Abstract
PP2A is composed of a scaffolding subunit (A), a catalytic subunit (C) and a regulatory subunit (B) that is classified into four families including B, B′, B′′ and B′′′/striatin. Here, we found that a distinct PP2A complex regulates NF‐κB signalling by dephosphorylation of IKKβ, IκBα and RelA/p65. The PP2A core enzyme AC dimer and the holoenzyme AB′′′C trimer dephosphorylate IKKβ, IκBα and RelA, whereas the ABC trimer dephosphorylates IκBα but not IKKβ and RelA in cells. In contrast, AB′C and AB′′C trimers have little effect on dephosphorylation of these signalling proteins. These results suggest that different forms of PP2A regulate NF‐κB pathway signalling through multiple steps each in a different manner, thereby finely tuning NF‐κB‐ and IKKβ‐mediated cellular responses.
Keywords: NF‐kappa B, protein phosphatase, protein phosphatase 2 A (PP2A)
Abbreviations
IκB, inhibitor of kappa B
IKKβ, inhibitor of kappa B kinase β
IKKβKN, kinase‐negative IKKβ mutant
NF‐κB, nuclear factor kappa B
PPPs, protein phosphatases
Nuclear factor kappa B (NF‐κB) is a critical transcription factor that regulates many cellular and organismal processes including immune and inflammatory responses, cellular growth and cell survival. The NF‐κB signalling pathway is regulated by the phosphorylation of several proteins including inhibitor of kappa B (IκB) kinase β (IKKβ), an inhibitory protein IκBα and an essential subunit of NF‐κB RelA/p65 1, 2, 3. Binding of cytokines such as TNF‐α to respective receptors leads to the conjugation of ubiquitin to several signalling components and recruits adaptor proteins and kinases 3, 4, 5, 6. These chains of reaction induce IKKβ activation through phosphorylation of two critical serine residues, Ser177 and Ser181, at its activation loop 7, 8. IKKβ in turn phosphorylates IκBα at the N‐terminal serines, Ser32 and Ser36, which leads to ubiquitination at lysine residues Lys21 and Lys22 and IκBα degradation by the ubiquitin‐proteasome system. These reactions result in the nuclear translocation of NF‐κB and binding to its cognate κB sites in the promoters/enhancers of its target genes. IKKβ also phosphorylates serine residues, Ser468 and Ser536, in the C‐terminal transactivation domain of RelA and promotes gene expression through the association of RelA with a transcriptional coactivator, CREB‐binding protein 9, 10, 11.
Aberrant activation of NF‐κB is linked to various diseases such as inflammatory disorders and cancer. Thus, there are numerous regulatory mechanisms at multiple levels to ensure the tight control of NF‐κB activity 1, 2, 3, 4, 5, 6. In particular, dephosphorylation of its protein represents the most important mechanism to downregulate NF‐κB activity. There are three families of protein serine/threonine phosphatases: protein phosphatases (PPPs), metal‐dependent protein phosphatases (PPMs) and DxDxT phosphatases 12, 13. PPP constitutes the largest family, containing several members including protein phosphatase 1 (PP1), PP2A, PP2B and PP5, whereas the PPM family includes PP2C. Several of these, such as PP1 14, PP2A 7, 15, 16, PP2C 17 and PP5 18, are required for dephosphorylation of the activation loop serines in IKKβ, which is a critical step to downregulate NF‐κB. In addition, protein phosphatases dephosphorylate downstream signalling proteins as well. Specifically, it has been reported that PP2B dephosphorylates IκBα 19 and PP2A 20 and PP2C 21 dephosphorylate RelA.
Among these phosphatases, PP2A is the most abundant, constituting approximately 1% of total cellular proteins 22, 23, 24. PP2A is a heterotrimeric complex consisting of a scaffolding (A), regulatory (B) and catalytic (C) subunit. The A and C subunits each are comprised of two possible variants, α and β. The monomeric C subunit is unstable and requires binding to the A subunit to exist as a stable core enzyme AC dimer. The B subunit family consists of four classes including B (B55/PR55), B′ (B56/PR61), B′′ (PR48/PR72/PR130) and B′′′(PR93/PR110)/striatin (Strn). In turn, there are four isoforms of the B class including α, β, γ and δ, five of the B′ class including α, β, γ, δ and ε, three of the B′′ class including α, β and γ, and three isoforms of the B′′′/Strn class including Strn, Strn3 and Strn4. These B subunits associate with the AC dimer to form the holoenzyme trimer. Crystal structure analysis revealed that the B subunits bind proximal to the C subunit active site in the holoenzyme trimer and determine the specificity for substrate proteins 25, 26.
A previous RNAi screen revealed that PP2A plays an important role in the regulation of NF‐κB signalling and identified the core enzyme subunits, Aα, Aβ, Cα and Cβ, as negative regulators of the NF‐κB signalling pathway in mouse astrocytes 16. However, this screen did not clarify whether the B subunit is involved in regulation of NF‐κB activity, nor identify the specific B subunits regulating each NF‐κB signalling step. It is plausible that, as cells usually express multiple isoforms of each B family protein, this failure was a result of complementation with other isoforms of the same family following the simple RNAi screen. In this study, we expressed B subunits together with A and C subunits in cells and investigated the role of these subunits on TNF‐α‐, IKKβ‐ and RelA‐induced NF‐κB activation. This assay revealed that distinct B subunits regulate specific steps of NF‐κB signalling. Dephosphorylation of IKKβ and RelA is mediated by the AC dimer and AB′′′C trimer, whereas dephosphorylation of IκBα is mediated by the AC dimer, ABC trimer and AB′′′C trimer.
