Abstract
A well-controlled microtubule organization is essential for intracellular transport, cytoskeleton maintenance, and cell development. KN motif and ankyrin repeat domain-containing protein 1 (KANK1), a member of KANK family, recruits kinesin family member 21A (KIF21A) to the cell cortex to control microtubule growth via its C-terminal ankyrin domain. However, how the KANK1 ankyrin domain recognizes KIF21A and whether other KANK proteins can also bind KIF21A remain unknown. Here, using a combination of structural, site-directed mutagenesis, and biochemical studies, we found that a stretch of ∼22 amino acids in KIF21A is sufficient for binding to KANK1 and its close homolog KANK2. We further solved the complex structure of the KIF21A peptide with either the KANK1 ankyrin domain or the KANK2 ankyrin domain. In each complex, KIF21A is recognized by two distinct pockets of the ankyrin domain and adopts helical conformations upon binding to the ankyrin domain. The elucidated KANK structures may advance our understanding of the role of KANK1 as a scaffolding molecule in controlling microtubule growth at the cell periphery.
Keywords: cell adhesion, cell signaling, kinesin, structural biology, X-ray crystallography, ankyrin domain
Introduction
Precisely controlled microtubule (MT)4 organization is essential for intracellular transport (1), skeleton maintenance, and neuronal development (2). The MTs, after reaching the cell cortex, change their directions to extend in the direction parallel to the cell periphery (3, 4). Interaction of the MTs with the cell cortex plays important roles in inducing cortical polarity, remodeling the cytoskeleton, and promoting focal adhesion turnover (5, 6). A cortical attachment complex, consisting of KANK1, KIF21A, liprin-α1, and liprin-β1, acts to restrict the MT overgrowth at the cell cortex (7).
KANK1 and its close homologs KANK2–4, belong to the KANK family (8), and they all consist of an N-terminal KN motif, followed by a coiled-coil domain and an ankyrin domain (9). KANK1 was first identified as the tumor suppressor in renal cell carcinoma (10) and was found to be essential for the function of podocyte by regulating the Rho GTPase activity (11, 12). As a scaffold protein, KANK1 mediates cytoskeleton construction by affecting polymerization of actin (13). It has been reported that KANK1 contributes to the talin-based molecular clutch to mediate force transmission by interaction with talin, liprin-β1, and KIF21A via the KN motif, the middle coiled-coil domain, and the C-terminal ankyrin domain, respectively (7, 13, 14). KANK2, another member of the KANK family, was reported to induce the adhesion sliding by promoting the turnover of the integrin-ligand complexes (15). In contrast, the functions of KANK3 and KANK4 remain largely unknown. VAB-19, the only KANK homolog in Caenorhabditis elegans, was found to mediate the cell adhesion by affecting F-actin organization (16). Mutations of KANK proteins cause neuronal and developmental disorders, such as nephrotic syndrome and cerebral palsy (12, 17).
Among the KANK1 binding partners, KIF21A is of special interest because it is a member of the kinesin-4 family and possesses the plus-end–directed motor activity (18). In addition, the mutations of KIF21A lead to congenital fibrosis of extraocular muscles type 1, a neurological disease (19). KIF21A consists of an N-terminal kinesin domain, a middle coiled-coil region, and a C-terminal WD40 domain (18). It has been reported that KIF21A is recruited by KANK1 to restrict the MT growth at the cell periphery via a stretch of about 40 amino acids in the middle region (7).
Akhmanova et al. (7) reported that the KANK1 ankyrin domain is responsible for interacting with KIF21A. The molecular mechanism of the recognition of KIF21A by the KANK1 ankyrin domain remains elusive. As one of the most versatile protein-protein binding modules, ankyrin repeats are usually formed by linear arrays of tandem copies of a 30–40-aa motif (20), which adopts a canonical helix-loop-helix conformation (21). Ankyrin repeats containing proteins are involved in many important biological processes, such as signal transduction (22), gene transcription (23), and neural development (24). In the past few years, we and other groups have determined several ankyrin-peptide complexes by X-ray crystallography, and those complexes display distinct binding modes (22, 23, 25–27), which is consistent with the diverse functions of the ankyrin repeats proteins in vivo.
