Abstract
Breast cancer is the most common cancer in women around the world. So far, many attempts have been made to treat this disease, but few effective treatments have been discovered. In this work, we reviewed the related articles in the limited period of time, 2000–2016, through search in PubMed, Scopus database, Google Scholar, and psychology and psychiatry literature (PsycINFO). We selected the articles about the correlation of microRNAs (miRNAs) and breast cancer in the insight into therapeutic applicability from mentioned genetics research databases. The miRNAs as an effective therapy for breast cancer was at the center of our attention. Hormone therapy and chemotherapy are two major methods that are being used frequently in breast cancer treatment. In the search for an effective therapy for breast cancer, miRNAs suggest a promising method of treatment. miRNAs are small, noncoding RNAs that can turn genes on or off and can have critical roles in cancer treatment; therefore, in the near future, usage of these biological molecules in breast cancer treatment can be considered a weapon against most common cancer-related concerns in women. Here, we discuss miRNAs and their roles in various aspects of breast cancer treatment to help find an alternative and effective way to treat or even cure this preventable disease.
Keywords: Breast cancer, chemotherapy, hormone therapy, microRNA, treatment
INTRODUCTION
Breast cancer strikes one million women around the world every year and is the most common cancer among American women. According to the January 2013 statistics of the World Health Organization, breast cancer is the most common malignancy among women, and it is the second leading cause of death for women around the world, accounting for 450,000 deaths annually. The incidence of this disease is increasing so that it is replacing heart disease as the first cause of death. Therefore, breast cancer can be considered a major cause of death among all cancer types.[1] Despite progressions in early diagnosis and treatment, breast cancer has remained the most common cancer-related cause of death among women living in developed countries.[2] Breast cancer is not a single disease, but rather this phenotype consists of a spectrum of distinct tumors. Although these tumors have different treatments and therapeutic outcomes, they are developed in a single organ.[3]
This has led to some research in the field of combining biological markers for improvement in disease diagnosis and prognosis.
There are six major subtypes of breast cancer including luminal A, luminal B, tumor enriched with human epidermal growth factor receptor 2 (HER2), triple-negative, normal-like, and claudin-low subtype.[4]
Triple-negative breast cancer (which does not express estrogen receptor [ER], progesterone receptor [PR], and HER-2) has a higher disease incidence rate and a higher risk of death as compared with other breast cancer subtypes.[5] Often, this cancer leads to death through metastasis.
Metastasis is a multistep process through which cancer cells invade distant tissues. In this process, primary tumor cells attack peripheral tissues and enter the blood and lymphatic systems. These cells can eventually lead to secondary tumor development in the destination tissue.[6]
One of the phenomena metastasis needs is vasculation, a crucial process that leads to new vessel development. This process is necessary for tumor growth, invasion, and metastasis.
Despite all progressions in cancer research, the molecular basis of malignancy has remained unknown as one of the most challenging aspects of the disease.[1]
Although early diagnosis of breast cancer has caused a decrease in death rates, prevention and treatment are still considered a general concern.
Each factor that can interfere with paths involved in breast cancer development would have at least a role in the improvement of disease treatment and the outcome which cancer patients receive. One such factor is microRNA (miRNA) molecule which can control many cancer development pathways.
MICRORNAS
miRNA biogenesis is a multistep process that begins in the cell nucleus. Pri-miRNA molecule is coded by RNA polymerase II enzyme, and Pri-miRNA expression occurs in distinctive genomic regions. There are many intergenic miRNAs with independent promoters. However, other miRNAs are clustered in poly cistronic transcripts. Specific miRNAs are located in the host intronic regions. This suggests that miRNAs’ biogenesis can be regulated by the host's gene promoters. Pri-miRNAs are cut down to 70–85 nucleotide molecules called pre-miRNAs. Pre-miRNAs have impaired stem-loop structure. This molecule is transported to the cytoplasm, where it is cut down more to transient double-strand miRNAs. One of these strands is mature miRNA sequence, with about 22 nucleotides in length. Another one is the complementary strand (usually shown with a star sign).
microRNP (miRNA plus ribonucleoprotein) complex separates the two strands from each other. One of these strands turns into functionally active miRNA while the other one degrades. miRNAs demonstrate high conserved evolution in different species. The regulatory function of miRNAs can be inferred from this issue that miRNAs have different levels of expression in various types of cells. miRNAs play their role through one of the two following ways: (1) through mRNA degradation process or (2) through protein expression suppression process. Most of the animal miRNAs bind to the 3’UTR of the targeted mRNA. However, miRNAs can be effective through binding to coding sequences or the 5’UTR of targeted mRNA. Complete or nearly complete binding to mRNAs can lead to gene silencing. However, most of the times, this binding is noncomplete. Therefore, this leads just to translation suppression and eventually to a decrease in protein level. There is a specific region in miRNA sequence with two to eight nucleotides in length located in 5’ miRNA terminal that is called “seed sequence.” This region of miRNA sequence is important for binding to mRNA molecules. Computer software has predicted that every miRNA has the potential to regulate 200 mRNAs. Hence, miRNAs are responsible for the regulation of at least one-third of human mRNAs.[7,8,9] Figure 1 depicts the process which miRNAs pass to be produced.