Materials and methods
Cell cultures, plasmids and transfection
cDNA‐encoding Aα, Aβ, Cα, Cβ, Bα, Bβ, Bγ, Bδ, B′α, B′β, B′γ, B′δ, B′ε, B′′β, Strn and Strn3 subunits of PP2A were amplified from a human cDNA library by PCR. The cDNAs were inserted into pRK‐HA and pRK‐Flag expression vectors. Expression plasmids encoding IKKβ, kinase‐negative IKKβ mutant (IKKβKN) and IκBα have been reported previously 27. Constitutive active IKKβEE mutant of which phosphorylation sites Ser177/181 in the activation loop were exchanged to Glu, the ubiquitination‐resistant IκBαRR mutant of which ubiquitination sites lysine residues Lys21/22 were exchanged to Arg, and the phosphatase‐inactive CαD85N mutant of which Asp 85 was exchanged to Gln were generated using the KOD‐Plus Mutagenesis kit (TOYOBO). Plasmids were transfected into GP2‐293 cells in Opti‐MEM (Invitrogen) using Lipofectamine Plus (Invitrogen) following the manufacturer's instructions. GP2‐293 cells were obtained from Clontech. Cells were cultured in DMEM supplemented with 10% FBS containing 2 mm L‐glutamine at 37 °C.
Luciferase assay
Nuclear factor kappa B activity was estimated by Dual‐Luciferase Reporter Assay System (Promega) using pNF‐κB and pRL‐TK Luciferase reporter plasmids. After transfection of 0.01 μg NF‐κB reporter plasmids with or without 0.01 μg expression plasmids of various PP2A subunits, IKKβ, IκBα and/or RelA into GP2‐293 cells in collagen‐coated 96‐well dishes, cells were incubated in the presence or absence of 50 ng·mL−1 TNF‐α for 18 h. After lysing cells with buffer from the assay system, luciferase activity was analysed following the manufacturer's instructions.
Immunoblotting
GP2‐293 cells in collagen‐coated 12‐well dishes were transfected with 0.2 μg expression plasmids of various PP2A subunits, IKKβ, IκBα and/or RelA. After 24‐h transfection, cells were washed with PBS and solubilized with buffer A consisting of 20 mm Tris/Cl, pH 7.5, 150 mm NaCl, 1 mm EGTA, 10 mm MgCl2, 60 mm β‐glycerophosphate, 1 mm Na3VO4, 1 mm 4‐amidino phenyl methyl sulfonyl fluoride, 50 KIU·mL−1 aprotinin, 20 μg·mL−1 pepstatin, 20 μg·mL−1 leupeptin, 2 mm DTT and 1% Triton X‐100. After centrifugation at 16 000 × g for 20 min at 4 °C, the supernatants were used as cell lysates. The cell lysates were subjected to SDS/PAGE and then gel‐separated proteins were transferred to PVDF membranes (Millipore) and subjected to immunoblotting using the SuperSignal West Pico Chemiluminescence System (Pierce). Antibodies used were as follows: anti‐Flag (M2) (Sigma, F‐3165), anti‐HA (Roche, 04‐902), anti‐IκBα (Santa Cruz Biotechnology, SC‐371), anti‐RelA/p65 (Santa Cruz Biotechnology, SC‐372), anti‐phospho‐IκBα (Cell Signaling Technology, 2859), anti‐phospho‐RelA/p65(S468) (Cell Signaling Technology, 3039), anti‐phospho‐RelA/p65(S536) (Cell Signaling Technology, 3033), anti‐PP2A (Cell Signaling Technology, 2039), anti‐PP2A (Epitomics, 1512‐1), anti‐IKKβ (Cell Signaling Technology, 2684) and anti‐phospho‐IKKα/β (Cell Signaling Technology, 2697).
In vitro dephosphorylation assay by PP2A
For the dephosphorylation assay of IKKβ, GP2‐293 cells in 10‐cm dishes were transfected with expression plasmids encoding Flag‐IKKβ, and then phosphorylated IKKβ was recovered from cell lysates via anti‐Flag (M2) agarose beads (Sigma). IKKβ proteins were eluted in a buffer containing 20 mm Tris/Cl, pH 7.5, 150 mm NaCl, 2 mm DTT, 1% Triton X‐100 and 3 × Flag peptides (Sigma) from anti‐Flag‐agarose beads with ultra‐free Centrifugal Filter Units (Millipore). For the dephosphorylation assay of IκBα and RelA, cells were transfected with plasmids encoding Flag‐RelA, HA‐IκBαRR and IKKβEE, and then phosphorylated IκBαRR and RelA protein complexes were purified from cell lysates via anti‐Flag (M2) agarose beads. PP2A proteins were purified from GP2‐293 cells following transfection with plasmids encoding the Flag‐tagged Aα subunit and HA‐tagged Cα subunit, together with or without HA‐tagged B subunits including Bα, Bβ, Bγ, Bδ, B′α, B′β, B′γ, B′δ, B′ε, B′′β, Strn and Strn3. PP2A core enzyme composed of Aα and Cα, and PP2A holoenzyme composed of Aα, Cα and various B subunits were purified from cell lysates via anti‐Flag (M2) agarose beads with ultra‐free Centrifugal Filter Units in the previously described buffer. Phosphorylated IKKβ proteins (0.001 μg) or protein complexes (0.001 μg) of phosphorylated IκBαRR and RelA were incubated with purified PP2A (0.001 μg) for 30 min at 37 °C, and then reaction mixtures were subjected to SDS/PAGE. Gel‐separated proteins were transferred to PVDF membranes and subjected to immunoblotting using anti‐phospho‐IKKα/β, anti‐phospho‐IκBα and anti‐phospho‐RelA antibodies, respectively.
Results
The PP2A core enzyme downregulates NF‐κB signalling
PP2A is composed of three subunits scaffolding (A), regulatory (B) and catalytic (C) subunit (Fig. 1A). Cells were transfected with expression plasmids of α and β isoforms of the A and C subunits, and the expression of PP2A proteins were analysed by immunoblotting (Fig. 1B). Then, the effects of expression of these proteins on NF‐κB activity were analysed by luciferase assay. Expression of the Cα and Cβ subunits and of the C subunits together with Aα or Aβ subunits suppressed TNF‐α‐induced NF‐κB activation (Fig. 1C) and suppressed IKKβ and constitutive active IKKβEE‐induced NF‐κB activation (Fig. 1D,E). In contrast, expression of the PP2A did not attenuate RelA‐mediated NF‐κB activity (Fig. 1F). These results suggest that PP2A suppresses NF‐κB activity by suppressing IκBα and/or IKKβ phosphorylation. Expression of inactive PP2A core enzyme consisting of the CαD85N mutant did not suppress TNF‐α‐, IKKβ‐ and IKKβEE‐induced NF‐κB activation (Fig. 1G,H).