To understand how the KANK1 ankyrin domain recognizes KIF21A and whether other KANK family members also possesses KIF21A-binding ability, we mapped the KANK1-binding motif in KIF21A and determined the crystal structures of the KIF21A peptide in complex with either the ankyrin domain of KANK1 or the ankyrin domain of KANK2. By structural analysis, mutagenesis, and biochemical experiments, we identified the key residues of KIF21A and KANK1/2 involved in the complex formation. Overall, our work not only uncovers the common KIF21A recognition mode by KANK1 and KANK2 but also reflects a broader understanding of the roles of KANK1 in clustering the cortical complex to inhibit outgrowth of MTs.
Results
The ankyrin domains of KANK1 and KANK2 bind to KIF21A(1146–1167) peptide
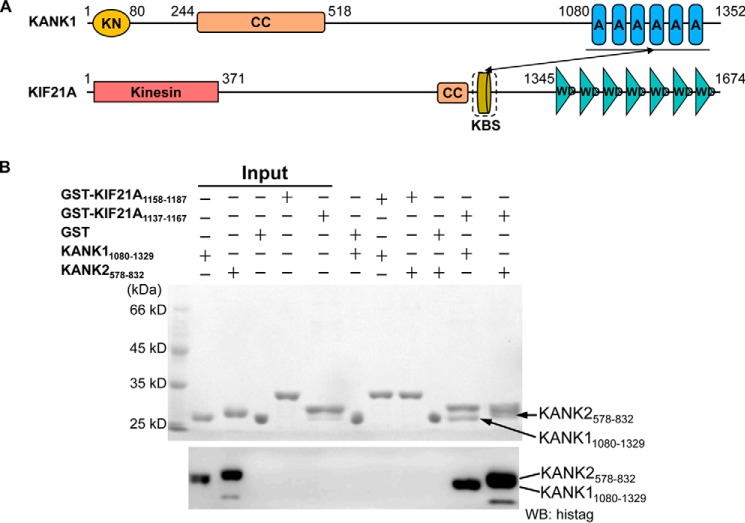
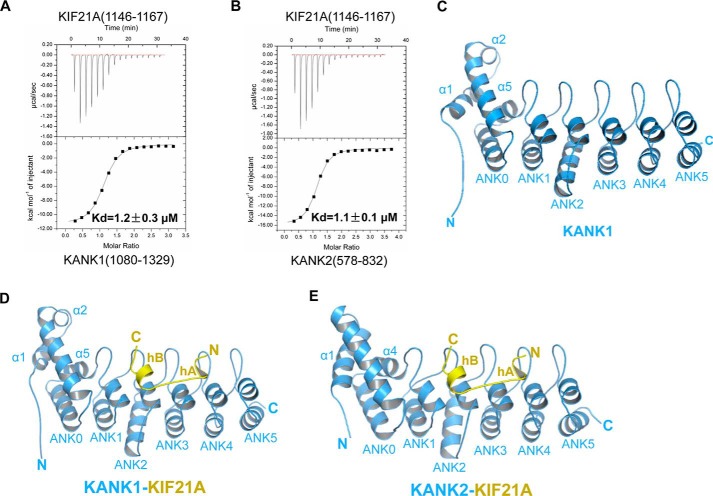
KANK1 contains several unique functional domains and has been found to associate with a stretch of a ∼40-aa motif (residues 1142–1180) of KIF21A via its C-terminal ankyrin domain (Fig. 1A and Fig. S1A) (7). We further narrow down the KANK1-binding site (KBS) in KIF21A to a ∼30-aa motif (residues 1137–1167) by a GST pull-down and Western blot experiment (Fig. 1B). Next we synthesized two peptides of KIF21A, 1138–1160 and 1146–1167, respectively, and tested their binding to the KANK1 ankyrin domain (residues 1080–1329) by isothermal titration calorimetry (ITC) binding experiments (Fig. S1B). We found that 1146–1167 of KIF21A, but not 1138–1160 of KIF21A, displayed KANK1-binding affinity, with a dissociation constant (Kd) of 1.2 μm (Fig. S1B and Table S1). We synthesized 1142–1167 of KIF21A and found that it bound to KANK1 only slightly stronger (Kd = 1.0 μm) (Fig. S2B and Table S1). KANK1 and KANK2 display similar domain organization, and their C-terminal ankyrin domains are highly homologous (>55% sequence identity) (Fig. S1A), which prompted us to test the binding of the KANK2 ankyrin domain (residues 578–832) to different KIF21A peptides. ITC binding data showed that the KANK2 ankyrin domain also bound to 1142–1167 and 1146–1167 of KIF21A, but not 1138–1160 of KIF21A, with Kd values of 0.5 and 1.0 μm, respectively (Fig. S1C and Table S1). Overall, our binding data indicated that 1146–1167 of KIF21A is sufficient for binding to the ankyrin domains of KANK1 and KANK2 (Fig. 2, A and B).
