Abstract
Chlamydia trachomatis is an obligate intracellular human pathogen responsible for the most prevalent sexually-transmitted infection in the world. For decades C. trachomatis has been considered an “energy parasite” that relies entirely on the uptake of ATP from the host cell. The genomic data suggest that C. trachomatis respiratory chain could produce a sodium gradient that may sustain the energetic demands required for its rapid multiplication. However, this mechanism awaits experimental confirmation. Moreover, the relationship of chlamydiae with the host cell, in particular its energy dependence, is not well understood. In this work, we are showing that C. trachomatis has an active respiratory metabolism that seems to be coupled to the sodium-dependent synthesis of ATP. Moreover, our results show that the inhibition of mitochondrial ATP synthesis at an early stage decreases the rate of infection and the chlamydial inclusion size. In contrast, the inhibition of the chlamydial respiratory chain at mid-stage of the infection cycle decreases the inclusion size but has no effect on infection rate. Remarkably, the addition of monensin, a Na+/H+ exchanger, completely halts the infection. Altogether, our data indicate that chlamydial development has a dynamic relationship with the mitochondrial metabolism of the host, in which the bacterium mostly depends on host ATP synthesis at an early stage, and at later stages it can sustain its own energy needs through the formation of a sodium gradient.
Keywords: ATP synthase, Chlamydia trachomatis, energy metabolism, host-pathogen interaction, mitochondrial metabolism, respiratory chain
Introduction
Chlamydia trachomatis is an obligate intracellular bacterial pathogen that produces two of the most prevalent human diseases, affecting millions of people worldwide. It is responsible for trachoma, the world leading cause of preventable blindness, with ∼21 million patients globally (1). C. trachomatis also causes the most common sexually-transmitted infection worldwide, with more than 130 million new cases diagnosed annually (2, 3). Most chlamydial genital tract infections are asymptomatic in women, leading to untreated chronic infections that can produce pelvic inflammatory disease, and may develop infertility and ectopic pregnancy (4, 5). Chlamydial infection can also increase the risk of HIV infection (6). Moreover, C. trachomatis infections have a high impact on public health with a yearly expenditure of more than $500 million in direct medical costs, by the American health care system alone (7).
C. trachomatis has a unique biphasic developmental cycle, consisting of the infectious elementary body (EB)4 and the metabolically-active reticulate body (RB). EBs have the ability to attach and enter the host's mucosal epithelia, forming an inclusion in about 2 h post-infection (hpi), and are differentiated into RBs. At ∼12 hpi, RBs mature and proliferate by binary fission and start to differentiate back to EBs after 18 hpi. The host cells are lysed, and EBs are released for another cycle of infection (8–11).
Extensive studies of the genome, cell biology, and pathogenesis have revealed important biological information about C. trachomatis (10, 12, 13); nevertheless, many aspects remain unknown, especially its energy metabolism and relationship with the host cell. For decades, chlamydiae species were considered “energy parasites” that entirely depended on the host cells to fulfill their energetic requirements (14–17), due to the apparent lack of flavoproteins and cytochromes, which are essential for the mitochondrial respiratory function. Moreover, the discovery of two ATP-ADP translocases (Npt1 and Npt2) (18), which allow ATP uptake from the host cell, further supported the energy parasite hypothesis. However, the genomic data indicate that Chlamydiae encode many pathways involved in energy metabolism, including glycolysis (12, 19, 20), an incomplete Krebs cycle (12), as well as the pentose–phosphate pathway (19). The expression of these enzymes is activated at mid-stage of the infection, at around 12 hpi (21, 22). Moreover, quantitative proteomics studies showed that the expression of glycolytic enzymes fluctuates dramatically between the EB and RB forms. For instance, the RBs have a higher expression of glycolytic enzymes compared with EBs (23), indicating that the two life stages have different energy strategies.
The genomic data have also shown that C. trachomatis contains the enzymes of a simplified respiratory chain that is completely different compared with the mitochondrial chain. The chlamydial oxidative phosphorylation system consists of the sodium-dependent NADH dehydrogenase (Na+-NQR), succinate dehydrogenase, cytochrome bd oxidase, and an A1–A0-ATPase (12, 24, 25). Furthermore, C. trachomatis seems to use menaquinone (26), instead of ubiquinone, which is used by mitochondria (27). Na+–NQR is a unique respiratory complex that is analogous to the mitochondrial complex I, incorporating the electrons from NADH into ubiquinone, feeding the lower part of the respiratory chain (28). However, in contrast with complex I, which is a H+ pump, Na+–NQR specifically pumps sodium across the plasma membrane, producing a sodium gradient (28–30). This gradient is critical in the physiology of other pathogenic bacteria, driving many homeostatic processes, such as nutrient transport and pH regulation (31, 32). Moreover, it is used to support the efflux of drugs in antibiotic-resistant strains (31). Remarkably, Na+–NQR is the only sodium pump found in the C. trachomatis genome (12), and the sodium gradient produced by this enzyme might be linked directly to the synthesis of ATP, through the A1–A0-ATPase. The A1–A0-ATPase is closely related to the V1–V0-ATPase but seems to fulfill the same role as the F1–F0-ATP synthase (33–36). In bacterial species such as Enterococcus hirae, Streptococcus pyogenes, and Treponema pallidum, this enzyme is able to drive ATP synthesis using the sodium gradient (29). Although a detailed biochemical characterization of this complex has not been carried out, subunit K of the chlamydial A1–A0-ATPase contains a sodium-binding motif (37, 38). Hence, the evidence suggests that C. trachomatis could use a sodium-linked respiratory chain to produce ATP.
Despite all the information obtained from genomic data, most of these hypotheses have not been corroborated experimentally at the protein/enzyme, metabolic, or cellular levels. Moreover, the role of these pathways in different stages of the infection cycle has not been elucidated. Some of the technical difficulties that have limited the studies on C. trachomatis include the fragility of the bacterial cells and the contamination of EB and RB preparations with host-cell mitochondria, due to their similar density (22). Moreover, the lack of a cell-free culture system for Chlamydiae cultivation also obstructs the study of energy metabolism. Although a glucose 6-phosphate-based host cell-free medium has been recently described to cultivate C. trachomatis (39), the metabolic state of the cells under this condition might not represent those found in the intracellular milieu. Nevertheless, important aspects of Chlamydia metabolism have been elucidated in preparations of isolated EBs and RBs. For example, Chlamydia psittaci's RBs show an active ATP-ADP exchange mechanism and ATP-dependent lysine transport (16). Moreover, exogenously added ATP can sustain protein synthesis by C. psittaci and C. trachomatis RBs (39, 40). In contrast, protein synthesis by EBs is stimulated specifically by glucose 6-phosphate (39). In addition, it has been shown that several glycolytic enzymes are active in purified C. trachomatis RBs (19).
In this work, we have addressed two critical questions regarding Chlamydiae biology, the functionality of the respiratory metabolism and the energetic relationship with the host cell. The respiratory activity of C. trachomatis was studied in situ, in intact infected HeLa cells, and in permeabilized infected cells. Our data show, for the first time, that C. trachomatis is able to sustain an active oxidative metabolism that is resistant to the inhibitors of the mitochondrial respiratory chain but sensitive to HQNO (2-heptyl-4-hydroxyquinoline-N-oxide), an inhibitor of Na+–NQR (41–45). In addition, we show that the chlamydial respiratory activity, in digitonin-permeabilized infected HeLa cells, is stimulated by ADP, supporting the operation of an active oxidative phosphorylation by this bacterium. To study C. trachomatis energy dependence of the host cells, in particular whether or not the respiratory chain is functional and can actively produce ATP, infected HeLa cells were treated with mitochondrial or chlamydial respiratory inhibitors and ionophores to disrupt proton and sodium gradients. The treatments were made at two stages of the life cycle, immediately after infection (1 hpi) and when the EBs are differentiated into RBs and start division (12 hpi) (11, 21, 23). Treatment with oligomycin A, which inhibits the mitochondrial ATP synthesis (46), at an early stage of infection almost completely abolished the infection rate, stopped inclusion development, and decreased the protein content of C. trachomatis. At 12 hpi, the inhibitory effects were reduced, and only a decrease in the inclusion size was noticed. The data indicate that the EBs have a strong dependence on the mitochondrially-produced ATP, especially during the early stages of the infection. The role of C. trachomatis respiratory chain in infection and growth was studied by testing the effects of HQNO. At 1 and 12 hpi, HQNO decreased the chlamydial protein level and inclusion size but not the infection rate. This indicates that Na+–NQR is particularly important to sustain the aerobic metabolism required for RB growth. Moreover, the Na+/H+ exchanger monensin (47, 48) was used to determine whether a Na+ gradient could be used by C. trachomatis to energize its membrane to support physiological processes. A drastic inhibition of infection, inclusion size, and chlamydial protein content was observed in the presence of this drug. Taken together, these results indicate that C. trachomatis generates a Na+ gradient to energize its membrane, which is essential for its infection and growth and that its energy dependence on the host cell is only partial.
Results
Respiratory activity of C. trachomatis–infected HeLa cells
To elucidate the ability of C. trachomatis to synthesize its own ATP, and to clarify whether C. trachomatis expresses the components of a functional respiratory chain, as suggested by the genomic data, the respiratory activities of intact non-infected and C. trachomatis–infected HeLa cells were measured with a Clark-type oxygen electrode (49). The respiratory activities were measured in 24-hpi–infected cells, which carry mostly RBs (10), in the presence of inhibitors of the mitochondrial oxidative phosphorylation system. The inhibitors tested were as follows: 1) rotenone (1 μm), which inhibits complex I (Ki = 4 nm) (50, 51); 2) antimycin A (1 μm), which inhibits complex III (Ki = 38 nm) (52); 3) KCN (1 mm), an inhibitor of complex IV (Ki = 0.2 μm) (53); and 4) oligomycin A (1 μm), which targets the F0 subunit of F1–F0-ATP synthase (54, 55) and inhibits mitochondrial ATP synthesis (Ki = 0.1 μm) (56). The experiments were also carried out in the presence of HQNO, which at low concentrations (1–2 μm) can inhibit specifically Na+–NQR (Ki = 0.3–0.5 μm) (41–45). However, at high concentrations (>20 μm) it can also inhibit the mitochondrial complexes I, II, and III (57–59). Moreover, the respiratory activity was measured in the presence of the protonophore CCCP (1 μm) and the Na+/H+ exchanger monensin (1 μm) (60, 61).