Figure 1.
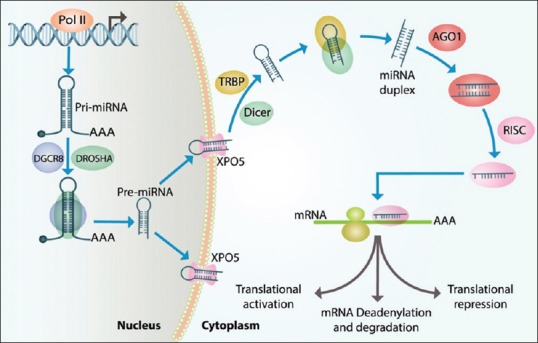
The process of miRNA expression and function: miRNA-encoding genes transcribed by RNA polymerase II enzyme in nucleus and led to production of a hairpin-formed molecule called “Pri-miRNA”. Pri-miRNA molecule turns into a precursor molecule called “Pre-miRNA” molecule by DROSHA and DGCR8 molecules’ action. Then, pre-miRNA enters cytoplasm through a nuclear export receptor called “Exportin 5.” In cytoplasm, this precursor is cut by Dicer-TRBP to produce a double-strand molecule known as “miRNA dublex.” After this process, one of these two strands remains as an active strand which can repress or even activate the target downstream genes in transcription or translation level. miRNA: microRNAs
miRNAs can be extracted from some body fluids such as serum, plasma, urine, and cells and tissues.[10]
miRNAs are degraded hardly, due to their small size. These small biological molecules remain in pristine in paraffin-embedded tissues for 10 years. Some miRNAs, which are present in blood circulation, can be measured in peripheral blood and serum easily.[11]
miRNA-encoding genes constitute one percent of the whole genome, while they control the expression of 30% of genes.[8] These molecules can increase and decrease the expression level of various genes.[12] MiRNAs may regulate the function of more than one gene, and one gene can also be regulated by the function of more than one miRNA.
So far, 474 mature miRNA-encoding genes are known in the human genome, while the computerized prediction for this number was more than 1000.[13]
Through gene regulation, miRNAs play critical roles in the normal biological processes such as differentiation, proliferation, and apoptosis. The relationship of miRNAs with disease severity, response to treatment, and outcome in several types of cancer has been established.[14] Although miRNAs’ roles in a wide range of cancer types are identified formerly, about breast cancer, there is a missed ring that should be found to create a link which would be scrutinized below.
CHANGED LEVEL OF EXPRESSION OF MIRNAS IN CANCER
Half of the known human miRNAs are related to cancer.[15] The expression levels of most of the miRNAs change in cancer. This changed expression level has effects on disease progression, severity, remission, and all aspects of disease.
The change of miRNA expression level has been previously established in a range of cancers, including lungs, prostate, thyroid, hepatocellular, pancreatic, gastric, colorectal, testicular, glioblastoma, and lymphatic cancers.
Additionally, each tissue has a specific amount of miRNAs called miRNA profile. This amount can vary from a few to millions. Also, a specific tumor has a different and unique miRNA profile from another one; therefore, miRNA levels in tumors are useful tool to determine metastasis resource and cancer subgrouping.[16]
Most of the studies focus on a decrease in miRNAs in breast cancer, while these biological molecules can also increase in a cancer patient's blood compared to a healthy subject's blood. miRNA profile has been checked in several breast cancer patients while their tumors were not resected yet. After surgery and excision of tumor out of patients’ breast tissue, the miRNA profile has been measured again for each patient. Outstanding decreases were seen in patients when these two miRNA profiles were compared with each other. The amount of these molecules has been dropped in patients’ blood after tumor resection. This issue provides that these molecules might be released from tumorous tissue to blood through exosome, microvesicles, and apoptotic bodies. However, the precise process through which these molecules are released to blood is yet to be answered.[17,18]
Some studies have accentuated that just specific miRNAs increase in cancer patients’ blood.[19]
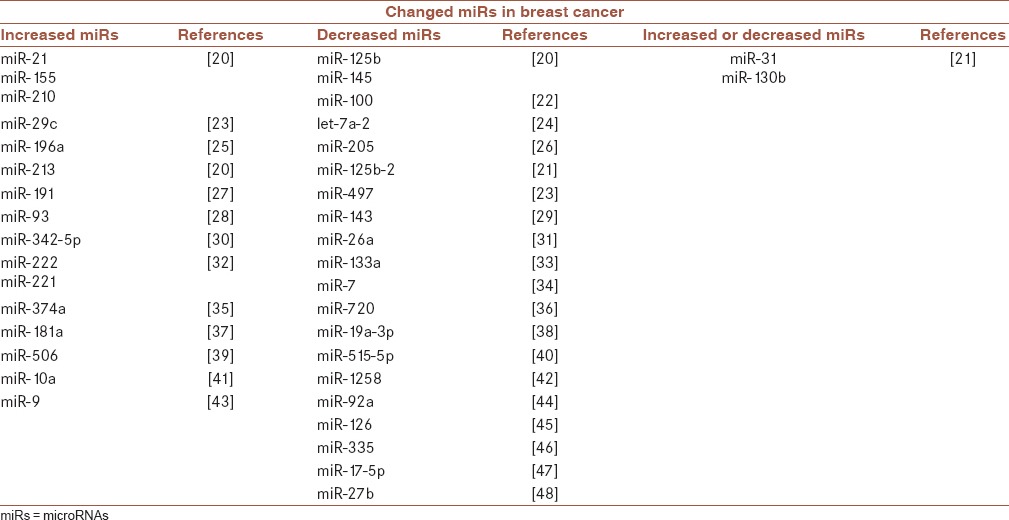
In breast cancer, there are also obvious differences in miRNA expression levels between normal and tumorous tissues. Several miRNAs always incur an obvious change in their expression levels in breast cancer. These miRNAs are shown in Table 1.