Figure 1.
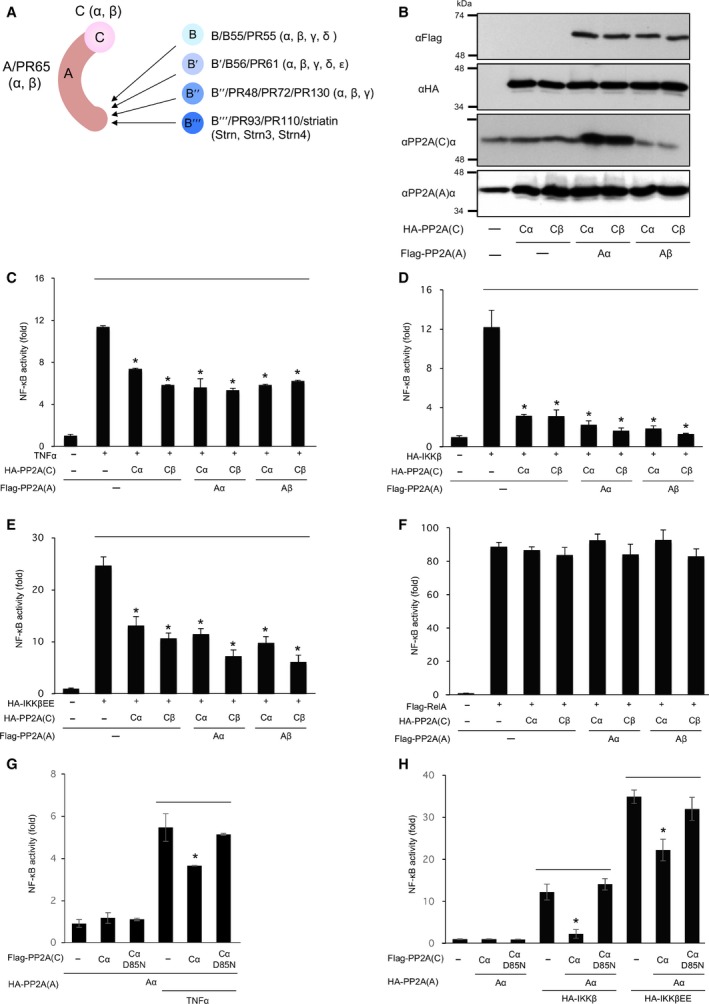
PP2A core enzymes suppress NF‐κB activity. (A) Schematic model of PP2A core enzyme and holoenzyme. (B) Expression of PP2A subunits in GP2‐293 cells. Cells were transfected with plasmids encoding HA‐PP2A Cα and Cβ, together with Flag‐PP2A Aα and Aβ, and expression of these subunits was analysed by immunoblotting. (C) Cells were transfected with NF‐κB reporter plasmids together with plasmids encoding PP2A core enzyme subunits, then stimulated with TNF‐α for 18 h. (D‐F) Cells were transfected with NF‐κB reporter plasmids and plasmids encoding PP2A core enzyme subunits together with plasmids encoding HA‐IKKβ (D), HA‐IKKβEE (E) or Flag‐RelA (F). (G) Cells were transfected with NF‐κB reporter plasmids and PP2A plasmids encoding Cα or CαD85N, and were incubated for 20 h. Then, cells were stimulated with TNF‐α for 4 h. (H) Cells were transfected with NF‐κB reporter plasmids and PP2A plasmids encoding Cα or CαD85N, together with plasmids encoding HA‐IKKβ and HA‐IKKβEE. Cells were incubated for 18 h, and NF‐κB activity was estimated by luciferase assay. *Statistically significance at P < 0.01.
Distinct PP2A holoenzymes downregulate NF‐κB signalling
To investigate the involvement of the PP2A in regulation of NF‐κB signalling, we constructed expression plasmids encoding each subunit of the B, B′′, B′′ and B′′′/Strn family (Fig. 2A). Among isoforms of B′′ and B′′′/Strn families, we used expression plasmids encoding B′′β for the B′′ family, along with Strn and Strn3 for the B′′′/Strn family, as B′′α, B′′γ and B′′′/Strn4 hardly express in cells. Cells were transfected with NF‐κB reporter plasmids and plasmids encoding Aα, Cα and each B subunit, and NF‐κB activity was investigated following TNF‐α treatment by luciferase assay. Expression of the AC holoenzyme, the ABC holoenzyme including A(Bα)C, A(Bβ)C, A(Bγ)C and A(Bδ)C, and the AB′′′C holoenzyme including A(Strn)C and A(Strn3)C suppressed TNF‐α‐induced NF‐κB activity, whereas the AB′C holoenzyme including A(B′α)C, A(B′β)C, A(B′γ)C, A(B′δ)C and A(B′ε)C and the AB′′C holoenzyme including A(B′′β)C had little effect on TNF‐α‐induced NF‐κB activation (Fig. 2B). Then, we investigated the effects of distinct B subunits on NF‐κB signalling. NF‐κB activity was analysed in cells transfected with plasmids encoding IKKβ or IKKβEE together with Aα, Cα and each B subunit. A luciferase assay demonstrated that expression of AC, ABC and AB′′′C attenuated IKKβ‐ and IKKβEE‐induced NF‐κB activity, whereas AB′C and AB′′C had little effect on NF‐κB activation (Fig. 2C,D). These results suggest that AC, ABC and AB′′′C suppress NF‐κB activity downstream of IKKβ.
Figure 2.
