Abstract
Tumor metastasis involves cancer cell invasion across basement membranes and interstitial tissues. The initial invasion step consists of adherence of the tumor cell to the extracellular matrix (ECM), and this binding transduces a variety of signals from the ECM to the tumor cell. Accordingly, it is critical to establish the mechanisms by which extracellular cues influence the intracellular activities that regulate tumor cell invasion. Here, we found that invasion of the basal-like breast cancer cell line BT-549 is enhanced by the ECM component chondroitin sulfates (CSs). CSs interacted with and induced proteolytic cleavage of N-cadherin in the BT-549 cells, yielding a C-terminal intracellular N-cadherin fragment that formed a complex with β-catenin. Of note, the cleavage of N-cadherin increased cytoplasmic and nuclear β-catenin levels; induced the matrix metalloproteinase 9 (MMP9) gene, a target of β-catenin nuclear signaling; and augmented the invasion potential of the cells. We also found that CS-induced N-cadherin proteolysis requires caveolae-mediated endocytosis. An inhibitor of that process, nystatin, blocked both the endocytosis and proteolytic cleavage of N-cadherin induced by CS and also suppressed BT-549 cell invasion. Knock-out of chondroitin 4-O-sulfotransferase-1 (C4ST-1), a key CS biosynthetic enzyme, suppressed activation of the N-cadherin/β-catenin pathway through N-cadherin endocytosis and significantly decreased BT-549 cell invasion. These results suggest that CSs produced by C4ST-1 might be useful therapeutic targets in the management of basal-like breast cancers.
Keywords: β-catenin (B-catenin), breast cancer, cadherin, cell invasion, chondroitin sulfate, extracellular matrix, glycobiology, glycosaminoglycan, proteoglycan, sulfotransferase
Introduction
Breast cancer comprises heterogeneous tumors with different clinical characteristics, disease courses, and responses to specific treatments. Tumor-intrinsic features, including classical histological and immunopathological classifications, separate breast tumors into multiple groups. The presence of specific markers in breast cancer has long been recognized to define subtypes with differential overall prognosis and to identify tumors susceptible to targeted treatments. The chief markers assessed are estrogen receptor (ER),2 progesterone receptor (PR), and human epidermal receptor 2 (HER2). Combinations of these markers allow for the assignment of individual cases to specific categories, namely ER+ (ER+/HER2−), HER2+ (ER−/HER2+), triple negative (ER−/PR−/HER2−), and triple positive (ER+/PR+/HER2+). Basal or basal-like subtype broadly corresponds to the ER−/PR−/HER2− cohort (1, 2), and these subtypes are linked to the worst prognosis among the subtypes (3). Thus, a better understanding of the specific nature of the basal-like subtype, especially its highly invasive property, is required.
N-cadherin is a cell–cell adhesion molecule that contributes to the invasive/metastatic phenotype and is also known as an epithelial–mesenchymal transition marker. Up-regulation of N-cadherin is more likely to be observed in breast tumors with a basal-like phenotype (4). The expression of N-cadherin correlates with both invasion and cell motility, and the forced expression of N-cadherin in non-invasive cells produces invasive cells (5). It has been reported that N-cadherin increases cell motility and invasive potential mediated by FGF receptor signaling (5). FGF receptor signaling dramatically up-regulates the expression of the matrix metalloproteinase MMP-9, which may endow cancer cells with a greater ability to degrade matrix protein barriers (6). Another report showed that the cleavage of N-cadherin by the disintegrin metalloproteinase ADAM10 regulates β-catenin signaling and plays a significant role during tumor invasion (7). These studies indicate that N-cadherin is responsible for invasive/metastatic properties and have examined N-cadherin-dependent intracellular events. However, which extracellular cues control tumor cell properties via N-cadherin has remained unclear.
Chondroitin sulfate (CS), a type of glycosaminoglycan, is present on the cell surface and in the extracellular matrix (ECM). There is ample evidence for a pro-tumorigenic role of CS in enhancement of cell proliferation, cell motility, and metastasis (8–11). CSs in the ECM act on tumor cells to promote tumor progression. For example, CS interacts with tumor cells via the multifunctional transmembrane protein CD44 (12, 13). The interaction of CS with CD44 triggers CD44 cleavage and thereby may be involved in tumor progression (14). Furthermore, it has been reported that the tumor-associated glycocalyx that includes CSs plays a key role in promoting and regulating breast cancer progression and metastasis (15).
CS chains consist of repeating disaccharide units [(-4GlcAβ1-3GalNAcβ1-)n] and are covalently linked to specific serine (Ser) residues in any of the core proteins via the so-called glycosaminoglycan–protein linkage region (GlcAβ1-3Galβ1-3Galβ1-4Xylβ1-O-Ser) (16, 17). The biosynthesis of the repeating disaccharide units is catalyzed by a combination of six homologous glycosyltransferase-chondroitin (Chn) synthases-1, -2, and -3, Chn-polymerizing factor, and Chn GalNAc transferases-1 and -2. The resulting backbones of CS chains are subsequently modified with sulfation and uronate epimerization (16). Based on the substrate preferences of Chn sulfotransferases identified to date, the biosynthetic scheme for CS-type sulfation can be classified into initial “4-O-sulfation” and “6-O-sulfation” pathways. In the initial step, the non-sulfated O-unit [GlcA-GalNAc] serves as a common acceptor substrate for two types of sulfotransferases, chondroitin 4-O-sulfotransferases (C4ST-1 and C4ST-2) (18–20) and chondroitin 6-O-sulfotransferse-1 (C6ST-1), forming monosulfated A-unit (GlcA-GalNAc(4-O-sulfate)) and C-unit (GlcA-GalNAc(6-O-sulfate)), respectively (see Fig. 7A, panel b). Subsequent sulfation of A- and C-units can also occur via GalNAc 4-sulfate 6-O-sulfotransferase (GalNAc4S-6ST) or CS-specific uronyl 2-O-sulfotransferase, resulting in the formation of disulfated disaccharide E-unit (GlcA-GalNAc(4,6-O-disulfate)) and D-unit (GlcA(2-O-sulfate)-GalNAc(6-O-sulfate)), respectively (see Fig. 7A, panel b) (16). Of these CS biosynthetic enzymes, it has been reported that the expression of GalNAc transferase-1, C4ST-1, and GalNAc4S-6ST is up-regulated in breast cancer cells (15). Moreover, the gene expression of C4ST-1 has been shown to be positively correlated with progression of breast cancers (6), and CSs produced by C4ST-1 function as a ligand of P-selectin in aggressive breast cancer cells (9). These data suggest that tumor cells drive a change in CS biosynthesis to acquire metastatic capacity. Here, we show that binding of CS to N-cadherin triggers endocytosis-dependent activation of the N-cadherin/β-catenin pathway to enhance the metastatic properties of a basal-like breast cancer cell line BT-549. In addition, a particular sulfation pattern of CS is important for regulation of the invasion potential of BT-549 cells through the N-cadherin/β-catenin pathway, because N-cadherin specifically recognized the CS E-unit, a specific sulfate epitope synthesized by C4ST-1 and GalNAc4S-6ST.
Figure 7.
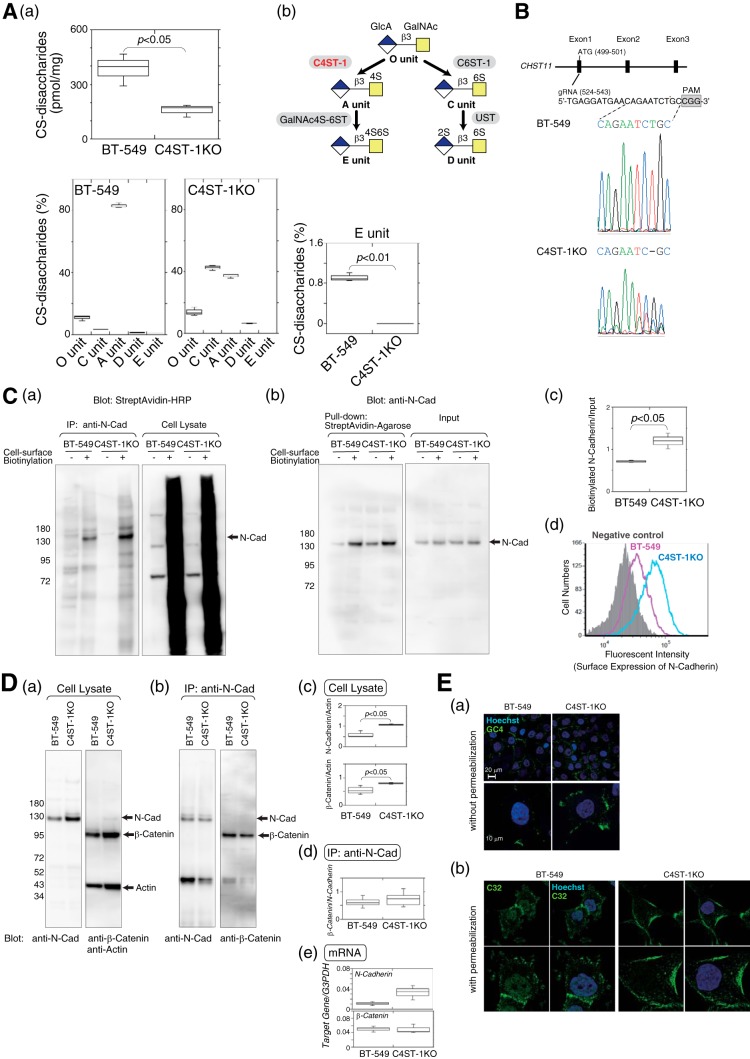
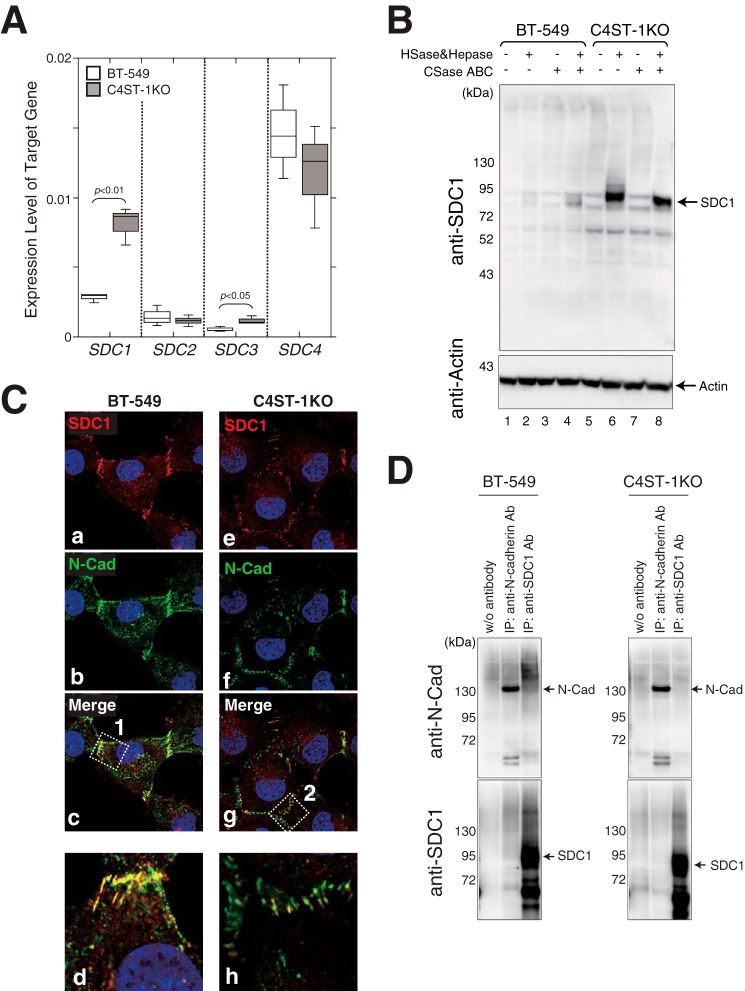
Surface expression level of N-cadherin was elevated due to inhibition of N-cadherin endocytosis and subsequent cleavage of N-cadherin, β-catenin signaling, and cell invasive activity was suppressed in C4ST-1KO cells compared with BT-549 cells. A, panel a, sulfated CS chains isolated from parental BT-549 cells and C4ST-1KO cells were analyzed using HPLC to measure the total amount and the composition of CS-disaccharides (n = 3). Panel b, outline of sulfation pathways. The C4-position of the GalNAc residue in the O-unit is sulfated by C4ST-1 to form an A-unit. Subsequently, the A-unit is converted to an E-unit by GalNAc4S-6ST. B, gRNAs targeting the CHST11 gene were designed in the 20 nucleotides upstream of the protospacer adjacent motif (PAM) sequence as shown in the drawing at top. The DNA sequences of the gRNA target site in parental BT-549 cells and C4ST-1KO cells were sequenced as shown at bottom. C, panels a and b, cell-surface proteins labeled with EZ-Link Sulfo-NHS-Biotin were immunoprecipitated with anti-N-cadherin antibody (panel a) or pulldown assay using StreptAvidin-agarose beads (panel b) and analyzed by immunoblotting with horseradish peroxidase-conjugated StreptAvidin (panel a) and anti-N-cadherin, respectively (panel b). Panel c, box-and-whisker plots of the data from panel b are shown. The relative level of biotinylated N-cadherin, standardized against the total N-cadherin (input), were quantified (n = 3). Statistical significance was assessed using a Student's t test. Panel d, surface N-cadherin expression on BT-549 cells (magenta) and C4ST-1KO cells (cyan) were compared by FACS analysis. The shaded area indicates cells treated without anti-N-cadherin antibody (negative control). D, panel a, total cell lysates from BT-549 and C4ST-1KO cells were analyzed by immunoblotting to examine the expression levels of N-cadherin and β-catenin. Panel b, amount of β-catenin associated with N-cadherin in BT-549 and C4ST-1KO cells was measured by immunoprecipitation (IP) with an anti-N-cadherin antibody followed by immunoblotting of co-precipitated β-catenin. Panel c, box-and-whisker plots of the data from panel a are shown (n = 3). Statistical significance was assessed using a Student's t test. Panel c, box-and-whisker plots of the data from panel b are shown (n = 3). Panel e, expression levels of N-cadherin and β-catenin in BT-549 and C4ST-1KO cells were analyzed using real-time PCR (n = 3). E, panel a, BT-549 and C4ST-1KO cells were stained without permeabilization using an antibody against the N-terminal domain of N-cadherin (GC4). Magnified images are shown below. Panel b, BT-549 and C4ST-1KO cells were permeabilized and then stained with an antibody against the C-terminal domain of N-cadherin (C32). Schematic representation of the binding domains of the anti-N-cadherin antibodies used in panel a and b is shown in Fig. 6A.
