Abstract
ATP-binding cassette (ABC) transporters help export various substrates across the cell membrane and significantly contribute to drug resistance. However, a recent study reported an unusual case in which the loss of an ABC transporter in Candida albicans, orf19.4531 (previously named ROA1), increases resistance against antifungal azoles, which was attributed to an altered membrane potential in the mutant strain. To obtain further mechanistic insights into this phenomenon, here we confirmed that the plasma membrane–localized transporter (renamed CDR6/ROA1 for consistency with C. albicans nomenclature) could efflux xenobiotics such as berberine, rhodamine 123, and paraquat. Moreover, a CDR6/ROA1 null mutant, NKKY101, displayed increased susceptibility to these xenobiotics. Interestingly, fluorescence recovery after photobleaching (FRAP) results indicated that NKKY101 mutant cells exhibited increased plasma membrane rigidity, resulting in reduced azole accumulation and contributing to azole resistance. Transcriptional profiling revealed that ribosome biogenesis genes were significantly up-regulated in the NKKY101 mutant. As ribosome biogenesis is a well-known downstream phenomenon of target of rapamycin (TOR1) signaling, we suspected a link between ribosome biogenesis and TOR1 signaling in NKKY101. Therefore, we grew NKKY101 cells on rapamycin and observed TOR1 hyperactivation, which leads to Hsp90-dependent calcineurin stabilization and thereby increased azole resistance. This in vitro finding was supported by in vivo data from a mouse model of systemic infection in which NKKY101 cells led to higher fungal load after fluconazole challenge than wild-type cells. Taken together, our study uncovers a mechanism of azole resistance in C. albicans, involving increased membrane rigidity and TOR signaling.
Keywords: ABC transporter, membrane transport, mTOR complex (mTORC), multidrug transporter, yeast, Azole resistance, CDR6, TOR signaling
Introduction
Candida albicans is the most common human fungal pathogen; causes vaginal, oral and systemic diseases; and contributes to 9–12% of all hospital-acquired infections, with mortality rates approaching 50% (1, 2). Azoles, polyenes, and echinocandins are three major categories of antifungals used to combat invasive fungal infections (3). However, prolonged use of antifungals increases the frequency of the generation of multidrug resistance (MDR)2 strains, which show resistance to more than one class of drugs. Target alteration and the overexpression of its gene products are the most common strategies accounting for MDR phenomena in Candida species (3, 4). Among the various mechanisms of MDR, enhanced drug expulsion by MDR strains of Candida species represents a prominent strategy. Several studies report low levels of drug accumulation in C. albicans drug-resistant clinical isolates. This occurs predominantly due to overexpression of plasma membrane (PM)–localized drug-efflux transporter proteins Cdr1 and Cdr2 of the ATP-binding cassette (ABC) superfamily and Mdr1 of the major facilitator superfamily (5–7).
Apart from enhanced efflux of incoming drugs, which impacts drug susceptibility, the influx of drugs across the PM is also considered to play an important part in the development of drug tolerance. The PM acts as a partition between the extracellular and intracellular environment, and previous studies have shown that alterations in the physical state of the PM could impact intracellular drug accumulation and susceptibility of C. albicans cells to azoles (8–10). Interestingly, in yeast, the PM tension activates TORC2 (target of rapamycin complex 2), which promotes sphingolipid biosynthesis (11). Sphingolipids are important constituents of the detergent-insoluble microdomain of the PM known as lipid rafts, which play an important role as a platform for various signaling mechanisms (12–14). In C. albicans, the PM-bound transporter Cdr1 has been shown to be specifically localized within the raft region (15), and any imbalance in raft lipid constituents, such as sphingolipids or ergosterol, results in altered drug susceptibility (16). The occurrence of some of the transporter proteins in raft domains emphasizes their additional role in cell signaling.
In fungi, Hsp90 and calcineurin are two important regulators of cellular response to combat drug-induced stress. Hsp90 acts on various downstream effectors and is required for stability of calcineurin (17). In Saccharomyces cerevisiae, Tor1 (target of rapamycin 1) affects the Hsp90 activity in a positive manner by suppressing the expression of Kns1 kinase which phosphorylates Ckb1 (a regulatory subunit of CK2 kinase) and modulates its function (18, 19). CK2 kinase phosphorylates Hsp90 and represses its activity in S. cerevisiae and C. albicans (20, 21). A recent study has shown that inhibition of Tor1 leads to inhibition of Hsp90 activity, resulting in hypersensitivity to azoles in S. cerevisiae and C. albicans (22). Thus, hyperactivation of TOR signaling can be an important mechanism used by C. albicans to bypass azole toxicity.
The present study deals with a poorly characterized ORF (orf19.4531) of C. albicans belonging to the PDR subfamily, which has earlier been observed to be up-regulated in a number of drug-resistant clinical isolates (23, 24). A recent report by Jiang et al. (25), which appeared during the course of the present study, suggested that this orf19.4531-encoded protein is localized on a punctuated compartment adjacent to the vacuolar membrane and that its absence results in selective resistance to azoles, and these authors designated it as ROA1 (regulator of azole sensitivity). However, the study could not provide any mechanistic insight into this surprising phenotype.
We demonstrate that orf19.4531 actually codes for a PM-localized protein, which could efflux antifungal berberine (BER), the fluorescent dye rhodamine 123 (R123), and the herbicide paraquat (PQ). The first member of the ABC superfamily characterized in C. albicans was designated CDR1 (Candida drug-resistant 1). The other members of the PDR subfamily, CDR2, CDR3, CDR4, and CDR5/CDR11, are partially characterized (26, 27). Considering the fact that orf19.4531 is a member of the PDR subfamily and we show that it plays a role in transport of and resistance to various xenobiotics, we designated it as CDR6 (Candida drug-resistant 6). However, because orf19.4531 is designated as ROA1 in the Candida Genome Database (CGD), we refer it as CDR6/ROA1. Interestingly, we observed that Cdr6/Roa1, in addition to being a PM-localized exporter of selected compounds, also impacts lipid homeostasis and the physical state of the PM, resulting in reduced accumulation of azoles in the cell. The decreased intracellular azole concentration results in enhanced resistance to azoles. The increased resistance to azoles could be well-demonstrated in vitro as well as in an in vivo mouse model. In addition, we observed that the TOR signaling was elicited in the CDR6/ROA1 null mutant NKKY101 and led to azole resistance by stabilizing calcineurin via activation of Hsp90. This cascade induces ribosome biogenesis and suppresses sphingolipid biosynthesis. Taken together, we uncovered in C. albicans the impact of Cdr6/Roa1 on drug resistance, transport, and cell signaling.
Results
Topology prediction by the software TOPOCONS suggested Cdr6/Roa1 to be similar to Cdr1 and Cdr2, both of which possess two transmembrane domains, each composed of six transmembrane helices, which are preceded by two well-conserved nucleotide-binding domains (Fig. 1A, NBD1 and NBD2). However, the phylogenetic analysis of Cdr6 together with all members of the PDR subfamily from C. albicans and S. cerevisiae showed that Cdr6 stands out as being in a distinct cluster (Fig. S1).
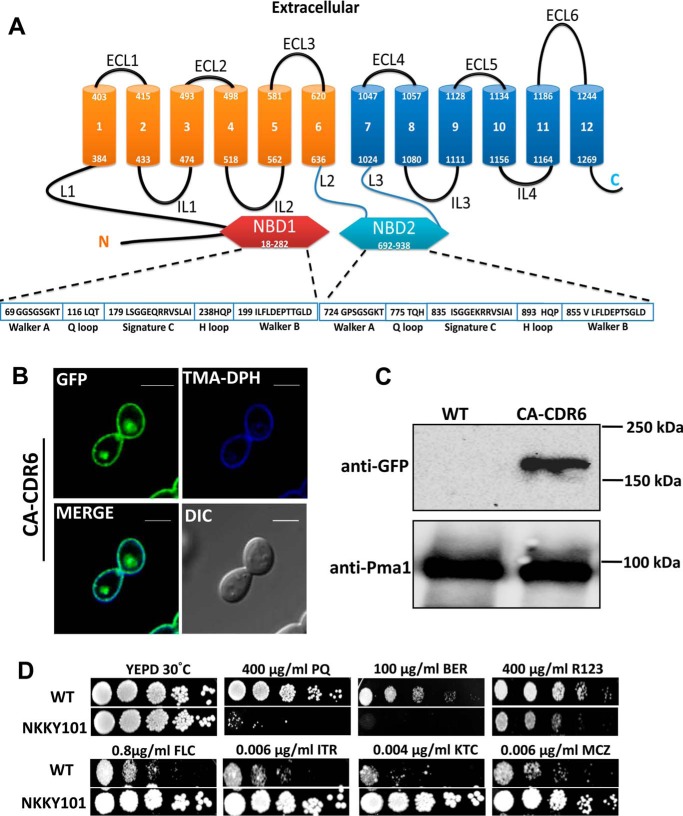
Figure 1.
Localization of Cdr6/Roa1 transporter in C. albicans and its role in drug resistance. A, topology prediction of Cdr6/Roa1 protein by TOPCONS software and pictorial representation showing typical domain arrangement of PDR subfamily with conserved nucleotide-binding domain (NBD). B, fluorescence imaging by confocal microscopy showing PM localization of Cdr6/Roa1-GFP with corresponding differential interference contrast (DIC) images, TMA-DPH (PM-specific stain) staining, and merged images. CA-CDR6 cells expressing Cdr6/Roa1-GFP were grown until midlog phase in YEPD medium and labeled with TMA-DPH (1.0 μm) for 10 min at 25 °C in the dark. Cells were washed with PBS buffer and imaged using the confocal microscope. Scale bar, 10 μm. C, immunoblot showing expression of Cdr6/Roa1-GFP protein. PMs were isolated using the sucrose gradient method, and equal amounts of proteins (80 μg) were resolved by SDS-PAGE (8% gel) and then probed with anti-GFP antibody. After probing, the membrane was striped and reprobed with anti-Pma1p polyclonal antibody. Pma1 is used as a control for the PM fraction and for loading. D, a comparison of susceptibilities by spot dilution assays between WT and CDR6/ROA1 null strain NKKY101. A 5-fold serial dilution of each strain was spotted onto PQ, BER, R123, FLC, ITR, KTC, and MCZ at the indicated concentrations in YEPD agar plates and grown for 48 h at 30 °C.
Cdr6 localizes to the plasma membrane
The bioinformatics analysis using the Wolf PSORT (http://www.genscript.com/wolf-psort.html)3 and CELLO version 2.5 (http://cello.life.nctu.edu.tw/)3 tools predicted that the C. albicans CDR6-encoding protein could be a PM-localized transporter. To validate this prediction, we cloned the orf19.4531 as a GFP-tagged protein (as described under “Experimental procedures”) in the WT C. albicans strain SC5314 at the RPS1 locus. The resulting strain, expressing N-terminal GFP-tagged Cdr6/Roa1 under the control of the pTDH3 promoter, was designated CA-CDR6. The expression and localization of the recombinant Cdr6/Roa1-GFP protein in the CA-CDR6 strain were confirmed by confocal microscopy, where green fluorescence of the GFP-tagged Cdr6/Roa1 distinctly showed a rimmed appearance on the PM (Fig. 1B). To verify the localization on the PM, trimethylaminodiphenyl-1,3,5-hexatriene (TMA-DPH) was used, which stains the PM specifically (28). As depicted in Fig. 1B in the merge channel, Cdr6/Roa1-GFP overlapped with that of the TMA-DPH stain, indicating the PM localization of Cdr6/Roa1-GFP. Cdr6/Roa1-GFP localization at the PM was further confirmed by Western blotting of the PM fraction using anti-GFP monoclonal antibody (Fig. 1C). Pma1 was used as a control for the PM fraction and loading. It is pertinent to mention that orf19.4531 (designated here as CDR6/ROA1) was earlier designated as ROA1 (regulator of azole sensitivity 1) and shown to be localized in small punctuated compartments adjacent to the vacuolar membrane (25).
Next, we further confirmed the localization of Cdr6/Roa1 by expressing it in a heterologous overexpression system, the AD1–8U− strain of S. cerevisiae, which has been widely used to express C. albicans and other yeast ABC transporter proteins (29–31). In the same manner, we cloned CDR6/ROA1 ORF in pABC3-GFP vector and integrated at the PDR5 locus in the AD1–8U− strain. Our confocal microscopy data for GFP-tagged Cdr6/Roa1 and Western blot analysis also confirmed its localization to the PM (data not shown).
Cells lacking CDR6/ROA1 display altered susceptibility to selected xenobiotics
A homozygous null of CDR6/ROA1 was constructed, where both of the alleles of diploid C. albicans WT strain SC5314 were deleted using a targeted gene deletion strategy (32). The deletion was confirmed by Southern blotting (data not shown). The resulting CDR6/ROA1 homozygous null strain was designated NKKY101 and subjected to extensive phenotypic analysis. The growth analysis under a variety of stress conditions, including thermal stress (20, 37, and 42 °C), non-fermentable carbon sources (xylulose, mannitol, glycerol, sorbitol, ethanol, and sucrose), metal stresses (CaCl2, MnCl2, CsCl, AgNO3, CuSO4, and NiSO4), oxidative stress (H2O2, PQ, and menadione), cell wall–perturbing agents (caspofungin, calcofluor white, SDS, caffeine, hygromycin B, and Congo red), cell membrane–perturbing agents (EDTA and NaCl), fluorescent dyes (rhodamine 6G, rhodamine B, and R123), polyenes (nystatin), azoles (fluconazole (FLC), itraconazole (ITR), miconazole (MCZ), and ketoconazole (KTC)), and other compounds (BER and curcumin), was performed. Most of the above-mentioned stresses, in comparison with WT cells, had no significant impact on growth of NKKY101 (Fig. S2), except in cases of PQ, BER, and R123, where increased susceptibility of NKKY101 strain was observed (Fig. 1D) accompanied with collateral high resistance to azoles (Fig. 1D; discussed below).