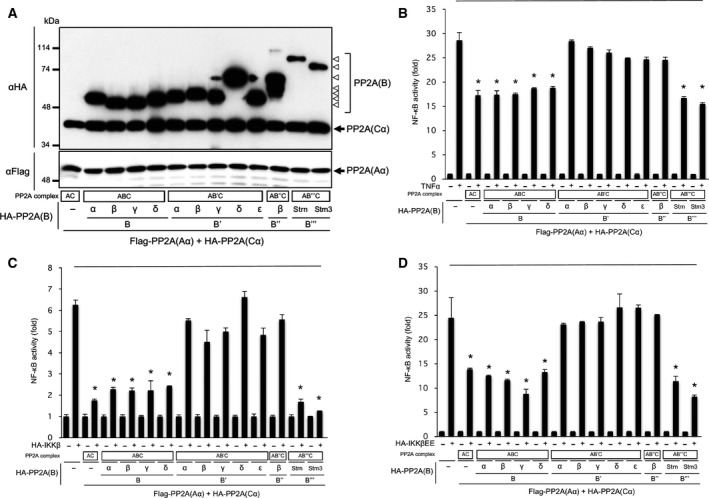
PP2A core enzyme AC and holoenzymes ABC and AB′′′C suppress TNF‐α‐ and IKKβ‐induced NF‐κB activity. (A) GP2‐293 cells were transfected with plasmids encoding PP2A core enzyme subunits Aα and Cα together with subunits of B, B′, B′′ and B′′′/Strn families. Following incubation for 24 h, cells were lysed and expression of these subunits was analysed by immunoblotting. (B) Cells were transfected with NF‐κB reporter plasmids and plasmids encoding PP2A subunits, and then stimulated with TNF‐α for 18 h. NF‐κB activity was estimated by luciferase assay. (C, D) Cells were transfected with NF‐κB reporter plasmids and plasmids encoding PP2A subunits together with plasmids encoding IKKβ (D) and IKKβEE (E). Cells were incubated for 18 h, and NF‐κB activity was estimated by luciferase assay. *Statistically significance at P < 0.01.
AC and AB′′′C dephosphorylate IKKβ and AC, ABC and AB′′′C dephosphorylate IκBα in cells
We next investigated the effects of distinct B subunits on phosphorylation of the IKKβ activation loop. Cells were transfected with expression plasmids encoding IKKβ and plasmids encoding Aα, Cα and each B subunit, and phosphorylation of the activation loop was investigated by immunoblotting. Expression of the AC core enzyme markedly suppressed IKKβ phosphorylation in a phosphatase activity‐dependent manner (Fig. 3A). The AC core enzyme and the AB′′′C holoenzyme markedly suppressed IKKβ phosphorylation, whereas three types of holoenzyme including ABC, AB′C and AB′′C had little effect (Fig. 3B). These lines of evidence indicate that AC and AB′′′C attenuate TNF‐α‐induced NF‐κB activity, at least in part, through dephosphorylation of IKKβ.
Figure 3.
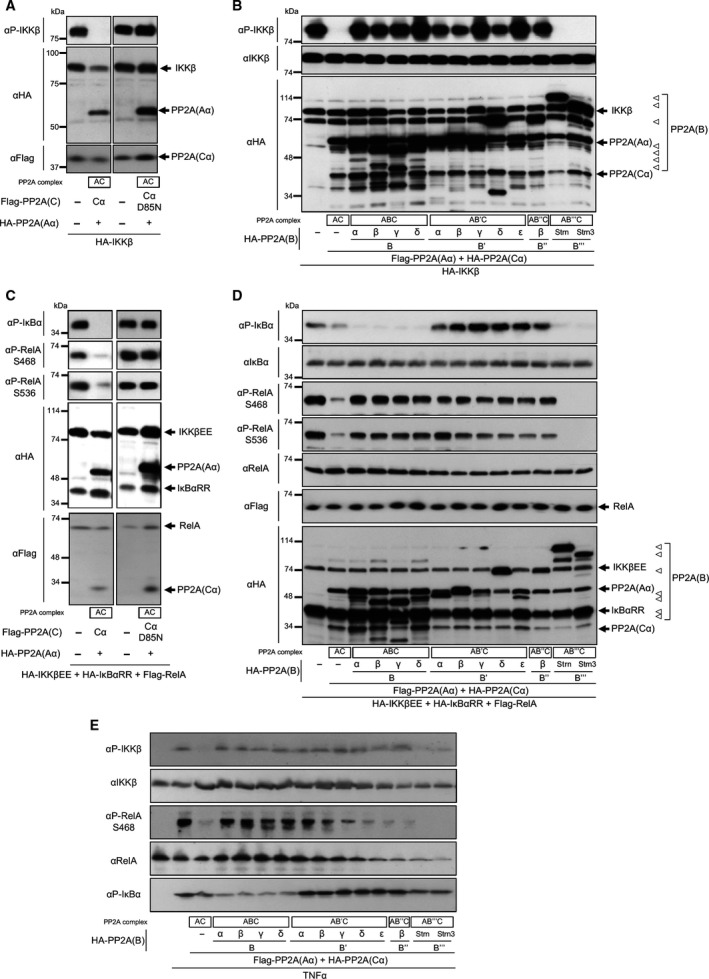
PP2A enzyme AC and AB′′′C suppress IKKβ and RelA phosphorylation, and AC, ABC and AB′′′C suppress IκBα phosphorylation in vivo. (A) GP2‐293 cells were transfected with plasmids encoding IKKβ and PP2A plasmids encoding Aα and Cα or phosphatase‐inactive CαD85N. (B) Cells were transfected with plasmids encoding IKKβ and PP2A plasmids encoding Aα, Cα and each subunit of B, B′, B′′ and B′′′/Strn families. (C) Cells were transfected with plasmids encoding IKKβEE, IκBαRR, RelA and PP2A plasmids encoding Aα and Cα or CαD85N. (D) Cells were transfected with plasmids encoding IKKβEE, IκBαRR, RelA and PP2A plasmids encoding Aα, Cα, and each subunit of B, B′, B′′ and B′′′/Strn families. Following incubation for 24 h, cells were lysed, and phosphorylation of proteins was estimated by immunoblotting. (E) GP2‐293 cells were transfected with expression plasmids of PP2A, and incubated for 24 h. Cells were stimulated with TNF‐α for 5 min, and phosphorylation of proteins was analysed by immunoblotting.