Table 1.
microRNAs that have changed expression level in breast cancer
In the screening of breast cancer samples based on different expression levels of miRNAs in tumorous and pri-tumorous tissues, it was found that miR-139-5p will drop in human breast cancer tumors.[49]
Some miRNAs in breast cancer have a different expression level in tumorous tissues compared to normal tissues. MiRNAs can also be useful in determining ER, PR (like miR-30), HER-2 status for breast cancer tumors, and for tumor subgrouping.[20]
Zhang et al. found a correlation between a decreased level of expression of miR-125b and poor survival rates in patients. They also found that upregulation of miR-125b leads to proliferation suppression and colonization suppression in cell lines.[50]
In the past years, the relevance between downregulation of miR-126 and miR-126* with poor prognosis has been established. In some studies, more than 15 miRNAs have been reported to be changed to develop potential of metastasis in breast cancer cell lines.[51,52]
Breast tumors consist of different cell populations, some of which are considered stem cells. Due to stem cell's abilities to self-proliferate and self-innovate, they may contribute to tumor development.[53]
miRNAs have the ability to regulate the expression of several genes simultaneously, which makes these molecules appropriate candidates for regulating self-proliferation process and determining cell destiny in stem cells. There are documents proving that specific miRNAs have a specific amount of expression in stem cells.[54]
miRNAs are necessary for G1/S cell cycle transition in stem cells, so manipulating such molecules can lead to facilitating the regulation of stem cells’ population.[55] Over the past 8 years, studies about miRNAs have led to information about the molecular basis of the disease, a tool for molecular subgrouping of disease, detecting new predisposing genes to breast cancer, and new biomarkers with diagnosis and prognosis application.[56]
MICRORNAS AS CANCER-CONTROLLING GENES
Various miRNAs have important roles in different stages of cancer [Figure 2].
Figure 2.
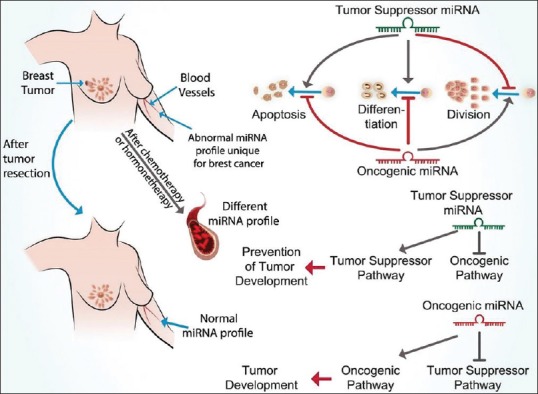
Assortment of miRNAs based on their functions and different roles in breast tumors: MicroRNAs can be divided into two general groups as tumor suppressor and oncogenic miRNAs. Each of these groups act with their specific effects on different pathways in cell as shown in the figure. Alterations in the amount of these miRNAs constitute a specific profile as microRNA profile, which is changed after resection of tumor, chemotherapy and hormonetherapy
Calin et al. found that half of known miRNAs are located in fragile or cancer-related genomic regions, so the role of these biological molecules in cancer is reinforced.[57]
miRNAs can have either an oncogenic or a tumor suppressor role in cancer development and the role they play depends on the path they activate or suppress. miRNAs play role in different biological events, including cell proliferation, tumor development, and migration.[58]
Therefore, they regulate gene expression in the process of tumor development and metastasis. Even it has been hypothesized that miRNAs are useful tools for suppression of oncogenic-miRNAs and for induction of miRNAs with tumor suppressor role.[11]
There are some documents verifying that miRNAs have role in embryogenesis and organogenesis in fetals.[59]
It has recently been demonstrated that miRNAs can suppress cell proliferation and induce apoptosis through their roles as a tumor suppressor.[60]
ONCOGENIC MICRORNAS
Oncogenic miRNAs are referred to all miRNAs which can contribute to help tumor cells to develop and remain. Every oncogenic miRNA exerts its role through one or more than one cancer-controlling pathways.