Figure 1.
Human KANK1(1080–1329) and KANK2(578–832) ankyrin domains specifically recognize the KIF21A(1137–1167) peptide. A, domain organizations of human KANK1 and KIF21A. KN, KANK N-terminal (KN) motif; CC, coiled-coil domain; AR, ankyrin repeat; Kinesin, kinesin motor domain; KBS, KANK1-binding site; WD, WD40 repeat. The interaction between the KANK1 ankyrin domain and the KANK1-binding site is denoted by the solid black arrow. B, pull-down binding assay between GST fusion peptides (KIF21A(1158–1187) and KIF21A(1137–1167)) and His-tagged ankryin domains of KANK1 and KANK2. GST protein is used as the control. His-tagged recombinant KANK1 and KANK2 were further detected by Western blotting.
Figure 2.
KIF21A(1146–1167) is sufficient for binding to KANK1/2 and overall structures of the KANK1/2 ankyrin domains with the KIF21A peptide. A, ITC binding between the ankyrin domain of KANK1 and the KIF21A(1146–1167) peptide. B, ITC binding between the ankyrin domain of KANK2 and the KIF21A(1146–1167) peptide. C, structure of the KANK1 ankyrin domain alone, shown in blue, with ANK0–5 and three extra N-terminal helices labeled. D, overall structure of the KANK1-KIF21A complex, with protein and peptide shown in blue and yellow, respectively. ANK0–5 and three extra N-terminal helices of KANK1, as well as hA and hB of KIF21A, are also labeled. E, overall structure of the KANK2-KIF21A complex, with protein and peptide shown in the same way as in Fig. 2D.
The structures of KANK1 ankyrin domain alone and in complex with the KIF21A(1146–1167) peptide
To provide insights into the molecular mechanisms of the KIF21A recognition by the KANK1 ankyrin domain, we set up crystal trials of the KANK1 ankyrin domain alone and with different KIF21A peptides encompassing 1146–1167. Finally, we successfully solved the crystal structure of the human KANK1 domain alone and in complex with the KIF21A peptide at 2.34 and 1.89 Å resolutions, respectively (Fig. 2 (C and D) and Table 1). We found that the structure of the ankyrin domain of KANK1 consists of one non-canonical ankyrin repeat (ANK0) and five canonical ankyrin repeats (ANK1–5), with folds of ANK1–5 characteristic of previously determined ankyrin structures (Fig. 2C and Fig. S1A) (22). Three extra helices at the N terminus of the KANK1 ankyrin domain, two preceding ANK0 (α1-α2) and one between ANK0 and ANK1 (α5), pack with ANK0 (α3-α4) to form a tight helix bundle (Fig. 2C and Fig. S1A). The helix bundle (α1–α5) and ANK1–5 are spaced apart and connected by five hairpin loops (Fig. 2C).
Table 1.
Data collection and refinement statistics
Values in parentheses are for the highest-resolution shell.