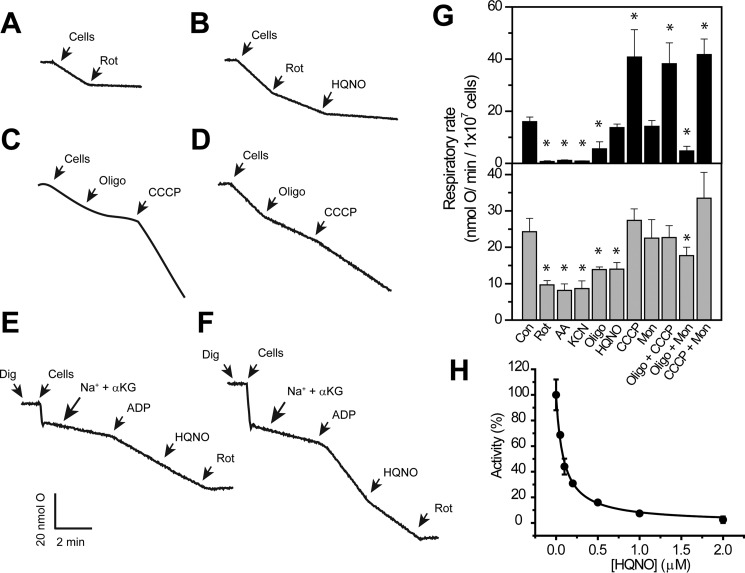
Oxygen consumption by non-infected HeLa cells follows the expected behavior for human cells, with >90% inhibition by all mitochondrial inhibitors (Fig. 1, A, C, and G) and an stimulatory effect of CCCP, which uncouples the respiratory chain and accelerates the rate of oxygen consumption, while inhibiting ATP synthesis (61). However, the respiratory activity of infected cells showed approximately a 40% resistance to all mitochondrial respiratory chain inhibitors (Fig. 1, B, D, and G), which is likely attributed to the oxygen consumption by intact chlamydial RBs. As shown in Fig. 1B, the rotenone-insensitive (non-mitochondrial) oxygen consumption is inhibited by a low concentration (1 μm) of the Na+–NQR inhibitor HQNO. To corroborate that HQNO acts specifically on chlamydial respiration, a titration of the rotenone-insensitive activity was performed (Fig. 1H). An inhibition constant (Ki) of 0.1 ± 0.03 μm was obtained, which is nearly identical to the Ki values obtained in other Na+–NQR complexes (41–45). These results show that C. trachomatis RBs have a highly active aerobic metabolism that might sustain endogenous ATP synthesis, corroborating the previous hypothesis. To clarify the functionality of C. trachomatis' oxidative phosphorylation system, experiments were carried out in permeabilized cells. Infected and non-infected HeLa cells were harvested and permeabilized with different concentrations of digitonin, and the respiratory activity was tested to find the optimal concentration. The concentration of digitonin used in this study (20 μg/ml/5 × 106 cells) allowed a complete permeabilization of the plasma membrane while keeping the mitochondrial and chlamydial membranes intact, judged by the stimulation of the respiratory activity with respiratory substrates. As shown in Fig. 1E, the respiratory activity of non-infected permeabilized cells with α-ketoglutarate is stimulated 3–4 times with ADP, indicating tightly coupled mitochondria. This activity is insensitive to HQNO and is completely inhibited by rotenone. However, the respiratory activity of infected cells is 40–50% more active with α-ketoglutarate, which is predicted as the main respiratory substrate of C. trachomatis (12, 22, 62). This activity is inhibited (50–60%) by HQNO, indicating that it is the result of chlamydial activity, and remarkably it is stimulated by ADP (Fig. 1F), showing state 3/state 4-like transitions, resembling mitochondria. This result corroborates the effects of monensin and CCCP over the respiratory activity of infected cells. Although monensin does not stimulate on its own the respiratory activity, the addition of CCCP and monensin increase the respiratory activity, indicating that it is coupled to energy production processes (Fig. 1G). These results strongly support that ATP synthesis through oxidative phosphorylation in C. trachomatis is indeed functional.
Figure 1.
Respiratory activities of non-infected and C. trachomatis–infected HeLa cells in the presence of respiratory inhibitors and ionophores. Representative traces of oxygen consumption of non-infected (A and C) and infected (B and D) intact HeLa cells in the presence of 1 μm HQNO, rotenone (Rot), oligomycin A (Oligo), or CCCP. Respiratory activities of non-infected (E) and infected (F) digitonin-permeabilized cells were in the presence of NaCl (20 mm), α-ketoglutarate (α-KG, 10 mm), and ADP (0.5 mm). G, oxygen consumption rates of non-infected (upper panel) and infected (lower panel) HeLa cells in the presence of 1 mm KCN or 1 μm of each of the following uncouplers or inhibitors: rotenone, antimycin A (AA), oligomycin A, CCCP, monensin (Mon), and HQNO. H, HQNO titration of rotenone-insensitive respiration of intact infected cells. *, p < 0.05 versus vehicle-treated control (Con).
Mitochondrial membrane energization in infected and non-infected cells
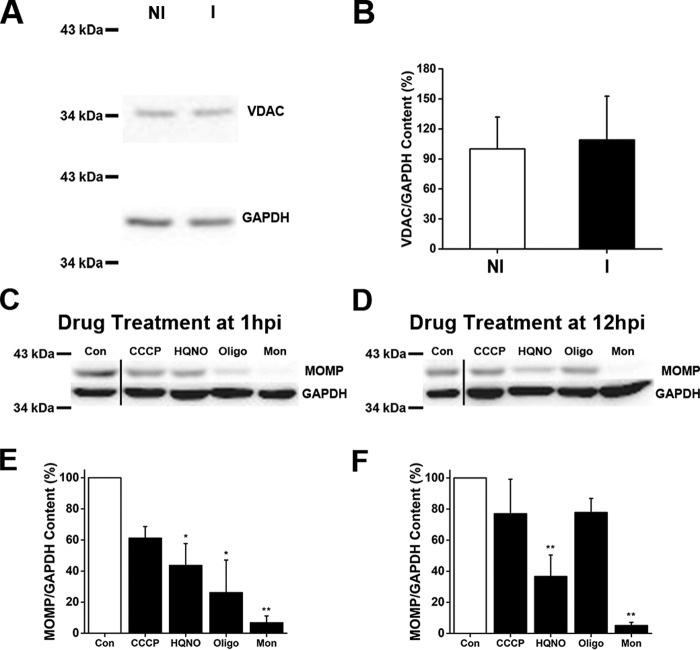
Our results show that the mitochondrial activity in infected cells is only partially sensitive to oligomycin A, and it is not stimulated by CCCP, in contrast with non-infected cells (Fig. 1, C and D), suggesting that the mitochondria from infected cells might not be coupled or might not sustain a large membrane potential. To address this question, experiments were performed using the fluorescent probe JC-1, which has been used widely to study mitochondrial membrane energization (63). Control cells exhibit JC-1 typical behavior, staining the mitochondrial network as bright red spots with a homogeneous cytosolic green fluorescence background (Fig. 2, A–C). Our results show that infected cells have a reduction of 50–65% in bright (highly energized) mitochondria compared with non-infected cells (Fig. 2), which could account for the small respiratory stimulation obtained by CCCP addition. In both infected and non-infected cells, the mitochondrial membrane potential (red JC-1 fluorescence) was collapsed by CCCP addition (Fig. 2A) and was resistant to both HQNO (Fig. 2B) and monensin (Fig. 2C). Western blot experiments using anti-VDAC (a typical marker of mitochondrial content (64)) antibodies showed no difference between infected and non-infected cells (Fig. 3, A and B). Thus, our data indicate that a smaller fraction of the mitochondria are capable of sustaining a significant membrane potential in infected cells. It is possible that the role of the mitochondria in these cells might not be limited to the synthesis of ATP. It should be pointed out that chlamydial membrane potential could not be studied using this probe, because the inclusion was not loaded after 30 min (or 1 h, not shown), appearing as empty areas devoid of green fluorescent signal (Fig. 2). This might be due to the specific membrane composition of the inclusion or to transporters that could eliminate the dye.
Figure 2.
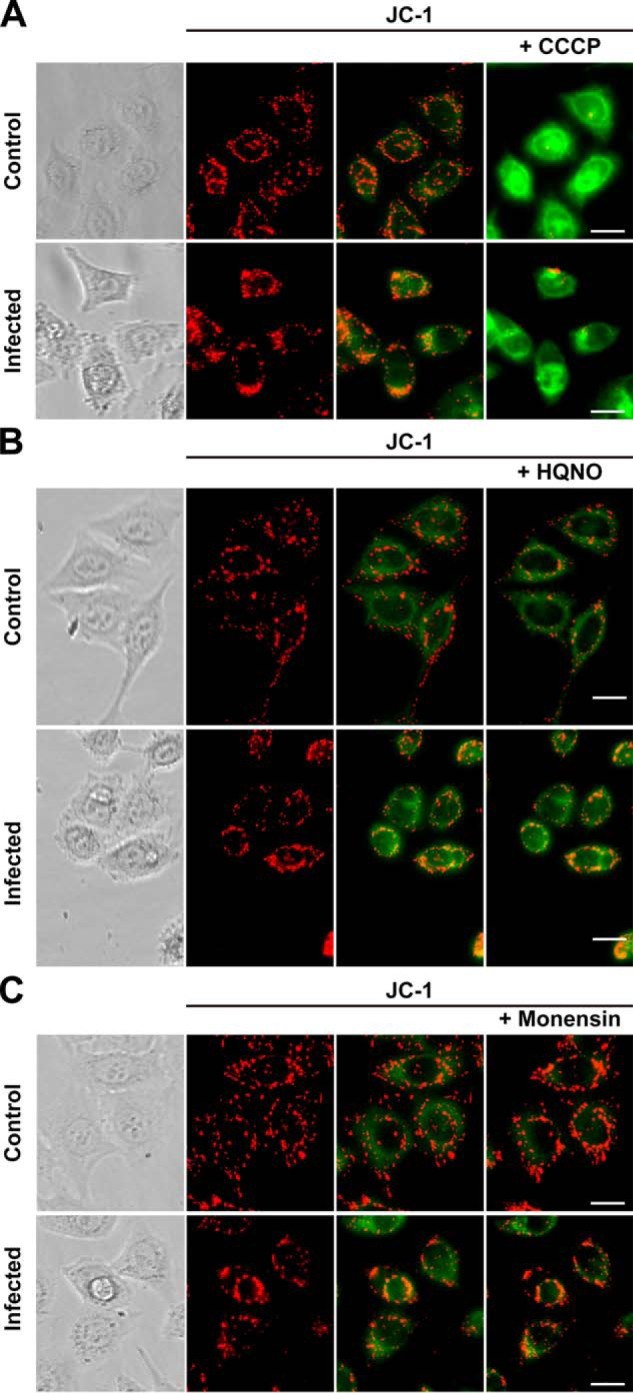
Effects of respiratory chain inhibitors or ionophores on mitochondrial membrane potential in Chlamydia-infected HeLa cells. The bright field images show non-infected cells (Control) or Chlamydia-infected cells (Infected). The mitochondrial membrane potential indicator JC-1 was used and visualized in red fluorescence (JC-1, left). The red/green fluorescent images (JC-1, middle) before and after the addition of the inhibitors (JC-1, right) are shown. A, CCCP (2 μm); B, HQNO (1 μm); and C, monensin (2 μm). Scale bar, 20 μm.