A single allele revertant strain was constructed by adding the CDR6/ROA1 allele at its native locus in a NKKY101 strain designated NKKY102. The revertant strain was confirmed by semiquantitative RT-PCR along with the WT and NKKY101 strains (Fig. S3). This revertant strain NKKY102 was used to test whether the observed changes in drug susceptibility could be rescued. For this, a serial dilution spot assay as well as a planktonic growth assay of WT, NKKY101, and NKKY102 strains on BER, R123, and PQ was performed. As depicted in Fig. 2A, the addition of a CDR6/ROA1 single allele in the NKKY101 strain was able to revert back the phenotype of NKKY101. The impact of Cdr6/Roa1 level on drug susceptibility was further assessed in a strain overexpressing untagged Cdr6/Roa1. This strain was constructed in C. albicans by integrating CDR6/ROA1 under the control of pTDH3 promoter at RPS1 locus in the SC5314 and designated NKKY103 (Fig. S3). Of note, as expected, the CDR6/ROA1-overexpressing strain NKKY103 showed higher resistance toward BER and PQ as compared with WT, NKKY101, and NKKY102 (Fig. 2A).
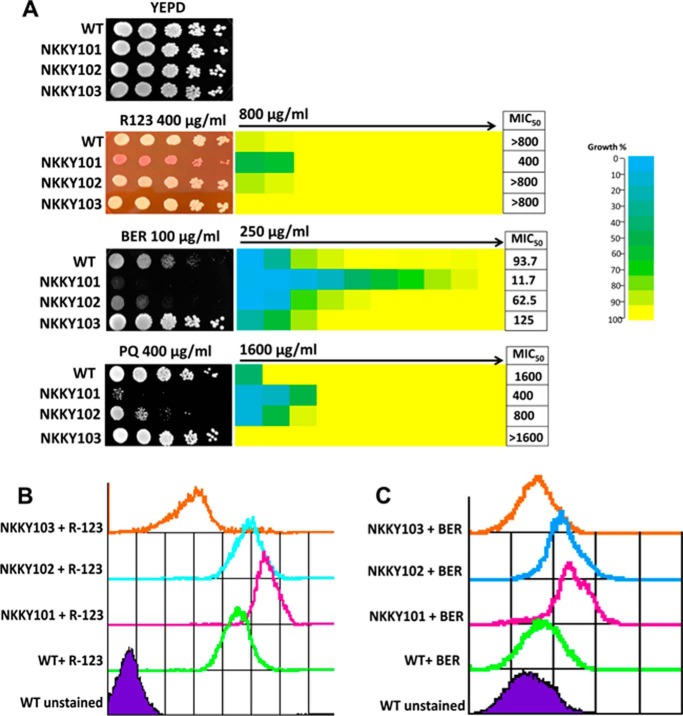
Figure 2.
R123, BER, and PQ are substrates of the Cdr6/Roa1 transporter. A, left, comparison of growth by spot dilution assays for C. albicans WT, CDR6/ROA1 null mutant NKKY101, CDR6/ROA1 revertant NKKY102, and CDR6/ROA1 overexpression strain NKKY103 cells onto R123, BER, and PQ at the indicated concentrations in YEPD agar plates. Right, MIC50 values for these strains, obtained by use of a broth microdilution assay, starting with the indicated concentration of xenobiotics in the first well in YEPD medium, as described under “Experimental procedures.” For each strain, optical densities were averaged for duplicate measurements, and relative growths (as compared with drug-free growth) were displayed with color as indicated with the color bar using three-color heat maps. B, R123 accumulation assay in WT, NKKY101, NKKY102, and NKKY103 strains cells using FACS. n = 10,000 cells for each sample. C, BER accumulation assay in WT, NKKY101, NKKY102, and NKKY103 strains cells using FACS. n = 10,000 cells for each sample.
Fluorescent BER and R123 are substrates of Cdr6/Roa1
Cdr6/Roa1 is a PM-localized PDR subfamily transporter; therefore, it was reasonable to speculate that it could also act as an exporter. Because the overexpression of Cdr6 leads to higher resistance to BER (Fig. 2A), we checked whether these fluorescent molecules (BER and R123) could act as substrates of Cdr6. FACS-based accumulation assays of R123 and BER for WT, NKKY101, NKKY102, and NKKY103 strains revealed that the CDR6/ROA1 null mutant NKKY101 accumulated higher amounts of R123 and BER as compared with other strains. As expected, the CDR6/ROA1-overexpressing NKKY103 strain showed the lowest amount of accumulation as compared with other strains (Fig. 2, B and C).
CDR6/NKKY101 null mutant shows reduced intracellular accumulation of azoles
As mentioned, our screen (Fig. 1D) revealed that whereas the CDR6/ROA1 null mutant NKKY101 displayed higher susceptibility to several xenobiotics, it was particularly resistant to azoles, such as triazoles (FLC and ITR) and imidazoles (MCZ and KTC) (Fig. 1D). This phenotype was quite surprising; therefore, further validation was done by employing multiple assays using different growth conditions (solid medium and liquid medium). As depicted in Fig. 3A, the spot assays revealed that the NKKY101 strain is indeed azole-resistant as compared with the WT, NKKY102, and NKKY103 strains. This was further confirmed by agar drug diffusion assays, where the size of the inhibitory zone (halo) indicates the extent of drug susceptibility. It was observed that the zone of inhibition was smallest in the NKKY101 strain as compared with the WT, NKKY102, and NKKY103 strains, pointing to the fact that the mutant strain is highly resistant to the tested drug (Fig. 3B). The liquid growth test further confirmed the resistant phenotype of the NKKY101 strain. It was evident from the growth analysis until 24 h that in the presence of FLC, the NKKY101 mutant strain showed significantly enhanced growth as compared with WT, NKKY102, and NKKY103 strains (Fig. 3C). During the course of our study, Jiang et al. (25) also reported azole resistance with deletion of orf19.4531; however, the study did not provide any explanation for the mechanism of resistance (25).
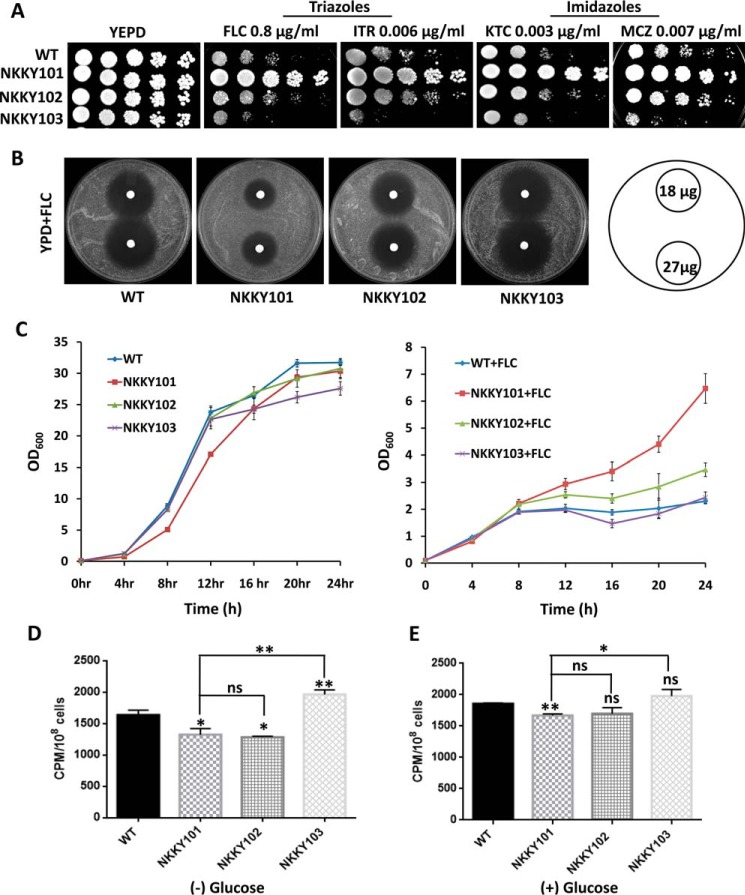
Figure 3.
CDR6/ROA1 deletion leads to resistance toward azoles and low accumulation of azoles. A, comparison of growth by spot dilution assays between WT, NKKY101, NKKY102, and NKKY103 strains. A 5-fold serial dilution of each strain was spotted on YEPD agar plates containing the triazoles (FLC and ITR) and imidazoles (KTC and MCZ) at the indicated concentrations and grown for 48 h at 30 °C. B, drug resistance profiles of WT, NKKY101, NKKY102, and NKKY103 strains on FLC by a filter disk assay as described under “Experimental procedures.” C, growth curve of WT, NKKY101, NKKY102, and NKKY103 strains in YEPD medium in the absence (left) and presence (right) of FLC. D, [3H]FLC accumulation levels in C. albicans WT, NKKY101, NKKY102, and NKKY103 strains were measured at 24 h in the absence of glucose after cells were carbon-starved for 3 h. E, [3H]FLC accumulation levels in C. albicans WT, NKKY101, NKKY102, and NKKY103 strains were measured at 24 h in the presence of 2% glucose after cells were carbon-starved for 3 h. All experiments were performed in biological triplicate, and the results are shown as means ± S.D. (error bars). A statistical significance value (**, p ≤ 0.01; *, p ≤ 0.05; ns, not significantly different) was employed using unpaired Student's t test.
The fact that resistance to azoles in the CDR6/ROA1 null mutant NKKY101 is observed in the absence of the transporter suggests that the transporter does not play a role as an azole exporter. We examined whether the import of FLC is affected in the NKKY101 strain, which may contribute to the observed resistance. For measuring drug accumulation in the CDR6/ROA1 null mutant NKKY101, radiolabeled [3H]FLC was used as described previously (33). The intracellular accumulation of radiolabeled [3H]FLC was quantitated in the WT, NKKY101, NKKY1012, and NKKY103 strains in the absence and presence of glucose. As depicted in Fig. 3 (D and E), a significantly lower level of FLC accumulated in the CDR6/ROA1 null mutant NKKY101 as compared with the WT and NKKY103 strains. The CDR6/ROA1-overexpressing strain NKKY103 showed the highest accumulation of FLC and highest susceptibility to azoles (Fig. 3, D and E). The difference in FLC accumulation was intensified between the WT and CDR6/ROA1 null mutant NKKY101 when the assay was performed in the absence of the carbon source glucose (Fig. 3D). The absence of glucose inhibits ATP-dependent FLC efflux transporters, which may otherwise mask the drug accumulation measurements during the assay.
Deletion of CDR6/ROA1 leads to higher PM rigidity
Because changes in the physical state of the PM have earlier been shown to impact drug diffusion and susceptibility of C. albicans cells (8–10, 16), we explored whether the intracellular drug accumulation observed could be correlated to any change in PM fluidity. To examine this possibility, we used fluorescence recovery after photobleaching (FRAP)-based lateral mobility analysis by employing the FAST-DiI dye, which we have used earlier for measuring the PM fluidity in yeast cells (16). As described under “Experimental procedures,” WT, NKKY101, and NKKY102 cells were incubated with the lipid probe FAST-DiI dye. A particular region of FAST-DiI dye–stained PM was selected and photobleached. The same region was imaged over time, and recovery of FAST-DiI dye fluorescence was measured. Recovery is due to lateral movement of the FAST-DiI dye from the unbleached region of PM to the bleached region (Fig. 4A). The time to recover the fluorescence in the bleached area is dependent on the physical state of the PM. For instance, higher membrane fluidity would lead to faster fluorescence recovery (16). Fig. 4B depicts the representative fluorescence recovery experiment images of WT, NKKY101, and NKKY102 strains. As shown in the overlapping recovery plot, the WT shows the fastest recovery, followed by the NKKY102 and NKKY101 strains, respectively (Fig. 4C).
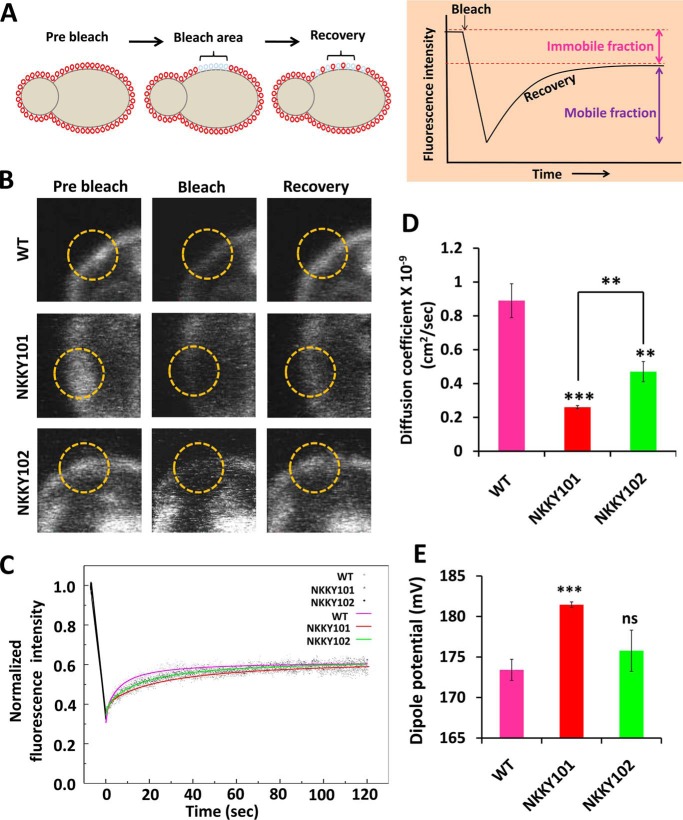
Figure 4.