| Parameters | KANK1(1080–1329) | KANK1(1080–1329)-KIF21A(1146–1167) | KANK2(578–832)-KIF21A(1146–1167) |
|---|---|---|---|
| Data collection | |||
| Radiation wavelength (Å) | 0.9796 | 0.9796 | 0.9796 |
| Space group | P212121 | P212121 | P1 |
| Cell dimensions | |||
| a, b, c (Å) | 42.90, 47.60, 137.75 | 37.65, 51.62, 137.61 | 45.96, 52.83, 53.48 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 | 73.02, 89.67, 84.91 |
| Resolution (Å) | 44.99–2.34 (2.38–2.34) | 48.33–1.89 (1.96–1.89) | 34.47–2.12 (2.20–2.12) |
| Rmerge | 0.111 (0.610) | 0.053 (0.92) | 0.092 (0.72) |
| I/σI | 28.1 (3.2) | 29.3 (3.5) | 10.2 (2.1) |
| Completeness (%) | 99.7 (98.9) | 99.5 (99.3) | 92.9 (93.4) |
| Redundancy | 5.7 (4.4) | 13.9 (14.2) | 3.8 (3.9) |
| Refinement | |||
| Resolution (Å) | 44.99–2.34 | 48.33–1.89 | 34.47–2.12 |
| No. of reflections (used/free) | 12363/584 | 22149/1997 | 25270/2559 |
| Rwork/Rfree | 0.212/0.242 | 0.222/0.253 | 0.188/0.235 |
| No. of atoms (non-hydrogen) | 1897 | 2061 | 4121 |
| Protein | 1864 | 1839 | 3658 |
| Peptide | NAa | 127 | 250 |
| Solvent | 33 | 95 | 213 |
| B-factors (Å2) | 59.2 | 49.6 | 43.8 |
| Protein | 59.1 | 49.3 | 43.1 |
| Peptide | NA | 55.8 | 55.4 |
| Solvent | 70.0 | 45.6 | 41.8 |
| RMSDs | |||
| Bond lengths (Å) | 0.011 | 0.010 | 0.010 |
| Bond angles (degrees) | 1.2 | 1.2 | 1.3 |
| Ramachandran plot favored/outliers (%) | 97.6/0.00 | 98.5/0.00 | 97.8/0.00 |
a NA, not applicable.
By comparing the apo-structure of the KANK1 ankyrin domain and that of the KANK1-KIF21A complex, we found that the structure of KANK1 did not change much upon KIF21A binding, with the root mean square deviation (RMSD) of the Cα atoms of the two KANK1 structures only 0.72 Å (calculated by PyMOL). In the KANK1-KIF21A complex structure, 1152–1166 of the KIF21A peptide were modeled into electron density, whereas 1146–1151 and 1167 of KIF21A were not visible (Fig. S2A). The KIF21A peptide binds to the inner groove of the KANK1 ankyrin domain constituted by the inner helix and the inter-repeat linkers of ANK1–5 (Fig. 1E). Unexpectedly, one 310-helix (1153ARR1155) and one α-helix (1160QMELL1164) are formed at the N- and C-terminal ends of the KIF21A peptide, respectively. The two helices, designated as hA and hB, respectively, are connected by a short linker (1156RTTT1159) (Fig. 2D). By CD spectroscopy, we found that the conformation of the KIF21A peptide alone lacks regular secondary structures (Fig. S3), thereby confirming that helical conformation of the KIF21A peptide was induced upon KANK1 binding.
Interactions between the ankyrin domains of KANK1/2 and KIF21A(1146–1167)
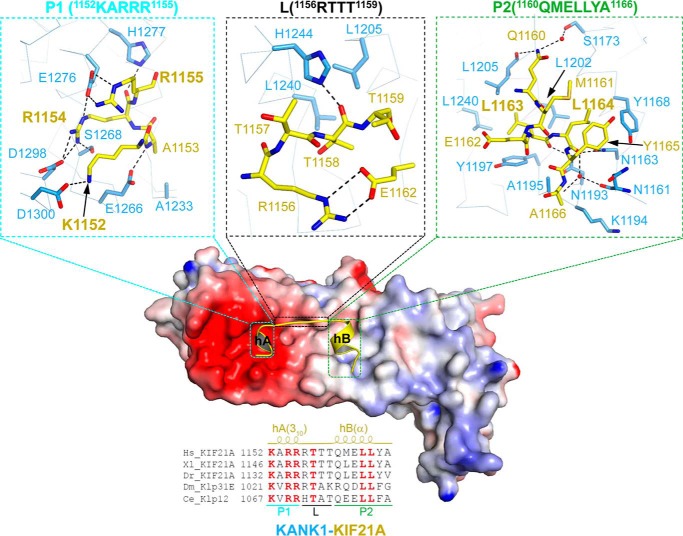
The KIF21A peptide was recognized by the KANK1 ankyrin domain via an acidic patch and a hydrophobic groove, designated as P1 and P2, respectively (Fig. 3). hA is recognized specifically by P1 and hB by P2, whereas the linker connecting hA and hB only makes few contacts with KANK1 (Fig. 3).