Figure 3.
Mitochondrial content and chlamydial major outer membrane protein content in Chlamydia-infected HeLa cells. A, mitochondrial content of non-infected HeLa cells (NI) and Chlamydia-infected HeLa cells (I) were analyzed by Western blot analysis, using antibodies against VDAC on mitochondrial outer membrane and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the loading control. B, protein levels of VDAC were normalized to levels of GAPDH. The plot shows the percentage of the VDAC/GAPDH ratio. Results were expressed as mean ± S.D., using three independent experiments. C and D, cell cultures were treated with respiratory chain inhibitors or ionophores at 1 hpi (C) or 12 hpi (D), and protein levels were analyzed by Western blotting using an anti-chlamydial MOMP antibody. GAPDH is shown as a loading control. E and F, graphical display of MOMP/GAPDH content expressed as ratio of MOMP/GAPDH band intensity percentage at 1 hpi (E) or 12 hpi (F) treatment with respect to the control (Con). n = 3–4. *, p < 0.05; **, p < 0.005. The vertical line between control and CCCP bands represents a splice junction (C and D).
Effect of respiratory chain inhibitors on the infectivity and growth of C. trachomatis
To characterize the host-cell energy contribution to chlamydial metabolism, a pharmacological approach was used, measuring the effects of the inhibition of the bacterial and mitochondrial respiratory chain over the infection rate and chlamydial replication (estimated as the chlamydial inclusion size), and the chlamydial protein content, by Western blot analysis against the chlamydial major outer membrane protein (MOMP) (65).
HeLa cells were inoculated with C. trachomatis EBs and treated with different inhibitors. The experiments were performed at two critical time points in the developmental cycle of the bacteria: at an early-stage (1 hpi), when Chlamydia cells are in the form of EBs, and at mid-stage infection (12 hpi), characterized by the differentiation of EBs into RBs (11). Inhibitor-treated cell cultures were fixed and stained at 36 hpi by HEMA 3 staining, to identify chlamydial inclusions in the host cells. Additionally, results obtained were corroborated by immunostaining, because some of the drugs used in this study decreased dramatically the size of the inclusion, resulting in possible underestimation of the infection rate by HEMA 3 (66). Immunofluorescence experiments were carried out with anti-MOMP antibodies that specifically recognize C. trachomatis MOMP (67, 68). MOMP is a membrane protein highly expressed in both EBs and RBs that provides structural support and regulates the permeability of the chlamydial cell membrane (65, 69). The inhibitors used in this part of the study were oligomycin A and HQNO, which allowed us to understand the role of the mitochondrial ATP synthesis and the chlamydial respiratory chain on infection and growth. Other respiratory inhibitors, such as rotenone and antimycin A, were also tested but proved too toxic for HeLa cells, and thus were not included in this study. Although a relatively high concentration of HQNO and oligomycin A was used (10 μm), their final “free” concentration could be significantly smaller, due to the ability of albumin (found in the fetal bovine serum) to bind highly hydrophobic molecules, especially because oligomycin and HQNO both contain the reported chemical moieties required for albumin binding (70, 71). Because of the uncertainty in the final effective inhibitor concentration reached in these assays, titrations were carried out. As shown in Fig. S1, the inhibitor concentrations used in this study provide maximal effects over chlamydial infection, with little to no toxicity toward non-infected control cells (Fig. S2).
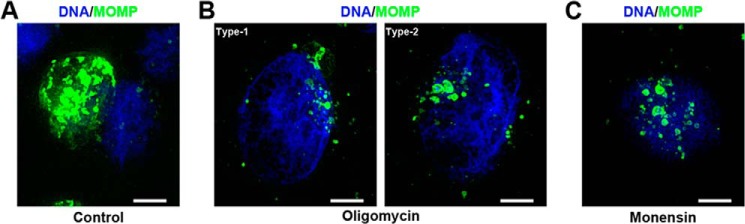
The treatment with the inhibitors had different effects depending on the time at which the inhibitor was added to the culture. At 1 hpi, oligomycin A produced a significant decrease of 76–84% in the chlamydial inclusion size (Fig. 4, A and C, and Fig. S3), and a 69% decrease in the infection rate (Fig. 4, A and D, and Fig. S3). This decrease was only evident with HEMA staining, and the immunostaining showed no effects. Immunostained cells were analyzed with the ×100 microscope objective lens (Fig. 5, A and B), and we discovered that the cells were indeed infected, but the chlamydial inclusion was not fully formed, thus explaining the discrepancy in the results with the two types of staining. At 12 hpi the inclusion size was reduced only by 39–42% (Fig. 4, B and E, and Fig. S4), and no effects were found in the rate of infection (Fig. 4, B and F, and Fig. S4). These results clearly indicate that the ATP production by the host-cell mitochondria supports the early stages of the infection process by the EBs.
Figure 4.
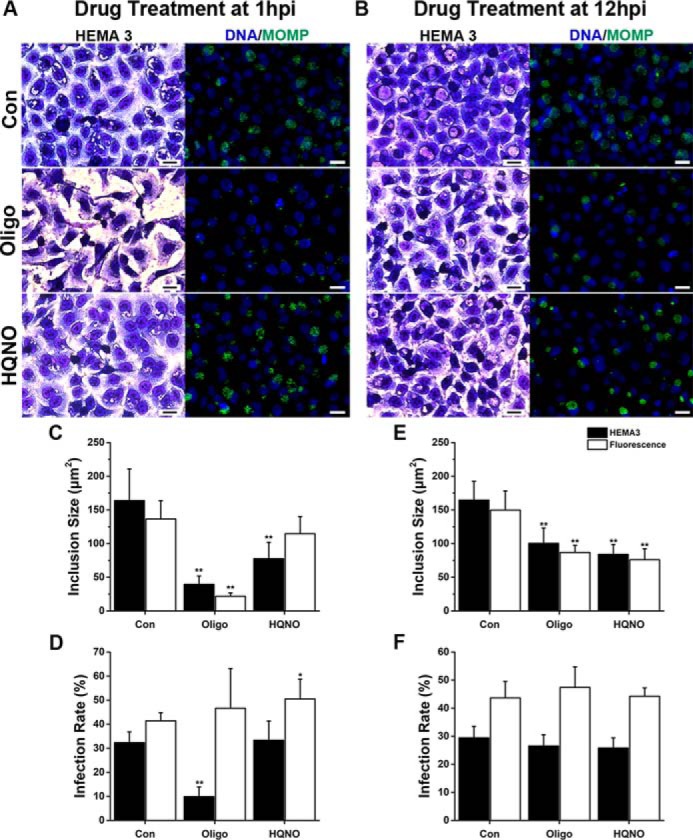
Effect of respiratory chain inhibitors on chlamydial infection in cell culture. A and B, representative images of C. trachomatis–infected HeLa cultures treated with 10 μm oligomycin A (Oligo) or 10 μm HQNO, at 1 or 12 hpi, and stained with HEMA 3 staining or immunofluorescence with anti-MOMP antibodies (green) at 36 hpi. In the fluorescent images, DNA is visualized with Hoechst 33342 (blue). Scale bars represent 20 μm. C and E, size of the chlamydial inclusion was quantified by measuring the area of the inclusions with the inhibitors added at 1 hpi (C) or 12 hpi (E). 110–340 inclusions were measured per condition per experiment using ImageJ. D and F, chlamydial infection rate was determined by quantifying the percentage of inclusion-positive cells with the inhibitors added at 1 hpi (D) or 12 hpi (F). More than 200 cells were quantified per condition per sample. Results were collected from 6 to 8 separate experiments and are expressed as mean ± S.D. Asterisks denote the significance from the vehicle-treated control (Con). *, p < 0.05; **, p < 0.005.
Figure 5.
Detail of Chlamydia-disrupted inclusion under oligomycin A or monensin treatment at 1 hpi. Representative images under ×100 objective lens of C. trachomatis–infected HeLa cells immunostained with anti-MOMP antibodies (green) at 36 hpi. A, control cells (vehicle-treated). B, 10 μm oligomycin A treatment. In type 1, small inclusions are present, and a main inclusion is still formed or it is reminiscent. In type 2, only small chlamydial inclusions are observed. C, 2 μm monensin treatment. Individual chlamydial inclusions are not fused in a single inclusion or it is disrupted. DNA is visualized with Hoechst 33342 (blue). Scale bar, 5 μm. Images are full focus images created after acquisition with BZ-X software merging 0.1 μm Z-stacks. Haze reduction was applied to optimize inclusion appearance.
The addition of HQNO to the cell cultures at 1 hpi did not reduce the infection rate (Fig. 4, A and D). However, as in the previous case, HEMA staining showed a decrease of 53% in the inclusion size (Fig. 4, A and C, and Fig. S3), which was not observed by the more sensitive immunostaining method. At 12 hpi, the inclusion size decreased by 49% (Fig. 4, B and E, and Fig. S4), and no effects on the infection rate were evident (Fig. 4, B and F, and Fig. S4). The effects of HQNO on the inclusion size (an indicator of chlamydial replication) suggest that Na+–NQR, the first complex of the respiratory chain (43, 72), is essential for the growth of C. trachomatis RBs, allowing the cells to produce its own ATP, but it has no important role early in the infection process.