Results
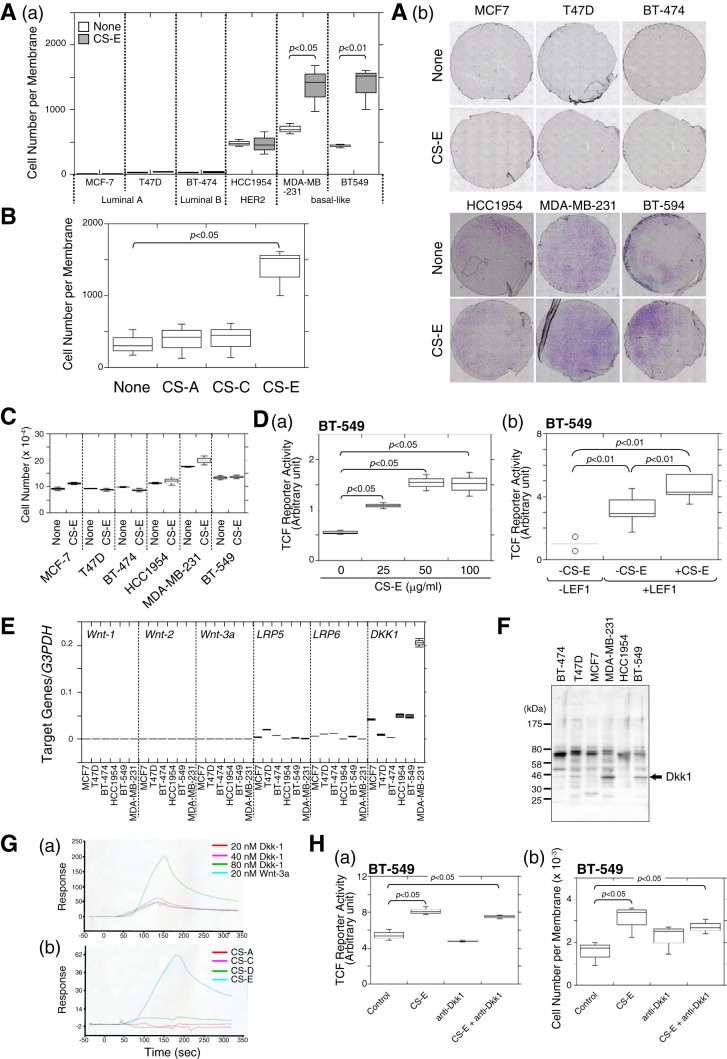
The most common sites of breast cancer metastasis are the bone, liver, and lung. In particular, the basal-like subtype of breast cancer preferentially metastasizes to the liver and brain (21). We speculated that the structural differences in CSs could affect the invasive potential of basal-like breast cancer cells and permit site-specific metastasis. Of the six breast cancer cell lines shown in Fig. 1A, panel a, HCC1954 (HER2 subtype), MDA-MB-231, and BT-549 cells (basal-like subtype) showed higher invasive potential than the other subtypes (Fig. 1A, panels a, open bars, and b). Interestingly, the invasive activity of the basal-like subtypes (MDA-MB-231 and BT-549 cells) was elevated by treatment with chondroitin sulfate E (CS-E), whereas invasion of the HER2 subtype (HCC1954 cells) was not affected by CS-E (Fig. 1A, panels a, closed bars, and panel b). CS consists of repeating disaccharide units of N-acetylgalactosamine (GalNAc) and glucuronic acid, and it has various structural modifications resulting from sulfation of different positions in the sugar residues. There are at least five different types of CSs (CS-A, CS-B, CS-C, CS-D, and CS-E), as defined by the sulfation pattern of the repeating disaccharide moieties. Major component disaccharide units of CS-A, CS-C, and CS-E are the A-unit, the C-unit, and the E-unit, respectively. Although CS-E enhanced the invasive activity of BT-549 cells, CS-A and CS-C did not (Fig. 1B). These results indicated that CS-E specifically activated the invasion potential of BT-549 cells. In addition, cell proliferation activities were not affected by CS-E (Fig. 1C).
Figure 1.
Invasion activity of human basal-like subtype breast cancer cells was enhanced by CS-E. A, panel a, invasion potential of six human breast cancer cell lines was measured using an in vitro invasion assay in the absence (open boxes) or presence (closed boxes) of CS-E. Three independent experiments were carried out, and box-and-whisker graph was plotted using data combined from these experiments. Statistical significance was assessed using a Student's t test. Panel b, invaded cells on the lower surface of the membrane in the invasion assay were stained with Giemsa stain, photographed under a light microscope, and counted. B, invasion activity of BT-549 cells treated with CS-A, CS-C, or CS-E was examined (n = 3). C, effect of CS-E on cell proliferation of six human breast cancer cell lines was investigated. Cells were treated with or without CS-E, and the cell number was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (n = 3). D, panel a, canonical Wnt signaling activity in BT-549 cells was measured using the pTCF7wt-luc reporter vector. The relative luciferase activity in the presence of different concentrations of CS-E was measured in duplicate, and the data from two independent experiments are presented with the S.D. (error bars). Panel b, pTCF7wt-luc reporter vector was co-transfected with the LEF expression vector (catalog no. SC114370, ORIGENE) into BT-549 cells, and the cells were treated with or without CS-E as indicated. The relative luciferase activity was measured (n = 6). E, mRNA expression levels of Wnt signal pathway-related genes in six breast cancer cell lines were measured using quantitative real-time PCR (n = 3). F, expression of Dkk-1 in six human breast cancer cell lines was examined by immunoblotting using anti-Dkk-1 antibody. G, panel a, concentration-dependent binding of Dkk-1 to CS-E was investigated by SPR. Wnt-3a is reported as a CS-E binding protein. Panel b, interactions between Dkk-1 and four isotyped of CSs were examined by SPR. H, panel a, Wnt signaling activities were measured after BT-549 cells were treated with either CS-E or anti-Dkk1 antibody, or both (n = 3). Panel b, invasion activity of BT-549 cells was measured after the cells were treated with either CS-E or anti-Dkk1 antibody or both (n = 3).
We first focused on the previously described link between canonical Wnt signaling activation and mammary gland tumors that was shown in the mouse (22) to investigate the mechanism underlying CS-E-dependent up-regulation of the invasion potential of BT-549 cells. In addition, frequent alterations in expression of β-catenin have been reported in breast cancer cell lines (23). We therefore examined whether CS-E could activate the canonical Wnt-signaling pathway, in which β-catenin functions as a central player. β-Catenin functions as a transcription cofactor in a complex with T cell factor/lymphoid enhancer factor (TCF/LEF) to regulate target gene expression. Canonical Wnt signaling can be measured using the reporter vector, pTcf7wtluc, which carries seven repeats of the TCF-binding consensus sequence upstream of the IFN-β basal promoter followed by the luciferase gene (24). TCF reporter activities in BT-549 cells were increased by treatment with CS-E in a dose-dependent manner (Fig. 1D, panel a). The effect of CS-E on TCF reporter activities was not canceled by overexpression of LEF1 (Fig. 1D, panel b). These results suggest that CS-E controls the expression level of β-catenin in these cells. We next examined the mRNA expression levels of genes related to the Wnt-signaling pathway in the six breast cancer cell lines using quantitative real-time PCR. The mRNAs for the Wnt ligands, Wnt-1, Wnt-2, and Wnt-3a, were very weakly expressed, and the mRNAs for the receptors for the canonical Wnt-signaling pathway, LRP5 and LRP6, were expressed at low levels in all six human breast cancer cell lines (Fig. 1E). The mRNA for the Dickkopf canonical WNT signaling pathway inhibitor 1 (Dkk1), which antagonizes canonical Wnt signaling by inhibiting LRP5/6 interaction with Wnt ligands, was expressed at various levels in the six cell lines (Fig. 1E), and Dkk1 proteins were highly expressed in BT-549 and MDA-MB-231 cells (Fig. 1F). We examined the possibility that Dkk1 might be a target of CS-E in BT-549 cells, because surface plasmon resonance (SPR) analysis demonstrated that Dkk1 specifically binds to CS-E (Fig. 1G). Neutralization of Dkk1 by anti-Dkk1 antibodies did not affect TCF reporter activities in BT-549 cells (Fig. 1H, panel a). In addition, CS-E could activate TCF reporter activities even when Dkk1 was blocked by its antibodies (Fig. 1H, panel a). Furthermore, anti-Dkk1 antibodies had no effects on the invasion activities of BT-549 cells, and did not abrogate the increased invasion induced by CS-E (Fig. 1H, panel b). These results suggested that CS-E up-regulated β-catenin-dependent transcriptional activities in BT-549 cells, but that the effect of CS-E was not mediated via the canonical Wnt-signaling pathway.
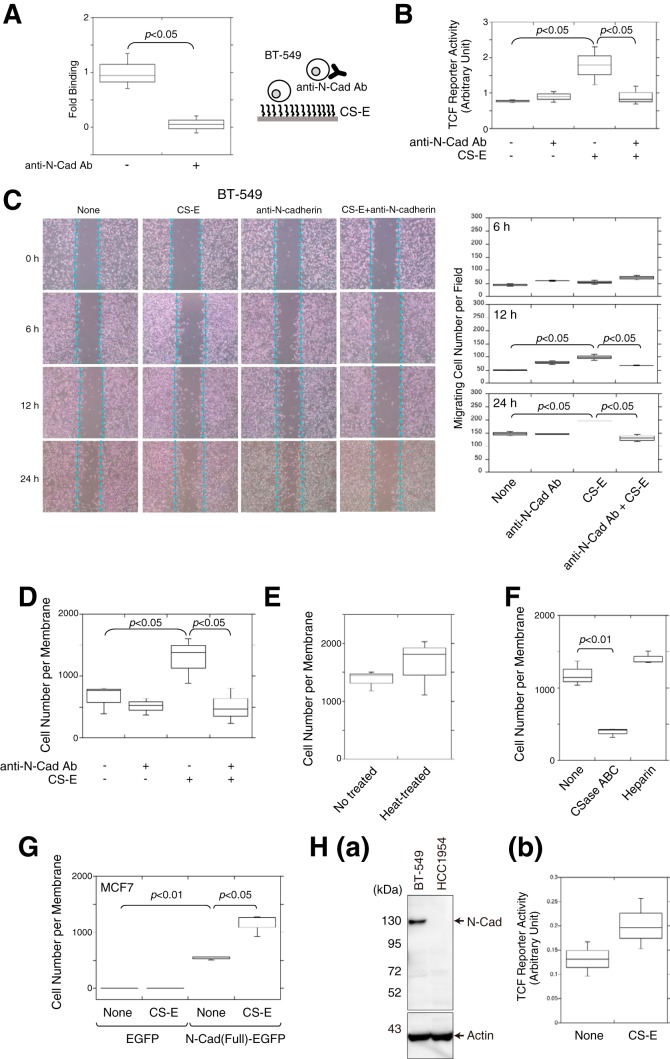
β-Catenin is not only a key transcription factor in canonical Wnt signaling but also a structural adaptor protein linking cadherins to the actin cytoskeleton in cell–cell adhesion (25). The cadherin-bound β-catenin can be released and made available for signaling. We therefore examined whether CS-E elevates the invasion potential of BT-549 cells by acting on cadherins, thereby increasing β-catenin-mediated signaling. It was previously reported that BT-549 cells highly express N-cadherin (5) and that CS-E binds to N-cadherin with high affinity (26). Consistent with these reports, adhesion of BT-549 cells to immobilized CS-E was significantly inhibited by anti-N-cadherin antibodies (Fig. 2A). In addition, blocking of the binding of CS-E to N-cadherin by anti-N-cadherin antibodies suppressed β-catenin-mediated transcriptional activities (Fig. 2B). These results suggested that CS-E mainly utilizes N-cadherin as a receptor and controls β-catenin signaling via N-cadherin in BT-594 cells. Next, we investigated whether up-regulation of β-catenin signaling by CS-E via N-cadherin could enhance cell migration and invasion. A scratch assay indicated that anti-N-cadherin antibodies completely abrogated the increased migration that was induced by CS-E at 24 h (Fig. 2C). Cells treated solely with anti-N-cadherin antibodies migrated slightly faster at 12 h compared with untreated cells, probably because cell–cell adhesion was reduced by dissociation of homophilic interactions between N-cadherin molecules. In addition, the invasion of BT-549 cells into Matrigel that was enhanced by CS-E was completely blocked by treatment with anti-N-cadherin antibodies (Fig. 2D). These combined data suggested that the binding of CS-E to N-cadherin up-regulates the migration and invasion of BT-549 cells. Moreover, as shown in Fig. 2E, the activities of CS-E were unaffected by heat treatment of CS-E at 94 °C for 20 min, excluding the possibility that up-regulation of the invasion was caused by protein components contaminated in CS-E preparations. Furthermore, heparin did not enhance the invasive activity of BT-549 cells, although highly sulfated CS like CS-E likely behaves as a heparin mimetic (Fig. 2F). In addition, removal of endogenous CSs from BT-549 cells by pre-treatment with chondroitinase ABC remarkably diminished the invasive potential of BT-549 cells (Fig. 2F).
Figure 2.
N-cadherin could function as a receptor for CS-E and enhanced the invasive activity of BT-549 cells. A, cells pre-treated with (+) or without (−) anti-N-cadherin antibody were added to the CS-E-coated wells, and cells bound to the CS-E on the plate were estimated using cellular lactate dehydrogenase activity as an indicator (n = 3). Statistical significance was assessed using a Student's t test. B, TCF reporter activity in the absence (−) or presence (+) of anti-N-cadherin antibody and/or CS-E was measured (n = 3). C, BT-549 cells cultured in a monolayer were scratched and subsequently cultured for the indicated periods in the presence of CS-E, anti-N-cadherin antibody, or both. Dashed lines show the edge of the gap newly created by scratching. D, invasive activity of BT-549 cells was measured in the absence or presence of CS-E and/or anti-N-cadherin antibody (Ab) (n = 3). E, BT-549 cells were stimulated with CS-E untreated (No treated) or pre-treated at 94 °C for 20 min (Heat-treated), and their invasive activities were measured (n = 3). F, invasive activities of BT-549 cells digested with chondroitinase ABC or pre-treated with heparin (50 μg/ml) were measured (n = 3). G, either EGFP- or N-Cad(Full)-EGFP-expressing MCF7 cells were treated with or without CS-E, and their invasive activities were measured (n = 3). H, panel a, cell lysates of BT-549 and HC1954 cells were analyzed by immunoblotting using an anti-N-cadherin antibody. Panel b, β-catenin transcriptional activities of HCC1954 cells in the presence or absence of CS-E were measured using TCF reporter vector (n = 3).