miR-21 as an oncogene is upregulated in breast cancer, especially in breast cancer with a high proliferation rate and severe phenotype, like pregnancy-associated breast cancer, and through anti-apoptosis function which leads to cell proliferation. Another reason verifies that miR-21 has an oncogenic role is that miR-21 suppresses two tumor suppressor genes called TPM1 and PDCD4.[61,62,63]
Downregulation of this miRNA causes tumor growth suppression. There is also an inverse relationship between miR-21 expression and PTEN gene expression. Therefore, PTEN is one of the target genes for miR-21.[63,64]
miR-200a is one of the miR-200 family members that has a role in breast cancer progression and distant metastasis.[65]
Nassirpour et al. found that miR-221 is upregulated in triple-negative breast cancer, and if this miRNA is knocked out, cell cycle suspension, apoptosis, inhibition of cell proliferation, and tumor growth inhibition will be induced.[66]
miR-155 also has an oncogenic role in triple-negative breast cancer and downregulates a tumor suppressor gene called VHL (Von Hippel–Lindau) through which it leads to angiogenesis in tumor. This miRNA contributes to migration and cell invasiveness.[61,67]
miR-9 plays a role as metastasis regulator through increasing tumor invasiveness and angiogenesis.[20]
In addition, some other oncogenic miRNAs are shown in Table 2. miRNAs are so necessary for the metastasis process that recently, the term “metasta miRNA” was coined to emphasize this role of miRNAs in migration, invasiveness, and their correlation with poor prognosis in cancer.[58]
Table 2.
Tumor suppressor microRNAs and oncogenic microRNAs and an excerpt of their roles in breast cancer

TUMOR SUPPRESSOR MICRORNAS
Tumor suppressor miRNAs are all miRNAs which act against tumor cells through different pathways. Every tumor suppressor has its own spectrum of strategies.
miR-205 acts as a tumor suppressor and is downregulated in triple-negative breast cancer. This miRNA normally is upregulated by the PTEN gene.[107]
miR-125b inhibits tumor growth, colony development, and proliferation.[108]
Upregulation of miR-125a/b decreases HER-2 in transcriptional and protein level and eventually leads to decreased cell mobility and invasiveness. miR-125a inhibits cell growth and decreases migration and cell proliferation in healthy status. Altogether, miR-125b has an antiproliferative role.[77,109]
As the expression level of miR-125a/b and miR-205 declines, the expression level of HER-2 gene rises and leads to invasiveness.[77]
miR-126 and miR-126* suppress metastasis and angiogenesis in breast cancer, so this miRNA is downregulated in this common cancer.[51,52]
Tavazoie et al. demonstrated that induction of silenced metastasis-related miRNAs in malignant cells can suppress metastasis to lungs and bones in metastatic breast cancer. miR-126 and miR-335 are downexpressed, and sometimes complete silencing of miR-335 is reported in breast cancer. Induction of miR-126 decreases the tumor proliferation rate and induction of miR-335 leads to suppress migration, invasiveness, and metastasis. Therefore, these miRNAs are known as metastasis suppressors.[52,110]
miR-200c has a role in the inhibition of cell proliferation, so this miRNA is known as a tumor suppressor.[111] Upregulation of miR-200c sensitizes cells to radiotherapy through inhibition of cell proliferation, increase in apoptosis, and development of double-strand DNA breakage.[112]
This miRNA decreases the metastasis rate by suppression of metastasis-associated proteins.[69]
Reinforcement of miR-200 family expression has resulted in the formation of “inhibition of metastasis” hypothesis.[113]
Another miR-200 family function is containment of EMT (epithelial–mesenchymal transition),[51] and its proof is downregulation of miR-200 family in EMT event in cell culture model.[114]
miRNAs can be inoculated in tumorous tissues, which prevents disease progression and even leads to tumor shrinkage.
miR-127 is downregulated in 75% of cancer patients while this miRNA increases after treatment. Downregulation of this miRNA correlates to overexpression of a proto-oncogene called BCL6. Thus, miR-127 has a protective role in breast cancer and can be considered a new treatment strategy.[64,115]
Another tumor suppressor miRNA is miR-206. This miRNA can prevent cell growth, migration, and invasiveness and is eventually able to inhibit metastasis. This miRNA also has role in apoptosis, and its downregulation leads to advanced stage of disease and shorter overall survival.[52]
miR-497 is a tumor suppressor miRNA whose upregulation can decrease cell mobility and proliferation and restrain cell cycle progression in G1 phase.[60]
miR-17-5p is known as a possible tumor suppressor since this miRNA is downregulated in breast cancer. miR-429 and miR-141 through suppression of metastasis-associated proteins lead to a reduced metastasis rate.[47]
miR-31 is downregulated in breast cancer and can limit metastasis through suppression of invasiveness and colonization. Recently, it has been reported that miR-31 can induce apoptosis and sensitize cells to anticancer therapy through suppression of the BCL2 gene.