To reveal the effects of each B subunit on the NF‐κB/IκBα complex, we transfected cells with expression plasmids encoding IκBαRR, RelA and IKKβEE, together with plasmids encoding Aα, Cα and each B subunit. Then, the effects of PP2A on IKKβEE‐mediated phosphorylation of IκBαRR and RelA were investigated by immunoblotting. Expression of the AC core enzyme suppressed phosphorylation of IκBαRR and RelA in a phosphatase activity‐dependent manner (Fig. 3C). AC, ABC and AB′′′C reduced IκBα phosphorylation efficiently, whereas AB′C and AB′′C had little effect on dephosphorylation of IκBαRR (Fig. 3D). In turn, AC and AB′′′C dephosphorylated RelA efficiently, whereas ABC, AB′C and AB′′C had little effect on dephosphorylation of RelA. These results suggest that AC, ABC and AB′′′C attenuate NF‐κB activity, at least in part, by dephosphorylation of IκBα.
Then, we investigated the effects of distinct B subunits on phosphorylation of endogenous NF‐κB signalling proteins in TNF‐α‐stimulated cells. Cells were transfected with expression plasmids of PP2A, and then stimulated with TNF‐α. AC and AB′′′C suppressed TNF‐α‐induced phosphorylation of IKKβ and RelA, whereas ABC, AB′C and AB′′C had little effect (Fig. 3E). AC, ABC and AB′′′C reduced TNF‐α‐induced IκBα phosphorylation, whereas AB′C and AB′′C had little effect. These results suggest that functional features of B subunits have physiological significance in NF‐κB signalling.
PP2A core enzyme and holoenzyme effectively dephosphorylate IKKβ, IκBα and RelA in vitro
Dephosphorylation of IKKβ, IκBαRR and RelA was analysed by using purified PP2A enzyme in vitro. Expression plasmids encoding Flag‐tagged Aα, HA‐tagged Cα and HA‐tagged B subunits were transfected in cells, and PP2A complexes of various isoforms of B subunit family were purified from cells by using Flag‐agarose beads (Fig. 4A). Phosphorylated IKKβ was purified from cells transfected with Flag‐IKKβ expression plasmid (Fig. 4B). Following incubation of phosphorylated IKKβ with various purified PP2A core enzyme and holoenzyme, dephosphorylation of IKKβ was analysed by immunoblotting. This assay revealed that purified PP2A complexes of different B subunits effectively dephosphorylate IKKβ, and these complexes showed similar levels of activity (Fig. 4C). Then, we purified phosphorylated complexes of IκBαRR and RelA from cells transfected with plasmids of Flag‐RelA, HA‐IκBαRR and IKKβEE by using Flag‐agarose beads (Fig. 4D). Following incubation of phosphorylated IκBαRR and RelA protein complexes with various purified PP2A core enzyme and holoenzyme, dephosphorylation of IκBαRR and RelA was analysed by immunoblotting. Purified PP2A complexes of different B subunits effectively dephosphorylate IκBαRR and RelA (Fig. 4E). These results indicated that, although all PP2A complexes have an ability to dephosphorylate IKKβ, IκBα and RelA in vitro, AC and AB′′′C preferentially dephosphorylate IKKβ and RelA, and AC, ABC and AB′′′C preferentially dephosphorylate IκBα in vivo, suggesting that intracellular mechanisms may relate to the preference of AC, ABC and AB′′′C to dephosphorylate IKKβ, IκBα and RelA.
Figure 4.
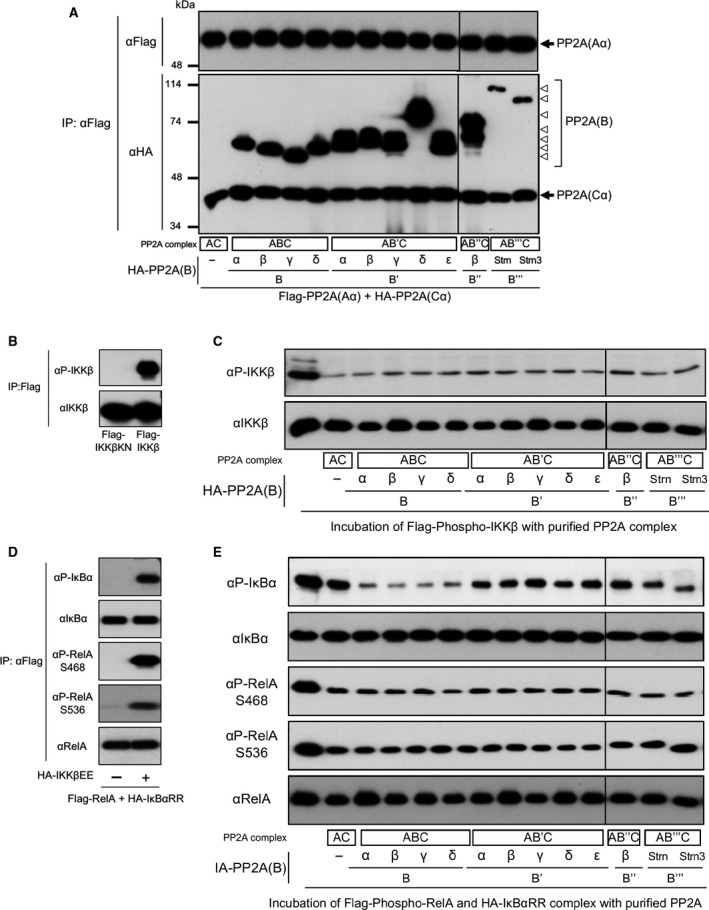
Dephosphorylation of IKKβ, IκBα and RelA by PP2A in vitro. (A) Cells were transfected with plasmids encoding PP2A core enzyme subunits Aα and Cα together with subunits of B, B′, B′′ and B′′′/Strn families. Following incubation for 24 h after transfection, PP2A enzymes were purified using anti‐Flag (M2) agarose beads. Purified proteins (0.01 μg) were subjected to SDS/PAGE and analysed by immunoblotting. (B) Cells were transfected with plasmids of Flag‐IKKβ and kinase‐inactive Flag‐IKKβKN. Phosphorylated IKKβ proteins were purified by using anti‐Flag (M2) agarose beads. (C) Phosphorylated IKKβ was incubated with purified PP2A, and dephosphorylation levels of IKKβ were analysed by immunoblotting. (D) Cells were transfected with plasmids encoding RelA, κBαRR and IKKβEE. Phosphorylated RelA and IκBα complexes were purified using anti‐Flag (M2) agarose beads. (E) Phosphorylated RelA and IκBα proteins were incubated with purified PP2A, and dephosphorylation levels of each protein were analysed by immunoblotting.