We further confirmed that N-cadherin could function as a receptor of CS-E to elevate the invasive activities. A previous report showed that non-invasive MCF7 cells acquired an invasive phenotype by overexpression of N-cadherin (6). As shown in Fig. 2G, N-cadherin-expressing MCF7 cells showed an invasive property, and their invasive activities were enhanced by CS-E. In contrast, although HCC1954 cells showed an invasive phenotype, their invasive activities were not enhanced by CS-E (see Fig. 1A). Remarkably, HCC1954 cells expressed no N-cadherin (Fig. 2H, panel a), and β-catenin transcriptional activities were not affected in the presence of CS-E (Fig. 2H, panel b). These results suggest that the cell-surface expression levels of N-cadherin control the CS-E-induced invasive properties of breast cancer cells.
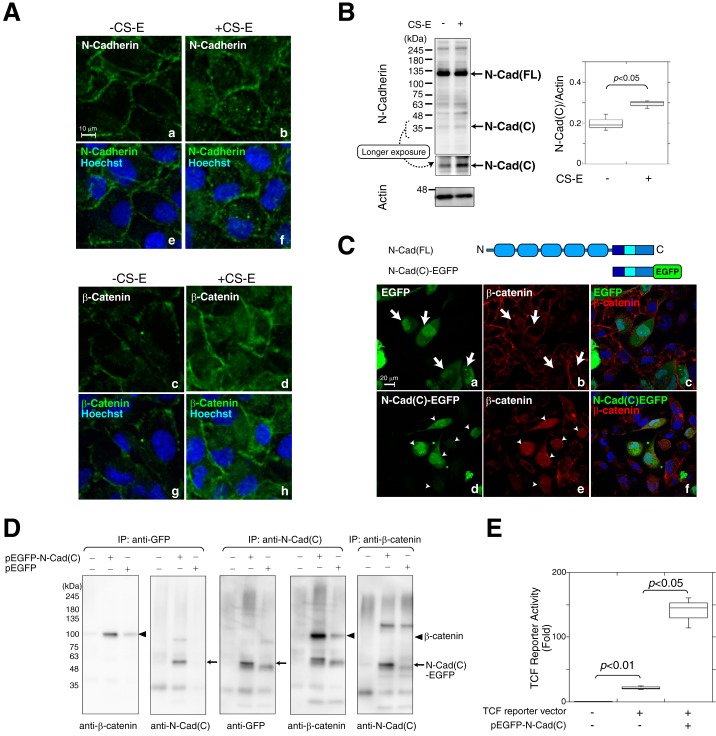
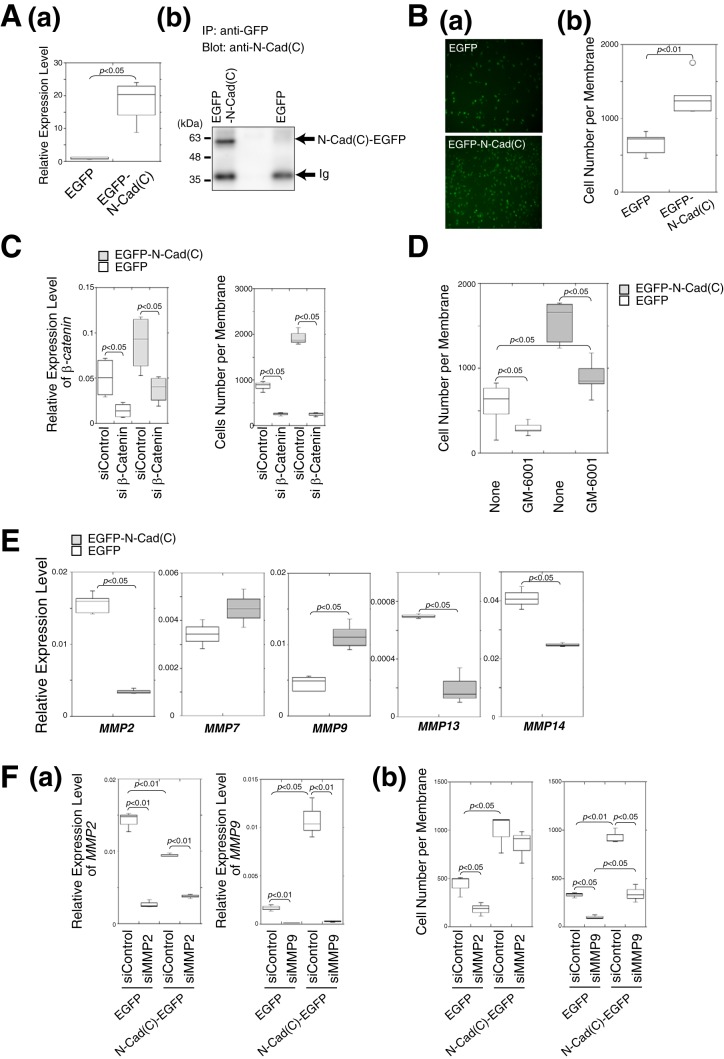
Next, we examined the cellular localization of N-cadherin and β-catenin in BT-549 cells in the presence or absence of CS-E. In the absence of CS-E, N-cadherin was mainly expressed at the plasma membrane, but small amounts of intracellular vesicular staining could also be seen scattered throughout the periphery of the cells (Fig. 3A, panels a and e). Treatment of the cells with CS-E increased the intracellular distribution of N-cadherin (Fig. 3A, panels b and f), and vesicular N-cadherin staining was observed in a perinuclear position. In parallel with the altered localization of N-cadherin from the plasma membrane to the cytoplasm, the intracellularly localized pool of β-catenin also increased with CS-E treatment (Fig. 3A, panels c–h). Previous studies have indicated that matrix metalloproteinase- and γ-secretase-mediated N-cadherin cleavage leads to the generation of intracellular C-terminal fragments and modulates signal transduction through influencing the cytoplasmic β-catenin pool (7). We therefore investigated whether N-cadherin was cleaved in the presence of CS-E using immunoblotting. In the absence of CS-E, N-cadherin was detected mostly as the full-length form, whereas a proteolytic fragment appeared in the presence of CS-E, albeit at a low level (Fig. 3B). These results suggested that N-cadherin is cleaved in a CS-E-dependent manner. The intracellular C-terminal fragments of N-cadherin are considered to be involved in signal transduction (7, 27). Therefore, to investigate the function of the C-terminal cytoplasmic domain released by proteolytic cleavage of N-cadherin, an EGFP-fused cytoplasmic domain of N-cadherin (N-Cad(C)-EGFP) was transiently expressed in BT-549 cells (Fig. 3C). When EGFP alone was expressed, β-catenin was distributed not in the nucleus but in the plasma membrane and cytoplasm (Fig. 3C, panels a–c, the cells indicated by white arrows in panel a and b). In contrast, the forced expression of N-Cad(C)-EGFP resulted in the translocation of β-catenin from the plasma membrane into the nucleus (Fig. 3C, panels d–f, the cells indicated by white arrowheads in d and e). Immunoprecipitation followed by immunoblotting analysis revealed a direct interaction between N-Cad(C)-EGFP and β-catenin (Fig. 3D). Immunoprecipitates of N-Cad(C)-EGFP obtained using either anti-EGFP antibody or anti-N-Cad(C) antibody contained β-catenin, whereas N-Cad(C)-EGFP was co-immunoprecipitated by an anti-β-catenin antibody (Fig. 3D). The combined data suggested that N-Cad(C) remains complexed with β-catenin and translocates into the nucleus. We next examined whether the expression of N-Cad(C) could enhance β-catenin-dependent transcriptional activities. As shown in Fig. 3E, the expression of N-Cad(C)-EGFP significantly increased TCF reporter activities. These results suggested the possibility that the N-Cad(C)–β-catenin complex is involved in the regulation of β-catenin-dependent transcriptional activities. To identify the target gene(s) induced by the N-Cad(C)–β-catenin complex, we established BT-549 cells that stably expressed N-Cad(C)-EGFP, and we checked their expression levels of N-Cad(C)-EGFP at both the mRNA and protein levels (Fig. 4A). Cell invasion activities were significantly increased in N-Cad(C)-EGFP-expressing cells compared with EGFP-expressing cells (Fig. 4B), suggesting that β-catenin-dependent transcriptional induction was accelerated by the expression of N-Cad(C)-EGFP. Consistent with this result, knockdown of β-catenin using siRNA decreased the invasion activities of N-Cad(C)-EGFP-expressing cells to the same level as the control cells expressing EGFP (Fig. 4C). In addition, the high invasion activities of N-Cad(C)-EGFP-expressing cells were decreased by treatment with the matrix metalloprotease (MMPs) inhibitor, GM-6001 (Fig. 4D). We therefore measured the gene expression levels of some MMPs that have been reported as targets of β-catenin. Of the five MMPs tested, only MMP9 mRNA was significantly up-regulated in N-Cad(C)-EGFP-expressing BT-549 cells (Fig. 4E). To confirm that N-Cad(C)-EGFP enhancement of the invasion of BT-549 cells is mediated by transcriptional induction of MMP9, the effect of knockdown of MMP9 on the invasion potential of N-Cad(C)-EGFP-expressing BT-549 cells was investigated. siRNA suppression of the expression of MMP9 significantly decreased the invasion activities of both N-Cad(C)-EGFP- and EGFP-expressing BT-549 cells (Fig. 4F, panel b). In contrast, knockdown of MMP2 decreased the invasion of EGFP-expressing BT-549 cells but did not affect the invasive activities of N-Cad(C)-EGFP-expressing BT-549 cells (Fig. 4F, panel b). These results suggested that the C-terminal fragments of N-cadherin led to the transcriptional induction of MMP9 in cooperation with β-catenin and thereby to enhancement of cell invasion. However, it is considered that β-catenin-targeted genes other than MMP9 are also involved in the invasion activities accelerated by the expression of N-Cad(C)-EGFP, because the inhibitor of MMPs, GM-6001, and the knockdown of MMP9 could not completely decrease the invasion activities of N-Cad(C)-EGFP-expressing cells to the same extent as the control cells expressing EGFP (Fig. 4, D and F).
Figure 3.
C-terminal fragments of N-cadherin generated by cleavage of N-cadherin in response to CS-E formed a complex with β-catenin, translocated to the nucleus, and activated N-cadherin/β-catenin signaling. A, BT-549 cells were incubated in the presence (+CS-E) or absence (−CS-E) of CS-E and stained with anti-N-cadherin antibody (green; panels a, b, e, and f) or anti-β-catenin antibody (green; panels c, d, g, and h). Hoechst was used for nuclear counter-staining (blue). B, total cell lysates from BT-549 cells incubated in the presence (+CS-E) or absence (−CS-E) of CS-E were subjected to immunoblotting using anti-N-cadherin antibody. N-Cad(FL) and N-Cad(C) indicate the full-length and the C-terminal fragment of N-cadherin, respectively. Densitometry analysis was performed to determine the relative level of N-Cad(C) proteins normalized against actin used as a loading control (n = 3). Statistical significance was assessed using a Student's t test. C, top, schematic representation of the full-length N-cadherin and the C-terminal fragment of N-cadherin fused to EGFP (N-Cad(C)-EGFP). After EGFP or N-Cad(C)-EGFP was expressed in BT-549 cells, immunofluorescence of EGFP (green) and β-catenin (red) was analyzed. White arrows (panels a and b) show EGFP-expressing cells, and white arrowheads (panels d and e) indicate the cells expressing N-Cad(C)-EGFP. Merged images are shown in panels c and f in which nuclei were counterstained with Hoechst (blue). D, BT-549 cells were transfected with (+) or without (−) the pEGFP-N-Cad(C) vector or the pEGFP empty vector. N-Cad(C)-EGFP and β-catenin were immunoprecipitated (IP) using anti-GFP antibody, anti-N-Cad(C) antibody, or anti-β-catenin antibody and were then analyzed by immunoblotting using anti-GFP antibody, anti-N-Cad(C) antibody, or anti-β-catenin antibody as indicated. E, BT-549 cells were transfected without (−) or with (+) the TCF luciferase reporter vector and/or the pEGFP-N-Cad(C) vector (n = 3). Statistical significance was assessed using a Student's t test.
Figure 4.
Expression of EGFP-N-Cad(C) increased cellular invasive potential through transcriptional induction of MMP9. A, panel a, expression level of the mRNA encoding the EGFP-labeled C-terminal domain of N-cadherin in BT-549 cells stably expressing either EGFP-N-Cad(C) or EGFP was examined using real-time PCR (n = 3). Data were combined from two independently performed experiments. Statistical significance was assessed using a Student's t test. Panel b, EGFP-N-Cad(C) protein was immunoprecipitated from the cells in panel a using anti-GFP antibody and was then analyzed by immunoblotting using an anti-N-cad(C) antibody. The band denoted as Ig represents immunoglobulin from the anti-GFP antibody used for immunoprecipitation (IP). B, panel a, BT-549 cells stably expressing either EGFP-N-Cad(C) or EGFP were subjected to a Matrigel invasion assay (n = 5). GFP fluorescent images of the invading cells are shown. Panel b, box-and-whisker plots of the data from panel a are shown (n = 5). C, EGFP-N-Cad(C)-expressing (shaded boxes) or EGFP-expressing BT-549 cells (open boxes) were transfected with either β-catenin siRNA (siβ-Catenin) or control siRNA (siControl), and then the gene expression level of β-catenin (n = 3, left) and the invasive activities (n = 3, right) of the two cell lines were examined. Two independent experiments were carried out. D, invasive activities of EGFP-N-Cad(C)-expressing (shaded boxes) or EGFP-expressing BT-549 cells (open boxes) were measured in the presence or absence of GM-6001, an inhibitor of MMPs (n = 6). E, expression levels of five MMP genes in EGFP-N-Cad(C)-expressing (open boxes) or EGFP-expressing BT-549 cells (shaded boxes) were analyzed using real-time PCR (MMP2 and MMP9, n = 6; MMP7, MMP14, and MMP13, n = 3). F, EGFP-N-Cad(C)- or EGFP-expressing BT-549 cells, were transfected with either MMP2 siRNA (siMMP2), MMP9 siRNA (siMMP9), or control siRNA (siControl), and then the gene expression level of MMP2 and MMP9 (n = 3) (panel a) and invasive activities (n = 3) (panel b) of the two cell lines were examined.