miR-7 suppresses mobility and invasiveness and represses the potential of connection-free growth in breast cancer cells. The ability of stem-like cells in metastasis to the brain in the animal breast cancer model is suppressed by this miRNA. The expression of this miRNA was found to be reduced in brain metastasis.
miR-15b/16 has an anti-angiogenic effect.[10,52,77,109]
Hormone receptors and epidermal growth factor receptors are known today as diagnostic indicators, and several miRNAs are recognized as their regulator.[47]
For example, Cui et al. demonstrated that miR-133a may have a tumor suppressor role in breast cancer, since this miRNA, through targeting epidermal growth factor receptor, can regulate cell cycle and cell proliferation.[33]
Leivonen et al. found that miR-342-5p prevents the growth of HER-2-positive cells in vitro.[30]
Recently, it has been demonstrated that miR-26 is downregulated in breast cancer and has a regulatory role in cell proliferation and apoptosis.[31]
Zhang et al. found that miR-30a can interfere with migration and invasiveness in breast cancer, and the expression of this miRNA has an inverse relationship with metastasis to lymphatic nodes and lungs. As a result, loss of this miRNA can lead to the beginning of metastasis.[116]
Induction of miR-720 noticeably causes suppression of invasiveness and migration of breast cancer cells in vitro and in vivo.[38] Several other miRNAs are also shown in Table 2.
miRNAs can be considered a promising treatment target in the near future for their roles as tumor suppressors. The treatment method of the expression level of tumor suppressor miRNAs is called “alternative miRNA therapy.”
MICRORNAS AS PROGNOSTIC AND DIAGNOSTIC TOOLS
miRNAs are also recognized as predictors for disease development and progression.[9]
Using miRNAs as such predictors seems beneficial because this method has the advantage of being noninvasive. miRNAs could be biological markers in cancer for the following reasons:
MICRORNAS AND BREAST CANCER SUBGROUPING
There are three major subgroups in breast cancer:
ER and PR positive
HER-2 positive
Triple negative (ER, PR, and HER-2 negative).
However, using the micro array method for determining miRNAs profile, this subgrouping is extended to more precise one, including:
Luminal A (ER positive with low grade)
Luminal B (ER positive with high grade)
HER-2 positive
Basal like (is almost equal to triple-negative status).[4]
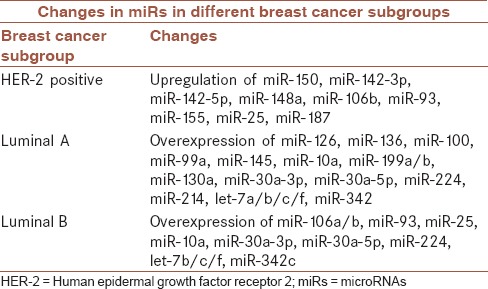
There are several miRNAs that are used for breast cancer subgrouping, as shown in Table 3.
Table 3.
microRNAs that are used for breast cancer subgrouping[74]
miRNAs can be used even as diagnostic tools to check whether a tumor is metastatic
miR-21 and miR-155 increased in metastatic tumors
miR-200 family decreased in metastatic tumors [118]
miR-21 has also been seen, especially in invasive breast cancer.[64]
Furthermore, Li et al. found that miR-720 downregulation is seen in breast cancer, and this decrease is especially seen in metastatic cells in this common cancer among women.[36]
There is an inverse relationship between miR-206 and ER alpha in ER-negative breast cancer and this miRNA upregulates in this form of breast cancer. Two binding sites for this miRNA have been found within 3’UTR of ER alpha gene, which can be one of the mechanisms leading to ER alpha gene silencing. On the other hand, inoculation of ER alpha agonist leads to miR-206 reduction. However, 64 other binding sites have been found in ER alpha gene for a variety of miRNAs, so each of these miRNAs can regulate this gene.[71]
The ability of miRNAs profiling in breast tumor subgrouping can be used currently. Therefore, this ability can help select cancer patients to receive adjuvant therapies. miRNA profiling can also be useful in discovering the molecular basis of breast cancer subgroups, and can therefore be effective in determining new therapy targets.
MICRORNAS AND ASSORTMENT OF PATIENTS BASED ON RESPONSE TO TWO CURRENT THERAPIES
One of the most important applications of miRNAs is patients’ assortment based on response to therapies.
There is a mutual relevance between miRNAs and anticancer agents. miRNAs can regulate the amount of sensitivity to therapy agents in cancer cells. Furthermore, anticancer agents can have an effect on miRNA expression levels.[119]
Thus, miRNAs seem to be appropriate tools for selecting a cancer treatment method.
RESPONSE TO CHEMOTHERAPY
Chemotherapy is the current therapeutic method that is being used for breast cancer, especially for triple-negative form, but the patients do not usually receive a desirable outcome. miRNAs can suggest a new alternative therapeutic method for yielding better breast cancer chemotherapy outcome.
Studies have shown that the expression level of miRNAs can relate to patients’ response to chemotherapy. For example, upregulation of miR-663 occurs in breast cancer and relates to chemoresistance.[120]
miRNAs can have a role in drug resistance. MiR-326 causes a reduction of drug resistance and is downregulated in progressed breast cancer.