Disruption of CDR6/ROA1 causes high PM rigidity. A, schematic of the FRAP. PM-bound FAST-DiI was photobleached. The same region was imaged over time, and recovery of FAST-DiI dye fluorescence was measured. Recovery was due to lateral diffusion of FAST-DiI dye from the unbleached region of PM to the bleached region. The time to recover the fluorescence in the bleach area is dependent on the physical state of the PM; higher membrane fluidity would lead to faster fluorescence recovery. B, images of FAST-DiI dye–labeled PM of WT, NKKY101, and NKKY103 cells (left column, t = 0 s, prebleach). A region of interest (ROI; yellow circle) was photobleached, and cells were imaged immediately thereafter (second column images, bleach) and after 120 s postbleach (third column, recovery). C, a qualitative estimate of rate of recovery can be obtained by comparing the slopes of the overlapped recovery curve, demonstrating slowest recovery in CDR6/ROA1 null mutant NKKY101 in comparison with WT and CDR6/ROA1 revertant NKKY102 strains. D, diffusion coefficients calculated for WT, NKKY101, and NKKY103 strains from quantitative FRAP experiments as described under “Experimental procedures” and plotted as a histogram. The experiment was performed in biological triplicates, and values are the means ± S.E. (error bars). A statistical significance value (***, p ≤ 0.001; **, p ≤ 0.01) was employed using Student's t test. E, PM dipole potential measured using di-8-ANEPPS. Change in dipole potential was measured utilizing di-8-ANEPPS after incorporation into the PM of WT, NKKY101, and NKKY103 strains. Dipole potential values were calculated from the fluorescence ratio (R), defined as the ratio of fluorescence intensity at an excitation wavelength of 420 nm to that at 520 nm (emission at 670 nm in both cases; see “Experimental procedures”). Experiments were performed at least in triplicate, and results are shown as means ± S.D. (error bars). ***, p ≤ 0.001; ns, not significantly different, calculated using an unpaired Student's t test.
Next, using different recovery data sets, we calculated the diffusion coefficient (as described under “Experimental procedures”). As evident from Fig. 4D, NKKY101 mutant showed the lowest diffusion coefficient value (0.26 ± 0.01 × 10−9 cm2/s), whereas the WT strain showed the maximum diffusion coefficient value (0.89 ± 0.10 × 10−9 cm2/s). Interestingly, the single allele revertant strain NKKY102 showed an intermediate diffusion coefficient value (0.47 ± 0.06 × 10−9 cm2/s). Further, we calculated the mobile fraction as described under “Experimental procedures.” Notably, the mobile fraction of the dye was similar for the WT (47.6 ± 2.1), NKKY101 (47.7 ± 3.3), and NKKY102 (49.5 ± 2.2) strains. This indicated that fluorescence recovery is comparable in WT, NKKY101, and NKKY102 strains; however, the rate of recovery is significantly different. We conclude from our FRAP experiments that the Cdr6/Roa1 protein is required to maintain the PM fluidity, and its absence makes the PM more rigid, resulting in reduced movement of azole drugs across the PM.
CDR6/ROA1 null mutant shows high PM dipole potential
Membrane dipole potential is the electrostatic potential difference within the membrane due to the nonrandom arrangement of amphiphile dipoles and solvent (water) molecules at the interface. As the origin of dipole potential is due to the “nonrandom” (restricted) orientation of lipid and solvent dipoles, a decrease in membrane fluidity could result in an increase in population of nonrandom dipoles, leading to an increase in membrane dipole potential. To explore the effect of CDR6/ROA1 deletion on the PM dipole potential of C. albicans, we stained the PM fractions of WT, NKKY101, and NKKY102 strains using the voltage-sensitive probe pyridinium, 4-(2-(6-dioctylamino)-2-naphthalenyl)ethenyl)-1-(3-sulfopropyl)-inner salt (di-8-ANEPPS) and measured the PM dipole potential as described under “Experimental procedures.” We observed that the NKKY101 strain had significantly higher PM dipole potential as compared with the WT and NKKY102 strains (Fig. 4E).
Disruption of CDR6/ROA1 perturbs lipid homoeostasis
Lipids are an essential component of the PM, and any changes in lipid homeostasis can alter the PM fluidity status. It has been reported earlier that the level of several ABC transporters in yeast and other organisms also impacts lipid homeostasis (30, 34, 35). The major fungal plasma membrane sterol, ergosterol, is an important lipid species that usually provides rigidity to the PM (9, 36). It is observed that the susceptibility to azole drugs that target the ergosterol biosynthesis pathway is affected by PM ergosterol levels (16, 28, 37).
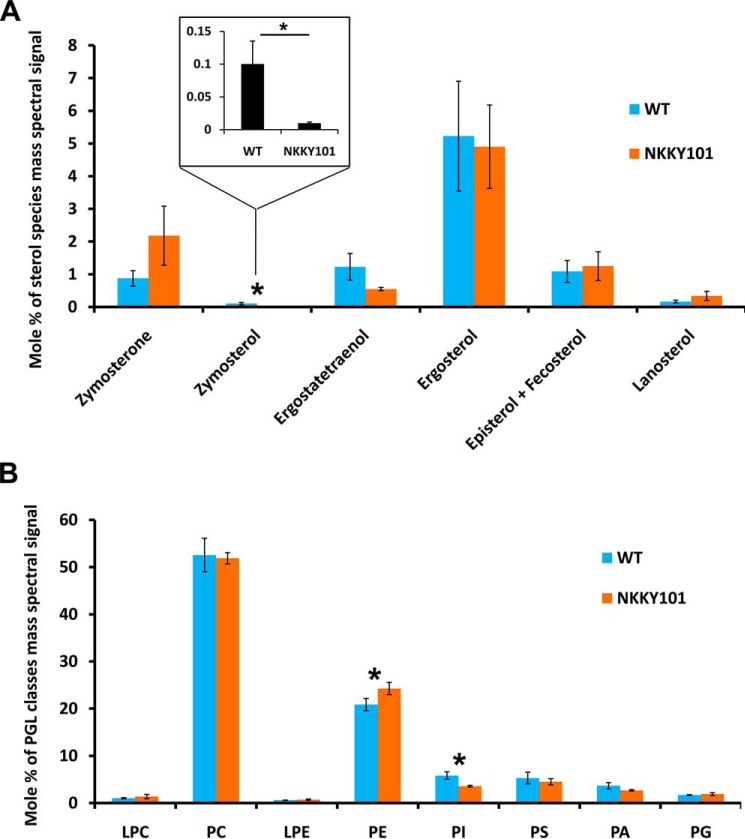
Our observed changes in intracellular drug accumulation and increased resistance to azoles in NKKY101 cells could be the result of imbalances in lipid homeostasis. This possibility was explored by performing high-throughput MS-based lipidome analysis using ESI-MS/MS. Our lipidome analysis revealed no major change in ergosterol and other sterol species content in NKKY101 strain as compared with WT (Fig. 5A). The zymosterol level was found to be significantly low in the case of the NKKY101 strain (Fig. 5A). However, a decrease in the level of zymosterol does not contribute to azole resistance. Together, the sterol analysis overruled the possibility of a role of sterols in influencing membrane fluidity and the phenotype of NKKY101 strain.
Figure 5.
Lipid profile of CDR6/ROA1 null NKKY101 mutant as compared with WT. A, sterol species level in WT and NKKY101 strain; data represent the percentage of total sterol mass spectral signal after normalization to internal standards. B, levels of PGL classes in WT and NKKY101 strain; the data represent the percentage of total PGL mass spectral signal after normalization to internal standards. Experiments were performed in biological quadruplet, and results are shown as means ± S.E. (error bars). A statistical significance value of p ≤ 0.05 (*) was calculated using Student's t test. The raw data for the graphs are available as Table S1. LPC, lysophosphatidylcholine; PC, phosphatidylcholine; LPE, lysophosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol; PA, phosphatidic acid; PG, phosphatidylglycerol.
The major phosphoglycerides (PGLs), which include lysophosphatidylcholine, phosphatidylcholine, lysophosphatidylethanolamine, phosphatidylethanolamine (PE), phosphatidylserine, phosphatidylinositol, phosphatidic acid, and phosphatidylglycerol, were detected in our analysis. No major variation between WT and NKKY101 strain was observed. However, as depicted in Fig. 5B, PE content was significantly high, whereas phosphatidylinositol was significantly low in the NKKY101 strain as compared with the WT strain. Even small changes in the levels of these PGLs can play an important role in the PM fluidity, particularly PE which has been assigned as a key regulator of the PM fluidity (38).
Inactivation of CDR6/ROA1 results in up-regulation of ribosome biogenesis genes
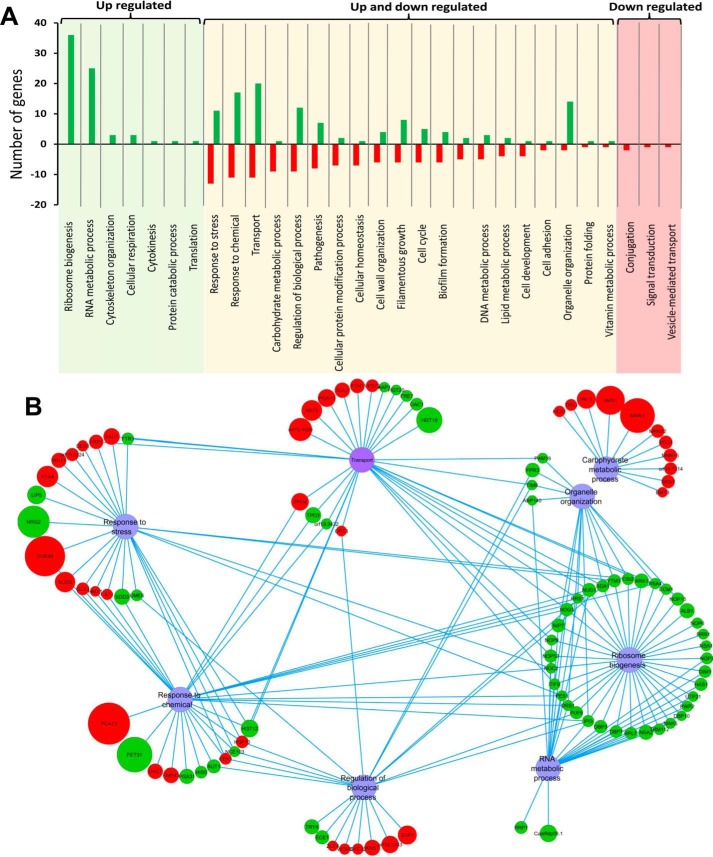
We performed a microarray-based genome-wide transcriptome analysis of WT and NKKY101 strains to assess the molecular basis of observed azole resistance. The comparative transcriptome analysis revealed that none of the known mechanisms of azole resistance involving expression of major drug exporters and ERG genes were significantly changed between the NKKY101 and WT strains. However, the comparative transcriptomic profile showed significant (≥2-fold) down-regulation and up-regulation of 135 and 116 genes, respectively, in the NKKY101 strain as compared with WT (Table S2). The transcription profile was further analyzed using CGD Go-slim mapper and was annotated on the basis of biological processes. Specifically, in the NKKY101 strain, expression of genes encoding proteins involved in ribosome biogenesis, RNA metabolic process, organelle organization, and filamentous growth was increased, whereas expression of carbohydrate metabolic process, cellular protein modification process, and cellular homeostasis was generally decreased (Fig. 6A). The genes belonging to the transport, response to chemicals, regulation of biological process, response to stress, and pathogenesis categories were differentially regulated (up- and down-regulated) in the NKKY101 strain as compared with WT (Fig. 6A).
Figure 6.
The comparative transcriptomic profile of CDR6/ROA1 null mutant NKKY101. A, the genes with ≥2-fold change are annotated on the basis of biological processes using CGD Go-slim mapper. The x axis indicates the biological functional categories, and the y axis indicates the number of genes. Left column, categories of genes that are only up-regulated; middle column, categories of genes that are up- and down-regulated; right column, the categories of genes that are only down-regulated. B, interaction map of the CDR6/ROA1 null mutant NKKY101 using five major categories from up- and down-regulated genes. Clustering was based on gene involvement in biological functions using Cytoscape software. Red and green circles, genes down- and up-regulating, respectively. The size of the circle is proportional to -fold change (the higher the -fold change, the bigger the size of the circle).
For a better understanding of the roles of genes belonging to more than one biological process, five major categories from up- and down-regulated genes were selected to make an interaction map (based on the biological processes in which they are involved) using Cytoscape software. As depicted in Fig. 6B, except for the carbohydrate metabolic process, other categories shared many common genes. Interestingly, the major portion (∼31% of up-regulated genes; 35 genes) among up-regulated genes belongs to ribosome biogenesis, which shared most of the genes with the RNA metabolic process category (Fig. 6, A and B).