Next, we aimed to visualize the movement of transfected full-length N-cadherin (N-Cad(FL)-EGFP) within cells using GFP fluorescence before and after the addition of CS-E. First, we expressed N-Cad(FL)-EGFP in BT-549 cells; however, overexpression of N-Cad(FL)-EGFP caused CS-E-independent intracellular relocation of N-Cad(FL)-EGFP. This phenomenon was probably due to the fact that endogenous N-cadherin is expressed at high levels in BT-549 cells (5). When N-Cad(FL)-EGFP was expressed in HeLa cells, which express N-cadherin at lower levels compared with BT-549 cells, we could observe CS-E-dependent intracellular relocation of N-Cad(FL)-EGFP. We therefore used HeLa cells to visualize N-Cad(FL)-EGFP endocytosis induced by CS-E. When N-Cad(FL)-EGFP was expressed in confluent cells that form stable cell–cell contacts, it was mainly localized on the plasma membrane (Fig. 5A, panels a and b). In contrast, treatment of the cells with CS-E enhanced the internalization of N-Cad(FL)-EGFP, and consequently N-Cad(FL)-EGFP disappeared from the cell surface (Fig. 5A, panels c and d). These effects of CS-E on the endocytosis of N-cadherin were specific, and other differently structured chondroitin sulfates, CS-A and CS-C, could barely induce internalization of N-Cad(FL)-EGFP (Fig. 5B). Because it was previously shown that N-cadherin specifically binds to CS-E with high affinity (26), it was considered that the interaction between N-cadherin and CS-E stimulates internalization of N-cadherin. Immunofluorescence analysis indicated that intracellular N-Cad(FL)-EGFP was partially co-localized with GM130, a marker of cis-Golgi (Fig. 5C, panels u and u′), but showed little merging with LAMP-1 or EEA-1, markers of lysosomes and early endosomes, respectively (Fig. 5C, panels o, o′, r, and r′). In addition, the intracellular localization of N-Cad(FL)-EGFP accorded well with that of caveolin-1 (Fig. 5C, panels x and x′). These results suggested that CS-E stimulates the internalization of N-cadherin, which is mediated via caveolin-positive vesicles.
Figure 5.
N-cadherin was translocated from the plasma membrane to the intracellular compartment in response to CS-E. A, HeLa cells transiently expressing N-Cad(FL)-EGFP were incubated in the presence (panels c and d) or absence (panels a and b) of CS-E and were then fixed and permeabilized for immunofluorescence analysis of EGFP (green). Nuclei were counterstained with Hoechst (blue). B, HeLa cells transiently expressing N-Cad(FL)-EGFP were treated overnight with CS-A (panels b and b′), CS-C (panels c and c′), or CS-E (panels d and d′). The areas within the dotted white squares (panels b–d) are magnified and shown in panels b′–d′. Arrowheads indicate N-Cad(FL)-EGFP translocated from the cell surface into the intracellular compartment. C, HeLa cells transfected with the expression vector of N-Cad(FL)-EGFP were incubated in the presence (+CS-E) or absence (−CS-E) of 25 μg/ml CS-E overnight. Cells were stained with anti-LAMP-1 antibody (panels b and m), anti-EEA-1 antibody (panels e and q), anti-GM130 antibody (panels h and t), or anti-caveolin-1 antibody (panels k and w) and Alexa594-labeled second antibody (red). GFP florescence is shown in the left columns, and merged images in which nuclei were counterstained with Hoechst (blue) are shown at right. Magnified images of the areas within the white boxes in panels o, r, u, and x are shown in panels o′, r′, u′, and x′, respectively.
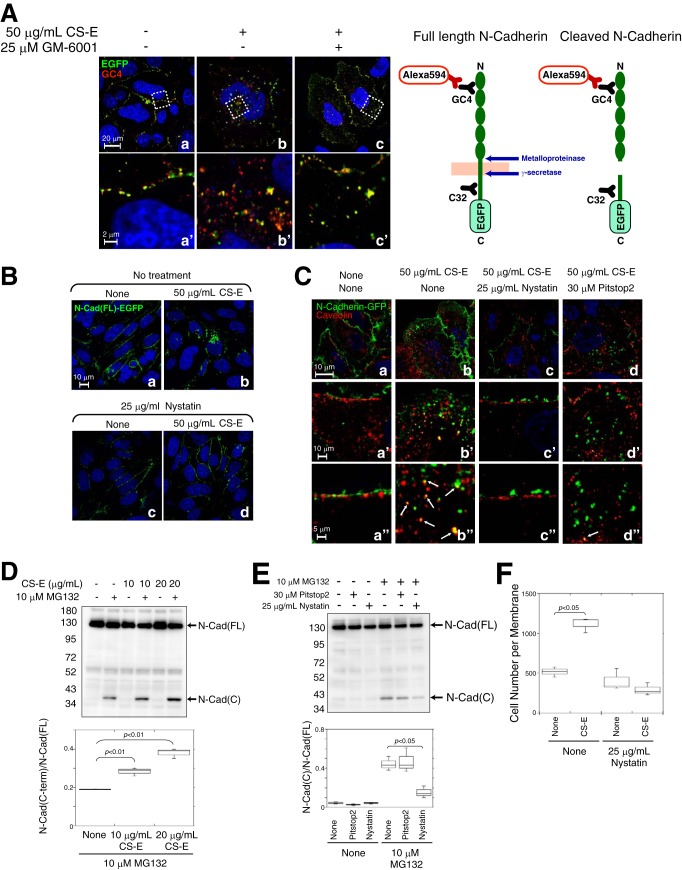
Based on the data shown in Figs. 3 and 4, it was considered that proteolytic release of the cytoplasmic domain of N-cadherin is important for β-catenin-dependent transcriptional induction. We therefore investigated where in the cell the proteolysis of N-cadherin that is triggered by CS-E occurs. It is generally accepted that N-cadherin is cleaved on the cell surface by multiple metalloproteases containing a disintegrin domain (ADAMs), MMPs, or other transmembrane proteases (7, 28–30). We analyzed the effect of the broad-range MMP inhibitor GM-6001, which is a potent, reversible inhibitor of zinc-containing proteases, including MMPs and ADAMs, on N-Cad(FL)-EGFP cleavage induced by CS-E. For this purpose, antibodies against the N-terminal domain of N-cadherin (clone GC-4) that were detected with Alexa594-labeled second antibodies in immunofluorescence were used in conjunction with EGFP fluorescence for detection of the cleavage of N-cadherin (Fig. 6A, right panel). Thus, if N-Cad(FL)-EGFP is cleaved and its C terminus is released into the cytosol, then the signals of the EGFP that are fused to the C terminus of N-cadherin will not co-localize or merge with the signals of GC-4 staining. In the absence of CS-E, N-Cad(FL)-EGFP was localized in the plasma membrane, where the signals derived from EGFP (green signal) and GC-4 (red signal) merged (Fig. 6A, panels a and a′). Following treatment with CS-E, some internalized N-Cad(FL)-EGFP molecules were labeled with both EGFP and GC-4, although others were detected as either a red or a green signal only (Fig. 6A, panels b and b′). When cells were treated with CS-E in the presence of GM-6001, internalized N-Cad(FL)-EGFP molecules were detected as a full-length N-cadherin protein that was labeled with both EGFP and GC-4 (Fig. 6A, panels c and c′). These results raise the possibility that N-cadherin is cleaved by zinc-containing proteases, including MMPs and ADAMs, after internalization into the cells, probably in their journey from the plasma membrane to a caveolin-positive intracellular compartment. However, further studies are needed to clarify the underlying mechanism.
Figure 6.
Caveolae-mediated endocytosis and subsequent cleavage of N-cadherin was enhanced by CS-E. A, left panel, HeLa cells transiently expressing N-Cad(FL)-EGFP were treated overnight with CS-E and/or GM6001 and stained with anti-N-cadherin antibody (GC4) and Alexa594-labeled second antibody. Nuclei were counterstained with Hoechst (blue). Magnified images of the areas within the white dotted boxes in panels a–c are shown in panels a′–c′, respectively. Right panel, schematic representation of the binding domains of anti-N-cadherin antibody (GC4 and C32) and the N-cadherin cleavage sites is shown. B, HeLa cells transiently expressing N-Cad(FL)-EGFP were treated with (panels c and d) or without (panels a and b) nystatin for 5 h, and then CS-E was added (panels b and d) or not (panels a and c) to the cells and incubated overnight. Nuclei were counterstained with Hoechst (blue). C, HeLa cells transiently expressing N-Cad(FL)-EGFP were incubated at 4 °C for 30 min and were then treated without (panel a) or with CS-E (panel b) or with CS-E together with nystatin (panel c) or Pitstop2 (panel d) for 1 h. Cells were then stained with anti-caveolin-1 antibody and Alexa594-labeled second antibody (red). Nuclei were counterstained with Hoechst (blue). Magnified images of panels a–d are shown in panels a′–d′ and a"–d". Arrows indicate co-localization of N-Cad(FL)-EGFP and caveolin-1. D, BT-549 cells were treated with or without CS-E and/or MG132 for 2 h and were then subjected to immunoblotting analysis using an anti-N-cadherin antibody (n = 3). E, BT-549 cells were treated with or without Pitstop2 or nystatin for 2 h in the presence or absence of MG132 and were then subjected to immunoblotting analysis using an anti-N-cadherin antibody (n = 3). D and E, the band intensity of N-Cad(C) was normalized to that of N-Cad(FL). F, invasion potential of BT-549 cells treated with or without nystatin in the presence or absence of CS-E was measured using an in vitro invasion assay (n = 3).
We next examined whether endocytosis is required for the proteolysis of N-cadherin induced by CS-E. Because N-Cad(FL)-EGFP molecules incorporated within the cells were mostly co-localized with caveolin-1 (Fig. 5D, panels x and x′), we inhibited endocytosis of N-cadherin using nystatin, an inhibitor of the caveolae-mediated endocytosis pathway (31, 32). Pre-treatment of cells with nystatin inhibited the internalization of N-Cad(FL)-EGFP that was induced by CS-E, and consequently N-Cad(FL)-EGFP was retained on the cell surface even in the presence of CS-E (Fig. 6B, panel d). In addition, N-Cad(FL)-EGFP that was internalized following CS-E treatment was co-localized with caveolin, a component of the caveolae membranes (Fig. 6C, panels b, b′, and b"). Interestingly, although nystatin blocked the transport of N-Cad(FL)-EGFP into the cells (Fig. 6C, panels c, c′, and c"), Pitstop2, a potent inhibitor of clathrin-mediated endocytosis (33), had little effect on the endocytosis of N-Cad(FL)-EGFP (Fig. 6C, panels d, d′, and d"). We next tried to detect the C-terminal cleavage fragments of N-cadherin in the presence of CS-E, but we failed to do so. Therefore, to detect these C-terminal cleavage fragments, we incubated the cells in the presence or absence of MG132 (benzyloxycarbonyl-Leu-Leu-Leu-al), an inhibitor of protein degradation by the proteasome, because it has been shown that some nuclear transcription factors can be detected by blocking their rapid proteasome-mediated degradation. Pre-treatment of BT-549 cells with MG132 allowed the C-terminal fragments of N-cadherin (N-Cad(C)) to be visualized by Western blotting (Fig. 6D), suggesting that cleavage of N-cadherin occurred in a cell-autonomous manner in BT-549 cells without the addition of exogenous CS-E (probably by endogenous CS-E). This phenomenon is considered to contribute to the high invasive activities of BT-549 cells. In addition, exogenously added CS-E enhanced the cleavage of N-cadherin (Fig. 6D). We next investigated whether the cleavage of N-cadherin could be affected by blocking the endocytosis of N-cadherin. As shown in Fig. 6E, nystatin inhibited the production of N-Cad(C), whereas Pitstop2 could not (Fig. 6E), suggesting that caveolae-mediated endocytosis is required for proteolysis of N-cadherin. Furthermore, the invasive activities of BT-549 cells were not increased even in the presence of CS-E when the caveolae-mediated endocytosis pathway was suppressed by nystatin (Fig. 6F).
As shown in Fig. 2F, digestion of endogenous CSs of BT-549 cells with chondroitinase ABC significantly reduced the invasive activities of BT-549 cells. In addition, a small but significant proportion of N-cadherin was constitutively cleaved in BT-549 cells (Fig. 6D). It is considered that endocytosis and subsequent proteolysis of N-cadherin occur in a cell-autonomous manner, because BT-549 cells endogenously express E-units, albeit at low levels (Fig. 7A, panel a, and Table 1). We therefore hypothesized that BT-549 cells that were deficient for E-units would display a non-invasive phenotype. As shown in Fig. 7A, panel b, the E-unit is biosynthesized by the successive action of C4ST-1 and GalNAc4S-6ST using the O-unit as a precursor. Therefore, to generate BT-549 cells that were deficient for E-units, we established C4ST-1–knock-out BT-549 cells (C4ST-1KO) using the CRISPR/Cas9 system. The sequences of the guide RNA (gRNA) target sites were analyzed, and the sites cleaved by the Cas9 nuclease were confirmed (Fig. 7B). As shown in Fig. 7A, panel a, E-units were completely depleted, A-units were decreased, and the C- and D-units were increased by the loss of C4ST-1. C4ST-1 is dominantly involved in the synthesis of E-units, as reported previously (34). Thus, C4ST-1KO cells are defective for the biosynthesis of E-units. Residual A-units in C4ST-1KO cells are synthesized by other sulfotransferases involved in the 4-O-sulfation of GalNAc residues in CS/dermatan sulfate, C4ST-2, and D4ST-1. In addition, the amounts of CS-disaccharides were decreased in C4ST-1KO cells (Fig. 7A, panel a), consistent with previous reports that C4ST-1 regulates CS content (34, 35).
Table 1.