Suppression of miR-21 leads to sensitize cancer cells to chemotherapy, and upregulation of this miRNA increases resistance to treatment with Taxol in breast cancer.
miR-125b causes increased drug resistance through suppression of a pro-apoptotic protein. miR-27 and transfection of doxorubicin-treated breast cancer cells with miR-451 lead to increased sensitivity to this drug. This is a good example for amending breast cancer patients’ responses to treatment through miRNA expression control. Hence, miR-451 and miR-27 lead to doxorubicin resistance while miR-200b and miR-200c increase sensitivity to doxorubicin. Thus, miR-200c relates to increased doxorubicin sensitivity and miR-326 regulates MRP1 (multidrug-resistant associated protein 1).[93,121,122]
Induction of miR-30c causes sensitivity to doxorubicin in vitro and in vivo. It is assumed that this miRNA acts as a tumor suppressor in doxorubicin-resistant breast tumors.[123]
miR-222 and miR-29a can contribute to docetaxel and adriamycin resistance development.[124]
miR-21 has an oncogenic role in breast cancer, so suppression of this miRNA can postpone tumor growth. On the other hand, induction of miRNAs with tumor suppressor role can lead to tumor shrinkage or prevent tumor progression. By the same token, in a study performed by Si et al. using anti-miR-21, the amount of this miRNA was reduced, and through inhibition of cell proliferation and promotion of apoptosis, led to decreased cell growth.[64]
Therefore, anti-miR-21 in combination with chemotherapy can reduce tumor growth. Considering that this miRNA (miR-21) has an oncogenic role, suppression of this molecule can sensitize tumor cells to anticancer therapies.
RESPONSE TO HORMONE THERAPY
By targeting a specific miRNA, there is a possibility of regulating different pathways involved in the development of hormone therapy resistance in breast cancer. In this process, the miRNAs that are silenced in breast cancer will be induced, and the miRNAs that are induced in breast cancer will also be suppressed.[14]
Tamoxifen is a drug used in ER-positive breast cancer and plays a role as an estrogen antagonist in the signal transduction process. miR-30c and miR-30a-3p have roles as predictors for tamoxifen efficacy in progressed breast cancer.[125,126]
miR-222, miR-221, and downregulation of miR-301 confer tamoxifen resistance, while induction of miR-15a and miR-16 can lead to sensitize cells to tamoxifen. Upregulation of miR-30c and miR-30a-3p contribute to an increased response to hormone therapy and increased disease progression-free survival. miR-375, miR-342, and miR-451 develop tamoxifen sensitivity while miR-210, miR-181a, miR-101, miR-30c, and miR-26a develop tamoxifen resistance.[127,128,129]
Trastuzumab is a drug against HER-2-positive breast cancer. Several miRNAs can target the HER-2 gene in HER-2-positive breast cancer patients. For example, miR-125b binds to the HER-2 gene 3’UTR through which prevents HER-2 overexpression and eventually leads to an increased response to trastuzumab drug in these patients. This miRNA declines in HER-2-positive breast cancer. Therefore, induction of this miRNA can be useful for HER-2-positive breast cancer treatment.[20,61,77]
Upregulation of miR-125b causes HER-2 protein reduction through which reduces migration, invasiveness, and tumor growth in breast cancer. MiR-331-3p also suppresses HER-2 expression and its downstream signal transduction. However, there are several other miRNAs that can regulate HER-2 expression.[77,130]
miR-221 and miR-222 are expressed two times as often in HER-2-positive breast cancer than normal status. Upregulation of miR-21 in HER-2-positive breast cancer patients causes trastuzumab resistance.[131]
CONCLUSION
miRNAs are tools with a wide scope of roles through affecting on genes with various applications. By manipulating these biological molecules, there is a possibility of influencing different processes involved in treatment, diagnosis, and prognosis of diseases. In particular, cancers have been mentioned as diseases in which miRNAs have many roles in their development and treatment. Since one of the most common cancers among women is breast cancer, miRNAs can also exert improvements in breast cancer treatment. There are also some advantages in miRNA-based therapy. Nowadays, we know that, using of miRNAs in different clinically grounds of cancers is simple and noninvasive, but the best thing is that, this technology does not have other methods’ side effects. So, in the near future, using of these smart and controller molecules would be turned into a dominant method to treat cancers, especially breast cancer, but it needs still researches to reach this goal and to be made applicable.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The figure and graphical abstract of this review article were designed by FB Scientific Art Design.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2013. Ann Oncol. 2013;24:792–800. doi: 10.1093/annonc/mdt010. [DOI] [PubMed] [Google Scholar]
- 3.Bertos NR, Park M. Breast cancer – One term, many entities? J Clin Invest. 2011;121:3789–96. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.D’Ippolito E, Iorio MV. MicroRNAs and triple negative breast cancer. Int J Mol Sci. 2013;14:22202–20. doi: 10.3390/ijms141122202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB, et al. MiR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8:e66502. doi: 10.1371/journal.pone.0066502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100:9779–84. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–13. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 13.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Iguchi H, Kosaka N, Ochiya T. Secretory microRNAs as a versatile communication tool. Commun Integr Biol. 2010;3:478–81. doi: 10.4161/cib.3.5.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 20.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 21.Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: Clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17:313–9. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 25.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, et al. MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–78. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 26.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Adams BD, Guttilla IK, White BA. Involvement of microRNAs in breast cancer. Semin Reprod Med. 2008;26:522–36. doi: 10.1055/s-0028-1096132. [DOI] [PubMed] [Google Scholar]