Hyperactivation of Tor1 leads to up-regulation of ribosome biogenesis in NKKY101 strain
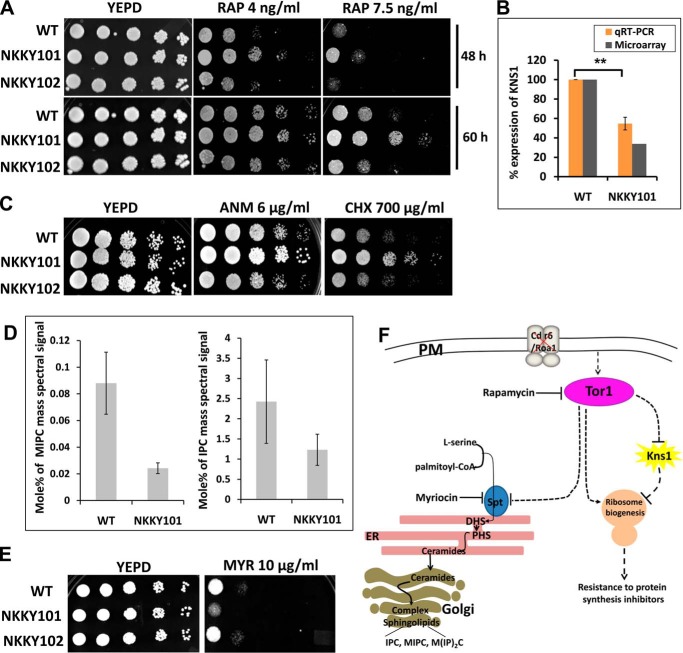
Our microarray data revealed that 35 genes related to ribosome biogenesis were among the most up-regulated gene category in the NKKY101 strain (Fig. 6). It is an established fact that Tor1 (target of rapamycin 1) regulates ribosome biogenesis in a positive manner, and Tor1 function is inhibited by rapamycin (39, 40). We compared the growth of WT, NKKY101, and NKKY102 strains in the presence of rapamycin using a spot assay. Fig. 7A shows that the NKKY101 strain showed a resistant phenotype on rapamycin as compared with WT, and this phenotype could be rescued in the revertant strain NKKY102. The resistance to rapamycin pointed to hyperactivation of Tor1 in the NKKY101 strain. To validate further that the observed up-regulation of the ribosome biogenesis gene in the NKKY101 strain is because of hyperactive Tor1, we treated the NKKY101 cells with 4 and 8 ng/ml concentrations of rapamycin (an inhibitor of Tor1) and checked the expression of some of the randomly selected ribosome biogenesis genes by quantitative RT-PCR. As depicted in Fig. S4A, the expression of selected genes was lower in rapamycin-treated cells in a dose-dependent manner in comparison with untreated cells. This suggests that the up-regulation of ribosome biogenesis genes in NKKY101 strain is linked to the hyperactivation of Tor1.
Figure 7.
Hyperactivation of Tor1 leads to up-regulation of ribosome biogenesis in the absence of CDR6/ROA1. A, analysis of Tor1 activation by comparison of growth in the presence of rapamycin at the indicated concentrations by spot dilution assays for WT, CDR6/ROA1 null strain NKKY101, and CDR6/ROA1 revertant strain NKKY102. A 5-fold serial dilution of each strain was spotted onto rapamycin at the indicated concentrations on YEPD agar plates and grown for 48–60 h at 30 °C. The resistant phenotype of the NKKY101 strain suggests hyperactivation of Tor1 in comparison with WT and NKKY102 strains. B, validation of microarray data of KNS1 expression by qRT-PCR in NKKY101 mutant compared with WT. Data (mean ± S.E. (error bars) of biological replicate with technical quadruplets) were normalized to an internal ACT1 mRNA control and represented as percentage relative expression in the mutant cultures compared with the wild-type cells. Statistical analysis was performed using Student's t test (**, p ≤ 0.01). C, a comparison of growth in the presence of protein synthesis inhibitors by spot dilution assays between WT, NKKY101, and NKKY102 strains. A 5-fold serial dilution of each strain was spotted onto ANM and CHX at the indicated concentrations on YEPD agar plates and grown for 48 h at 30 °C. D, left, MIPC level in WT and NKKY101 strains; data represent the percentage of total MIPC mass spectral signal after normalization to internal standards. Right, IPC level in WT and NKKY101 strains; data represent the percentage of total IPC mass spectral signal after normalization to internal standards. E, WT, CDR6/ROA1 null strain NKKY101, and CDR6/ROA1 revertant strain NKKY102 were analyzed for growth on YEPD agar plates in the presence of myriocin at the indicated concentrations and incubated at 30 °C for 48 h. F, model for CDR6/ROA1 deletion effects on ribosome biogenesis and complex sphingolipid biosynthesis via TOR signaling. In the absence of Cdr6/Roa1 protein, the Tor1 becomes active, which down-regulates the KNS1 transcript and also induces ribosome biogenesis, thereby enabling cells to survive in the presence of protein synthesis inhibitors. Tor1 suppresses complex sphingolipid biosynthesis by inhibition of the SPT enzymatic step.
Considering that Kns1 kinase in S. cerevisiae negatively regulates the ribosome biogenesis and Tor1 relieves the impact of Kns1 from ribosome biogenesis by negatively regulating KNS1 transcript expression (19), our observed ∼3-fold down-regulation of orf19.4979, a homologue of S. cerevisiae Kns1 kinase in NKKY101, strongly indicates that the impact of hyperactive Tor1 on Kns1 kinase expression is conserved in C. albicans (Fig. 7B). The negative impact of Tor1 on KNS1 transcript in the NKKY101 strain was further validated by the quantification of the expression of the KNS1 transcript in rapamycin-treated conditions, where inhibition of Tor1 by rapamycin induced the expression of KNS1 (Fig. S4B).
Next, we performed spot assays of the WT, NKKY101, and NKKY102 strains in the presence of protein synthesis inhibitors anisomycin (ANM) and cycloheximide (CHX) (41, 42). Our spot assay showed that growth of the NKKY101 strain was much better as compared with WT and NKKY102 strains in the presence of protein synthesis inhibitors (Fig. 7C). Of note, the induction of ribosome biogenesis encourages protein synthesis, thus overcoming the growth inhibition by protein synthesis inhibitors (43, 44). Finally, we concluded from this experiment that deletion of CDR6/ROA1 hyperactivates Tor1, which ultimately drives ribosome biogenesis (Fig. 7F).
Tor1 hyperactivation in NKKY101 mutant inhibits the complex sphingolipid biosynthesis
Recent studies have suggested that Tor1 negatively regulates the complex sphingolipids synthesis by inhibiting the Orm phosphorylation (45, 46). In S. cerevisiae, it has been shown that Orm proteins negatively regulate the synthesis of sphingolipids by inhibiting the serine palmitoyltransferase (SPT) enzyme, and phosphorylation of Orm proteins relieves their inhibitory activity (45, 47, 48). Because we observed hyperactivation of Tor1 in the NKKY101 strain, the complex sphingolipid level in our lipidome data for NKKY101 and WT strains was analyzed. As expected, the inhibitory effect of hyperactive Tor1 on sphingolipid synthesis was evident, which was reflected in the lower amount of complex sphingolipids (IPC and MIPC) in NKKY101 as compared with WT strain (Fig. 7D).
To validate whether in C. albicans the observed Tor1 inhibitory effect on sphingolipids synthesis is because of SPT inhibition, we performed spot assays of WT, NKKY101, and NKKY102 strains on myriocin (a specific inhibitor of SPT) (49). Growth in the presence of myriocin is strongly dependent on the SPT level, and inhibition of SPT makes cells susceptible to myriocin. As illustrated in Fig. 7E, the NKKY101 strain showed growth impairment in the presence of myriocin as compared with WT and NKKY102 strain. This validates that in the NKKY101 strain of C. albicans, Tor1 hyperactivation represses the SPT activity, resulting in decreased sphingolipid synthesis (Fig. 7F).
Hsp90 activation occurs via Tor1 in NKKY101 strain
Hsp90 is a molecular chaperone that plays a crucial role in antifungal drug resistance (50). Impairment of Hsp90 function converts azoles from fungistatic to fungicidal (50, 51). Hsp90 stabilizes the calcineurin protein, and its activation is essential for cells to survive in the presence of azoles (52–54). CK2 kinase phosphorylates Hsp90 and represses its activity in C. albicans and S. cerevisiae (20, 21). S. cerevisiae Kns1 kinase activates the CK2 kinase by phosphorylation of Ckb1 (a regulatory subunit of CK2 kinase), and Tor1 negatively regulates the expression of Kns1 kinase (18, 19). Thus, active Tor1 decreases CK2 kinase activity, which in turn activates Hsp90.
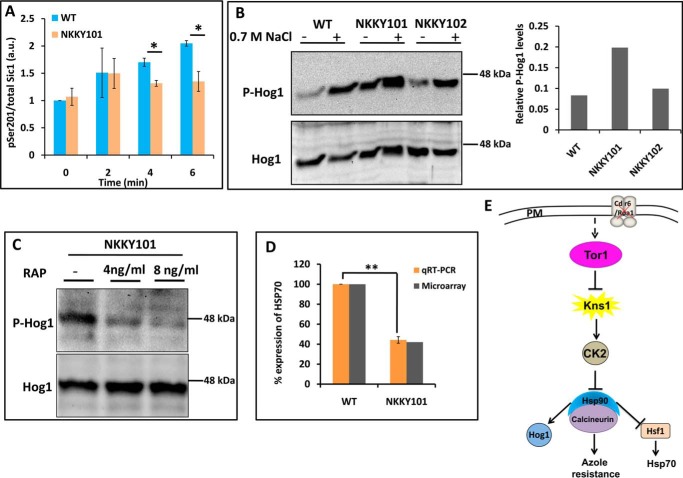
The observed down-regulation of Kns1 in the NKKY101 strain (Fig. 7B) prompted us to analyze whether it has any impact on CK2 kinase activity. For monitoring the CK2 kinase activity, the phosphorylation of Sic1, which is phosphorylated by the CK2 kinase at serine 201, was quantitated (55). Notably, CDR6/ROA1 null mutant NKKY101 showed significantly lowered phosphorylation of Sic1 at 4 and 6 min (Fig. 8A). This implied that, similar to S. cerevisiae (18), the low level of Kns1 kinase activity also has a negative impact on CK2 kinase activity in C. albicans.
Figure 8.
Tor1 activation in NKKY101 strain causes Hsp90 activation via CK2 kinase. A, CDR6/ROA1 deletion suppresses CK2 activity. CK2 activity was monitored in an in vitro assay using protein lysates from C. albicans WT and NKKY101 strains, using recombinant Sic1 as a substrate. The assay was performed in biological duplicates with technical replicates. Error bars, S.D.; *, p < 0.05 (Student's t test). B, the CDR6/ROA1 deletion causes Hog1 activation. NaCl was used as a positive control to detect the active phosphorylated form of Hog1. The phosphorylated Hog1 level is low in the WT and CDR6/ROA1 revertant NKKY102 strains under basal conditions and induced in the presence of NaCl. However, the phosphorylated Hog1 level is high in the CDR6/ROA1 null strain NKKY101 even in basal conditions. The addition of NaCl increased the phosphorylated Hog1 in the null strain. Bands were quantified using Bio-Rad Image Lab software, and the ratio of phosphorylated Hog1 to total Hog1 was plotted for WT, NKKY101, and NKKY102 strains for basal conditions (without NaCl induction). A similar trend of phosphorylated Hog1 was observed in biological triplicate experiments for basal conditions (Fig. S5). C, Hog1 activation in CDR6/ROA1 null strain NKKY101 is Tor1-dependent. Rapamycin was used as a Tor1 inhibitor. The phosphorylated Hog1 level decreased in CDR6/ROA1 null strain NKKY101 when treated with rapamycin (4 and 8 ng/ml) in comparison with untreated conditions. D, validation of microarray data of HSP70 expression by qRT-PCR in NKKY101 mutant compared with WT. Data (mean ± S.E. (error bars) of biological replicate with technical triplicates) were normalized to an internal ACT1 mRNA control and represent percentage relative expression in the mutant cultures compared with the wild-type cells. Statistical analysis was performed using Student's t test (**, p ≤ 0.01). E, model for Hsp90 activation. Deletion of CDR6/ROA1 activates the Tor1, which down-regulates the KNS1 transcript, resulting in decreased CK2 kinase activity. This decrease in CK2 kinase activity causes activation of Hsp90 and its client proteins.
We observed decreased CK2 kinase activity in the NKKY101 strain, implying that Hsp90 function may be induced in this strain. To determine whether decreased CK2 kinase activity in the NKKY101 strain activates Hsp90, we evaluated the activity of mitogen-activated protein kinase Hog1 (21, 56) and heat shock transcription factor Hsf1, which are downstream effectors of Hsp90 (57, 58).
Hog1 requires active Hsp90 to warrant its activation by phosphorylation (21, 56). As expected, we observed a high amount of phosphorylated Hog1 in the NKKY101 strain as compared with WT and NKKY102 strains (Fig. 8B). The results were confirmed by experiments in biological triplicate (Fig. S5). The Tor1-dependent high amount of Hog1 phosphorylation in NKKY101 strain was further validated by measuring the Hog1 phosphorylation in NKKY101 strain after treatment with rapamycin as described under “Experimental procedures.” As expected, the inhibition of Tor1 by rapamycin resulted in a decrease in Hog1 phosphorylation in the treated sample in comparison with the untreated sample (Fig. 8C).
We further validated the Hsp90 activation in NKKY101 strain by monitoring the repression of the HSP70 gene, for which basal level expression requires active Hsf1 TF (59). Hsp90 exerts a suppressive effect on Hsf1; thus, activation of Hsp90 represses Hsf1-dependent genes. As expected, in our microarray data, we also observed ∼2.4-fold down-regulation of HSP70 in NKKY101 strain as compared with WT (Fig. 8D). These results indicate that Hsp90 is present in a hyperactive state in CDR6/ROA1 null mutant NKKY10.