Disaccharide composition of CS chains from parental BT-549 cells and C4ST-1 KO cells
Values are expressed as picomoles of disaccharide per mg of dried homogenates of these cells and the means ± S.D. of three determinations
| Parental BT-549 cells | C4ST-1 KO cells | |
|---|---|---|
| ΔDi-0Sa | 41.48 ± 6.77 (10.9) | 19.14 ± 2.37 (13.9) |
| ΔDi-6S | 13.47 ± 2.66 (3.5) | 18.86 ± 1.16 (42.5) |
| ΔDi-4S | 321.86 ± 78.28 (83.1) | 126.49 ± 4.95 (37.3) |
| ΔDi-diSD | 5.81 ± 1.42 (1.50) | 1.24 ± 0.16 (6.4) |
| ΔDi-diSE | 3.59 ± 1.11 (0.92) | ND |
| ΔDi-triS | NDb | ND |
| Total | 386.22 ± 87.66 | 158.72 ± 34.73 |
a The abbreviations used are as follows: ΔDi-0S, ΔHexAα1-3GalNAc; ΔDi-6S, ΔHexAα1-3GalNAc(6-O-sulfate); ΔDi-4S, ΔHexAα1-3GalNAc(4-O-sulfate); ΔDi-diSD, ΔHexA(2-O-sulfate)α1-3GalNAc(6-O-sulfate); ΔDi-diSE, ΔHexAα1-3GalNAc(4-,6-O-sulfate); ΔDi-triS: ΔHexA(2-O-sulfate)α1-3GalNAc(4-,6-O-sulfate).
b ND means not detected.
As shown in Fig. 6, the cell surface-expressed N-cadherin is transported into the cells in response to the binding of CS-E and is cleaved to release its cytoplasmic domain, which forms a complex with β-catenin. We therefore hypothesized that N-cadherin might be retained on the cell surface in the C4ST-1KO cells that were defective for CS-E expression. As expected, a cell-surface biotinylation assay demonstrated that the cell-surface expression levels of N-cadherin were increased in C4ST-1KO cells compared with the parental BT-549 cells (Fig. 7C, panels a–c). Consistent with this result, flow cytometric analysis also showed an increased expression level of N-cadherin on the cell surface in C4ST-1KO cells (Fig. 7C, panel d). In addition, we checked the total expression level of N-cadherin and β-catenin by Western blotting. Both N-cadherin and β-catenin were more highly expressed in C4ST-1KO cells than in BT-549 cells (Fig. 7D, panels a and c), probably because N-cadherin is degraded together with β-catenin in BT-549 cells. The binding of β-catenin to N-cadherin was not altered in the absence of CS-E (Fig. 7D, panel b and d). Moreover, the gene expression levels of N-cadherin and β-catenin were not significantly different between BT-549 cells and C4ST-1KO cells (Fig. 7D, panel e). We further compared the cellular localization of N-cadherin between BT-549 cells and C4ST-1KO cells using immunofluorescence. In non-permeabilized cells, the cell-surface distribution of N-cadherin was visualized using GC4, a monoclonal anti-N-cadherin antibody that reacts with the N-terminal half of the extracellular domain of N-cadherin. In this experiment, more N-cadherin molecules were distributed on the cell surface in C4ST-1KO cells, compared with BT-549 cells (Fig. 7E, panel a). The plasma membrane and intracellular localization of N-cadherin was examined in permeabilized cells using C32, a monoclonal anti-N-cadherin antibody that reacts with the C-terminal half of the intracellular domain of N-cadherin (see Fig. 6). In BT-549 cells, N-cadherin was localized in the cytoplasm and nucleus as well as in the plasma membrane, and in C4ST-1KO cells a large fraction of N-cadherin resided on the cell surface (Fig. 7E, panel b).
We next examined which CS-PGs interact with N-cadherin. Syndecan (SDC) family is well-known as cell-surface PGs modified with HS and CS. Thus, we checked the gene expression levels of four members of the SDC family. As shown in Fig. 8A, the gene expression levels of SDC1 and SDC3 were significantly higher in C4ST-1KO cells than parental BT-549 cells. Consistent with this result, the expression of SDC1 core proteins increased in C4ST-1KO cells more than BT-549 cells (Fig. 8B). Interestingly, SDC1 core proteins were hardly modified with CS chains in C4ST-1KO cells (Fig. 8B, lanes 6–8). We then investigated the cellular localization of N-cadherin and SDC1 in BT-549 and C4ST-1KO cells (Fig. 8C). N-cadherin was mostly co-localized with SDC1 at the plasma membrane of BT-549 cells (Fig. 8C, panels c and d). In contrast, the localization of N-cadherin and SDC1 did not merge well on the plasma membrane of C4ST-1KO cells (Fig. 8C, panels g and h). Furthermore, the interaction between SDC1 and N-cadherin was investigated. As shown in Fig. 8D, N-cadherin was co-immunoprecipitated with SDC1 in BT-549 cells, albeit at low efficiency, whereas no bands of N-cadherin interacting with SDC1 could be detected in C4ST-1KO cells. These results suggest the possibility that CS chains on the SDC1 could interact with N-cadherin.
Figure 8.
Modification of SDC1 core proteins with glycosaminoglycan chains and the interaction between N-cadherin and SDC1. A, gene expression levels of SDC1, SDC2, SDC3, and SDC4 in BT-549 (open boxes) and C4ST-1KO cells (shaded boxes) were analyzed by real-time PCR (n = 3). Statistical significance was assessed using a Student's t test. B, after the cell lysates of BT-549 and C4ST-1KO cells were digested with heparitinase and heparinase (HSase&Hepase) and/or chondroitinase ABC (CSase ABC) as indicated, the digests were subjected to immunoblotting using anti-SDC1 (Prestige Antibodies®, catalog no. HPA006185, Sigma) or anti-actin antibodies. C, BT-549 and C4ST-1KO cells were immunostained with anti-SDC1 and anti-N-cadherin antibodies. Magnified images of the areas within white boxes 1 (panel c, BT-549) and 2 (panel g, C4ST-1KO) are shown in panels d and h, respectively. D, cell lysates digested with heparitinase, heparinase, and chondroitinase ABC were immunoprecipitated in the presence or absence (w/o antibody) of anti-N-cadherin (IP, anti-N-cadherin antibody (Ab)) or anti-SDC1 antibodies (IP, anti-SDC1 antibody). Immunoprecipitates were analyzed by immunoblotting using the indicated antibodies.
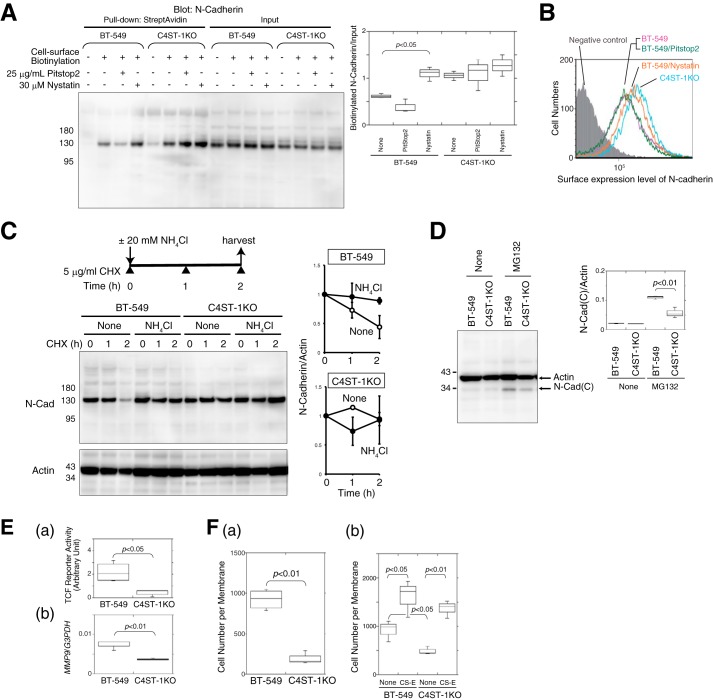
To confirm that a difference in endocytosis gave rise to this difference in cellular localization of N-cadherin, the surface expression levels of N-cadherin were analyzed using both cell-surface biotinylation and FACS analysis in the presence of endocytosis inhibitors. As expected, the surface expression of N-cadherin in parental BT-549 cells was increased to the level of that in C4ST-1KO cells by blocking endocytosis of N-cadherin using nystatin (Fig. 9, A and B). Furthermore, a cycloheximide chase analysis demonstrated that the half-life of the full-length N-cadherin expressed in BT-549 cells was ∼2 h (Fig. 9C, open circle). In contrast, the level of N-cadherin in BT-549 cells remained relatively constant over the 2-h chase when endocytosis was blocked using NH4Cl (Fig. 9C, closed circle). N-cadherin expressed in C4ST-1KO cells was relatively stable both in the absence and presence of NH4Cl (Fig. 9C). Finally, we compared the production of the C-terminal fragment of N-cadherin by BT-549 and C4ST-1KO cells using Western blotting. As shown in Fig. 9D, the amount of the C-terminal fragment of N-cadherin was significantly decreased in C4ST-1KO cells compared with BT-549 cells. As shown in Fig. 9E, β-catenin-dependent transcriptional activities, which were measured using the TCF reporter vector, were significantly suppressed in C4ST-1KO cells (Fig. 9E, panel a), and thus the expression level of the β-catenin-target gene MMP9 was reduced (Fig. 9E, panel b). These results indicated that the N-cadherin/β-catenin pathway was depressed in C4ST-1KO cells due to a lack of CS-E. As expected, C4ST-1KO cells showed lower invasive activities compared with the parental BT-549 cells (Fig. 9F, panel a). In addition, exogenously added CS-E restored the invasive activities of C4ST-1KO cells to the same level as those of BT-549 cells stimulated by CS-E (Fig. 9F, panel b), suggesting that CS-E directly promotes tumor invasion. Furthermore, these data suggested that molecules involved in tumor invasion, except for C4ST-1, are fully functional in C4ST-1KO cells. Based on the combined data, it is concluded that tumor cells drive invasive activity by taking advantage of both tumor- and tissue-derived CSs.
Figure 9.
CSs synthesized by C4ST-1 controlled endocytosis of N-cadherin, cleavage of N-cadherin, β-catenin signaling, and cell invasive activity in BT-549 cells. A, after BT-549 and C4ST-1KO cells were treated with or without Pitstop2 or nystatin for 2 h, the cell-surface proteins were labeled with Ez-Link sulfo-NHS-Biotin (2nd to 4th, 6th to 8th, 10th to 12th, and 14th to 16th lanes), which was followed by a pulldown assay using StreptAvidin-agarose beads. The pulldown complex was analyzed by immunoblotting using anti-N-cadherin. The box-and-whisker graph at right indicates the cell-surface expression level of N-cadherin, normalized against the total expression level of N-cadherin (input), in BT-549 and C4ST-1KO cells (n = 3). Statistical significance was assessed using a Student's t test. B, surface N-cadherin expression on BT-549 cells (magenta), BT-549 cells treated with Nystatin (orange) or with Pitstop2 (green), and on C4ST-1KO cells (cyan) were compared by FACS analysis. The shaded area indicates cells treated without anti-N-cadherin antibody (negative control). C, disappearance rate of the full-length N-cadherin in BT-549 and C4ST-1KO cells during a cycloheximide chase in the presence or absence of an inhibitor of endocytosis, NH4Cl, was analyzed by Western blotting. In the graph at right the presence or absence of NH4Cl is indicated by closed and open circles, respectively. Two independent experiments were carried out for each data set. Error bars represent standard deviation (S.D.). D, amount of the C-terminal fragment, N-Cad(C), generated in BT-549 and C4ST-1KO cells in the presence or absence of 10 μm MG132 was analyzed by Western blotting. In the graph at right, the quantified expression level of N-Cad(C) was compared between BT-549 and C4ST-1 cells. Three independent experiments were carried out for each data set. E, panel a, activation level of β-catenin signaling in BT-549 and C4ST-1KO cells was estimated using the TCF reporter vector (n = 3). Panel b, gene expression level of MMP9 in BT-549 and C4ST-1KO cells was examined using real-time PCR (n = 3). F, panel a and b, invasive activity of BT-549 and C4ST-1KO cells was measured in a Matrigel invasion assay in the absence or presence of CS-E (n = 3).
Discussion
Extensive evidence supports the importance of tumor-associated CSs in promoting aggressive and metastatic behavior of malignant cells (9, 36). In addition, CSs present in the ECM of normal tissue affect tumor cells (14). A previous report showed that CD44, an ECM-binding molecule of tumor cells, is proteolytically cleaved by interaction with certain ECM components such as hyaluronan and CSs and that CD44-dependent motility is thereby up-regulated (14). This finding indicates that tumor cells utilize the glycocalyx containing CSs to invade into normal tissue. We found here that CSs could significantly increase the invasion potential of two basal-like cancer cell lines, BT-549 and MDA-MB-231 (Fig. 1A, panel a). However, the molecular mechanism by which CSs contribute to tumor cell invasion has remained unclear. We previously found that CS is a receptor for N-cadherin in osteoblasts (26). In addition, N-cadherin is expressed in highly invasive tumor cell lines, including BT-549 cells, and contributes to the invasive phenotype (5, 6). These findings raised the possibility that CSs transduce signals into breast cancer cells through N-cadherin to up-regulate the tumor invasion potential. As expected, the invasion activity of basal-like subtype BT-549 cells that was elevated by stimulation with CSs was inhibited by pre-treatment with an anti-N-cadherin antibody (Fig. 2D).
What happens to N-cadherin after the binding of CSs to N-cadherin? Classical cadherins are proteolytically cleaved by multiple proteases in a context-dependent manner (30). It is generally accepted that proteolysis of cadherins proceeds in two steps. This proteolysis process is well-known to be a regulated intramembrane proteolysis, by which transmembrane proteins are cleaved to release cytoplasmic domains that often enter the nucleus to regulate gene transcription (37). In the first step, the extracellular region of N-cadherin is cleaved by multiple proteases, including ADAMs, MMPs, and other transmembrane proteases. Subsequently, a second cleavage occurs within the transmembrane segment by another protease to liberate the cytoplasmic C-terminal fragment (CTF). The CTFs of classical cadherins can also influence intracellular signaling events by interacting with molecules involved in multiple pathways. For example, the N-cadherin CTF can interact with β-catenin. This complex enters the nucleus, leads to β-catenin-dependent transcriptional activation, and can promote cell migration and invasion (7). Thus, proteolysis of N-cadherin is considered to be a regulated step in N-cadherin/β-catenin nuclear signaling; however, what regulates the proteolysis of N-cadherin has remained unclear. We have shown here that binding of CSs with N-cadherin triggers endocytosis-associated proteolysis of N-cadherin to activate β-catenin nuclear signaling. To date, it has been reported that N-cadherin is cleaved by ADAM10, MMP-7, MT5-MMP, and presenilin 1 (7, 28, 29, 38, 39) in intracellular compartments as well as at the cell surface. In addition, ADAM10 is predominantly localized in the Golgi, and L1 adhesion molecule ectodomain cleavage occurs in the Golgi as well as at the plasma membrane (40). Therefore, endocytosis is probably necessary to deliver N-cadherin cargo to a subcellular compartment where proteases are active. Consistent with this notion, previous studies showed that proteolytic processing of protocadherin (Pcdh) is tightly linked to subcellular trafficking that is mediated by the endosomal sorting complex required for transport (ESCRT complex)-0, -I, -II, and -III (41). In early brain development or in undifferentiated cell lines, rapid endocytosis of Pcdh facilitates its proteolytic processing to generate an intracellular fragment that may enter the nucleus and regulate gene expression. As development progresses or cells differentiate, Pcdh intracellular fragments are no longer produced probably because of inhibition of endocytosis due to homophilic interactions between Pcdh molecules. In addition, the amount and the structure of CSs are tightly regulated during brain development (42). These findings suggest that sugar signals control cadherin homophilic interactions, endocytosis of cadherin, and its subsequent cleavage. Furthermore, as described above, the regulation of a regulated intramembrane proteolysis is thought to occur at the level of the first proteolysis step. The first ectodomain shedding of N-cadherin is mediated by MMPs. MMP7 is reported to be one of the proteases involved in the first cleavage of N-cadherin (28) and is activated by CS-E (43). Thus, CS-E might also enhance N-cadherin proteolysis through activation of the proteases involved in N-cadherin cleavage.