Effect of ionophores on the infection and growth of C. trachomatis
CCCP is a proton ionophore that dissipates the H+ gradient, producing mitochondrial uncoupling that hinders the production of ATP (73). Our results with the staining techniques indicate that the infection rate decreased 22–39% after the addition of CCCP at 1 hpi (Fig. 6, A and C, and Fig. S5). However, the inclusion size remained unchanged (Fig. 6, A and D, and Fig. S5). The infection rate and inclusion size were not modified by the treatment with CCCP at 12 hpi (Fig. 6, B, E and F, and Fig. S6). Although the general behavior corroborates that C. trachomatis requires a functional mitochondrial metabolism early on, the effects of CCCP seemed attenuated compared with oligomycin A. This suggests that the concentration of CCCP that was used might not be enough to fully uncouple cell mitochondria in culture. Higher concentrations of CCCP (5 and 10 μm) were tested but proved too toxic for HeLa cells (data not shown).
Figure 6.
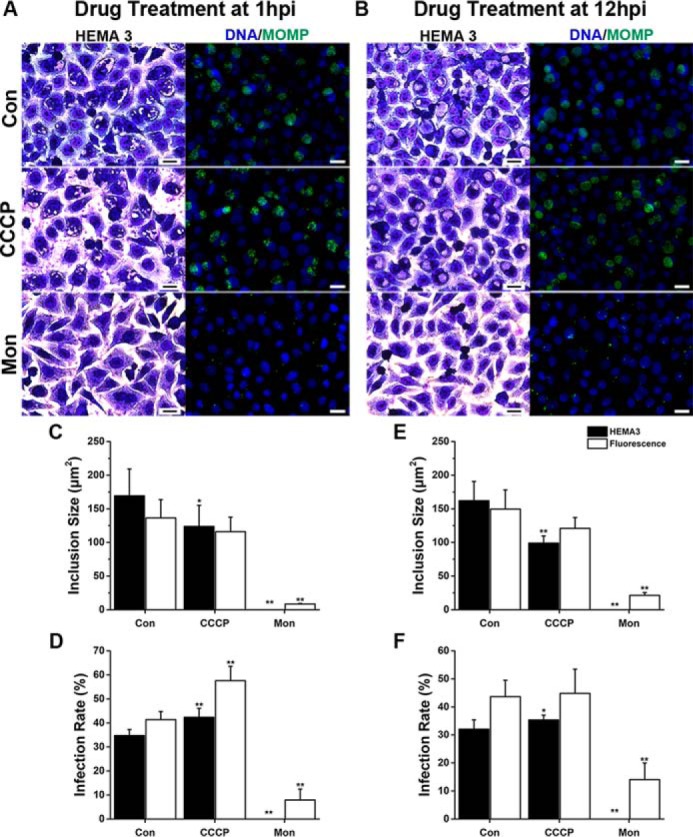
Effect of ionophores on chlamydial infection in cell culture. A and B, HeLa cells infected with C. trachomatis were treated with 2 μm monensin (Mon) or CCCP at 1 or 12 hpi and stained with HEMA 3 staining or immunofluorescence using anti-chlamydial antibodies (green) at 36 hpi. Hoechst was used for DNA labeling (blue in fluorescent images). Scale bars, 20 μm. C and E, area of the chlamydial inclusion was measured in >110 inclusions per condition, in six separate experiments using ImageJ. D and F, chlamydial infection rate represents the percentage of infected cells when treatment was applied at 1 hpi (D) or 12 hpi (F). >200 cells were quantified per condition of six to eight different experiments. Error bars represent standard deviation of the mean. Asterisks denote the significance from vehicle-treated control (Con). *, p < 0.05; **, p < 0.005.
As discussed previously, the inhibition of Na+–NQR has a significant effect over chlamydial development, and is particularly important in RB physiology, which indicates that the cells have an active aerobic metabolism that might be dependent on the sodium gradient. To assess this hypothesis, the effect of monensin was tested over chlamydial growth. Monensin acts as a Na+/H+ exchanger, which dissipates the Na+ gradient while maintaining the membrane potential (60), and thus it specifically acts on the Na+-driven processes. Remarkably, inclusions were not detected by HEMA 3 staining when 2 μm monensin was added to the infected cell culture, regardless of the time of addition (Fig. 6). Nonetheless, small inclusions were detected by immunofluorescence at 1 hpi (8.41 ± 0.94 μm2) (Figs. 5, A and C, and 6, A and C, and Fig. S5) and at 12 hpi (21.18 ± 2.89 μm2) (Fig. 6, B and E, and Fig. S6), representing a 94 and 86% reduction, respectively. The infection rate was also reduced by 81 and 68% when the ionophore was added at 1 and 12 hpi, respectively (Fig. 6, A, B, D, and F, and Figs. S5 and S6). These data show that the sodium gradient is crucial in the chlamydial infection process at an early stage, and its disruption eliminates the infection at middle stage of the infection, when the inclusions are already formed. This result highlights the paramount importance of sodium-energized membranes to sustain C. trachomatis infection process and growth. Taken together, the data suggest that a sodium gradient, produced by the Na+–NQR complex, is essential for the infection and growth of C. trachomatis, likely sustaining the ability to produce energy through the aerobic metabolism. Moreover, the sodium gradient produced by Na+–NQR can also be used by the cell to carry other essential homeostatic processes, such as pH regulation, through the Na+-H+ antiporter NhaD (74), as well as nutrient transport, which seems to be carried mostly by Na+-dependent transporters (12, 75).
Expression of major outer membrane protein of C. trachomatis
Chlamydial inclusion size is an indirect indicator of cell replication, growth, and maturity. In some instances, such as in the presence inhibitors of the glucose 6-phosphate transporter, which blocks cell replication entirely, a normal inclusion size is observed (68). The two staining techniques that we used provide semi-quantitative measurements of the inhibitor effects. To confirm that the change in inclusion size measured by immunofluorescence is accompanied by a corresponding change in chlamydial load, we determined whether the respiratory chain inhibitors and ionophores had an effect on chlamydial protein content, quantifying the levels of MOMP by Western blot analysis (Fig. 3, C–F). Oligomycin A decreased the content of MOMP (versus loading control GAPDH) by ∼74% at 1 hpi. MOMP/GAPDH content was also reduced in the presence of HQNO, by 56 and 63%, at 1 and 12 hpi, respectively. These results indicate that the MOMP content correlates with the immunofluorescence data, further confirming that the ATP production from the host cell sustains the chlamydial growth at an early stage of the developmental cycle, which switches to a high energy production by the chlamydial cells when the EBs are differentiated to RBs. Monensin dramatically reduces the content of MOMP/GAPDH (95% approximately) regardless of the time of addition, confirming the staining data. These data further substantiate the evidence indicating that the sodium gradient is important to produce the energy necessary for C. trachomatis infection and growth.
Discussion
C. trachomatis energy metabolism
C. trachomatis is an obligate intracellular pathogen that had been long considered an energy parasite, depending entirely on the host cell to fulfill its energy needs (17). Early studies with preparations obtained from infected hen eggs showed that C. psittaci did not contain flavoproteins or the types of cytochromes normally found in human mitochondria (14, 15), which led to the conclusion that Chlamydiae metabolism was inactive and depended completely on the host cell to fulfill its energy requirements. However, these experiments were carried out on EBs, which are not metabolically active. Moreover, it was later described that Chlamydiae express ATP-ADP antiporters, which allows the net transport of high-energy phosphate from the host cell (16), and it has been demonstrated that the host-free protein synthesis in RBs can be entirely supported by exogenous ATP (40), further supporting the energy parasite hypothesis.
The advent of the genomic data provided new tools to understand Chlamydiae biology. In particular, it was described that the genome of C. trachomatis encodes many enzymes of key energy metabolism pathways, including glycolysis, the Krebs cycle, and a simplified respiratory chain (12, 19, 20). However, it has remained unknown whether these pathways are actually functional. C. trachomatis genome contains all glycolysis genes except for hexokinase (12, 23, 25). Glucose 6-phosphate seems to be the physiological substrate of glycolysis, which is captured through the UhpC transporter (62, 76). Previous studies have shown that the glycolytic enzymes are indeed expressed by both RBs and EBs (23) and that their contents are higher in RBs (21, 23, 25). Although C. trachomatis could sustain its own energetic demand through glycolysis, it has not been experimentally tested whether the role of this pathway is the synthesis of ATP or whether it is used to synthesize glycogen (via gluconeogenesis) or other biosynthetic intermediates.