It is a well-established fact that Hsp90 stabilizes the catalytic subunit of the calcineurin protein by interacting with it, and calcineurin provides resistance against azoles by activating the calcineurin-dependent pathway (52, 60). Inhibition of Hsp90 activity using Hsp90-specific inhibitors also shows a negative impact on calcineurin function (22). Our results suggest that in the NKKY101 strain, hyperactive Tor1 influences the Hsp90 activity in a positive manner by modulating the CK2 kinase activity via effector kinase Kns1 and active Hsp90, leading to an activation of the calcineurin protein, thus contributing to azole resistance in the NKKY101 strain (Fig. 8E).
Cdr6/Roa1 is not required for virulence
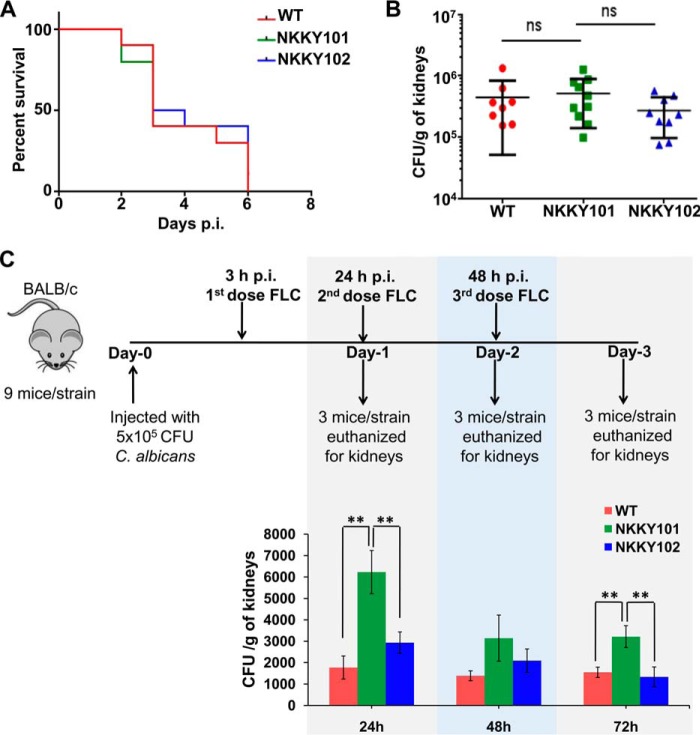
In our recent studies, we reported that various transporter proteins of C. albicans perform diverse functions and are also required to maintain the virulence (30, 61). Whether Cdr6/Roa1 played any role in virulence was examined in a mouse model. To test the infection capacity, mouse survival and kidney fungal burden assays were performed in immunocompetent BALB/c mice by injecting them with WT, NKKY101, and NKKY102 strains. It was observed that all of the C. albicans strains have similar virulence, and in all of the cases, no mice survived beyond 6 days (Fig. 9A). There was also no significant difference in overall kidney fungal burdens in mice infected with WT, NKKY101, and NKKY102 strains (Fig. 9B). These experiments ruled out any role of Cdr6/Roa1 in virulence of C. albicans.
Figure 9.
Absence of CDR6/ROA1 hampers in vivo azole therapy. A, the survival curve of mice infected with C. albicans WT, NKKY101, and NKKY102 shows similar patterns; all of the mice succumbed to infection within 6 days postinfection. B, fungal kidney burden that was examined from moribund mice was not statistically different between WT-, NKKY101-, and NKKY102-infected mice. C, on average, cfu recovered from kidneys after a short FLC therapy (20 mg/kg intraperitoneally once after 3 h postinfection, once on day 1 and once on day 2 postinfection) are significantly higher in NKKY101-infected mice compared with WT and NKKY102 strains, as determined at the indicated times (24, 48, and 72 h). Statistically significant differences in the cfu levels between WT, NKKY101, and NKKY102 strains are indicated. **, p ≤ 0.01 (Student's t test).
Absence of Cdr6/Roa1 hampers in vivo azole efficacy
To evaluate the in vivo significance of the observed in vitro azole resistance in CDR6/ROA1 null mutant NKKY101, we employed a mouse model of candidiasis. Immunocompetent female BALB/c mice were infected with the WT, NKKY101, and NKKY102 mutant cells via tail vein injection described under “Experimental procedures.” After 3 h of infection, FLC treatment was given intraperitoneally. The FLC treatment was followed for the next 2 days. The experimental regimen is depicted in Fig. 9C. Each day, three mice from each group were euthanized and enumerated for C. albicans burden in kidneys. Notably, as mentioned above, our in vivo data revealed that CDR6/ROA1 null mutant NKKY101 did not affect virulence in the mouse model (Fig. 9, A and B); however, when infection was challenged with FLC, interesting correlations were observed. Higher kidney fungal burden was observed from kidneys of mice infected with CDR6/ROA1 null mutant NKKY101 as compared with WT and CDR6/ROA1 revertant NKKY102 strains (Fig. 9C). Of note, the fungal burden was significantly higher after 24 and 72 h in mice infected with CDR6/ROA1 null mutant NKKY101 in comparison with WT (p < 0.001 for 24 and 72 h) and CDR6/ROA1 revertant NKKY102 strain (p < 0.001 for 24 and 72 h). Together, the increased fungal burdens from kidneys of mice infected with CDR6/ROA1 null mutant point to the role of Cdr6/Roa1 in azole resistance in vivo during FLC treatment.
Discussion
Cdr1 (Candida drug resistance 1) belongs to the largest ABCG/PDR subfamily of transporters in C. albicans and is well characterized as a multidrug exporter imparting MDR. A close homologue of Cdr1, designated Cdr2, has also been shown to impact clinical drug resistance (7). However, the role of other members of this subfamily remains poorly described. For instance, none of the members are proven transporters. A few of the ABCG/PDR members have been shown to translocate phosphoglycerides between the two monolayers of the lipid bilayer and maintain membrane lipid asymmetry (27). The present study deals with a poorly characterized PDR subfamily member, orf19.4531, which encodes a PM-localized protein. In a recent report (25), this particular protein has been shown to be localized on a punctuated compartment adjacent to the vacuolar membrane. However, the authors could not detect any protein expression in their Western blot. We speculate that proper expression of the protein could not be achieved with the system used in that study. Our present data from confocal microscopy and Western blotting confirm that localization of Cdr6/Roa1 is in the PM. Our functional analysis revealed that, similar to PM-localized CDR proteins, orf19.4531 also functions as a multidrug exporter and particularly effluxes BER, R123, and PQ. We therefore designated orf19.4531 as CDR6/ROA1.
It was observed here that homozygous deletion of CDR6/ROA1 leads to an increased resistance to azoles. This observation is consistent with a previous report (25). The [3H]FLC accumulation data confirmed here a low intracellular accumulation of the drug in CDR6/ROA1 null mutant NKKY101. Additionally, high-throughput lipidomics and FRAP data suggested that the CDR6/ROA1 deletion resulted in lipid imbalances and increased rigidity of the PM, thereby providing a plausible explanation for the observed reduced entry of azole into the CDR6/ROA1 null mutant. High-throughput lipid analysis further revealed no significant imbalance in ergosterol levels. However, PE content was significantly higher in the CDR6/ROA1 null mutant. According to recent reports, PE could also act as a key regulator of PM fluidity, wherein an increase in its level leads to high PM rigidity (38). Our lipidome data support the conclusions of Jiang et al. (25), who have recently suggested a probable role of orf19.4531 in the regulation of transport and redistribution of lipids across different membranes, resulting in an impact on the plasma membrane potential. The observed high PM rigidity in CDR6/ROA1 null could be a plausible reason for the increased membrane potential observed by Jiang et al. (25). Additionally, this high PM rigidity could result in an increase in the population of nonrandom dipoles, leading to an increase in membrane dipole potential. Our dipole potential measurements showed that the NKKY101 strain has significantly higher PM dipole potential as compared with WT and NKKY102 strains (Fig. 4E).
Together, an increase in PE content in the absence of Cdr6/Roa1 in PM results in an increase in membrane rigidity, which impacts membrane potential and PM dipole potential, resulting in a decrease in drug entry into the cells and enhanced azole resistance. These observations are also consistent with the earlier reports that the physical state of the PM impacts intracellular drug accumulation and drug susceptibility of C. albicans cells (8–10, 16).
We explored the cause of observed enhanced resistance to azoles of the CDR6/ROA1 null mutant by performing whole-genome transcriptomic analysis. These data confirmed the absence of ERG gene regulation and also of the azole drug transporters (Cdr1, Cdr2, and Mdr1) involved in clinical drug resistance. Notably, genes involved in ribosome biogenesis and RNA processing were up-regulated categories in the CDR6/ROA1 null mutant NKKY101 as compared with WT (Fig. 6). It is worth mentioning that well-conserved rapamycin-sensitive Tor protein kinases regulate ribosome biogenesis and various other processes (62). In S. cerevisiae, two Tor proteins (Tor1 and Tor2) have been characterized, whereas in C. albicans, a single Tor homolog is present (63). S. cerevisiae Tor1 negatively regulates expression of Kns1 kinase and relieves its negative impact from the ribosome biogenesis (19). Our growth analysis in the presence of rapamycin demonstrates that TOR is hyperactivated in the CDR6/ROA1 null mutant NKKY101 and induces ribosome biogenesis. This was well supported by the repressed expression of orf19.4979 (a homologue of the S. cerevisiae Kns1 kinase) transcript (Fig. 7B). Finally, enhanced growth of CDR6/ROA1 null mutant NKKY101 as compared with WT in the presence of protein synthesis inhibitors (CHX and ANM) confirmed the impact of induced ribosome biogenesis on translation. To the best of our knowledge, this is the first study to demonstrate a role of the TOR signaling cascade in azole resistance in a pathogenic yeast drug transporter null mutant (Fig. 10).
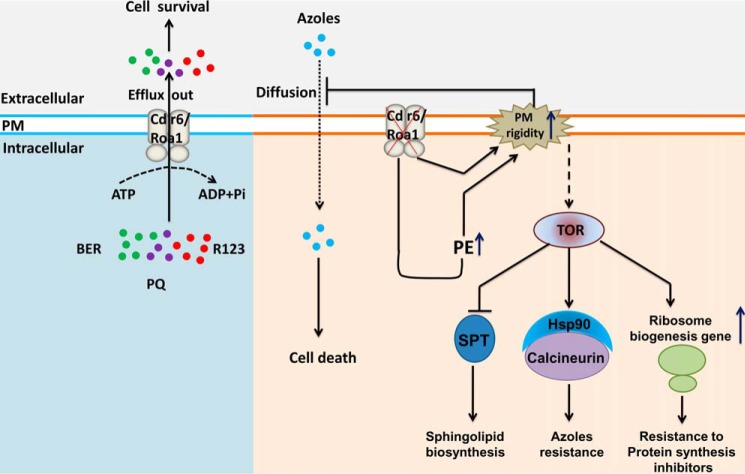
Figure 10.
Proposed model for C. albicans Cdr6/Roa1. Cdr6/Roa1 is a PM-localized transporter that effluxes BER (green circles), R123 (violet circles), and PQ (red circles). Deletion of CDR6/ROA1 leads to increased PE levels and high plasma membrane rigidity, resulting in reduced entry of azoles (sky blue circles) inside the cells, and provides resistance against azoles. The absence of Cdr6/Roa1 activates Tor1, which affects multiple cellular functions, including (i) induction of ribosome biogenesis, which provides resistance to protein synthesis inhibitors (CHX and ANM); (ii) Hsp90 activation, which acts on the client protein calcineurin and provides resistance to azoles; and (iii) inhibition of the SPT enzymatic step, which results in suppression of complex sphingolipid biosynthesis.
Tor1 negatively regulates complex sphingolipid synthesis, which was evident from the observed decrease in complex sphingolipid biosynthesis in the CDR6/ROA1 null mutant. This was further supported by the growth analysis in the presence of myriocin (a specific inhibitor of the sphingolipid biosynthesis rate-limiting enzyme SPT), where the NKKY101 strain showed a growth defect in comparison with WT. These observations indicated that Tor1 activation in C. albicans may suppress the biosynthesis of complex sphingolipids by inhibiting the SPT enzyme. In the case of S. cerevisiae, it is well established that Tor1 negatively regulates the complex sphingolipid synthesis by inhibiting Orm phosphorylation, where the non-phosphorylated form of Orm proteins negatively regulates the synthesis of sphingolipids by inhibiting the SPT enzyme (45, 46). The S. cerevisiae Tap42-Sit4-PP2A protein phosphatase complex activates the Npr1 kinase, which in turn phosphorylates Orm proteins. Tor1 imposes inhibition on the Tap42-Sit4-PP2A complex, thus inactivating Npr1 kinase (45, 46), preventing Orm phosphorylation, resulting in inhibition of SPT and repression of sphingolipids synthesis. Inhibition of Tor1 by rapamycin showed increased phosphorylated Orm proteins in S. cerevisiae, which can induce de novo synthesis of complex sphingolipids (46). Together, our study shows that Tor1 regulates complex sphingolipid biosynthesis, and the pathway appears to be evolutionarily conserved in C. albicans.