Distinct B subunits regulate the binding of PP2A to the substrate proteins in vivo
To resolve the discrepancies in the dephosphorylation of IKKβ and RelA between in vitro and in vitro, we investigated intracellular mechanisms of the preference of AC, ABC, AB′′C and AB′′′C to dephosphorylate NF‐κB signalling proteins. The effects of B subunits on the interaction between PP2A and IKKβ were analysed by immunoprecipitation assay. Cells were transfected with plasmids encoding Flag‐tagged CαD85N, HA‐tagged Aα, HA‐tagged B subunits and HA‐tagged IKKβ. PP2A complexes were immunoprecipitated by using Flag‐agarose beads from cells, and the association of IKKβ proteins to the PP2A complexes was analysed by immunoblotting (Fig. 5). This assay revealed that IKKβ associates to PP2A, and B, B′ and B″ subunits block the association. These results suggest that the preference of AC, ABC, AB′′C and AB′′′C to dephosphorylate NF‐κB signalling proteins is regulated by the binding features of B subunits to the substrate proteins in cells.
Figure 5.
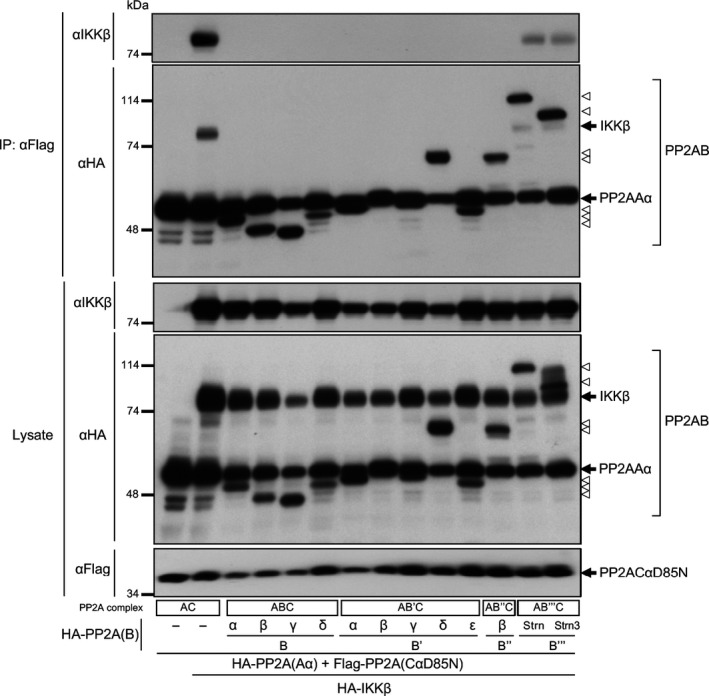
PP2A B, B′ and B′′ subunits suppress the association of PP2A AC to IKKβ in vivo. Cells were transfected with plasmids encoding Aα, CαD85N mutant and subunits of B families together with IKKβ plasmids. Following incubation for 24 h after transfection, PP2A complexes were immunoprecipitated and the association of IKKβ proteins to the PP2A complexes was analysed by immunoblotting.
Discussion
Many studies have revealed important and complicated roles of PP2A in IKKβ regulation. For example, it has been reported that PP2A suppresses IKKβ activity in vitro and in vivo 7, 15, 16. However, another contradictory study shows that PP2A forms a stable complex with IKKβ and promotes its kinase activity in vivo 28. We found here that AC and AB′′′C dephosphorylate IKKβ in cells, whereas ABC, AB′C and AB′′C do not. These results suggest the possibility that ABC, AB′C and AB′′C complexes positively regulate IKKβ by competing against AC and AB′′′C complexes. The data in this study may help to reconcile the seemingly contradictory observations that PP2A both downregulates and promotes IKKβ activity. To confirm the specific regulation by distinct PP2A complexes, we tried to knockdown of multiple isoforms of each B family protein by RNAi. However, RNAi screen in GP2‐293 cells did not clarify distinct functions of each isoform of B family protein, because expression levels of B and B‴ family proteins are very low in GP2‐293 cells, and these cells express multiple isoforms of B′ and B″ family proteins.
NF‐κB signalling is not uniformly regulated by PP2A but rather is subjected to specific regulation by distinct PP2A complexes at multiple steps. In contrast to AC and AB′′′C, which dephosphorylate IKKβ, IκBα and RelA, ABC dephosphorylates IκBα but not IKKβ and RelA in cells. These results indicate that ABC suppresses NF‐κB activity without inhibition of IKKβ. A recent study revealed that IKKβ not only activates NF‐κB through the phosphorylation of IκBα but also regulates many cellular functions by phosphorylating various proteins in an NF‐κB‐independent manner 29. For example, IKKβ regulates autophagy 30, mRNA stability 31, apoptosis 32, angiogenesis 33 and cellular vesicle trafficking 34 by phosphorylating various proteins that are unrelated to IκBα. Thus, ABC potentially suppresses NF‐κB without inhibiting these NF‐κB‐independent functions of IKKβ, thereby ensuring that PP2A regulates NF‐κB activity and IKKβ‐mediated cellular responses independently.