C. trachomatis respiratory chain is highly simplified, consisting of Na+–NQR, succinate dehydrogenase, cytochrome bd oxidase, and an A1–A0-ATPase (12, 24). These enzymes constitute a unique respiratory chain that could potentially produce ATP linked to the production of sodium gradient, because Na+–NQR is a primary sodium pump (77) and the A-type ATPase has been reported to catalyze ATP synthesis using the sodium gradient (35). Eukaryotic mitochondria and most types of bacteria contain a respiratory chain that is able to build a proton gradient across the membrane, which sustains ATP synthesis. This gradient is also used to energize other primary functions, such as nutrient transport and pH regulation (78). However, different types of pathogenic bacteria, such as γ-proteobacteria, bacteroidetes, and Chlamydiae (e.g. Vibrio cholerae (79), Klebsiella pneumoniae (80), Haemophilus influenzae (81), and Bacteroides fragilis (32), etc.) can substitute (or supplement) transmembrane proton gradients for sodium gradients and encode a variety of sodium pumps, including the Na+–NQR complex (24, 31, 32, 82, 83). Na+–NQR is a respiratory enzyme that catalyzes the transfer of electrons from NADH to ubiquinone and is the entry site of redox equivalents, produced by the primary and intermediary metabolism, into the respiratory chain (30, 43, 72, 84). Na+–NQR fulfills the same function as mitochondrial complex I, but in contrast with the latter, it acts as a sodium-specific ion pump (77, 86). The genome of C. trachomatis contains Na+–NQR, as part of the respiratory chain, and interestingly also contains an A1–A0-ATPase (12, 13, 22), which could carry a proton-independent sodium-driven ATP synthesis (12, 35). In this work, we demonstrate for the first time that the intact C. trachomatis RBs in situ have a very active oxidative metabolism, comparable with the mitochondrial activity. The chlamydial respiratory activity is insensitive to all mitochondrial inhibitors, but it is sensitive to low concentrations of HQNO. The high sensitivity toward this inhibitor and the low inhibition constant (sub-micromolar range) obtained indicates that Na+–NQR is indeed active in the RBs. Moreover, our results indicate that the sodium gradient is absolutely essential in the physiology of C. trachomatis, because monensin blocked completely the infection, growth, and protein expression of the bacteria. To corroborate this hypothesis, experiments were carried out in permeabilized cells, which allow a detailed examination of the metabolic properties of organelles (87–89) and in this case RBs. Experiments were performed using α-ketoglutarate, which is predicted as the main respiratory substrate for C. trachomatis (62). Our results show that the HQNO-sensitive respiratory activity is stimulated by ADP, closely resembling the state 3 to state 4 transitions of mitochondria, strongly suggesting that the respiratory activity of this pathogen is coupled to the synthesis of ATP. Thus, Na+–NQR is not only functional, but its sodium-pumping activity is critical in the physiology of the bacterial cell. Our data agree with previous reports indicating that Na+–NQR is expressed very early in the infection (1–3 hpi) (25). A recent report has also shown that C. trachomatis infectivity is drastically reduced by the treatment with a novel Na+–NQR inhibitor PEG-2 (90). Na+–NQR activity is not only important for energy production, and the gradient of sodium seems to fulfill many other important functions, including the transport of nutrients and other intermediate metabolites, which explains the lethal effect of monensin on bacterial infection. Indeed, C. trachomatis genome contains a variety of sodium-coupled amino acid transporters (12, 22, 91) and a sodium-dicarboxylate translocator (22) that may feed the incomplete chlamydial Krebs cycle with glutamate or α-ketoglutarate (62). It should be pointed out that it has been suggested that the main role of the A1–A0-ATPase could be the formation of the sodium membrane potential using the host's ATP as an energy source (35). However, the presence of a fully functional and highly active respiratory chain and the stimulation of the respiration by ADP indicate that the physiologic role of the A-type ATPase is the synthesis of ATP.
Effects of mitochondrial inhibitors and uncouplers
In this study, we explored the energy dependence of C. trachomatis and its ability to produce ATP endogenously, employing different respiratory inhibitors and ionophores to test their effect on chlamydial infection, growth, and protein expression, in a HeLa cell-based culture system.
The results found in this work offer the first clues to understand the functionality of Chlamydiae aerobic metabolism and its relationship with the host cell, and they indicate that C. trachomatis indeed requires ATP from the host cell to support its own growth, as originally proposed (17), but this relationship is dynamic. Our results demonstrate that the addition of oligomycin A at an early stage of infection produces a nearly complete inhibition of chlamydial growth, reducing the infection rate, inclusion size, and protein content by depriving EBs from the host-cell mitochondrial ATP. Thus, C. trachomatis EBs rely mostly on the mitochondrial ATP production during the early stages of the infection to carry out the chlamydial infection process (internalization, inclusion forming, and differentiation to RBs). However, the data indicate that there is a shift of energy dependence during the chlamydial developmental cycle. The addition of HQNO, which inhibits Na+–NQR and the entire chlamydial respiratory function, decreases the chlamydial inclusion size and chlamydial content when the EBs have been differentiated into RBs, suggesting that the RBs have the ability to produce their own ATP and that the energy demand is supplemented by the host cell. This is in agreement with the temporal expression of some of the genes involved in chlamydial energy metabolism. For instance, adt mRNA, encoding an ATP-ADP translocase, is detected all through the developmental cycle of C. trachomatis (25), but sdhA and pky transcripts, encoding succinate dehydrogenase and pyruvate kinase, respectively, are expressed at mid-stage of the developmental cycle (11). Moreover, a recent report has shown that the NADH and NADPH content increases steadily in the chlamydial inclusion from 12 to 24 hpi, being initially distributed at the inner border of the inclusion and later on localized homogeneously throughout the inclusion, indicating an active metabolism. Similarly, the free/protein-bound NAD(P)H ratio decreases in the same time frame (92) suggesting that C. trachomatis uptakes NAD(P)H from the host cell to support the activity of Na+–NQR, which is mostly active during mid-stage infection, corroborating our data.
It should be noted that the effects of HQNO were smaller compared to monensin, which may be due to a relatively low concentration of HQNO reaching C. trachomatis cells, due to the presence of albumin, or because both EBs and RBs are enclosed in an inclusion, which might decrease the permeability of this inhibitor. Alternatively, HQNO might not inhibit C. trachomatis Na+–NQR as effectively as V. cholerae Na+–NQR, considering that the ubiquinone-binding site (where HQNO is bound) has a 70% identity between these two bacteria (27). Also, the function of an Na+/H+ antiporter (NhaD) (74) might maintain, to a certain extent, the sodium gradient across the C. trachomatis membrane (24, 31, 93). The relevance of this antiporter is supported by the change in the pH in the chlamydial inclusion as the infection progresses, as it increases over time from pH 6.26 at 4 hpi to 6.60 (at 12 hpi) and finally 7.25 at 20 hpi (94, 95), indicating an active influx of H+ to the RBs.
To corroborate that the effects of the inhibitors were specific and were not due to toxicity, the viability of C. trachomatis-infected HeLa cells was evaluated in the presence of the inhibitors (Fig. S2). The data indicate that most of the inhibitors (at the concentrations used in this report) do not produce a decrease in cell viability, except for oligomycin A, which has a modest effect (24%). Even though some toxicity over the host cell was evident (and expected), this parameter does not correlate with the effects over chlamydial growth, i.e. monensin has no toxicity over the HeLa cells and abolishes completely C. trachomatis growth. Thus, the pharmacological strategy used in this study seems suitable to characterize chlamydial energetics.
Energetic relationship of C. trachomatis with the host
The main findings of this work are summarized in Fig. 7. During the initial phase of the infection, the EBs mostly depend on host-cell energy, but at mid-stage of the developmental cycle characterized by the presence of RBs, they switch and depend to a limited extent on the host cells, because RBs are able to produce their own energy. A Na+ gradient is fundamental in the chlamydial infection process and growth. The treatment with monensin produced the biggest reductions on infection rate, inclusion size, and protein synthesis, indicating that a Na+ gradient is paramount as an energy source for C. trachomatis and is necessary in both the early and mid-stage developmental cycle. Therefore, a sodium-based metabolism of C. trachomatis could be a new target for drug design. Multidrug-resistant C. trachomatis strains have been reported to all antibiotics recommended for treatment (96, 97) and even to the alternative treatments, including macrolides (98, 99) and fluoroquinolones (100, 101). Moreover, C. trachomatis can spread multidrug resistance through horizontal transfer (102). Therefore, there is an urgent need to understand chlamydial metabolism and identify new suitable targets to treat infections.
Figure 7.
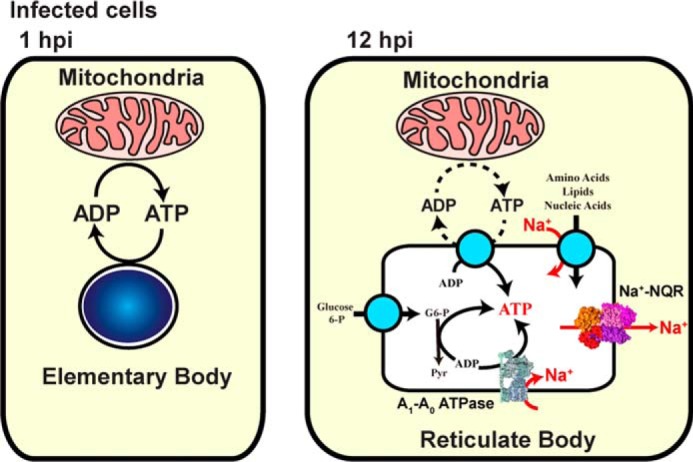
Scheme of Na+-based chlamydial energy metabolism.
Experimental procedures
Cell culture
The human cervical cancer cell line HeLa 229 was maintained in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin 100 units/ml and 100 μg/ml) at 37 °C in a humidified atmosphere (5% CO2 and 95% air). Confluent cultures were treated with 0.05% trypsin/EDTA for 3 min at 37 °C. Trypsin was inactivated with culture media containing 10% FBS. Cells were plated at the indicated densities and fed every 2 days.
Purification of Chlamydia trachomatis elementary bodies
Elementary bodies of C. trachomatis (serovar L2b) were prepared according to Scidmore (103). Briefly, HeLa cell monolayers, 6.6 × 104 cells/cm2, were infected with Chlamydia trachomatis EBs at a multiplicity of infection of 10 in the absence of antibiotics. The infected cells were collected at 44 hpi and were lysed in Hanks' balanced salt solution (HBSS), passing the suspension through a 18-gauge metal cannula attached to a sterile syringe. The sample was cleared at low speed centrifugation (500 × g) for 15 min at 4 °C. Subsequently, the EBs were pelleted by centrifugation at 30,000 × g for 30 min and resuspended in sucrose/phosphate/glutamate buffer (SPG) (103).
Oxymetric measurements
After 1 day in vitro, HeLa cells grown on 10-cm dishes in the absence of antibiotics at a density of 6.6 × 104 cells/cm2 were infected with C. trachomatis EBs, using 0.3–0.35 infection-forming units (IFU) per cell. The respiratory activity was measured in non-infected and C. trachomatis infected HeLa cells (24 hpi), using a Clark-type oxygen electrode (YSI Inc.), in a custom-made glass chamber of 2 ml. HeLa cells were harvested by trypsinization, washed, and resuspended in HBSS. The respiratory activity of intact cells was measured in HBSS buffer at 37 °C. In addition, the respiratory activity was measured in digitonin-permeabilized cells. Harvested cells were pelleted, and washed twice with KHE buffer (150 mm KCl, 50 mm HEPES, 1 mm EDTA, pH 7.5). Permeabilization was performed in the oxymetric chamber using the same buffer, by adding 20 μg/ml/5 × 106 cells of digitonin. The inhibitors and ionophores used in these experiments were 1 μm rotenone, 1 μm HQNO, 1 μm antimycin A, 1 mm KCN, 1 μm oligomycin A, 1 μm CCCP, or 1 μm monensin.