Hsp90 coordinates cellular circuitry essential for responses to drug-induced stress. Its post-translational modifications, such as phosphorylation, regulate its ability to interact and stabilize client proteins (17, 20, 64). Calcineurin, a protein phosphatase, is a client protein of Hsp90 that acts as a key mediator of Hsp90-dependent azole resistance in yeast (50, 52). Our results illustrate the role of Hsp90-dependent activation of calcineurin in azole resistance in the NKKY101. For instance, based on analysis of the Hsp90 client proteins, including the protein kinase Hog1 and heat shock transcription factor Hsf1, we observed that Hsp90 is present in the hyperactive state in the NKKY101. This particular state of Hsp90 is a consequence of hyperactive Tor1 in the NKKY101, which modulates the CK2 kinase activity via effector kinase Kns1. Thus, we demonstrate a dual mechanism in the NKKY101, where a high PM rigidity results in reduced intracellular accumulation of azoles and Tor1-dependent activation of Hsp90 and its client protein calcineurin, both leading to increase azole resistance. This was also well supported by an in vivo mouse model, where cells infected with the NKKY101 strain displayed higher fungal burden in the presence of fluconazole (Fig. 9C).
Our results show increased activity of Tor1 in the CDR6/ROA1 null mutant NKKY101. How TOR activity is regulated in CDR6/ROA1 null mutant NKKY101 remains to be elucidated. We speculate that the alteration in PM tension in the CDR6/ROA1 null mutant may be causing Tor1 activation, as it has been shown that the PM stress, due to membrane stretch or inhibition of sphingolipid synthesis, could activate the TOR signaling in S. cerevisiae (11). Mechanistic details and physiological relevance of Tor1 hyperactivity in CDR6/ROA1 null mutant will be interesting to further investigate, as the absence of none of the other ABC transporters has shown such a phenotype. Further, a recent study by Bastidas et al. (65) has shown that inhibition of Tor1 by rapamycin leads to an induced expression (∼34-fold) of CDR6/ROA1 transcripts in C. albicans, implying that Tor1 and Cdr6/Roa1 may impose a reciprocal impact on each other.
In summary, we present the characterization of a PM-localized ABC drug transporter, Cdr6/Roa1, and provide evidence that CDR6/ROA1 deletion leads to hyperactivation of Tor1, thus affecting multiple cellular functions, including Hsp90 activation, induction of ribosome biogenesis, and suppression of complex sphingolipid biosynthesis (Fig. 10). Our result highlights that Cdr6/Roa1 is an exporter of xenobiotics, and it also adopts multiple strategies to display collateral increased resistance to azoles.
Experimental procedures
Materials
The growth media YEPD (yeast extract/peptone/dextrose) and LB broth were purchased from Himedia (Mumbai, India). CuSO4, NiSO4, CaCl2, MnCl2, CsCl, AgNO3, NaCl, H2O2, and SDS were obtained from Qualigens (Mumbai, India). The antifungal compounds PQ, caspofungin, calcofluor white, caffeine, hygromycin B, Congo red, rhodamine 6G, rhodamine B, R123, MG, nystatin, ITC, MCZ, KTC, FLC, BER, RAP, MYR, ANM, CHX, curcumin, and the chemicals sucrose, Tris buffer, DMSO, sorbitol, EDTA, poly-l-lysine, and TMA-DPH were purchased from Sigma. FLC was generously provided by Ranbaxy Laboratories (Gurgaon, Haryana, India). Di-8-ANEPPS and FAST-DiI were purchased from Molecular Probes/Invitrogen (Eugene, OR). Radioactive fluconazole ([3H]FLC; 481 GBq/mmol, 13 Ci/mmol, 1 μCi/μl; 77 μm FLC) was custom-synthesized by Amersham Biosciences. The oligonucleotides used in the present study, as listed in Table S3, were obtained from Sigma Genosys (Bangalore, India).
Strains and culture conditions
The yeast strains used in the present study are listed in the Table S4. C. albicans strains were grown and maintained in YEPD medium. Strain stocks were made in 15% glycerol at −80 °C and freshly revived in YEPD before use. Bacterial strain Escherichia coli DH5α was used as a host for the construction and propagation of plasmids. E. coli cells were grown in LB medium containing 100 μg/ml ampicillin (Amresco) or 25 μg/ml chloramphenicol (Sigma-Aldrich) as required.
Plasmid constructions
To construct the CDR6/ROA1-specific knockout cassette, we ligated three fragments and cloned two fragments (500 bp upstream and 500 bp downstream of CDR6/ROA1) in pBluescript KS+ vector in a single cloning step. For this purpose, we amplified 500 bp upstream and 500 bp downstream of the CDR6/ROA1 ORF from C. albicans strain SC5314 by PCR using primer pairs CDR6-KpnI/CDR6-XhoI-NotI and CDR6-SacII-NotI/CDR6-SacI (Table S3). After digestion of PCR products (KpnI/NotI and NotI/SacI) and of the vector (KpnI/SacI), products were ligated to yield pNK1. Next the SAT1 flipper cassette was isolated from pSFS2A vector by XhoI and SacII digestion and cloned into pNK1 to give rise to pNK2.
For the CDR6/ROA1 revertant, the 500-bp upstream region of CDR6/ROA1 in pNK2 was replaced with the CDR6/ROA1 ORF (open reading frame) along with its promoter. The CDR6/ROA1 ORF was amplified along with its promoter from C. albicans SC5314 genomic DNA using CDR6-Rev-KpnI and CDR6-Rev-XhoI primers (Table S3). The PCR product was digested with KpnI and XhoI and cloned into pNK2 to yield pNK3.
To construct overexpression plasmids CIp-SAT-PTDH3-CDR6/ROA1 and CIp-SAT-PTDH3-GFP-CDR6/ROA1-SP, we first amplified the orf19.4531 open reading frame using primers 4531FP and 4531R (Table S3) and transferred the resulting PCR product into the pDONR207 vector using InvitrogenTM GatewayTM BP ClonaseTM as described previously (66). orf19.4531 was subsequently transferred from the resulting donor plasmid pDONR207::orf19.4531 into the destination vectors CIp-SAT-PTDH3-GTW and CIp-SAT-PTDH3-GFP-GTW-SP using InvitrogenTM GatewayTM LRClonaseTM (66). The CIp-SAT-PTDH3-GTW vector is a derivative of plasmid CIp10 that carries the sequence for integration at the RPS1 locus on C. albicans Chr1, the SAT1 marker for nourseothricin resistance selection (32, 67), and a GatewayTM cassette flanked by the attR sequences and preceded by the PTDH3 constitutive promoter. The CIp-SAT-PTDH3-GFP-GTW-SP vector is similar to CIp-SAT-PTDH3-GTW except that it harbors the GFP open reading frame between PTDH3 and the GatewayTM cassette and a platform for molecular barcode cloning and IlluminaTM sequencing. The resulting plasmids, CIp-SAT-PTDH3-CDR6/ROA1 and CIp-SAT-PTDH3-GFP-CDR6/ROA1, were then used to transform C. albicans SC5314 strain.
Yeast strain transformation
C. albicans strains were transformed by an electroporation method as reported previously (32), and selection was performed on YEPD plates containing nourseothricin.
C. albicans strain construction
CDR6 knockout strain construction
The CDR6/ROA1 knockout cassette was removed from the pNK2 plasmid by KpnI/SacI digestion; integration was performed in C. albicans (SC5314) using an electroporation method (32). Southern blots were performed to confirm the first allele deletion, and the strain was designated NKKY100. To perform the second allele deletion, we removed the selection marker (SAT1 cassette) from the transformants by growing them in yeast extract maltose medium as described previously (32). For deletion of the second allele, the integration was performed in the heterozygous null strain using the CDR6/ROA1 knockout cassette and confirmed by Southern blotting and semiquantitative RT-PCR. The CDR6/ROA1 homozygous null strain was designated as NKKY101.
CDR6/ROA1 revertant strain construction
The CDR6/ROA1 revertant cassette was removed from the plasmid pNK3 by KpnI/SacI digestion, and transformation was performed in the nourseothricin-sensitive CDR6/ROA1 homozygous null strain NKKY1011. The CDR6/ROA1 revertant (NKKY102) strain construction was validated by semiquantitative RT-PCR.
CDR6/ROA1 overexpression strain construction
To construct the CDR6/ROA1-overexpressing strain in C. albicans, we linearized the CIp-SAT-PTDH3-GFP-CDR6/ROA1-SP and CIp-SAT-PTDH3-CDR6/ROA1 plasmids with StuI and transformed C. albicans strain SC5314 to give rise to the GFP-tagged CDR6/ROA1 overexpression strain (CA-CDR6) and untagged CDR6/ROA1 overexpression strain (NKKY103), respectively.
Bioinformatic analysis
The phylogenetic tree was constructed using MEGA version 6 software (68). Cdr6/Roa1 protein topology was predicted using the TOPOCONS software (69).
Isolation of plasma membrane and immunodetection of Cdr6/Roa1-GFP protein
PMs were prepared from WT and Cdr6/Roa1-GFP–expressing C. albicans strains using sucrose density gradient centrifugation (70). The PM fractions (80 μg) were separated by 8% PAGE, blotted, and probed with HRP-conjugated anti-GFP monoclonal antibody (Santa Cruz Biotechnology, Inc., Dallas, TX) as described previously (70). Pma1 was used as a PM fraction control as well as a loading control; it was detected using anti-Pma1p polyclonal antibody (a gift from Professor Ramon Serrano, Universidad Politecnica de Valencia-CSIC, Valencia, Spain). Pma1p antibodies were detected with HRP-conjugated goat anti-rabbit antibody.
Semiquantitative and quantitative RT-PCR
cDNA synthesis was performed using the RevertAid H Minus First Strand cDNA synthesis kit (Thermo Scientific) following the protocol described by the manufacturer. The semiquantitative RT-PCR was performed using cDNA product (1 μl) for the PCR amplification reaction (25 μl) using gene-specific primers (Table S3). Quantitative RT-PCR was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) and Applied Biosystems® 7500 real-time PCR systems using gene-specific primers (Table S3). The ACT1 gene signal was used for normalization, and results were compared between the Ct values of WT and NKKY101 mutant.
Drug susceptibility assays
Spot assay
The spot assay was performed using 5-fold serial dilutions with 5 μl of cells from each dilution spotted on YEPD plates with and without drug (30).
Planktonic growth assay
The planktonic growth assay was performed using a broth microdilution method in YEPD medium with an inoculum of 1 × 104 cells/ml as described previously (71).
Agar drug diffusion assay
The assay was performed using an inoculum of 1 × 105 cells/ml as described previously (29).
BER and R123 accumulation assay
The accumulation of R123 and BER in C. albicans strains was measured by flow cytometry. Briefly, log phase cells were set to an A600 of 0.1 in PBS + 2% glucose. Then a 10 μm final concentration of R123 or BER was added to the culture, and the cells were incubated at 30 °C for 1 h. The cells were then washed twice with ice-cold PBS and analyzed with the FACSort flow cytometer (BD Biosciences) using filter FL2 and filter FL1 for R123 and BER, respectively. A total of 10,000 events were considered. The analysis was performed using CellQuest software.
[3H]FLC accumulation assay
For the import assay with WT, NKKY101, and NKKY102 strains, 200 μl of concentrated, glucose-starved (3-h starvation) cell culture was added to 14-ml round bottom tubes containing 250 μl of YNB with or without glucose as well as 50 μl of 0.77 μm [3H]FLC (freshly diluted 1:100 from stock). The resulting final [3H]FLC concentration in each sample was 77 nm (23.6 ng/ml), which is significantly below the MIC50 value for the strains. After a 24-h incubation with [3H]FLC, 200 μl of the treated cell solution was added to a fresh 14-ml round bottom tube containing 5 ml of stop solution (YNB + 20 mm (6 mg/liter) unlabeled FLC). The solution was then poured over prewetted glass fiber filters and dried. Another 5 ml of stop solution was poured over each filter to wash each sample again. The filters containing washed cells were transferred to 5-ml scintillation vials. 3 ml of scintillation mixture (Ecoscint XR, National Diagnostics, Atlanta, GA) was added, and the radioactivity associated with the filter was measured in a Beckman Coulter scintillation analyzer. Results were normalized to cpm per 1 × 108 cells. All experiments were performed as biological triplicates, and results are shown as means ± S.E. A statistical significance value of 0.05 was employed using an unpaired Student's t test.
Fluorescence imaging of labeled C. albicans and FRAP experiments
C. albicans staining with FAST-DiI was performed as described previously (16) with minor modifications. C. albicans culture was suspended at a density of 108 cells/ml in 1 m sorbitol, 0.1 m EDTA buffer and labeled using a final concentration of 10 μm FAST-DiI.
Confocal imaging was carried out on an inverted Zeiss LSM 510 Meta confocal microscope with a plan-apochromat ×100/1.4 numerical aperture oil-immersion objective using the 561-nm laser. Fluorescence emission was collected using the 575–630-nm bandpass filter with the confocal pinhole set at 1 Airy unit with a zoom factor of 7. The fluorescent periphery of the cell that represents the plasma membrane was selected for bleaching and monitoring recovery of fluorescence. The data on the diffusion coefficient (D) and mobile fraction (Mf) were calculated from quantitative FRAP experiments where just the bleached region was scanned to achieve improved temporal resolution. FRAP experiments were performed with Gaussian spot photobleaching and line-scanning mode with a circular region of interest of 1-μm radius. Data representing the mean fluorescence intensity in the membrane region (obtained using Zeiss LSM 510 software, version 3.2) within the bleached spot were corrected for background and analyzed. For a two-dimensional diffusion model, FRAP data were fitted to determine the characteristic diffusion time (τd) (72),
| (Eq. 1) |
where F(t) is the mean background-corrected and normalized fluorescence intensity at time t in the membrane region within the bleached spot, F(∞) is the recovered fluorescence at time t → ∞, and F(0) is the bleached fluorescence intensity at time t → 0. I0 and I1 are modified Bessel functions. The effective two-dimensional diffusion coefficient (D) is determined from the equation (73),
| (Eq. 2) |
where ω is the radius of the bleached spot. The mobile fraction (Mf) is calculated according to the equation,
| (Eq. 3) |
where F(p) is the mean background-corrected and normalized prebleach fluorescence intensity. Non-linear curve fitting of fluorescence recovery data to Equation 1 was carried out using GraphPad Prism software version 4.00 (La Jolla, CA).