Figure 3.
Detailed interactions between the KANK1 ankyrin domain (residues 1080–1329) and the KIF21A peptide (residues 1146–1167). Center, the electrostatic surface of the KANK1 ankyrin domain bound with the KIF21A peptide (yellow schematic); top, 1152KARR1155 of KIF21A bound to the acidic patch (P1 pocket) of KANK1; top center, 1156RTTT1159 OF KIF21A makes few contacts with the periphery of the acidic patch (L region) of KANK1; top right, 1160QMELLYA1166 bound to the hydrophobic pocket of KANK1 (P2 pocket). The residues involved in the protein-peptide interactions are labeled and shown in stick representation. Bottom, KIF21A orthologs aligned on 1152–1166 of human KIF21A, with the absolutely conserved residues colored in red. The 310- and α-helices in the KIF21A peptide are labeled at the top of the sequences as hA(310) and hB(α), respectively.
The N terminus of the KIF21A peptide (1152KARR1155) is fitted into the acidic patch (P1) of KANK1 and interacts with ANK3–5 of KANK1 primarily via electrostatic interactions (Fig. 2D). Specifically, the side chain of the KIF21A Lys1152 is hydrogen-bonded to Asp1300 of KANK1 (Fig. 3). The backbone amide and carbonyl groups of the KIF21A Ala1153 form one direct hydrogen bond with Glu1266 of KANK1 and one water-mediated hydrogen bond with Ser1243 of KANK1, respectively. Ala1153 of KIF21A also makes hydrophobic contacts with Ala1233 of KANK1 (Fig. 3 and Figs. S2B and S4). The side chain of the KIF21A Arg1154 is accommodated into a negatively changed pocket KANK1 by forming several intermolecular hydrogen bonds with the side chains of Ser1268, Glu1276, and Asp1298. Arg1155 of KIF21A is further hydrogen-bonded to Glu1276 and His1277 of KANK1 (Fig. 3 and Figs. S2B and S4).
The C terminus of KIF21A, including hB, binds to the hydrophobic groove (P2) of the KANK1 ankyrin domain formed by ANK1–3 (Figs. 2D and 3). Specifically, Leu1163 of KIF21A is accommodated into the hydrophobic pocket composed of Tyr1197, Leu1202, Leu1205, and Leu1240 of KANK1. Leu1164 of KIF21A makes hydrophobic contact with Tyr1168 and Leu1202 of the KANK1 (Fig. 3 and Figs. S2B and S4). Additional intermolecular hydrophobic interactions are also found between Lys1194 and Ala1195 of KANK1 and Ala-1166 of KIF21A (Fig. 3 and Fig. S4). Intramolecular hydrophobic interactions within KIF21A, such as those between Met1161, Leu1164, and Tyr1165, stabilize the peptide conformation (Fig. 3).
In addition to the hydrophobic interactions, hydrogen bonds are also involved in the interactions between the C terminus of KIF21A and P2 of KANK1 (Figs. S2B and S4). The side chain of Gln1160 of KIF21A forms one direct hydrogen bond and one water-mediated hydrogen bond, with the backbone carbonyl group of Leu1205 of KANK1 and the side chain of Ser1173 of KANK1, respectively (Fig. 3 and Fig. S4). The backbone carbonyl group of Gln1160 of KIF21A also forms one water-mediated hydrogen bond with the side chain of Ser1173 of KANK1 (Fig. 3 and Fig. S4). The backbone carbonyl groups of Leu1163 and Leu1164 of KIF21A are hydrogen-bonded to the side chains of Asn1193 and Asn1163 of KANK1, respectively (Fig. 3 and Fig. S4).