Among the four classes of B subunit families, the B′′′/Strn family proteins only facilitate dephosphorylation of three proteins including IKKβ, IκBα and RelA. Members of the B′′′/Strn family are evolutionarily conserved and have critical roles in biological processes such as development and cell growth 35, 36. B′′′/Strn family proteins form a large complex termed Striatin‐interacting phosphatase and kinase (STRIPAK) along with the germinal centre kinase family and many adaptor proteins, and recruit C subunits via A subunits of PP2A in the complexes. STRIPAK complexes have a critical role in protein dephosphorylation and act as important regulators of multiple vital signalling pathways, including the Hippo pathway, mitogen‐activated protein kinases and cytoskeleton remodelling. Recent studies suggest that the dysregulation of STRIPAK complexes correlates with human diseases including cancer 35, 36, 37. Regulation of NF‐κB signalling pathway by AB‴C may, therefore, be involved in STRIPAK complex‐related biological processes and cancer.
PP2A is a confirmed tumour suppressor protein that is genetically altered or functionally inactivated in many cancers 22, 23, 24. Its A and C subunits are reportedly inactivated through several mechanisms including somatic mutation, phosphorylation, methylation, and/or increased expression of PP2A inhibitors, and inactivation of PP2A is linked to cancer progression. Expression of Bα, Bβ and Bγ is also decreased in many cancers owing to deletion, point mutation and DNA hypermethylation 38, 39, 40, 41, 42, 43, 44. Furthermore, it has been revealed that IKKβ and NF‐κB signalling pathways are linked to inflammation and cancer 45. Therefore, the regulation of IKKβ and NF‐κB signalling by distinct PP2A complexes may be involved in multiple human diseases including cancer, and thus might serve as specific and novel targets for disease therapy.
Author contributions
HK and YT designed and performed the experiments, interpreted the study, and wrote the paper. KO, MK, YN, ET and TH performed the experiments.
Acknowledgements
This work was supported, in whole or in part, by Grants‐in‐Aid from the Japan Society for the Promotion of Science (JSPS) grant numbers #15H01406, #15H01382, and #23370062.
Edited by Kazuhiro Iwai
References
- 1. Hayden MS and Ghosh S (2008) Shared principles in NF‐kappaB signaling. Cell 132, 344–362. [DOI] [PubMed] [Google Scholar]
- 2. Vallabhapurapu S and Karin M (2009) Regulation and function of NF‐kappaB transcription factors in the immune system. Annu Rev Immunol 27, 693–733. [DOI] [PubMed] [Google Scholar]
- 3. Hinz M and Scheidereit C (2014) The IkappaB kinase complex in NF‐kappaB regulation and beyond. EMBO Rep 15, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rieser E, Cordier SM and Walczak H (2013) Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci 38, 94–102. [DOI] [PubMed] [Google Scholar]
- 5. Iwai K (2014) Diverse roles of the ubiquitin system in NF‐kappaB activation. Biochim Biophys Acta 1843, 129–136. [DOI] [PubMed] [Google Scholar]
- 6. Shimizu Y, Taraborrelli L and Walczak H (2015) Linear ubiquitination in immunity. Immunol Rev 266, 190–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E and Karin M (1997) A cytokine‐responsive IkappaB kinase that activates the transcription factor NF‐kappaB. Nature 388, 548–554. [DOI] [PubMed] [Google Scholar]
- 8. Delhase M, Hayakawa M, Chen Y and Karin M (1999) Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 284, 309–313. [DOI] [PubMed] [Google Scholar]
- 9. Sakurai H, Chiba H, Miyoshi H, Sugita T and Toriumi W (1999) IkappaB kinases phosphorylate NF‐kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem 274, 30353–30356. [DOI] [PubMed] [Google Scholar]
- 10. Schwabe RF and Sakurai H (2005) IKKbeta phosphorylates p65 at S468 in transactivaton domain 2. FASEB J 19, 1758–1760. [DOI] [PubMed] [Google Scholar]
- 11. Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L and Greene WC (2005) NF‐kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol 25, 7966–7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brautigan DL (2013) Protein Ser/Thr phosphatases–the ugly ducklings of cell signalling. FEBS J 280, 324–345. [DOI] [PubMed] [Google Scholar]
- 13. Shi Y (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. [DOI] [PubMed] [Google Scholar]
- 14. Li HY, Liu H, Wang CH, Zhang JY, Man JH, Gao YF, Zhang PJ, Li WH, Zhao J, Pan X et al (2008) Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat Immunol 9, 533–541. [DOI] [PubMed] [Google Scholar]
- 15. Fu DX, Kuo YL, Liu BY, Jeang KT and Giam CZ (2003) Human T‐lymphotropic virus type I tax activates I‐kappa B kinase by inhibiting I‐kappa B kinase‐associated serine/threonine protein phosphatase 2A. J Biol Chem 278, 1487–1493. [DOI] [PubMed] [Google Scholar]
- 16. Li S, Wang L, Berman MA, Zhang Y and Dorf ME (2006) RNAi screen in mouse astrocytes identifies phosphatases that regulate NF‐kappaB signaling. Mol Cell 24, 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prajapati S, Verma U, Yamamoto Y, Kwak YT and Gaynor RB (2004) Protein phosphatase 2Cbeta association with the IkappaB kinase complex is involved in regulating NF‐kappaB activity. J Biol Chem 279, 1739–1746. [DOI] [PubMed] [Google Scholar]
- 18. Chiang CW, Liu WK, Chiang CW and Chou CK (2011) Phosphorylation‐dependent association of the G4‐1/G5PR regulatory subunit with IKKbeta negatively modulates NF‐kappaB activation through recruitment of protein phosphatase 5. Biochem J 433, 187–196. [DOI] [PubMed] [Google Scholar]
- 19. Pons S and Torres‐Aleman I (2000) Insulin‐like growth factor‐I stimulates dephosphorylation of ikappa B through the serine phosphatase calcineurin (protein phosphatase 2B). J Biol Chem 275, 38620–38625. [DOI] [PubMed] [Google Scholar]