Measurement of membrane dipole potential
Dipole potential measurements were carried out by a dual-wavelength ratiometric approach using the voltage-sensitive fluorescence probe di-8-ANEPPS (74–77). PM fractions (10 μg/ml) of C. albicans WT, NKKY101, and NKKY102 strains were incubated with the di-8-ANEPPS probe (1 nm) in the dark for 30 min at 25 °C. Steady-state fluorescence measurements were performed with an RF-5301PC spectrofluorophotometer (Shimadzu, Kyoto, Japan) using 1-cm path length quartz cuvettes at room temperature (∼23 °C). Excitation and emission slits with a band pass of 5 nm were used for all measurements. Background intensities of samples were subtracted from each sample to cancel any contribution due to the solvent Raman peak. Fluorescence intensities were recorded at two excitation wavelengths (420 and 520 nm). The emission wavelength was fixed at 670 nm. The fluorescence ratio (R), defined as the ratio of fluorescence intensity at an excitation wavelength of 420 nm to that at 520 nm (emission at 670 nm in both cases), which is a measure of membrane dipole potential, was calculated (74). The choice of the emission wavelength (670 nm) at the red edge of the fluorescence spectrum has previously been shown to rule out membrane fluidity effects (78). Dipole potential (ψD) in mV was calculated from R using the linear relationship (74),
| (Eq. 4) |
Experiments were performed in triplicate, and results are shown as means ± S.D. Student's t test was used to calculate statistical significance value.
Lipidome analysis
Lipid extraction was performed as described previously (71). Lipidome analysis was performed at the Kansas Lipidomics Research Center Analytical Laboratory, using an ESI source on a triple quadrupole mass spectrometer (ESI-MS/MS) (API 4000, Applied Biosystems) (71). Experiments were performed in quadruplet, and results are shown as means ± S.E. A statistical significance value of 0.05 was employed using Student's t test.
Transcriptome analysis
Samples for transcriptome analysis were prepared as described previously (30). Microarray and data scanning was performed by Genotypic Technology Ltd. (Bangalore, India). The genes whose level varied ≥2-fold between NKKY101 and WT were considered significantly affected genes. The transcription profile was analyzed using the Go-Slim mapper and was annotated on the basis of biological processes (79). Cytoscape version 3.4.0 software was used to create an interacting map of the top five up- and down-regulated categories (80). The microarrays used in the present study along with complete raw transcriptome data can be accessed from the NCBI Gene Expression Omnibus database under accession number GSE70340.
CK2 kinase activity assay
The recombinant protein His6-Sic1 was expressed and purified from E. coli as reported previously (55). Briefly, His6-Sic1 was purified on Ni2+-nitrilotriacetic acid beads, as described by the manufacturer (QIAexpressionist Handbook, Qiagen), and eluted with 100 mm imidazole. The protein concentration was measured by the Bradford method using the Bio-Rad protein assay kit.
C. albicans strains were grown in YPD medium until early exponential phase (0.2–0.3 OD/ml), and crude extract was prepared as described previously (81). The CK2 activity of crude protein extracts (0.35 μg) was tested using the purified recombinant protein His6-Sic1 (7.5 μg) as a substrate in a reaction mix containing 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.1 mm ATP at 30 °C for the indicated times. Then CK2 phosphorylation on His6-Sic1 was analyzed by SDS-PAGE, blotted, and probed with anti-Sic1-pSer-201 antibody (1:2000 dilution) and with anti-His probe antibody (1:1000 dilution; Santa Cruz Biotechnology) as a control (82). Densitometric analysis of three independent experiments was performed by using the ImageJ program.
Hog1 Western blots
Overnight cultures of C. albicans WT (SC5314), NKKY101, and NKKY102 strains were diluted to A600 = 0.2 and grown at 30 °C for 6 h. Cells were pelleted down and immediately frozen in liquid nitrogen. Cell extract was prepared as described previously (81), and 60 μg of total cell extract protein was separated by 8% PAGE and blotted. The level of phosphorylated Hog1 protein was detected using anti-dually phosphorylated p38 antibody (Cell Signaling Technology), and total Hog1 protein was detected with anti-Hog1p antibody (Y-215; Santa Cruz Biotechnology). Band intensities were quantified through Image LabTM software (Bio-Rad), and Hog1 was used as a loading control.
Mouse survival and kidney fungal burden assay
All animal experiments performed in the absence of drug were executed at the University Hospital Center of Lausanne as described previously (30). Briefly, for all mouse experiments, female BALB/c mice (6 weeks old; Charles River France) were housed in ventilated cages with free access to food and water. Overnight secondary cultures grown in YEPD at 30 °C were washed twice and resuspended in 5 ml of PBS to the desired concentration. For survival and kidney fungal burden experiments in the absence of drug, groups of 10 mice were used. The mice were injected through the lateral tail vein with 250 μl of a cell suspension containing 2 × 106 cells/ml for survival and 8 × 105 cells/ml for fungal burden experiments. The postinfection day of natural death or euthanasia of moribund animals was recorded for each mouse. Statistical analyses of the survival data were performed using the log–rank (Mantel–Cox) test. Concerning the fungal burden experiment at 3 days postinfection, the kidneys were recovered, and the cfu were determined as described previously (83). Statistical analyses of the differences between cfu values were performed using the Mann–Whitney test.
The kidney burden study in the presence of FLC was performed at the Public Health Research Institute (Rutgers University). For the kidney burden study in the presence of FLC with single-strain infections, groups of 10 female mice were used. On day 0, female BALB/c mice were challenged with suspensions of C. albicans strains (WT, NKKY101, and NKKY102) at 2 × 105 cells/ml, administered by tail vein injection. At 3 h after infection, mice were treated with FLC (20 mg/kg) via intraperitoneal injection, and these treatments were administered once daily for 3 days after the first dose. Three mice from each strain group were euthanized at 24, 48, and 72 h postinfection, and kidneys were harvested and enumerated for C. albicans burdens.
Ethical statement
All survival assay animal experiments were performed at the University Hospital Center of Lausanne with approval through the Institutional Animal Use Committee, Affaires Vétérinaires du Canton de Vaud, Switzerland (authorization number 1734.3), according to decree 18 of the federal law on animal protection. All animal drug treatment experiments were conducted at the Public Health Research Institute, Rutgers University. All animals were housed and handled according to the guidelines set forth by the institutional animal care and use committee. Animal protocols (protocol number D-15138-A1) were reviewed and approved by the institutional animal care and use committee of Rutgers University. The animal experiments were conducted in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and were in full compliance with the United States Animal Welfare Act (Public Law 98-198).
Author contributions
N. K. K. and R. P. conceived of and designed the study. N. K. K. designed and performed selection experiments, drug-susceptibility assays, strain construction, R123 and BER transport assays, qRT-PCR, and Western blotting. R. P., A. C., N. K. K., and P. S. designed and N. K. K. and P. S. performed the FRAP experiment to measure the PM fluidity. B. D. E. and T. C. W. designed and performed the azole accumulation experiment. P. C. designed and performed CK2 kinase activity assays. R. P., N. K. K., and A. S. designed and performed the lipidomie analysis. N. C. designed and performed the C. albicans mouse study in the presence of azole. A. T. C. and D. S. designed and performed the virulence and kidney burden assay in the absence of drug. N. K. K. and M. G. designed and analyzed the transcriptome profile. M. C. and C. E. constructed the CDR6/ROA1 overexpression plasmid. A. K. M., R. P., and N. K. K. designed the Hog1 activity analysis, and N. K. K. performed the experiment. N. A. G. provided reagents for the rebuttal experiment. R. P., N. K. K., and M. G. wrote the paper with input from the other authors. All authors read and approved the final paper.
Supplementary Material
Acknowledgments
We acknowledge Dr. Sunil Shetty for valuable discussions and suggestions during the work. We acknowledge AIRF, JNU, for providing instrumental support and Ashok Kumar Sahu and Tripti Panwar for confocal microscopy. We thank Amarchand Kumawat for help with ultracentrifugation. We thank Kaushal Kumar Mahto, Mohd. Wasi, and Debasree Kundu for help during lipid preparation, bioinformatic analysis, and Hog1 Western blotting, respectively. The help of Dr. Rupinder Kaur in providing the facility for C. albicans strain growth during the FRAP experiment is greatly appreciated.
This work was supported, in part, by the Department of Biotechnology, Ministry of Science and Technology, Government of India, Grants BT/01/CEIB/10/III/02, BT/PR7392/MED/29/652/2012, and BT/PR14879/BRB10/885/2010 (to R. P.). Research in the laboratory of N. C. was supported by National Institutes of Health Grant R01AI124499. The research work in the laboratory of A. K. M. was supported by a DST PURSE grant. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The microarrays used in the present study along with complete raw transcriptome data can be accessed from the NCBI Gene Expression Omnibus database under accession number GSE70340.
This article contains Figs. S1–S5 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MDR
- multidrug resistance
- ABC
- ATP-binding cassette
- TOR
- target of rapamycin
- PM
- plasma membrane
- BER
- berberine
- R123
- rhodamine 123
- PQ
- paraquat
- CDR6
- Candida drug-resistant 6
- TMA-DPH
- trimethylaminodiphenyl-1,3,5-hexatriene
- CR
- congo red
- FLC
- fluconazole
- ITR
- itraconazole
- MCZ
- miconazole
- KTC
- ketoconazole
- FRAP
- fluorescence recovery after photobleaching
- PGL
- phosphoglyceride
- PE
- phosphatidylethanolamine
- SPT
- serine palmitoyltransferase
- CGD
- Candida Genome Database
- di-8-ANEPPS
- 4-(2-(6-dioctylamino)-2-naphthalenyl)ethenyl)-1-(3-sulfopropyl)-inner salt
- ESI
- electrospray ionization
- ANM
- anisomycin
- CHX
- cycloheximide
- IPC
- inositol phosphorylceramide
- MIPC
- mannosylinositol phosphorylceramide
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Pfaller M. A., and Diekema D. J. (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaoutis T. E., Argon J., Chu J., Berlin J. A., Walsh T. J., and Feudtner C. (2005) The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin. Infect. Dis. 41, 1232–1239 10.1086/496922 [DOI] [PubMed] [Google Scholar]
- 3. Cowen L. E. (2008) The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6, 187–198 10.1038/nrmicro1835 [DOI] [PubMed] [Google Scholar]
- 4. Sanglard D. (2016) Emerging threats in antifungal-resistant fungal pathogens. Front. Med. 3, 11 10.3389/fmed.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanglard D., Kuchler K., Ischer F., Pagani J. L., Monod M., and Bille J. (1995) Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39, 2378–2386 10.1128/AAC.39.11.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White T. C. (1997) Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41, 1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lyons C. N., and White T. C. (2000) Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44, 2296–2303 10.1128/AAC.44.9.2296-2303.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kohli A., Smriti, Mukhopadhyay K., Rattan A., and Prasad R. (2002) In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46, 1046–1052 10.1128/AAC.46.4.1046-1052.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mukhopadhyay K., Kohli A., and Prasad R. (2002) Drug susceptibilities of yeast cells are affected by membrane lipid composition drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46, 3695–3705 10.1128/AAC.46.12.3695-3705.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prasad T., Chandra A., Mukhopadhyay C. K., and Prasad R. (2006) Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob. Agents Chemother. 50, 3597–3606 10.1128/AAC.00653-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T. C., and Loewith R. (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14, 542–547 10.1038/ncb2480 [DOI] [PubMed] [Google Scholar]
- 12. Pike L. J. (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47, 1597–1598 10.1194/jlr.E600002-JLR200 [DOI] [PubMed] [Google Scholar]
- 13. Mollinedo F. (2012) Lipid raft involvement in yeast cell growth and death. Front. Oncol. 2, 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rella A., Farnoud A. M., and Del Poeta M. (2016) Plasma membrane lipids and their role in fungal virulence. Prog. Lipid Res. 61, 63–72 10.1016/j.plipres.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasrija R., Panwar S. L., and Prasad R. (2008) Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52, 694–704 10.1128/AAC.00861-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukhopadhyay K., Prasad T., Saini P., Pucadyil T. J., Chattopadhyay A., and Prasad R. (2004) Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 48, 1778–1787 10.1128/AAC.48.5.1778-1787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taipale M., Jarosz D. F., and Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- 18. Sanchez-Casalongue M. E., Lee J., Diamond A., Shuldiner S., Moir R. D., and Willis I. M. (2015) Differential phosphorylation of a regulatory subunit of protein kinase CK2 by target of rapamycin complex 1 signaling and the Cdc-like kinase Kns1. J. Biol. Chem. 290, 7221–7233 10.1074/jbc.M114.626523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J., Moir R. D., McIntosh K. B., and Willis I. M. (2012) TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell 45, 836–843 10.1016/j.molcel.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mollapour M., Tsutsumi S., Truman A. W., Xu W., Vaughan C. K., Beebe K., Konstantinova A., Vourganti S., Panaretou B., Piper P. W., Trepel J. B., Prodromou C., Pearl L. H., and Neckers L. (2011) Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol. Cell 41, 672–681 10.1016/j.molcel.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diezmann S., Michaut M., Shapiro R. S., Bader G. D., and Cowen L. E. (2012) Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet. 8, e1002562 10.1371/journal.pgen.1002562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shekhar-Guturja T., Gunaherath G. M. K. B., Wijeratne E. M. K., Lambert J.-P., Averette A. F., Lee S. C., Kim T., Bahn Y.-S., Tripodi F., Ammar R., Döhl K., Niewola-Staszkowska K., Schmitt L., Loewith R. J., Roth F. P., et al. (2016) Dual action antifungal small molecule modulates multidrug efflux and TOR signaling. Nat. Chem. Biol. 12, 867–875 10.1038/nchembio.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu T. T., Znaidi S., Barker K. S., Xu L., Homayouni R., Saidane S., Morschhäuser J., Nantel A., Raymond M., and Rogers P. D. (2007) Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 6, 2122–2138 10.1128/EC.00327-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhamgaye S., Bernard M., Lelandais G., Sismeiro O., Lemoine S., Coppée J.-Y., Le Crom S., Prasad R., and Devaux F. (2012) RNA sequencing revealed novel actors of the acquisition of drug resistance in Candida albicans. BMC Genomics 13, 396 10.1186/1471-2164-13-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang L., Xu D., Chen Z., Cao Y., Gao P., and Jiang Y. (2016) The putative ABC transporter encoded by the orf19.4531 plays a role in the sensitivity of Candida albicans cells to azole antifungal drugs. FEMS Yeast Res. 16, fow024 10.1093/femsyr/fow024 [DOI] [PubMed] [Google Scholar]