Membrane potential analysis
Mitochondrial membrane potential formation was determined using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1). Non-infected and C. trachomatis–infected HeLa cells were incubated with 5 μm JC-1 in HBSS buffer at 36 hpi for 30 min. Non-internalized dye was washed thoroughly, and live cell imaging was performed using Keyence BZ-X710 fluorescence microscope, and images were collected with a cooled monochrome CCD camera and BZ-X viewer software (version 01.03.00.05; Keyence). The objective lens were Nikon Plan Fluor ELWD 40X/0.60 Ph2 DM, ∞/0–2 WD 3.7–2.7 lens.
HEMA 3 staining and imaging
HeLa cells grown on glass coverslips in the absence of antibiotics at a density of 4 × 104 cells/cm2 were infected with C. trachomatis EBs, using 0.3 IFU/cell. At 1 or 12 hpi, EMEM culture media (supplemented with 10% FBS) with oligomycin A, HQNO, CCCP, or monensin was added to the HeLa cells cultures. Infected and non-infected HeLa cells were fixed at 36 hpi and stained with the differential staining HEMA 3 staining kit (Fisher), which has been used to identify the intracellular chlamydial vesicles (104, 105). Cells were visualized with a Leica DM IL LED Fluo microscope, and images were collected with a DFC450 C camera and Leica Application Suite 4.4.0 software. The objective lens used were Leica N PLAN L ×40/0.55, CORR Ph2, ∞/0–2/C lens. To optimize the intensity ranges of the images, contrast and brightness were adjusted through linear level adjustments, as needed, using Adobe Photoshop CS5.1 (Adobe Systems).
Immunohistochemistry and imaging
Non-infected and C. trachomatis–infected HeLa cell cultures were fixed with methanol at −20 °C for 10 min and washed with phosphate-buffered saline (PBS, pH 7.4). The cells were permeabilized with 0.04% Triton X-100 in PBS (PBS-TX) (75) for 15 min at room temperature and blocked with 10% normal donkey serum in PBS-TX for 1 h. Primary antibodies, anti-MOMP (1: 1,000, Pierce), were diluted in 1% normal donkey serum/PBS-TX and incubated overnight at 4 °C. Secondary antibodies, donkey anti-goat Alexa Fluor 488 (1:500, Life Technologies, Inc.) were incubated 1 h at room temperature, followed by Hoechst 33342 DNA staining (1 μg/ml). Coverslips were mounted with Fluoromount-G (anti-fade) solution (Southern Biotech). Immunofluorescent images were visualized with a Keyence BZ-X710 fluorescence microscope, and were collected with a cooled monochrome CCD camera and BZ-X viewer software (as described above). The objective lens were Nikon Plan Fluor ELWD ×40/0.60 Ph2 DM, ∞/0–2 WD 3.7–2.7 lens and Nikon Plan Apo λ ×100/1.45 oil, ∞/0.17 WD 0.13 lens. All images comparing non-infected and infected cells were acquired with the same exposure times and analyzed with BZ-X Analyzer software (version 1.3.05; Keyence). Contrast and brightness of images were adjusted through linear level adjustments, as needed, to optimize the intensity ranges of the images using Adobe Photoshop CS5.1.
Immunoblotting
Whole-cell lysates from infected and non-infected HeLa were prepared in lysis buffer containing 50 mm Tris, 1 mm EDTA, 0.1% SDS, and 1 mm PMSF, pH 7.4. Protein samples were run in a 15% SDS-PAGE and electroblotted onto a polyvinylidine fluoride membrane. Blots were blocked with 5% non-fat dry milk in TBS-T (150 mm NaCl, 20 mm Tris, 0.01% Triton X-100, pH 7.6) for 1 h at room temperature and incubated at 4 °C overnight with anti-MOMP, anti-GAPDH (Pierce, 1:50,000; 1:10,000), and anti-VDAC, Cell Signaling, 1:2000). Horseradish-peroxidase-conjugated secondary antibodies were incubated for 1 h at room temperature and detected with the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Chemiluminescence signals were detected and acquired by an Omega Lum G gel imaging system and acquisition software (version 1.0; Aplegen). Band intensities were quantified using ImageJ (85).
Author contributions
P. L., D. P., M. R. L., and K. T. performed the experiments. K. T., P. L., M. R. L., and O. J. designed the experiments. K. T., P. L., M. R. L., X. F., and O. J. analyzed the data and wrote the manuscript. All authors contributed to the final review of the manuscript.
Supplementary Material
Acknowledgments
We thank Ana Kaizer and Izabella Melo for their valuable contributions in different parts of this study. We thank Daniel A. Raba for critically reading the manuscript.
This work was supported in part by IIT startup funds (to O. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- EB
- elementary body
- RB
- reticulate body
- MOMP
- major outer membrane protein
- CCCP
- carbonyl cyanide 3-chlorophenylhydrazone
- HQNO
- 2-n-heptyl-4-hydroxyquinoline N-oxide
- Na+–NQR
- sodium-dependent NADH dehydrogenase
- hpi
- hours post-infection
- VDAC
- voltage-dependent anion channel
- HBSS
- Hanks' balanced salt solution
- IFU
- infection-forming unit.
References
- 1. Taylor H. R., Burton M. J., Haddad D., West S., and Wright H. (2014) Trachoma. Lancet 384, 2142–2152 10.1016/S0140-6736(13)62182-0 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization. (2012) Global incidence and prevalence of selected curable sexually transmitted infections–2008, World Health Organization, Geneva, Switzerland [Google Scholar]
- 3. Newman L., Rowley J., Vander Hoorn S., Wijesooriya N. S., Unemo M., Low N., Stevens G., Gottlieb S., Kiarie J., and Temmerman M. (2015) Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on Systematic Review and Global Reporting. PLoS ONE 10, e0143304 10.1371/journal.pone.0143304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malhotra M., Sood S., Mukherjee A., Muralidhar S., and Bala M. (2013) Genital Chlamydia trachomatis: an update. Indian J. Med. Res. 138, 303–316 [PMC free article] [PubMed] [Google Scholar]
- 5. Haggerty C. L., Gottlieb S. L., Taylor B. D., Low N., Xu F., and Ness R. B. (2010) Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 201, (suppl.) 134–155 10.1086/652395 [DOI] [PubMed] [Google Scholar]
- 6. Fleming D. T., and Wasserheit J. N. (1999) From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 75, 3–17 10.1136/sti.75.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Owusu-Edusei K. Jr., Chesson H. W., Gift T. L., Tao G., Mahajan R., Ocfemia M. C., and Kent C. K. (2013) The estimated direct medical cost of selected sexually transmitted infections in the United States, 2008. Sex. Transm. Dis. 40, 197–201 10.1097/OLQ.0b013e318285c6d2 [DOI] [PubMed] [Google Scholar]
- 8. Hammerschlag M. R. (2002) The intracellular life of chlamydiae. Semin. Pediatr. Infect. Dis. 13, 239–248 10.1053/spid.2002.127201 [DOI] [PubMed] [Google Scholar]
- 9. Abdelrahman Y. M., and Belland R. J. (2005) The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959 10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 10. Elwell C., Mirrashidi K., and Engel J. (2016) Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400 10.1038/nrmicro.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw E. I., Dooley C. A., Fischer E. R., Scidmore M. A., Fields K. A., and Hackstadt T. (2000) Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37, 913–925 10.1046/j.1365-2958.2000.02057.x [DOI] [PubMed] [Google Scholar]
- 12. Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., and Davis R. W. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 10.1126/science.282.5389.754 [DOI] [PubMed] [Google Scholar]
- 13. Vandahl B. B., Birkelund S., and Christiansen G. (2004) Genome and proteome analysis of Chlamydia. Proteomics 4, 2831–2842 10.1002/pmic.200400940 [DOI] [PubMed] [Google Scholar]
- 14. Allen E. G., and Bovarnick M. R. (1957) Association of reduced diphosphopyridine nucleotide cytochrome c reductase activity with meningopneumonitis virus. J. Exp. Med. 105, 539–547 10.1084/jem.105.6.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allen E. G., and Bovarnick M. R. (1962) Enzymatic activity associated with meningopneumonitis. Ann. N.Y. Acad. Sci. 98, 229–233 [DOI] [PubMed] [Google Scholar]
- 16. Hatch T. P., Al-Hossainy E., and Silverman J. A. (1982) Adenine nucleotide and lysine transport in Chlamydia psittaci. J. Bacteriol. 150, 662–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moulder J. W. (1991) Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55, 143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tjaden J., Winkler H. H., Schwöppe C., Van Der Laan M., Möhlmann T., and Neuhaus H. E. (1999) Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181, 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iliffe-Lee E. R., and McClarty G. (1999) Glucose metabolism in Chlamydia trachomatis: the “energy parasite” hypothesis revisited. Mol. Microbiol. 33, 177–187 10.1046/j.1365-2958.1999.01464.x [DOI] [PubMed] [Google Scholar]
- 20. Iliffe-Lee E. R., and McClarty G. (2002) Pyruvate kinase from Chlamydia trachomatis is activated by fructose-2,6-bisphosphate. Mol. Microbiol. 44, 819–828 10.1046/j.1365-2958.2002.02924.x [DOI] [PubMed] [Google Scholar]
- 21. Nicholson T. L., Olinger L., Chong K., Schoolnik G., and Stephens R. S. (2003) Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185, 3179–3189 10.1128/JB.185.10.3179-3189.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McClarty G. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity (Stephens R. S., ed) pp. 69–100, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 23. Skipp P. J., Hughes C., McKenna T., Edwards R., Langridge J., Thomson N. R., and Clarke I. N. (2016) Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PLoS ONE 11, 1–17 10.1371/journal.pone.0096696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dibrov P., Dibrov E., Pierce G. N., and Galperin M. Y. (2004) Salt in the wound: a possible role of Na+ gradient in chlamydial infection. J. Mol. Microbiol. Biotechnol. 8, 1–6 10.1159/000082075 [DOI] [PubMed] [Google Scholar]
