2.
Figure S6. Alignment of ASY4 (At2g33793) with ASY3 and mapping of T‐DNA insertion SAIL_886_D04 in asy4.
Table S1. ASY1 sample‐specific Brassica proteins with their putative Arabidopsis orthologues.
Table S2. Gene ontology enrichment analysis of ASY1 sample‐specific proteins.
Table S3. Functional grouping of ASY1 sample‐specific proteins.
Table S4. Summary of analysis of meiotic candidates.
Appendix S1. PPI network of ASY1 sample‐specific proteins in Cytoscape format.
Appendix S2. Supporting experimental procedures.