- 26. Prasad R., Banerjee A., Khandelwal N. K., and Dhamgaye S. (2015) The ABCs of Candida albicans multidrug transporter Cdr1. Eukaryot. Cell 14, 1154–1164 10.1128/EC.00137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smriti, Krishnamurthy S., Dixit B. L., Gupta C. M., Milewski S., and Prasad R. (2002) ABC transporters CdrLp, Cdr2p and Cdr3p of a human pathogen candida albicans are general phospholipid translocators. Yeast 19, 303–318 10.1002/yea.818 [DOI] [PubMed] [Google Scholar]
- 28. Abe F., and Hiraki T. (2009) Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1788, 743–752 10.1016/j.bbamem.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 29. Lamping E., Monk B. C., Niimi K., Holmes A. R., Tsao S., Tanabe K., Niimi M., Uehara Y., and Cannon R. D. (2007) Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot. Cell 6, 1150–1165 10.1128/EC.00091-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khandelwal N. K., Kaemmer P., Forster T. M., Singh A., Coste A. T., Andes D. R., Hube B., Sanglard D., Chauhan N., Kaur R., d'Enfert C., Mondal A. K., and Prasad R. (2016) Pleiotropic effects of the vacuolar ABC transporter MLT1 of Candida albicans on cell function and virulence. Biochem. J. 473, 1537–1552 10.1042/BCJ20160024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rawal M. K., Khan M. F., Kapoor K., Goyal N., Sen S., Saxena A. K., Lynn A. M., Tyndall J. D. A., Monk B. C., Cannon R. D., Komath S. S., and Prasad R. (2013) Insight into pleiotropic drug resistance ATP-binding cassette pump drug transport through mutagenesis of Cdr1p transmembrane domains. J. Biol. Chem. 288, 24480–24493 10.1074/jbc.M113.488353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuss O., Vik A., Kolter R., and Morschhäuser J. (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127 10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 33. Mansfield B. E., Oltean H. N., Oliver B. G., Hoot S. J., Leyde S. E., and White T. C. (2010) Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 6, e1001126 10.1371/journal.ppat.1001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prasad R., Khandelwal N. K., and Banerjee A. (2016) Yeast ABC transporters in lipid trafficking. Fungal Genet. Biol. 93, 25–34 10.1016/j.fgb.2016.05.008 [DOI] [PubMed] [Google Scholar]
- 35. Tarling E. J., de Aguiar Vallim T. Q., and Edwards P. (2013) A role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24, 342–350 10.1016/j.tem.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowen L. E., Sanglard D., Howard S. J., Rogers P. D., and Perlin D. S. (2014) Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 10.1101/cshperspect.a019752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kodedová M., and Sychrová H. (2015) Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10, e0139306 10.1371/journal.pone.0139306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawaliby R., Trubbia C., Delporte C., Noyon C., Ruysschaert J.-M., Van Antwerpen P., and Govaerts C. (2016) Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 291, 3658–3667 10.1074/jbc.M115.706523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loewith R., and Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heitman J., Movva N. R., and Hall M. N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- 41. Croons V., Martinet W., Herman A. G., Timmermans J.-P., and De Meyer G. R. Y. (2009) The protein synthesis inhibitor anisomycin induces macrophage apoptosis in rabbit atherosclerotic plaques through p38 mitogen-activated protein kinase. J. Pharmacol. Exp. Ther. 329, 856–864 10.1124/jpet.108.149948 [DOI] [PubMed] [Google Scholar]
- 42. Grollman A. P. (1967) Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 242, 3226–3233 [PubMed] [Google Scholar]
- 43. Rao S. S., and Grollman A. P. (1967) Cycloheximide resistance in yeast: a property of the 60s ribosomal subunit. Biochem. Biophys. Res. Commun. 29, 696–704 10.1016/0006-291X(67)90273-2 [DOI] [PubMed] [Google Scholar]
- 44. Sweeney R., Yao C. H., and Yao M. C. (1991) A mutation in the large subunit ribosomal RNA gene of Tetrahymena confers anisomycin resistance and cold sensitivity. Genetics. 127, 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu M., Huang C., Polu S. R., Schneiter R., and Chang A. (2012) Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J. Cell Sci. 125, 2428–2435 10.1242/jcs.100578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimobayashi M., Oppliger W., Moes S., Jenö P., and Hall M. N. (2013) TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell 24, 870–881 10.1091/mbc.E12-10-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., and Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 10.1038/nature08787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han S., Lone M. A., Schneiter R., and Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 10.1073/pnas.0911617107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Delgado A., Casas J., Llebaria A., Abad J. L., and Fabrias G. (2006) Inhibitors of sphingolipid metabolism enzymes. Biochim. Biophys. Acta 1758, 1957–1977 10.1016/j.bbamem.2006.08.017 [DOI] [PubMed] [Google Scholar]
- 50. Cowen L. E., and Lindquist S. (2005) Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185–2189 10.1126/science.1118370 [DOI] [PubMed] [Google Scholar]
- 51. Robbins N., Uppuluri P., Nett J., Rajendran R., Ramage G., Lopez-Ribot J. L., Andes D., and Cowen L. E. (2011) Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7, e1002257 10.1371/journal.ppat.1002257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sanglard D., Ischer F., Marchetti O., Entenza J., and Bille J. (2003) Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976 10.1046/j.1365-2958.2003.03495.x [DOI] [PubMed] [Google Scholar]
- 53. Imai J., and Yahara I. (2000) Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell Biol. 20, 9262–9270 10.1128/MCB.20.24.9262-9270.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumar R., Musiyenko A., and Barik S. (2005) Plasmodium falciparum calcineurin and its association with heat shock protein 90: mechanisms for the antimalarial activity of cyclosporin A and synergism with geldanamycin. Mol. Biochem. Parasitol. 141, 29–37 10.1016/j.molbiopara.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 55. Coccetti P., Rossi R. L., Sternieri F., Porro D., Russo G. L., di Fonzo A., Magni F., Vanoni M., and Alberghina L. (2004) Mutations of the CK2 phosphorylation site of Sic1 affect cell size and S-Cdk kinase activity in Saccharomyces cerevisiae. Mol. Microbiol. 51, 447–460 10.1046/j.1365-2958.2003.03836.x [DOI] [PubMed] [Google Scholar]
- 56. Hawle P., Horst D., Bebelman J. P., Yang X. X., Siderius M., and van der Vies S. M. (2007) Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p). Eukaryot. Cell 6, 521–532 10.1128/EC.00343-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Leach M. D., Budge S., Walker L., Munro C., Cowen L. E., and Brown A. J. P. (2012) Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 8, e1003069 10.1371/journal.ppat.1003069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Duina A. A., Kalton H. M., and Gaber R. F. (1998) Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273, 18974–18978 10.1074/jbc.273.30.18974 [DOI] [PubMed] [Google Scholar]
- 59. Nicholls S., Leach M. D., Priest C. L., and Brown A. J. P. (2009) Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 74, 844–861 10.1111/j.1365-2958.2009.06883.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cowen L. E. (2013) The fungal Achilles' heel: targeting Hsp90 to cripple fungal pathogens. Curr. Opin. Microbiol. 16, 377–384 10.1016/j.mib.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 61. Shah A. H., Singh A., Dhamgaye S., Chauhan N., Vandeputte P., Suneetha K. J., Kaur R., Mukherjee P. K., Chandra J., Ghannoum M. A., Sanglard D., Goswami S. K., and Prasad R. (2014) Novel role of a family of major facilitator transporters in biofilm development and virulence of Candida albicans. Biochem. J. 460, 223–235 10.1042/BJ20140010 [DOI] [PubMed] [Google Scholar]
- 62. De Virgilio C., and Loewith R. (2006) The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 38, 1476–1481 10.1016/j.biocel.2006.02.013 [DOI] [PubMed] [Google Scholar]
- 63. Cruz M. C., Goldstein A. L., Blankenship J., Del Poeta M., Perfect J. R., McCusker J. H., Bennani Y. L., Cardenas M. E., and Heitman J. (2001) Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob. Agents Chemother. 45, 3162–3170 10.1128/AAC.45.11.3162-3170.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trepel J., Mollapour M., Giaccone G., and Neckers L. (2010) Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bastidas R. J., Heitman J., and Cardenas M. E. (2009) The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5, e1000294 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cabral V., Chauvel M., Firon A., Legrand M., Nesseir A., Bachellier-Bassi S., Chaudhari Y., Munro C. A., and d'Enfert C. (2012) Modular gene over-expression strategies for Candida albicans. Methods Mol. Biol. 845, 227–244 10.1007/978-1-61779-539-8_15 [DOI] [PubMed] [Google Scholar]
- 67. Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., and Brown A. J. (2000) CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325–327 10.1002/1097-0061(20000315)16:4%3C325::AID-YEA538%3E3.0.CO%3B2-# [DOI] [PubMed] [Google Scholar]
- 68. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bernsel A., Viklund H., Hennerdal A., and Elofsson A. (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37, W465–W468 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shukla S., Saini P., Smriti, Jha S., Ambudkar S. V., Prasad R., and Ambudkar S. (2003) Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot. Cell 2, 1361–1375 10.1128/EC.2.6.1361-1375.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mahto K. K., Singh A., Khandelwal N. K., Bhardwaj N., Jha J., and Prasad R. (2014) An assessment of growth media enrichment on lipid metabolome and the concurrent phenotypic properties of Candida albicans. PLoS One 9, e113664, 10.1371/journal.pone.0113664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pucadyil T. J., Kalipatnapu S., Harikumar K. G., Rangaraj N., Karnik S. S., and Chattopadhyay A. (2004) G-protein-dependent cell surface dynamics of the human serotonin1A receptor tagged to yellow fluorescent protein. Biochemistry 43, 15852–15862 10.1021/bi0480887 [DOI] [PubMed] [Google Scholar]
- 73. Koppel D. E., Sheetz M. P., and Schindler M. (1980) Lateral diffusion in biological membranes. A normal-mode analysis of diffusion on a spherical surface. Biophys. J. 30, 187–192 10.1016/S0006-3495(80)85087-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Starke-Peterkovic T., Turner N., Vitha M. F., Waller M. P., Hibbs D. E., and Clarke R. J. (2006) Cholesterol effect on the dipole potential of lipid membranes. Biophys. J. 90, 4060–4070 10.1529/biophysj.105.074666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gross E., Bedlack R. S. Jr., and Loew L. M. (1994) Dual-wavelength ratiometric fluorescence measurement of the membrane dipole potential. Biophys. J. 67, 208–216 10.1016/S0006-3495(94)80471-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Haldar S., Kanaparthi R. K., Samanta A., and Chattopadhyay A. (2012) Differential effect of cholesterol and its biosynthetic precursors on membrane dipole potential. Biophys. J. 102, 1561–1569 10.1016/j.bpj.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Starke-Peterkovic T., Turner N., Else P. L., and Clarke R. J. (2005) Electric field strength of membrane lipids from vertebrate species: membrane lipid composition and Na+-K+-ATPase molecular activity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R663–R670 [DOI] [PubMed] [Google Scholar]
- 78. Clarke R. J., and Kane D. J. (1997) Optical detection of membrane dipole potential: avoidance of fluidity and dye-induced effects. Biochim. Biophys. Acta. 1323, 223–239 10.1016/S0005-2736(96)00188-5 [DOI] [PubMed] [Google Scholar]
- 79. Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., Cherry J. M., and Sherlock G. (2004) GO::TermFinder: open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sharma P., Meena N., Aggarwal M., and Mondal A. K. (2005) Debaryomyces hansenii, a highly osmo-tolerant and halo-tolerant yeast, maintains activated Dhog1p in the cytoplasm during its growth under severe osmotic stress. Curr. Genet. 48, 162–170 10.1007/s00294-005-0010-9 [DOI] [PubMed] [Google Scholar]
- 82. Tripodi F., Cirulli C., Reghellin V., Marin O., Brambilla L., Schiappelli M. P., Porro D., Vanoni M., Alberghina L., and Coccetti P. (2010) CK2 activity is modulated by growth rate in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 398, 44–50 10.1016/j.bbrc.2010.06.028 [DOI] [PubMed] [Google Scholar]
- 83. Vandeputte P., Ischer F., Sanglard D., and Coste A. T. (2011) In vivo systematic analysis of Candida albicans Zn2-Cys6 transcription factors mutants for mice organ colonization. PLoS One 6, e26962 10.1371/journal.pone.0026962 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.