Dynamic behavior of cadherin that is regulated by endocytic pathways in cell migration in physiological and pathological conditions has been reported, focusing particularly on neural development and cancer metastasis (44). N-cadherin endocytosis at neural synapses is regulated by β-catenin, an N-cadherin binding partner (45). Activation of NMDA receptors decreases tyrosine phosphorylation of β-catenin, increases binding of β-catenin to N-cadherin, and thus the rate of N-cadherin endocytosis is reduced (45). Thus, tyrosine phosphorylation of β-catenin regulates NMDA-dependent N-cadherin endocytosis. In addition, proteoglycans control the phosphorylation of β-catenin through activation of the PTP1B-like phosphatase (46). Furthermore, the Slit/Robo pathway, which requires proteoglycans bearing CS modifications for its activation (47), activates its downstream tyrosine kinase Abl that phosphorylates β-catenin, resulting in a decrease in the affinity of β-catenin for N-cadherin (48). Therefore, CSs might indirectly control N-cadherin endocytosis through regulation of β-catenin phosphorylation in neuronal systems. In cancer cells, cadherin endocytosis plays an important role in maintaining a rounded cell morphology to adopt mitosis-related cell rounding and to decrease cell–cell adhesion for control of cancer cell migration. Thus, cadherin internalization is a key requirement for cancer metastasis. In addition, cadherin endocytosis is also regulated by cancer-associated cell signaling, such as receptor tyrosine kinase signaling. Lu et al. (49) reported that EGF signaling enhanced internalization of E-cadherin to induce epithelial–mesenchymal transition in a breast carcinoma cell line. Furthermore, it has been reported that EGF signaling and CSs are reciprocally regulated. EGF signaling increases the production of CSs in astrocytes (50), heparin-binding–EGF molecules secreted into the tumor microenvironment bind to CS-E with high affinity (51), and CSs controlled EGF signaling (52). Therefore, we cannot exclude the possibility that CSs control cadherin endocytosis in an indirect manner through tyrosine kinase signaling, although we did examine whether N-cadherin endocytosis occurred via direct interactions between N-cadherin and CS-E in this study.
Why does the interaction of CSs with N-cadherin lead to endocytosis of N-cadherin? During the biogenesis of cell–cell contacts, N-cadherin shuttles between an intracellular and a plasma membrane pool, and the formation of the N-cadherin–dependent cell–cell contacts results from the recruitment of the intracellular pool of N-cadherin to the plasma membrane (53). It is considered that exogenously added CSs interact with cadherin and loosen the homophilic interaction between cadherin molecules. X-ray crystallography analysis has shown that homophilic interaction of cadherin is mediated by interaction between extracellular cadherin (EC)-1 (EC1) domains. These domains are present in an N-terminal cell-surface–exposed region of cadherin that is composed of tandemly repeated EC domains, EC1–EC5. The interfaces between EC1 domains have clusters of positively charged amino acids that can electrostatically interact with negatively charged CSs. Thus, binding of CSs to an EC1 domain might destabilize homophilic interactions between N-cadherins, leading to enhanced endocytosis of N-cadherin.
What is the pathophysiological significance of CS-E-induced proteolysis of N-cadherin? Breast cancer mainly metastasizes to the bony skeleton, the lungs, liver, and brain via the circulation. Liver metastasis in particular is a frequent occurrence in patients with breast cancer (54). If breast cancer liver metastasis is left untreated, the survival time is only 4–8 months. It is therefore necessary to understand the molecular mechanism that underlies liver metastasis in breast cancer patients. Although liver metastasis consists of multiple steps, liver-specific homing of breast cancer cells is an initial important step of metastasis, which means that it is necessary to understand the interaction between breast cancer cells and the liver microenvironment. Liver and bone express CSs and have a high content of CS-E. As shown in this study, breast cancer cells might bind through their N-cadherin to CS-E in the liver microenvironment to elevate their invasive potential via activation of the N-cadherin/β-catenin pathway (Fig. 10). In addition, CS-E that is expressed in cancer cells might form a positive feedback loop, thereby amplifying N-cadherin/β-catenin signaling. This notion is consistent with our finding that loss of CS-E led to a suppression of invasive potential in C4ST-1 knock-out cancer cells (Fig. 9F). Indeed, the genes encoding CS-E biosynthetic enzymes, C4ST-1 and GalNAc4S-6ST, are transcribed to a higher degree in breast carcinoma samples than in healthy tissue (55). Therefore, cell-autonomous activation of N-cadherin/β-catenin signaling by CS-E might contribute to breast cancer malignancy. Further studies are needed to elucidate whether cancer cell-derived CS-E promotes tumor growth and metastasis in vivo.
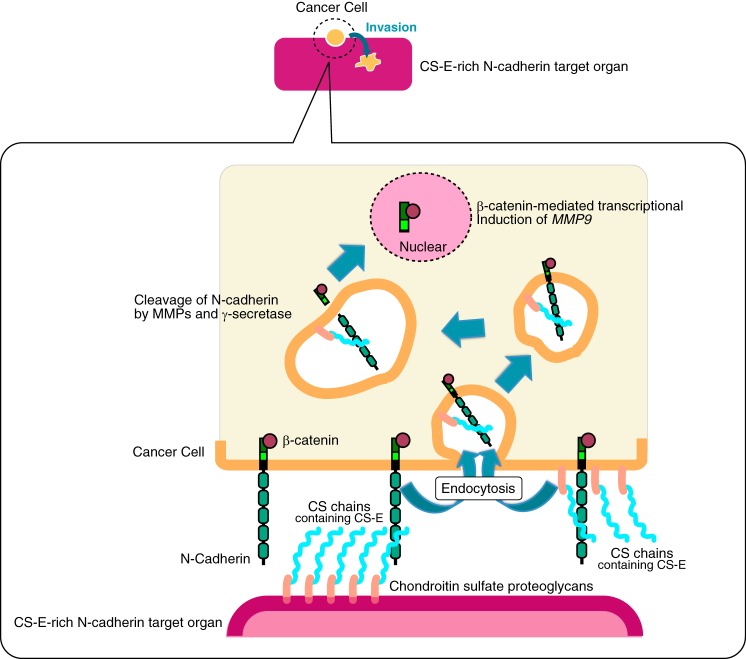
Figure 10.
Illustrated summary of how CS chains accelerate the invasive activity of a basal-like breast cancer cell line BT-549. First, CS chains expressed in a CS-E-rich target organ or in BT-549 cells themselves bind to N-cadherin. The binding of CSs to N-cadherin triggers endocytosis of N-cadherin, followed by cleavage of N-cadherin. As a result, the C-terminal domain of N-cadherin that is associated with β-catenin is released and is translocated into the nucleus, and transcriptional induction of MMP9 by β-catenin is increased, which enhances cancer cell invasion. CS chains expressed in the BT-549 cells themselves are considered to be utilized for cell-autonomous enhancement of invasive potential, whereas BT-549 cells might take advantage of CS chains expressed in a CS-E-rich N-cadherin target organ for metastasis.
Experimental procedures
Cell culture and stable transfection
The human breast cancer cell lines MCF7 (ATCC® HTB-22TM), T47D (ATCC® HTB-133TM), HCC1954 (ATCC® CRL-2338TM), and BT-549 (ATCC® HTB-122TM) and the human cervical cancer cell line HeLa (ATCC® CCL-2TM) were obtained from the American Type Culture Collection (ATCC). The MDA-MB-231 cell line (no. 92020424) was purchased from the European Collection of Cell Culture (Salisbury, UK). RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% l-glutamine was used as the culture medium for the human breast cancer cell lines. HeLa cells were grown in DMEM supplemented with 10% heat-inactivated FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% l-glutamine. All cell lines were maintained at 37 °C and 5% CO2.
The expression plasmids (pEGFP-N1-N-Cad(C) or pEGFP-N1) were transfected into BT-549 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfectants were cultured in the presence of 25 μg/ml G418. Colonies surviving in the presence of 25 μg/ml G418 were picked up and purified using a fluorescence-activated cell sorting (FACS)-based method (FACSAriaTM III, BD Biosciences).
Plasmid construction
A human N-cadherin ORF clone, pF1KB4700-human N-cadherin, was obtained from the Kazusa DNA Research Institute (Chiba, Japan). For expression of the EGFP-fused full-length or C-terminal region of N-cadherin, a polymerase chain reaction using the following primers and pF1KB4700-human N-cadherin as a template was performed: forward primer 1 for amplification of N-Cad(FL), 5′-CTAGCTAGCGCCATGTGCCGGATAGC-3′ (underline, NheI site; bold, start codon); forward primer 2 for amplification of N-Cad(C)), 5′-CTAGCTAGCGCCATGAAACGCCGGGATAAAG-3′ (underline, NheI site; bold, start codon); and reverse primer, 5′-CGGGATCCACGTCATCACCTCCACC-3′ (underline, BamHI site). pEGFP-N1-N-Cad(FL) and pEGFP-N1-N-Cad(C) were constructed by inserting the NheI–BamHI fragment of a PCR product into the NheI and BamHI sites of pEGFP-N1 (Clontech).
Real-time PCR
Total RNA was isolated from cells using the Maxwell® 16 LEV simplyRNA purification kit (Promega). For reverse transcription, 1 μg of total RNA was treated with Moloney murine leukemia virus reverse transcriptase (Invitrogen) using random primers (nonadeoxyribonucleotide mixture; pd(N)9) (Takara Bio Inc., Shiga, Japan). Quantitative real-time PCR was conducted using FastStart DNA master plus SYBR Green I in a LightCycler 1.5 (Roche Applied Science) according to the manufacturer's protocols. The housekeeping gene, GAPDH, was used as an internal control for quantification. The primers used for real-time PCR are shown in Table 2.
Table 2.
Primers used for real-time PCR
| Gene name | 5′-Primer | 3′-Primer |
|---|---|---|
| hWnt-1 | 5′-ACCGAGGCTGTCGAGAAACG-3′ | 5′-GCCGGTAGTCACACGTGCAG-3′ |
| hWnt-2 | 5′-GCTCACCCCCGAGGTCAACT-3′ | 5′-CCTGGCTAATGGCACGCATC-3′ |
| h&mWnt-3a | 5′-ACAACAATGAGGCTGGG-3′ | 5′-ATCTCCGAGGCACTGTCATA-3′ |
| hLRP5 | 5′-CAAGGTCGTCGGAACCAACC-3′ | 5′-AGAAGGCCTCAGGCACGATG-3′ |
| hLRP6 | 5′-GTGGATGCCCTGCCCACTAC-3′ | 5′-GGCTGTGGATGGGAAGGATG-3′ |
| hDkk-1 | 5′-GTGCGCAGAGGACGAGGAGT-3′ | 5′-GTGACGCATGCAGCGTTTTC-3′ |
| hN-cadherin | 5′-ATGAAGAAGGTGGAGGAGA-3′ | 5′-AGATCGGACCGGATACT-3′ |
| β-Catenin | 5′-AGCTTCCAGACACGCTATCAT-3′ | 5′-CGGTACAACGAGCTGTTCTAC-3′ |
| GFP | 5′-ACCACTACCTGAGCACCCAGTC-3′ | 5′-GTCCATGCCGAGAGTGATCC-3′ |
| hMMP2 | 5′-TCTCCTGACATTGACCTTGGC-3′ | 5′-CAAGGTGCTGGCTGAGTAGATC-3′ |
| hMMP7 | 5′-TGAGCTACAGTGGGAACAGG-3′ | 5′-TCATCGAAGTGAGCATCTCC-3′ |
| hMMP9 | 5′-TTGACAGCGACAAGAAGTGG-3′ | 5′-GCCATTCACGTCGTCCTTAT-3′ |
| hMMP13 | 5′-TCCCAGGAATTGGTGATAAAGTAGA-3′ | 5′-CTGGCATGACGCGAACAATA-3′ |
| hMMP14 | 5′-TTGGACTGTCAGGAATGAGG-3′ | 5′-GCAGCACAAAATTCTCCGTG-3′ |
Invasion assay
The invasion assay was carried out using Corning® BioCoatTM Matrigel® invasion chambers according to the manufacturer's instructions. Cells (5 × 104 cells/ml) were incubated in the presence or absence of 50 μg/ml CS isoforms (CS-A, CS-C, and CS-E were derived from whale cartilage, shark cartilage, and squid cartilage, respectively) (Seikagaku Corp., Tokyo, Japan) for 20 min at room temperature and were then added to the upper chamber and allowed to invade for 22 h at 37 °C in a CO2 incubator. In the case of treatment with MMP inhibitor, cells were pre-incubated for 20 min in the presence of 100 μm of the MMPs inhibitor, GM-6001 (BML-E1300, Enzo Life Science), before adding CS to the cells. Cells were transfected with the following siRNA corresponding to MMP2 and MMP9. Silencer® Select siMMP2 (catalog no. 4427038; Assay ID s8852) and MMP9 siRNA (catalog no. sc-29400) were purchased from Thermo Fisher Scientific and Santa Cruz Biotechnology, Inc., respectively. Twenty four hours after transfection, cells (5 × 104 cells/ml) were seeded on a Matrigel-coated membrane and cultured for 22 h at 37 °C. MCF7 cells were transfected with either pEGFP-N1 empty vector or pEGFP-N1-N-cadherin(full-length) vector and cultured for 12 h. GFP-positive cells (0.83 × 104 cells/ml) were inoculated on a Matrigel-coated membrane.