- 28.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, et al. P53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–50. [PubMed] [Google Scholar]
- 30.Leivonen SK, Sahlberg KK, Mäkelä R, Due EU, Kallioniemi O, Børresen-Dale AL, et al. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104. doi: 10.1016/j.molonc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Li L, Wu M, Liu M, Xie X, Guo J, et al. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One. 2013;8:e65138. doi: 10.1371/journal.pone.0065138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Cui W, Zhang S, Shan C, Zhou L, Zhou Z. MicroRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962–74. doi: 10.1111/febs.12398. [DOI] [PubMed] [Google Scholar]
- 34.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–79. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou X, et al. MiR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35:469–78. doi: 10.1093/carcin/bgt330. [DOI] [PubMed] [Google Scholar]
- 37.Zhang K, Zhang Y, Liu C, Xiong Y, Zhang J. MicroRNAs in the diagnosis and prognosis of breast cancer and their therapeutic potential (review) Int J Oncol. 2014;45:950–8. doi: 10.3892/ijo.2014.2487. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of fra-1 proto-oncogene. Oncogene. 2014;33:3014–23. doi: 10.1038/onc.2013.258. [DOI] [PubMed] [Google Scholar]
- 39.Arora H, Qureshi R, Park WY. MiR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One. 2013;8:e64273. doi: 10.1371/journal.pone.0064273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilam A, Edry L, Mamluk-Morag E, Bar-Ilan D, Avivi C, Golan D, et al. Involvement of IGF-1R regulation by miR-515-5p modifies breast cancer risk among BRCA1 carriers. Breast Cancer Res Treat. 2013;138:753–60. doi: 10.1007/s10549-013-2502-5. [DOI] [PubMed] [Google Scholar]
- 41.Hoppe R, Achinger-Kawecka J, Winter S, Fritz P, Lo WY, Schroth W, et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur J Cancer. 2013;49:3598–608. doi: 10.1016/j.ejca.2013.07.145. [DOI] [PubMed] [Google Scholar]
- 42.Tang D, Zhang Q, Zhao S, Wang J, Lu K, Song Y, et al. The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer. Clin Biochem. 2013;46:926–32. doi: 10.1016/j.clinbiochem.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 43.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–9. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene SB, Herschkowitz JI, Rosen JM. Small players with big roles: MicroRNAs as targets to inhibit breast cancer progression. Curr Drug Targets. 2010;11:1059–73. doi: 10.2174/138945010792006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang J, Ahmad A, Sarkar FH. The role of microRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci. 2012;13:13414–37. doi: 10.3390/ijms131013414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–8. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 49.Biagioni F, Bossel Ben-Moshe N, Fontemaggi G, Canu V, Mori F, Antoniani B, et al. MiR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–29. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wee EJ, Peters K, Nair SS, Hulf T, Stein S, Wagner S, et al. Mapping the regulatory sequences controlling 93 breast cancer-associated miRNA genes leads to the identification of two functional promoters of the Hsa-mir-200b cluster, methylation of which is associated with metastasis or hormone receptor status in advanced breast cancer. Oncogene. 2012;31:4182–95. doi: 10.1038/onc.2011.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells – Perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 54.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 55.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 56.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–24. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–62. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 60.Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, et al. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–9. [PubMed] [Google Scholar]
- 63.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 64.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY, et al. MiR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 65.Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389–99. doi: 10.1158/1078-0432.CCR-12-1959. [DOI] [PubMed] [Google Scholar]
- 66.Nassirpour R, Mehta PP, Baxi SM, Yin MJ. MiR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8:e62170. doi: 10.1371/journal.pone.0062170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–89. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–48. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A microRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 71.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 72.Liu H, Cao YD, Ye WX, Sun YY. Effect of microRNA-206 on cytoskeleton remodelling by downregulating cdc42 in MDA-MB-231 cells. Tumori. 2010;96:751–5. doi: 10.1177/030089161009600518. [DOI] [PubMed] [Google Scholar]
- 73.Negrini M, Calin GA. Breast cancer metastasis: A microRNA story. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 75.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szász AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 77.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC, et al. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 78.Zhai L, Ma C, Li W, Yang S, Liu Z. MiR-143 suppresses epithelial-mesenchymal transition and inhibits tumor growth of breast cancer through down-regulation of ERK5. Mol Carcinog. 2016;55:1990–2000. doi: 10.1002/mc.22445. [DOI] [PubMed] [Google Scholar]
- 79.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, et al. MiR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461–6. [PubMed] [Google Scholar]