- 25. Belland R. J., Zhong G., Crane D. D., Hogan D., Sturdevant D., Sharma J., Beatty W. L., and Caldwell H. D. (2003) Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483 10.1073/pnas.1331135100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barta M. L., Thomas K., Yuan H., Lovell S., Battaile K. P., Schramm V. L., and Hefty P. S. (2014) Structural and biochemical characterization of Chlamydia trachomatis hypothetical protein CT263 supports that menaquinone synthesis occurs through the futalosine pathway. J. Biol. Chem. 289, 32214–32229 10.1074/jbc.M114.594325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tuz K., Li C., Fang X., Raba D. A., Liang P., Minh D. D., and Juárez O. (2017) Identification of the catalytic ubiquinone-binding site of Vibrio cholerae sodium-dependent NADH dehydrogenase: a novel ubiquinone-binding motif. J. Biol. Chem. 292, 3039–3048 10.1074/jbc.M116.770982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juárez O., Morgan J. E., Nilges M. J., and Barquera B. (2010) Energy transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 12505–12510 10.1073/pnas.1002866107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Juárez O., Morgan J. E., and Barquera B. (2009) The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 284, 8963–8972 10.1074/jbc.M809395200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Juárez O., and Barquera B. (2012) Insights into the mechanism of electron transfer and sodium translocation of the Na+-pumping NADH:quinone oxidoreductase. Biochim. Biophys. Acta 1817, 1823–1832 10.1016/j.bbabio.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Häse C. C., Fedorova N. D., Galperin M. Y., and Dibrov P. A. (2001) Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65, 353–370 10.1128/MMBR.65.3.353-370.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reyes-Prieto A., Barquera B., and Juárez O. (2014) Origin and evolution of the sodium-pumping NADH: ubiquinone oxidoreductase. PLoS ONE 9, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruppert C., Wimmers S., Lemker T., and Müller V. (1998) The A1A0-ATPase from Methanosarcina mazei: cloning of the 5′ end of the aha operon encoding the membrane domain and expression of the proteolipid in a membrane-bound form in Escherichia coli. J. Bacteriol. 180, 3448–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McMillan D. G., Ferguson S. A., Dey D., Schröder K., Aung H. L., Carbone V., Attwood G. T., Ronimus R. S., Meier T., Janssen P. H., and Cook G. M. (2011) A1Ao-ATP synthase of Methanobrevibacter ruminantium couples sodium ions for ATP synthesis under physiological conditions. J. Biol. Chem. 286, 39882–39892 10.1074/jbc.M111.281675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pisa K. Y., Huber H., Thomm M., and Müller V. (2007) A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274, 3928–3938 10.1111/j.1742-4658.2007.05925.x [DOI] [PubMed] [Google Scholar]
- 36. Müller V., Lemker T., Lingl A., Weidner C., Coskun U., and Grüber G. (2005) Bioenergetics of archaea: ATP synthesis under harsh environmental conditions. J. Mol. Microbiol. Biotechnol. 10, 167–180 10.1159/000091563 [DOI] [PubMed] [Google Scholar]
- 37. Dzioba J., Häse C. C., Gosink K., Galperin M. Y., and Dibrov P. (2003) Experimental verification of a sequence-based prediction: F(1)F(0)-type ATPase of Vibrio cholerae transports protons, not Na+ ions. J. Bacteriol. 185, 674–678 10.1128/JB.185.2.674-678.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murata T., Yamato I., Kakinuma Y., Leslie A. G. W., and Walker J. E. (2005) Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science 308, 654–659 10.1126/science.1110064 [DOI] [PubMed] [Google Scholar]
- 39. Omsland A., Sager J., Nair V., Sturdevant D. E., and Hackstadt T. (2012) Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. U.S.A. 109, 19781–19785 10.1073/pnas.1212831109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatch T. P., Miceli M., and Silverman J. A. (1985) Synthesis of protein in host-free reticulate bodies of Chlamydia psittaci and Chlamydia trachomatis. J. Bacteriol. 162, 938–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakayama Y., Hayashi M., Yoshikawa K., Mochida K., and Unemoto T. (1999) Inhibitor studies of a new antibiotic, korormicin, 2-n-heptyl-4-hydroxyquinoline N-oxide and Ag+ toward the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biol. Pharm. Bull. 22, 1064–1067 10.1248/bpb.22.1064 [DOI] [PubMed] [Google Scholar]
- 42. Tokuda H., and Unemoto T. (1984) Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus. J. Biol. Chem. 259, 7785–7790 [PubMed] [Google Scholar]
- 43. Tuz K., Mezic K. G., Xu T., Barquera B., and Juárez O. (2015) The kinetic reaction mechanism of the Vibrio cholerae sodium-dependent NADH dehydrogenase. J. Biol. Chem. 290, 20009–20021 10.1074/jbc.M115.658773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strickland M., Juárez O., Neehaul Y., Cook D. A., Barquera B., and Hellwig P. (2014) The conformational changes induced by ubiquinone binding in the Na+-pumping NADH:ubiquinone oxidoreductase (Na+–NQR) are kinetically controlled by conserved glycines 140 and 141 of the NqrB subunit. J. Biol. Chem. 289, 23723–23733 10.1074/jbc.M114.574640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Juárez O., Neehaul Y., Turk E., Chahboun N., DeMicco J. M., Hellwig P., and Barquera B. (2012) The role of glycine residues 140 and 141 of subunit B in the functional ubiquinone binding site of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 287, 25678–25685 10.1074/jbc.M112.366088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Penefsky H. S. (1985) Mechanism of inhibition of mitochondrial adenosine triphosphatase by dicyclohexylcarbodiimide and oligomycin: relationship to ATP synthesis. Proc. Natl. Acad. Sci. U.S.A. 82, 1589–1593 10.1073/pnas.82.6.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Nevel C. J., and Demeyer D. I. (1977) Effect of monensin on rumen metabolism in vitro. Appl. Environ. Microbiol. 34, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bogachev A. V., Murtasina R. A., Shestopalov A. I., and Skulachev V. P. (1993) The role of protonic and sodium potentials in the motility of E. coli and Bacillus FTU. Biochim. Biophys. Acta 1142, 321–326 10.1016/0005-2728(93)90160-H [DOI] [PubMed] [Google Scholar]
- 49. Clark L. C., and Sachs G. (1968) Bioelectrodes for tissue metabolism. Ann. N. Y. Acad. Sci. 148, 133–153 10.1111/j.1749-6632.1968.tb20346.x [DOI] [PubMed] [Google Scholar]
- 50. Degli Esposti M. (1998) Inhibitors of NADH–ubiquinone reductase: an overview. Biochim. Biophys. Acta 1364, 222–235 10.1016/S0005-2728(98)00029-2 [DOI] [PubMed] [Google Scholar]
- 51. Ernster L., Dallner G., and Felice Azzone G. (1963) Differential effects of rotenone and amytal on mitochondrial electron and energy transfer. J. Biol. Chem. 238, 1124–1131 [Google Scholar]
- 52. Miyoshi H., Kondo H., Oritani T., Saitoh I., and Iwamura H. (1991) Inhibition of electron transport of rat liver mitochondria by unnatural (−)-antimycin A3. FEBS Lett. 292, 61–63 10.1016/0014-5793(91)80834-P [DOI] [PubMed] [Google Scholar]
- 53. Petersen L. C. (1977) The effect of inhibitors on the oxygen kinetics of cytochrome c oxidase. Biochim. Biophys. Acta 460, 299–307 10.1016/0005-2728(77)90216-X [DOI] [PubMed] [Google Scholar]
- 54. Macouillard-Poulletier de Gann, Belaud-Rotureau M. A., Voisin P., Leducq N., Belloc F., Canioni P., and Diolez P. (1998) Flow cytometric analysis of mitochondrial activity in situ: application to acetylceramide-induced mitochondrial swelling and apoptosis. Cytometry 33, 333–339 10.1002/(SICI)1097-0320(19981101)33:3%3C333::AID-CYTO7%3E3.0.CO%3B2-H [DOI] [PubMed] [Google Scholar]
- 55. Kalbácová M., Vrbacký M., Drahota Z., and Melková Z. (2003) Comparison of the effect of mitochondrial inhibitors on mitochondrial membrane potential in two different cell lines using flow cytometry and spectrofluorometry. Cytometry A. 52, 110–116 [DOI] [PubMed] [Google Scholar]
- 56. Soper W., Decker L., and Pedersen L. (1979) Mitochondrial ATPase complex. A dispersed, cytochrome-deficient, oligomycin-sensitive preparation from rat liver containing molecules with a tripartite structural arrangement. J. Biol. Chem. 254, 11170–111706 [PubMed] [Google Scholar]
- 57. Hacker B., Barquera B., Crofts A. R., and Gennis R. B. (1993) Characterization of mutations in the cytochrome b subunit of the bc1 complex of Rhodobacter sphaeroides that affect the quinone reductase site (Qc). Biochemistry 32, 4403–4410 10.1021/bi00067a033 [DOI] [PubMed] [Google Scholar]
- 58. Hägerhäll C. (1997) Succinate: quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320, 107–141 10.1016/S0005-2728(97)00019-4 [DOI] [PubMed] [Google Scholar]
- 59. Miyadera H., Shiomi K., Ui H., Yamaguchi Y., Masuma R., Tomoda H., Miyoshi H., Osanai A., Kita K., and Omura S. (2003) Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl. Acad. Sci. U.S.A. 100, 473–477 10.1073/pnas.0237315100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Inabayashi M., Miyauchi S., Kamo N., and Jin T. (1995) Conductance change in phospholipid bilayer membrane by an electroneutral ionophore, monensin. Biochemistry 34, 3455–3460 10.1021/bi00010a038 [DOI] [PubMed] [Google Scholar]
- 61. Heytler P. G. (1963) Uncoupling of oxidative phosphorylation by carbonyl cyanide phenylhydrazones. I. Some characteristics of m-CI-CCP action on mitochondria and chloroplasts. Biochemistry 2, 357–361 10.1021/bi00902a031 [DOI] [PubMed] [Google Scholar]
- 62. Mehlitz A., Eylert E., Huber C., Lindner B., Vollmuth N., Karunakaran K., Goebel W., Eisenreich W., and Rudel T. (2017) Metabolic adaptation of Chlamydia trachomatis to mammalian host cells. Mol. Microbiol. 103, 1004–1019 10.1111/mmi.13603 [DOI] [PubMed] [Google Scholar]
- 63. Reers M., Smith T. W., and Chen L. B. (1991) J-Aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry 30, 4480–4486 10.1021/bi00232a015 [DOI] [PubMed] [Google Scholar]