After the non-invading cells and Matrigel were removed from the upper surface of the membrane by scrubbing with a cotton swab, invading cells (those adhering to the bottom surface of the membrane) were stained with Giemsa (WAKO Pure Chemicals, Osaka, Japan). Alternatively, cells expressing EGFP or EGFP-N-Cad(C) were counted taking advantage of the fluorescence of EGFP. For this purpose, all invaded cells on a membrane were photographed using a Keyence BZ-8000 fluorescence microscope and counted.
It was checked whether the Matrigel-coated membrane might contain proteases using AmpliteTM universal fluorimetric protease activity assay kit (AAT Bioquest® Inc.). No activities of chymotrypsin, trypsin, thermolysin, protease K, protease XIV, and human leukocyte elastase were detected. In addition, the amounts of HS and CS in a Matrigel-coated membrane were measured. The membrane contained only HS (64.9 ± 10.5 pmol of HS-disaccharides).
Luciferase reporter assay
One day before transfection, cells were seeded on a 24-well culture plate. The reporter plasmid, pTCF7wt-luc (1 μg) (24), and the reference plasmid, pRL-TK (harboring the thymidine kinase promoter just upstream of Renilla luciferase; Promega) (0.1 μg), were transfected into cells using Lipofectamine 2000 (Invitrogen). After 12 h, the cells were washed once with PBS; fresh growth medium was added, and the cells were cultured for an additional 12 h with or without either 50 μg/ml CS-E or 200 ng/ml anti-Dkk1 antibody (H-120) (sc-25516, Santa Cruz Biotechnology). The cells were washed with PBS and lysed in 100 μl of Passive Lysis Buffer (Promega). Firefly luciferase and Renilla luciferase activities were measured using 5 μl of cell lysate with the Dual-Luciferase Reporter Assay System (Promega). The Lumat LB9507 luminometer (EG&G Berthold, Germany) was used for measurement in the linear range. “Relative activity” was defined as the ratio of firefly luciferase activity to Renilla luciferase activity.
Immunoblotting and immunoprecipitation
BT-549 cells stably expressing either N-Cad(C)-EGFP or EGFP were solubilized with lysis buffer (1% Nonidet P-40, 20 mm Tris-HCl (pH 7.5), 0.15 m NaCl, 1 mm EDTA, 10% glycerol, and protease inhibitor mixture (Nacalai Tesque)). Cell lysates were centrifuged at 9100 × g for 15 min, and the resulting supernatants were pre-cleared using 10 μl of protein G-Sepharose (GE Healthcare) for 2 h at 4 °C. Pre-cleared supernatants were incubated with anti-GFP antibody (dilution ratio 1:875) (Clone RQ1, Code No. D153-3, Medical & Biological Laboratories Co., Ltd.), anti-N-cadherin antibody (dilution ratio 1:437.5) (Clone 32/N-cadherin, Immunogen mouse N-cadherin amino acids 802–819, Material No. 610920, BD Transduction LaboratoriesTM), or anti-β-catenin antibody (dilution ratio 1:437.5) (Clone 14/β-catenin, Material No. 610514, BD Transduction LaboratoriesTM) at 4 °C overnight. After adding 10 μl of protein G-Sepharose, each sample was incubated for 2 h at 4 °C. The protein G-Sepharose beads recovered by centrifugation were washed four times with the lysis buffer. Each sample was resolved on 7.5% SDS-polyacrylamide gels, transferred to PVDF membranes, and incubated overnight with anti-GFP antibody (dilution ratio 1:1000) (Clone RQ1, Medical & Biological Laboratories Co., Ltd.), anti-N-cadherin antibody (dilution ratio 1:2000) (Clone 32/N-cadherin, BD Transduction LaboratoriesTM), or anti-β-catenin antibody (dilution ratio 1:2000) (Clone 14/β-catenin, BD Transduction LaboratoriesTM). The bound antibody was detected with anti-mouse IgG conjugated to horseradish peroxidase (GE Healthcare).
Immunofluorescence
Cells grown until 80–90% confluent on a 3.5-cm glass-bottomed dish (AGC Techno Glass Co. Ltd., Shizuoka, Japan) were transfected with 1 μg of pEGFP-N1-N-cadherin(FL) using Lipofectamine 2000 (Invitrogen). The following day, the cells were treated with or without 25 μg/ml CS-E and incubated for 20 h at 37 °C. Cells transiently expressing N-cadherin(FL) were fixed with PBS containing 4% paraformaldehyde for 20 min on ice. After washing with PBS, the cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min at room temperature. After blocking with PBS containing 2% BSA for 1 h at room temperature, cells were incubated overnight at 4 °C with the following antibodies: anti-N-cadherin antibody (dilution ratio 1:200) (Clone GC-4, catalog no. C3865, Sigma); anti-N-cadherin antibody (dilution ratio 1:200) (Clone 32, catalog no. 610920, BD Transduction LaboratoriesTM); anti-GM130 antibody (dilution ratio 1:50) (Clone 35/GM130, catalog no. 610822, BD Transduction LaboratoriesTM); anti-caveolin antibody (dilution ratio 1:100) (catalog no. C13630, BD Transduction LaboratoriesTM); anti-EEA1 antibody (dilution ratio 1:200) (Clone EEA1-C33, catalog no. E8034, Sigma); or anti-LAMP-1 antibody (dilution ratio 1:200) (Clone H4A3, Material no. 555798, BD Biosciences). Hoechst 33342 was used as a nuclear counterstain. Images were acquired with a Zeiss LSM700 confocal laser-scanning system (Carl Zeiss Inc., Oberkochen, Germany) equipped with an inverted Axio observer Z1 microscope.
Biacore analysis
The interaction between Dkk-1 and CSs was analyzed using BiacoreJ (GE Healthcare) as described previously (18).
Scratch assay
Cells were seeded in 6-well plates and cultured until confluent. Using a pipette tip, a straight scratch was made in the center, simulating a wound. After one wash of 1 ml/well of PBS, cells were cultured in 1 ml/well of RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% l-glutamine in the presence or absence of 5 μg/ml anti-N-cadherin antibodies (Clone GC-4) for 30 min at 37 °C, and then 2.5 μl/well of 20 μg/ml CS-E were added. After 0, 6, 12, and 24 h, the cells were imaged at ×10 magnification, and three fields of view were imaged along the wound edge. The number of cells migrating from the leading edge of the monolayer were counted.
Binding assay
Biotinylated CS-E was immobilized on a Nunc ImmobilizerTM streptavidin 96-well plate (Nalge Nunc International, Rochester, NY) as described previously (56, 57). Briefly, CS-E was dissolved in 100 mm MES-NaOH, pH 5.5, at a concentration of 1 mg/ml. To this solution were added 2.5 μg of biotin-LC-hydrazide freshly dissolved in dimethyl sulfoxide and 0.25 μl of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (Pierce). The mixture was incubated overnight at room temperature with continuous shaking. Excess biotinylating reagents were removed by centrifugal filtration using the Ultrafree®-MC (5000 NMWL filter unit) (Millipore, Billerica, MA) against several changes of phosphate-buffered saline. After the Nunc ImmobilizerTM streptavidin was washed with PBS containing 0.1% Tween 20 (PBST), 40 μg/ml biotinylated CS-E was added to each well, and incubated for 1 h at room temperature. After washing with PBST, each well was blocked with PBS containing 2% BSA for 30 min at room temperature. After three washes with 100 μl of PBS, 50,000 cells/well in RPMI 1640 medium containing 10 mm HEPES were added to each well and were then incubated for 30 min at 37 °C. After the cells were washed with PBS containing 0.03% BSA, cellular lactate dehydrogenase was released from the cells by adding 1 ml of 1% Triton X-100. The enzyme activity of cellular lactate dehydrogenase was measured as an indicator of cell viability using the Cyto Tox-ONE homogeneous membrane integrity assay (Promega).
Generation of C4ST-1 knock-out BT-549 cells
CRISPR/Cas9 plasmids targeting the human C4ST-1 gene were constructed using a GeneArt® CRISPR nuclease vector with orange fluorescent protein reporter kit (catalog no. A21174) (Thermo Fisher Scientific). Two single-stranded oligonucleotides specific for the C4ST-1 gene were annealed as follows: top oligonucleotide, 5′-GCGGAGGAACCCCTTCGGTGGTTTT-3′, and bottom oligonucleotide, 5′-CACCGAAGGGGTTCCTCCGCCGGTG-3′ (underlined sequences were complementary to the overhang sequence in the linearized CRISPR nuclease vector). The resulting double-stranded oligonucleotides encoding CRISPR RNA specific for the C4ST-1 gene were inserted into a GeneArt® CRISPR nuclease vector. The constructed CRISPR/Cas9 plasmids targeting the human C4ST-1 gene were transfected into BT-549 cells using Lipofectamine 2000 (Thermo Fisher Scientific). Forty eight hours after transfection, orange fluorescent protein-positive cells were isolated using the FACSAriaTM III cell sorter. These cells were cloned by limiting dilution analysis and were then analyzed by genome sequencing to check the accurate and efficient gene editing of the C4ST-1 gene using the following primers: forward sequencing primer for C4ST-1, 5′-TTTCTTCCTCCCTCCCAGAT-3′, and reverse sequencing primer for C4ST-1, CAGGAAAGGGCA.
Disaccharide analysis of GAGs from BT-549 cells
CSs isolated and purified from human breast cancer BT-549 cells were analyzed as described previously (18, 19, 58).
Cell-surface protein biotinylation assay
After washing with pre-chilled PBS containing 1 mm MgCl2 and 0.1 mm CaCl2 (PBS-CM), cells were incubated with PBS-CM containing 0.5 mg/ml Ez-Link® Sulfo-NHS-Biotin (Thermo Fisher Scientific) for 30 min at 4 °C with occasional shaking. After removing this reagent, PBS-CM containing 0.5 mg/ml Ez-Link® Sulfo-NHS-Biotin was added to the cells again and incubated for 30 min at 4 °C with occasional shaking. The cells were washed with PBS-CM and then incubated with quenching solution (4 mm glycine) for 10 min at 4 °C to neutralize uncross-linked reagents.
Flow cytometry
Cells (1 × 105 cells) were fixed with PBS containing 4% paraformaldehyde on ice for 30 min. After washing with PBS, the cells were incubated with PBS containing 2% BSA on ice for 30 min and were then stained with anti-N-cadherin antibody (dilution ratio 1:100) (Clone GC-4, catalog no. C3865, Sigma) on ice. After 1 h, the cells were washed and were then incubated with mouse IgG(H&L) antibody conjugated with DyLightTM 488 and pre-adsorbed (dilution ratio 1:200) (Rockland Immunochemicals Inc., Limerick, PA) on ice for 1 h. The cells were analyzed using the BD AccuriTM C6 flow cytometer (BD Biosciences).
Statistical analysis
Data are expressed as the mean ± S.D. of the mean. Statistical significance was determined using Student's t test.
Author contributions
S. N. and H. Kinouchi performed the research and analyzed the data. S. N. and H. Kitagawa conceived the idea, designed the research, and wrote the manuscript. H. Kitagawa coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are especially grateful to Prof. Kunitada Shimotohno (Chiba Institute of Technology, Chiba, Japan) and Assistant Prof. Makoto Hijikata (Institute for Virus Research, Kyoto University, Kyoto, Japan) for the kind gift of the reporter vector, pTcf7wt-luc. We thank Arisa Toda, Junko Yamada, Misa Akutagawa, Eri Nakamatsu, Setsuko Konoike, Manami Tachibana, and Naho Sakae for technical support.
This work was supported in part by Grants-in-aid for Scientific Research on Innovative Areas 23110003 (to H. K.), Scientific Research (B) 16H05088 (to H. K.), and (C) 25460080 (to S. N.), and the Supported Program for the Strategic Research Foundation at Private Universities, 2012–2016 (to H. K.), from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- estrogen receptor
- PR
- progesterone receptor
- ADAM
- disintegrin and metalloprotease
- CS
- chondroitin sulfate
- EC
- extracellular cadherin
- ECM
- extracellular matrix
- GalNAc4S-6ST
- N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase
- MMP
- matrix metalloprotease
- N-Cad(C)
- C-terminal of N-cadherin
- PG
- proteoglycan
- EGFP
- enhanced green fluorescent protein
- Chn
- chondroitin
- TCF
- T cell factor
- LEF
- lymphoid enhancer factor
- N-Cad(FL)
- full-length N-cadherin
- SDC
- syndecan
- CTF
- C-terminal fragment
- Pcdh
- protocadherin
- gRNA
- guide RNA
- SPR
- lymphoid enhancer factor
- HS
- heparan sulfate.