- 80.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, et al. LIFR is a breast cancer metastasis suppressor upstream of the hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–7. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–14. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 85.Rivas MA, Venturutti L, Huang YW, Schillaci R, Huang TH, Elizalde PV, et al. Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res. 2012;14:R77. doi: 10.1186/bcr3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu F, Deng H, Yao H, Liu Q, Su F, Song E, et al. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 88.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kato M, Paranjape T, Müller RU, Nallur S, Gillespie E, Keane K, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. Elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–24. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. P53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 91.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–48. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 92.Huang L, Dai T, Lin X, Zhao X, Chen X, Wang C, et al. MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochem Biophys Res Commun. 2012;425:127–33. doi: 10.1016/j.bbrc.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 93.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–9. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 94.Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen YY, et al. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ. 2011;18:16–25. doi: 10.1038/cdd.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2016;291:22855. doi: 10.1074/jbc.A110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 97.Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, et al. MiR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–74. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 98.Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34:1889–99. doi: 10.1093/carcin/bgt107. [DOI] [PubMed] [Google Scholar]
- 99.Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, et al. Analysis of miR-195 and miR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–30. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 101.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 102.Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–45. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang H, Wang P, Li X, Wang Q, Deng ZB, Zhuang X, et al. Restoration of miR17/20a in solid tumor cells enhances the natural killer cell antitumor activity by targeting Mekk2. Cancer Immunol Res. 2014;2:789–99. doi: 10.1158/2326-6066.CIR-13-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fan M, Sethuraman A, Brown M, Sun W, Pfeffer LM. Systematic analysis of metastasis-associated genes identifies miR-17-5p as a metastatic suppressor of basal-like breast cancer. Breast Cancer Res Treat. 2014;146:487–502. doi: 10.1007/s10549-014-3040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.He T, Qi F, Jia L, Wang S, Song N, Guo L, et al. MicroRNA-542-3p inhibits tumour angiogenesis by targeting angiopoietin-2. J Pathol. 2014;232:499–508. doi: 10.1002/path.4324. [DOI] [PubMed] [Google Scholar]
- 106.Moriarty CH, Pursell B, Mercurio AM. MiR-10b targets Tiam1: Implications for Rac activation and carcinoma migration. J Biol Chem. 2010;285:20541–6. doi: 10.1074/jbc.M110.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Joerger AC, Fersht AR. Structure-function-rescue: The diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–42. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. MiR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–62. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 109.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6:575–83. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–31. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ren Y, Han X, Yu K, Sun S, Zhen L, Li Z, et al. MicroRNA-200c downregulates XIAP expression to suppress proliferation and promote apoptosis of triple-negative breast cancer cells. Mol Med Rep. 2014;10:315–21. doi: 10.3892/mmr.2014.2222. [DOI] [PubMed] [Google Scholar]
- 112.Lin J, Liu C, Gao F, Mitchel RE, Zhao L, Yang Y, et al. MiR-200c enhances radiosensitivity of human breast cancer cells. J Cell Biochem. 2013;114:606–15. doi: 10.1002/jcb.24398. [DOI] [PubMed] [Google Scholar]
- 113.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 114.Castilla MÁ, Díaz-Martín J, Sarrió D, Romero-Pérez L, López-García MÁ, Vieites B, et al. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS One. 2012;7:e47709. doi: 10.1371/journal.pone.0047709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 116.Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, et al. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–28. doi: 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- 117.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 119.Bu Y, Lu C, Bian C, Wang J, Li J, Zhang B, et al. Knockdown of dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncol Rep. 2009;21:13–7. [PubMed] [Google Scholar]
- 120.Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2) J Biol Chem. 2013;288:10973–85. doi: 10.1074/jbc.M112.434340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79:817–24. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 122.Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang B, et al. MiRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 2013;7:1579–84. doi: 10.3892/mmr.2013.1403. [DOI] [PubMed] [Google Scholar]
- 123.Fang Y, Shen H, Cao Y, Li H, Qin R, Chen Q, et al. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res. 2014;47:60–9. doi: 10.1590/1414-431X20133324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 125.Sehdev S, Martin G, Sideris L, Lam W, Brisson S. Safety of adjuvant endocrine therapies in hormone receptor-positive early breast cancer. Curr Oncol. 2009;16(Suppl 2):S14–23. doi: 10.3747/co.v16i0.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105:13021–6. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cittelly DM, Das PM, Salvo VA, Fonseca JP, Burow ME, Jones FE, et al. Oncogenic HER2{Delta} 16 suppresses miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of breast tumors. Carcinogenesis. 2010;31:2049–57. doi: 10.1093/carcin/bgq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodríguez-González FG, Sieuwerts AM, Smid M, Look MP, Meijer-van Gelder ME, de Weerd V, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;127:43–51. doi: 10.1007/s10549-010-0940-x. [DOI] [PubMed] [Google Scholar]
- 130.Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. MiR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–24. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]