- 64. Craigen W. J., and Graham B. H. (2008) Genetic strategies for dissecting mammalian and Drosophila voltage-dependent anion channel functions. J. Bioenerg. Biomembr. 40, 207–212 10.1007/s10863-008-9146-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Byrne G. I. (2010) Chlamydia trachomatis strains and virulence: rethinking links to infection prevalence and disease severity. J. Infect. Dis. 201, S126–S133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chowdhary A., Malhotra V. L., Deb M., and Rai U. (1998) Screening for chlamydial infections in women with pelvic inflammatory diseases. J. Commun. Dis. 30, 163–166 [PubMed] [Google Scholar]
- 67. Bas S., and Vischer T. L. (1998) Chlamydia trachomatis antibody detection and diagnosis of reactive arthritis. Br. J. Rheumatol. 37, 1054–1059 10.1093/rheumatology/37.10.1054 [DOI] [PubMed] [Google Scholar]
- 68. Engström P., Bergström M., Alfaro A. C., Syam Krishnan K., Bahnan W., Almqvist F., and Bergström S. (2015) Expansion of the Chlamydia trachomatis inclusion does not require bacterial replication. Int. J. Med. Microbiol. 305, 378–382 10.1016/j.ijmm.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 69. Bavoil P., Ohlin A., and Schachter J. (1984) Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J. H., and Copeland L. (1974) Chemical modification of mitochondria. Uncoupler binding by mitochondria in different metabolic states. Arch. Biochem. Biophys. 162, 64–72 10.1016/0003-9861(74)90105-2 [DOI] [PubMed] [Google Scholar]
- 71. Takehara K., Yuki K., Shirasawa M., Yamasaki S., and Yamada S. (2009) Binding properties of hydrophobic molecules to human serum albumin studied by fluorescence titration. Anal. Sci. 25, 115–120 10.2116/analsci.25.115 [DOI] [PubMed] [Google Scholar]
- 72. Steuber J., Halang P., Vorburger T., Steffen W., Vohl G., and Fritz G. (2014) Central role of the Na+-translocating NADH:quinone oxidoreductase (Na+–NQR) in sodium bioenergetics of Vibrio cholerae. Biol. Chem. 395, 1389–1399 [DOI] [PubMed] [Google Scholar]
- 73. Ghoul M., Pommepuy M., Moillo-Batt A., and Cormier M. (1989) Effect of carbonyl cyanide m-chlorophenylhydrazone on Escherichia coli halotolerance. Appl. Environ. Microbiol. 55, 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Herz K., Vimont S., Padan E., and Berche P. (2003) Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of Vibrio cholerae in a saline environment. J. Bacteriol. 185, 1236–1244 10.1128/JB.185.4.1236-1244.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tuz K., Hsiao Y.-C., Juárez O., Shi B., Harmon E. Y., Phelps I. G., Lennartz M. R., Glass I. A., Doherty D., and Ferland R. J. (2013) The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions. J. Biol. Chem. 288, 13676–13694 10.1074/jbc.M112.420786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schwöppe C., Winkler H. H., and Neuhaus H. E. (2002) Properties of the glucose 6-phosphate transporter from Chlamydia pneumoniae (HPTcp) and the glucose 6-phosphate sensor from Escherichia coli (UhpC). J. Bacteriol. 184, 2108–2115 10.1128/JB.184.8.2108-2115.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Juárez O., Shea M. E., Makhatadze G. I., and Barquera B. (2011) The role and specificity of the catalytic and regulatory cation-binding sites of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 286, 26383–26390 10.1074/jbc.M111.257873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paroutis P., Touret N., and Grinstein S. (2004) The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215 10.1152/physiol.00005.2004 [DOI] [PubMed] [Google Scholar]
- 79. Barquera B., Hellwig P., Zhou W., Morgan J. E., Häse C. C., Gosink K. K., Nilges M., Bruesehoff P. J., Roth A., Lancaster C. R., and Gennis R. B. (2002) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41, 3781–3789 10.1021/bi011873o [DOI] [PubMed] [Google Scholar]
- 80. Bertsova Y. V., and Bogachev A. V. (2004) The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563, 207–212 10.1016/S0014-5793(04)00312-6 [DOI] [PubMed] [Google Scholar]
- 81. Hayashi M., Nakayama Y., and Unemoto T. (1996) Existence of Na+-translocating NADH-quinone reductase in Haemophilus influenzae. FEBS Lett. 381, 174–176 10.1016/0014-5793(96)00114-7 [DOI] [PubMed] [Google Scholar]
- 82. Häse C. C., and Barquera B. (2001) Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta 1505, 169–178 10.1016/S0005-2728(00)00286-3 [DOI] [PubMed] [Google Scholar]
- 83. Dibrov P. (2005) The sodium cycle in vibrio cholerae: riddles in the dark. Biochemistry 70, 150–153 [DOI] [PubMed] [Google Scholar]
- 84. Verkhovsky M. I., and Bogachev A. V. (2010) Sodium-translocating NADH:quinone oxidoreductase as a redox-driven ion pump. Biochim. Biophys. Acta 1797, 738–746 10.1016/j.bbabio.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 85. Abràmoff M. D., Magalhães P. J., and Ram S. J. (2004) Image processing with Image J. Biophotonics Int. 11, 36–42 [Google Scholar]
- 86. Juárez O., Athearn K., Gillespie P., and Barquera B. (2009) Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry 48, 9516–9524 10.1021/bi900845y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Juárez O., Guerra G., Martínez F., and Pardo J. P. (2004) The mitochondrial respiratory chain of Ustilago maydis. Biochim. Biophys. Acta 1658, 244–251 10.1016/j.bbabio.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 88. Divakaruni A. S., Rogers G. W., and Murphy A. N. (2014) Measuring mitochondrial function in permeabilized cells using the seahorse XF analyzer or a Clark-type oxygen electrode. Curr. Protoc. Toxicol. 60, 25.2.1–25.2.16 10.1002/0471140856.tx2502s.60 [DOI] [PubMed] [Google Scholar]
- 89. Vercesi A. E., Bernardes C. F., Hoffmann M. E., Gadelha F. R., and Docampo R. (1991) Digitonin permeabilization does not affect mitochondrial function and allows the determination of the mitochondrial membrane potential of Trypanosoma cruzi in situ. J. Biol. Chem. 266, 14431–14434 [PubMed] [Google Scholar]
- 90. Dibrov P., Dibrov E., Maddaford T. G., Kenneth M., Nelson J., Resch C., and Pierce G. N. (2017) Development of a novel rationally designed antibiotic to inhibit a nontraditional bacterial target. Can. J. Physiol. Pharmacol. 95, 595–603 10.1139/cjpp-2016-0505 [DOI] [PubMed] [Google Scholar]
- 91. Dean D. (2009) Chlamydia trachomatis today: treatment, detection, immunogenetics and the need for a greater global understanding of chlamydial disease pathogenesis. Drugs Today (Barc.) 45, Suppl. B, 25-31 [PMC free article] [PubMed] [Google Scholar]
- 92. Szaszák M., Steven P., Shima K., Orzekowsky-Schröder R., Hüttmann G., König I. R., Solbach W., and Rupp J. (2011) Fluorescence lifetime imaging unravels C. trachomatis metabolism and its crosstalk with the host cell. PLoS Pathog. 7, e1002108 10.1371/journal.ppat.1002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kalman S., Mitchell W., Marathe R., Lammel C., Fan J., Hyman R. W., Olinger L., Grimwood J., Davis R. W., and Stephens R. S. (1999) Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21, 385–389 10.1038/7716 [DOI] [PubMed] [Google Scholar]
- 94. Schramm N., Bagnell C. R., and Wyrick P. B. (1996) Vesicles containing Chlamydia trachomatis serovar L2 remain above pH 6 within HEC-1B cells. Infect. Immun. 64, 1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Grieshaber S., Swanson J. A., and Hackstadt T. (2002) Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell. Microbiol. 4, 273–283 10.1046/j.1462-5822.2002.00191.x [DOI] [PubMed] [Google Scholar]
- 96. Mourad A., Sweet R. L., Sugg N., and Schachter J. (1980) Relative resistance to erythromycin in Chlamydia trachomatis. Antimicrob. Agents Chemother. 18, 696–698 10.1128/AAC.18.5.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Somani J., Bhullar V. B., Workowski K. A., Farshy C. E., and Black C. M. (2000) Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J. Infect. Dis. 181, 1421–1427 10.1086/315372 [DOI] [PubMed] [Google Scholar]
- 98. Binet R., and Maurelli A. T. (2007) Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob. Agents Chemother. 51, 4267–4275 10.1128/AAC.00962-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Binet R., Bowlin A. K., Maurelli A. T., and Rank R. G. (2010) Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob. Agents Chemother. 54, 1094–1101 10.1128/AAC.01321-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dessus-Babus S., Bébéar C. M., Charron A., Bébéar C., and de Barbeyrac B. (1998) Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained in vitro. Antimicrob. Agents Chemother. 42, 2474–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Morrissey I., Salman H., Bakker S., Farrell D., Bébéar C. M., and Ridgway G. (2002) Serial passage of Chlamydia spp. in sub-inhibitory fluoroquinolone concentrations. J. Antimicrob. Chemother. 49, 757–761 10.1093/jac/dkf031 [DOI] [PubMed] [Google Scholar]
- 102. Suchland R. J., Sandoz K. M., Jeffrey B. M., Stamm W. E., and Rockey D. D. (2009) Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob. Agents Chemother. 53, 4604–4611 10.1128/AAC.00477-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Scidmore M. A. (2005) Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr. Protoc. Microbiol. 2005 Chapter 11:Unit 11A.1 10.1002/9780471729259.mc11a01s00 [DOI] [PubMed] [Google Scholar]
- 104. Vonck R. A., Darville T., O'Connell C. M., and Jerse A. E. (2011) chlamydial infection increases gonococcal colonization in a novel murine coinfection model. Infect. Immun. 79, 1566–1577 10.1128/IAI.01155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Uriarte S. M., Molestina R. E., Miller R. D., Bernabo J., Farinati A., Eiguchi K., Ramirez J. A., and Summersgill J. T. (2002) Effect of macrolide antibiotics on human endothelial cells activated by Chlamydia pneumoniae infection and tumor necrosis factor-α. J. Infect. Dis. 185, 1631–1636 10.1086/340575 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.