References
- 1. Nielsen T. O., Hsu F. D., Jensen K., Cheang M., Karaca G., Hu Z., Hernandez-Boussard T., Livasy C., Cowan D., Dressler L., Akslen L. A., Ragaz J., Gown A. M., Gilks C. B., van de Rijn M., and Perou C. M. (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 10, 5367–5374 10.1158/1078-0432.CCR-04-0220 [DOI] [PubMed] [Google Scholar]
- 2. Carey L. A., Dees E. C., Sawyer L., Gatti L., Moore D. T., Collichio F., Ollila D. W., Sartor C. I., Graham M. L., and Perou C. M. (2007) The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 13, 2329–2334 10.1158/1078-0432.CCR-06-1109 [DOI] [PubMed] [Google Scholar]
- 3. Nishimura R., and Arima N. (2008) Is triple negative a prognostic factor in breast cancer? Breast Cancer 15, 303–308 10.1007/s12282-008-0042-3 [DOI] [PubMed] [Google Scholar]
- 4. Sarrió D., Rodriguez-Pinilla S. M., Hardisson D., Cano A., Moreno-Bueno G., and Palacios J. (2008) Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68, 989–997 10.1158/0008-5472.CAN-07-2017 [DOI] [PubMed] [Google Scholar]
- 5. Nieman M. T., Prudoff R. S., Johnson K. R., and Wheelock M. J. (1999) N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 147, 631–644 10.1083/jcb.147.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hazan R. B., Phillips G. R., Qiao R. F., Norton L., and Aaronson S. A. (2000) Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J. Cell Biol. 148, 779–790 10.1083/jcb.148.4.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reiss K., Maretzky T., Ludwig A., Tousseyn T., de Strooper B., Hartmann D., and Saftig P. (2005) ADAM10 cleavage of N-cadherin and regulation of cell–cell adhesion and β-catenin nuclear signalling. EMBO J. 24, 742–752 10.1038/sj.emboj.7600548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Willis C. M., and Klüppel M. (2014) Chondroitin sulfate-E is a negative regulator of a pro-tumorigenic Wnt/β-catenin-collagen 1 axis in breast cancer cells. PLoS ONE 9, e103966, 10.1371/journal.pone.0103966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cooney C. A., Jousheghany F., Yao-Borengasser A., Phanavanh B., Gomes T., Kieber-Emmons A. M., Siegel E. R., Suva L. J., Ferrone S., Kieber-Emmons T., and Monzavi-Karbassi B. (2011) Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res. 13, R58 10.1186/bcr2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Price M. A., Neudauer C. L., Wilson C., Ferrone S., Xia H., Iida J., Simpson M. A., and McCarthy J. B. (2004) Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. J. Cell Biol. 165, 881–891 10.1083/jcb.200403174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang J., Price M. A., Li G. Y., Bar-Eli M., Salgia R., Jagedeeswaran R., Carlson J. H., Ferrone S., Turley E. A., and McCarthy J. B. (2009) Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer Res. 69, 7538–7547 10.1158/0008-5472.CAN-08-4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lesley J., Hyman R., and Kincade P. W. (1993) CD44 and its interaction with extracellular matrix. Adv. Immunol. 54, 271–335 10.1016/S0065-2776(08)60537-4 [DOI] [PubMed] [Google Scholar]
- 13. Kawashima H., Atarashi K., Hirose M., Hirose J., Yamada S., Sugahara K., and Miyasaka M. (2002) Oversulfated chondroitin/dermatan sulfates containing GlcAβ1/IdoAα1–3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J. Biol. Chem. 277, 12921–12930 10.1074/jbc.M200396200 [DOI] [PubMed] [Google Scholar]
- 14. Sugahara K. N., Hirata T., Tanaka T., Ogino S., Takeda M., Terasawa H., Shimada I., Tamura J., ten Dam G. B., van Kuppevelt T. H., and Miyasaka M. (2008) Chondroitin sulfate E fragments enhance CD44 cleavage and CD44-dependent motility in tumor cells. Cancer Res. 68, 7191–7199 10.1158/0008-5472.CAN-07-6198 [DOI] [PubMed] [Google Scholar]
- 15. Iida J., Dorchak J., Clancy R., Slavik J., Ellsworth R., Katagiri Y., Pugacheva E. N., van Kuppevelt T. H., Mural R. J., Cutler M. L., and Shriver C. D. (2015) Role for chondroitin sulfate glycosaminoglycan in NEDD9-mediated breast cancer cell growth. Exp. Cell Res. 330, 358–370 10.1016/j.yexcr.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 16. Mikami T., and Kitagawa H. (2013) Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830, 4719–4733 10.1016/j.bbagen.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 17. Nadanaka S., and Kitagawa H. (2008) Heparan sulphate biosynthesis and disease. J. Biochem. 144, 7–14 10.1093/jb/mvn040 [DOI] [PubMed] [Google Scholar]
- 18. Nadanaka S., Ishida M., Ikegami M., and Kitagawa H. (2008) Chondroitin 4-O-sulfotransferase-1 modulates Wnt-3a signaling through control of E disaccharide expression of chondroitin sulfate. J. Biol. Chem. 283, 27333–27343 10.1074/jbc.M802997200 [DOI] [PubMed] [Google Scholar]
- 19. Nadanaka S., Kinouchi H., Taniguchi-Morita K., Tamura J., and Kitagawa H. (2011) Down-regulation of chondroitin 4-O-sulfotransferase-1 by Wnt signaling triggers diffusion of Wnt-3a. J. Biol. Chem. 286, 4199–4208 10.1074/jbc.M110.155093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nadanaka S., Kinouchi H., and Kitagawa H. (2016) Histone deacetylase-mediated regulation of chondroitin 4-O-sulfotransferase-1 (Chst11) gene expression by Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 480, 234–240 10.1016/j.bbrc.2016.10.035 [DOI] [PubMed] [Google Scholar]
- 21. Smid M., Wang Y., Zhang Y., Sieuwerts A. M., Yu J., Klijn J. G., Foekens J. A., and Martens J. W. (2008) Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68, 3108–3114 10.1158/0008-5472.CAN-07-5644 [DOI] [PubMed] [Google Scholar]
- 22. Nusse R., and Varmus H. E. (1982) Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31, 99–109 10.1016/0092-8674(82)90409-3 [DOI] [PubMed] [Google Scholar]
- 23. Pierceall W. E., Woodard A. S., Morrow J. S., Rimm D., and Fearon E. R. (1995) Frequent alterations in E-cadherin and α- and β-catenin expression in human breast cancer cell lines. Oncogene 11, 1319–1326 [PubMed] [Google Scholar]
- 24. Ueda Y., Hijikata M., Takagi S., Takada R., Takada S., Chiba T., and Shimotohno K. (2002) Wnt/β-catenin signaling suppresses apoptosis in low serum medium and induces morphologic change in rodent fibroblasts. Int. J. Cancer 99, 681–688 10.1002/ijc.10418 [DOI] [PubMed] [Google Scholar]
- 25. Nelson W. J., and Nusse R. (2004) Convergence of Wnt, β-catenin, and cadherin pathways. Science 303, 1483–1487 10.1126/science.1094291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koike T., Izumikawa T., Tamura J., and Kitagawa H. (2012) Chondroitin sulfate-E fine-tunes osteoblast differentiation via ERK1/2, Smad3 and Smad1/5/8 signaling by binding to N-cadherin and cadherin-11. Biochem. Biophys. Res. Commun. 420, 523–529 10.1016/j.bbrc.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 27. Marambaud P., Wen P. H., Dutt A., Shioi J., Takashima A., Siman R., and Robakis N. K. (2003) A CBP binding transcriptional repressor produced by the PS1/ϵ-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114, 635–645 10.1016/j.cell.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 28. Williams H., Johnson J. L., Jackson C. L., White S. J., and George S. J. (2010) MMP-7 mediates cleavage of N-cadherin and promotes smooth muscle cell apoptosis. Cardiovasc. Res. 87, 137–146 10.1093/cvr/cvq042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Porlan E., Martí-Prado B., Morante-Redolat J. M., Consiglio A., Delgado A. C., Kypta R., López-Otín C., Kirstein M., and Fariñas I. (2014) MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 16, 629–638 10.1038/ncb2993 [DOI] [PubMed] [Google Scholar]
- 30. McCusker C. D., and Alfandari D. (2009) Life after proteolysis: exploring the signaling capabilities of classical cadherin cleavage fragments. Commun. Integr. Biol. 2, 155–157 10.4161/cib.7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh R. D., Puri V., Valiyaveettil J. T., Marks D. L., Bittman R., and Pagano R. E. (2003) Selective caveolin-1-dependent endocytosis of glycosphingolipids. Mol. Biol. Cell 14, 3254–3265 10.1091/mbc.E02-12-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Puri V., Watanabe R., Singh R. D., Dominguez M., Brown J. C., Wheatley C. L., Marks D. L., and Pagano R. E. (2001) Clathrin-dependent and -independent internalization of plasma membrane sphingolipids initiates two Golgi targeting pathways. J. Cell Biol. 154, 535–547 10.1083/jcb.200102084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von Kleist L., Stahlschmidt W., Bulut H., Gromova K., Puchkov D., Robertson M. J., MacGregor K. A., Tomlin N., Pechstein A., Chau N., Chircop M., Sakoff J., von Kries J. P., Saenger W., Kräusslich H. G., et al. (2011) Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell 146, 471–484 10.1016/j.cell.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 34. Uyama T., Ishida M., Izumikawa T., Trybala E., Tufaro F., Bergström T., Sugahara K., and Kitagawa H. (2006) Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J. Biol. Chem. 281, 38668–38674 10.1074/jbc.M609320200 [DOI] [PubMed] [Google Scholar]
- 35. Izumikawa T., Okuura Y., Koike T., Sakoda N., and Kitagawa H. (2011) Chondroitin 4-O-sulfotransferase-1 regulates the chain length of chondroitin sulfate in co-operation with chondroitin N-acetylgalactosaminyltransferase-2. Biochem. J. 434, 321–331 10.1042/BJ20101456 [DOI] [PubMed] [Google Scholar]
- 36. Theocharis A. D., Skandalis S. S., Tzanakakis G. N., and Karamanos N. K. (2010) Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 277, 3904–3923 10.1111/j.1742-4658.2010.07800.x [DOI] [PubMed] [Google Scholar]
- 37. Brown M. S., Ye J., Rawson R. B., and Goldstein J. L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 10.1016/S0092-8674(00)80675-3 [DOI] [PubMed] [Google Scholar]
- 38. Kohutek Z. A., diPierro C. G., Redpath G. T., and Hussaini I. M. (2009) ADAM-10-mediated N-cadherin cleavage is protein kinase Cα-dependent and promotes glioblastoma cell migration. J. Neurosci. 29, 4605–4615 10.1523/JNEUROSCI.5126-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jang C., Choi J. K., Na Y. J., Jang B., Wasco W., Buxbaum J. D., Kim Y. S., and Choi E. K. (2011) Calsenilin regulates presenilin 1/γ-secretase-mediated N-cadherin ϵ-cleavage and β-catenin signaling. FASEB J. 25, 4174–4183 10.1096/fj.11-185926 [DOI] [PubMed] [Google Scholar]
- 40. Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Gast D., Joumaa S., Zentgraf H., Fogel M., and Altevogt D. P. (2003) ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 17, 292–294 [DOI] [PubMed] [Google Scholar]
- 41. Buchanan S. M., Schalm S. S., and Maniatis T. (2010) Proteolytic processing of protocadherin proteins requires endocytosis. Proc. Natl. Acad. Sci. U.S.A. 107, 17774–17779 10.1073/pnas.1013105107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyata S., Komatsu Y., Yoshimura Y., Taya C., and Kitagawa H. (2012) Persistent cortical plasticity by upregulation of chondroitin 6-sulfation. Nat. Neurosci. 15, 414–422 10.1038/nn.3023 [DOI] [PubMed] [Google Scholar]
- 43. Ra H. J., Harju-Baker S., Zhang F., Linhardt R. J., Wilson C. L., and Parks W. C. (2009) Control of promatrilysin (MMP7) activation and substrate-specific activity by sulfated glycosaminoglycans. J. Biol. Chem. 284, 27924–27932 10.1074/jbc.M109.035147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawauchi T. (2012) Cell adhesion and its endocytic regulation in cell migration during neural development and cancer metastasis. Int. J. Mol. Sci. 13, 4564–4590 10.3390/ijms13044564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tai C. Y., Mysore S. P., Chiu C., and Schuman E. M. (2007) Activity-regulated N-cadherin endocytosis. Neuron 54, 771–785 10.1016/j.neuron.2007.05.013 [DOI] [PubMed] [Google Scholar]
- 46. Balsamo J., Leung T., Ernst H., Zanin M. K., Hoffman S., and Lilien J. (1996) Regulated binding of PTP1B-like phosphatase to N-cadherin: control of cadherin-mediated adhesion by dephosphorylation of β-catenin. J. Cell Biol. 134, 801–813 10.1083/jcb.134.3.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chanana B., Steigemann P., Jäckle H., and Vorbrüggen G. (2009) Reception of Slit requires only the chondroitin-sulphate-modified extracellular domain of Syndecan at the target cell surface. Proc. Natl. Acad. Sci. U.S.A. 106, 11984–11988 10.1073/pnas.0901148106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rhee J., Buchan T., Zukerberg L., Lilien J., and Balsamo J. (2007) Cables links Robo-bound Abl kinase to N-cadherin-bound β-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat. Cell Biol. 9, 883–892 10.1038/ncb1614 [DOI] [PubMed] [Google Scholar]
- 49. Lu Z., Ghosh S., Wang Z., and Hunter T. (2003) Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell 4, 499–515 10.1016/S1535-6108(03)00304-0 [DOI] [PubMed] [Google Scholar]
- 50. Smith G. M., and Strunz C. (2005) Growth factor and cytokine regulation of chondroitin sulfate proteoglycans by astrocytes. Glia 52, 209–218 10.1002/glia.20236 [DOI] [PubMed] [Google Scholar]
- 51. Deepa S. S., Umehara Y., Higashiyama S., Itoh N., and Sugahara K. (2002) Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J. Biol. Chem. 277, 43707–43716 10.1074/jbc.M207105200 [DOI] [PubMed] [Google Scholar]
- 52. Sirko S., von Holst A., Weber A., Wizenmann A., Theocharidis U., Götz M., and Faissner A. (2010) Chondroitin sulfates are required for fibroblast growth factor-2-dependent proliferation and maintenance in neural stem cells and for epidermal growth factor-dependent migration of their progeny. Stem Cells 28, 775–787 10.1002/stem.309 [DOI] [PubMed] [Google Scholar]
- 53. Mary S., Charrasse S., Meriane M., Comunale F., Travo P., Blangy A., and Gauthier-Rouvière C. (2002) Biogenesis of N-cadherin-dependent cell–cell contacts in living fibroblasts is a microtubule-dependent kinesin-driven mechanism. Mol. Biol. Cell 13, 285–301 10.1091/mbc.01-07-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma R., Feng Y., Lin S., Chen J., Lin H., Liang X., Zheng H., and Cai X. (2015) Mechanisms involved in breast cancer liver metastasis. J. Transl. Med. 13, 64 10.1186/s12967-015-0425-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Potapenko I. O., Haakensen V. D., Lüders T., Helland A., Bukholm I., Sørlie T., Kristensen V. N., Lingjaerde O. C., and Børresen-Dale A. L. (2010) Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol. Oncol. 4, 98–118 10.1016/j.molonc.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., and Kjellén L. (2008) Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 10.1073/pnas.0705807105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nadanaka S., Purunomo E., Takeda N., Tamura J., and Kitagawa H. (2014) Heparan sulfate containing unsubstituted glucosamine residues: biosynthesis and heparanase-inhibitory activity. J. Biol. Chem. 289, 15231–15243 10.1074/jbc.M113.545343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nadanaka S., Zhou S., Kagiyama S., Shoji N., Sugahara K., Sugihara K., Asano M., and Kitagawa H. (2013) EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J. Biol. Chem. 288, 9321–9333 10.1074/jbc.M112.416909 [DOI] [PMC free article] [PubMed] [Google Scholar]