- 20. Yang J, Fan GH, Wadzinski BE, Sakurai H and Richmond A (2001) Protein phosphatase 2A interacts with and directly dephosphorylates RelA. J Biol Chem 276, 47828–47833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chew J, Biswas S, Shreeram S, Humaidi M, Wong ET, Dhillion MK, Teo H, Hazra A, Fang CC, López‐Collazo E et al (2009) WIP1 phosphatase is a negative regulator of NF‐kappaB signalling. Nat Cell Biol 11, 659–666. [DOI] [PubMed] [Google Scholar]
- 22. Sangodkar J, Farrington CC, McClinch K, Galsky MD, Kastrinsky DB and Narla G (2016) All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J 283, 1004–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eichhorn PJ, Creyghton MP and Bernards R (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795, 1–15. [DOI] [PubMed] [Google Scholar]
- 24. Westermarck J and Hahn WC (2008) Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med 14, 152–160. [DOI] [PubMed] [Google Scholar]
- 25. Xing Y, Xu Y, Chen Y, Jeffrey PD, Chao Y, Lin Z, Li Z, Strack S, Stock JB and Shi Y (2006) Structure of protein phosphatase 2A core enzyme bound to tumor‐inducing toxins. Cell 127, 341–353. [DOI] [PubMed] [Google Scholar]
- 26. Cho US and Xu W (2007) Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57. [DOI] [PubMed] [Google Scholar]
- 27. Tsuchiya Y, Asano T, Nakayama K, Kato T Jr, Karin M and Kamata H (2010) Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV‐induced NF‐kappaB activation. Mol Cell 39, 570–582. [DOI] [PubMed] [Google Scholar]
- 28. Kray AE, Carter RS, Pennington KN, Gomez RJ, Sanders LE, Llanes JM, Khan WN, Ballard DW and Wadzinski BE (2005) Positive regulation of IkappaB kinase signaling by protein serine/threonine phosphatase 2A. J Biol Chem 280, 35974–35982. [DOI] [PubMed] [Google Scholar]
- 29. Chariot A (2009) The NF‐kappaB‐independent functions of IKK subunits in immunity and cancer. Trends Cell Biol 19, 404–413. [DOI] [PubMed] [Google Scholar]
- 30. Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S et al (2010) The IKK complex contributes to the induction of autophagy. EMBO J 29, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM et al (2011) The IkappaB kinase complex regulates the stability of cytokine‐encoding mRNA induced by TLR‐IL‐1R by controlling degradation of regnase‐1. Nat Immunol 12, 1167–1175. [DOI] [PubMed] [Google Scholar]
- 32. Yan J, Xiang J, Lin Y, Ma J, Zhang J, Zhang H, Sun J, Danial NN, Liu J and Lin A (2013) Inactivation of BAD by IKK inhibits TNFalpha‐induced apoptosis independently of NF‐kappaB activation. Cell 152, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC et al (2007) IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130, 440–455. [DOI] [PubMed] [Google Scholar]
- 34. Nair‐Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y, Banerjee M, Overholtzer M, Roche PA, Tampé R et al (2014) TLR signals induce phagosomal MHC‐I delivery from the endosomal recycling compartment to allow cross‐presentation. Cell 158, 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi Z, Jiao S and Zhou Z (2016) STRIPAK complexes in cell signaling and cancer. Oncogene 35, 4549–4557. [DOI] [PubMed] [Google Scholar]
- 36. Hwang J and Pallas DC (2014) STRIPAK complexes: structure, biological function, and involvement in human diseases. Int J Biochem Cell Biol 47, 118–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madsen CD, Hooper S, Tozluoglu M, Bruckbauer A, Fletcher G, Erler JT, Bates PA, Thompson B and Sahai E (2015) STRIPAK components determine mode of cancer cell migration and metastasis. Nat Cell Biol 17, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bluemn EG, Spencer ES, Mecham B, Gordon RR, Coleman I, Lewinshtein D, Mostaghel E, Zhang X, Annis J, Grandori C et al (2013) PPP2R2C loss promotes castration‐resistance and is associated with increased prostate cancer‐specific mortality. Mol Cancer Res 11, 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng Y, Liu W, Kim ST, Sun J, Lu L, Sun J, Zheng SL, Isaacs WB and Xu J (2011) Evaluation of PPP2R2A as a prostate cancer susceptibility gene: a comprehensive germline and somatic study. Cancer Genet 204, 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y et al ; METABRIC Group (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mosca L, Musto P, Todoerti K, Barbieri M, Agnelli L, Fabris S, Tuana G, Lionetti M, Bonaparte E, Sirchia SM et al (2013) Genome‐wide analysis of primary plasma cell leukemia identifies recurrent imbalances associated with changes in transcriptional profiles. Am J Hematol 88, 16–23. [DOI] [PubMed] [Google Scholar]
- 42. Muggerud AA, Musto P, Todoerti K, Barbieri M, Agnelli L, Fabris S, Tuana G, Lionetti M, Bonaparte E, Sirchia SM et al (2010) Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res 12, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ruvolo PP, Qui YH, Coombes KR, Zhang N, Ruvolo VR, Borthakur G, Konopleva M, Andreeff M and Kornblau SM (2011) Low expression of PP2A regulatory subunit B55alpha is associated with T308 phosphorylation of AKT and shorter complete remission duration in acute myeloid leukemia patients. Leukemia 25, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan J, Lee PL, Li Z, Jiang X, Lim YC, Hooi SC and Yu Q (2010) B55beta‐associated PP2A complex controls PDK1‐directed myc signaling and modulates rapamycin sensitivity in colorectal cancer. Cancer Cell 18, 459–471. [DOI] [PubMed] [Google Scholar]
- 45. DiDonato JA, Mercurio F and Karin M (2012) NF‐kappaB and the link between inflammation and cancer. Immunol Rev 246, 379–400. [DOI] [PubMed] [Google Scholar]