In contrast to hA and hB, the linker region of the KIF21A peptide (1156RTTT1159) only makes few contacts with the periphery region of the acidic patch of the KANK1 ankyrin domain (L) (Fig. 3). The side chain of Arg1156 of KIF21A points to the solvent and forms intramolecular hydrogen bonds with Glu1162 of KIF21A. The side chains of Thr1157 and Thr1159 of KIF21A also point to the solvent (Fig. 3). Thr1158 of KIF21A, which is hydrogen-bonded to His1244 of KANK1 and contacts Leu1205 and Leu1240 of KANK1 via hydrophobic interactions, is the only buried linker residue (Fig. 3 and Fig. S4). Collectively, the coordinated recognition of N- and C-terminal sequences of the KIF21A peptide by P1 and P2 pockets of KANK1 constitutes the unusual ankyrin-peptide recognition mode.
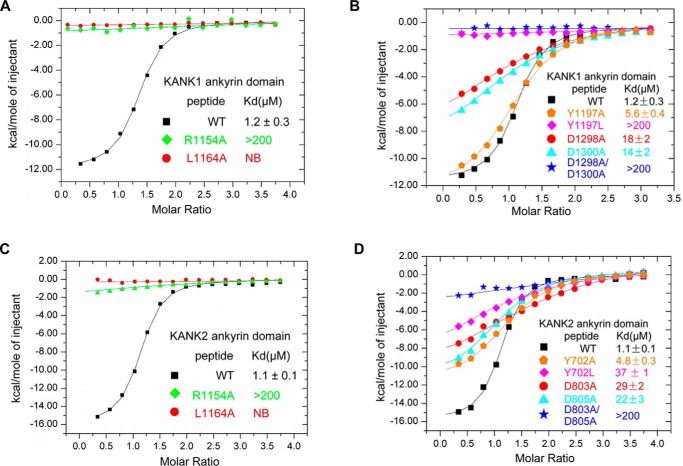
By aligning the sequence of the KIF21A peptide across species, we identified several residues that are absolutely conserved from C. elegans to humans, including Lys1152, Arg1154, Arg1155, Thr1157, Leu1163, and Leu1164 (Fig. 3). All of them interact with KANK1 directly except Thr1157, which is a putative phosphorylation site (https://www.phosphosite.org/proteinAction.action?id=14693)5 (28). We constructed two single mutants of the KIF21A peptide, R1154A and L1164A, and tested their binding to KANK1. We found by ITC that the mutants displayed no binding or very weak binding affinity toward the KANK1 ankyrin domain (Fig. 4A and Table S1).
Figure 4.
Comparison of the binding affinity measured by ITC between KANK1/2 and the KIF21A peptide with either that of KANK1/2 and mutant peptide or that of mutant protein and wild type peptide. A, comparison of the KANK1-binding affinities of wild-type and mutant KIF21A peptides. B, comparison of the KIF21A-binding affinities of KANK1 and its mutants. C, comparison of the KANK2-binding affinities of wild-type and mutant KIF21A peptides. D, comparison of the KIF21A-binding affinities of KANK2 and its mutants. NB, no detectable binding.
To verify the importance of the KANK1 residues in recognizing KIF21A, we chose to mutate Tyr1197, Asp1298, and Asp1300 because Asp1298 and Asp1300 of KANK1 constitute P1, and Tyr1197 is the P2 pocket residue (Fig. 2). We constructed D1298A, D1300A, Y1197A, and Y1197L and tested their KIF21A-binding affinities by ITC. The binding data show that Y1197A, D1298A, and D1300A diminished the KIF21A binding by ∼4.5-, ∼15-, and 10-fold, respectively (Fig. 4B). We further made the double mutant D1298A/D1300A and found that it displayed very weak binding affinity toward the KIF21A peptide (Kd > 200 μm) (Fig. 4B and Table S1). Y1197L also decreased the KIF21A binding severely (Kd > 200 μm) because replacement of Tyr1197 with a Leu in KANK1 would change the depth of the hydrophobic pocket and lead to close contacts with Leu1163 of KIF21A (Fig. 3). Together, the mutagenesis and binding experiments demonstrate that both P1 and P2 of the KANK1 ankyrin domain are required for binding to KIF21A.
KANK1 and KANK2 ankyrin domains share a common mechanism of KIF21A recognition
To investigate whether other KANK members also recognize KIF21A in a way similar to that observed in the KANK1-KIF21A complex, we further determined the 2.12 Å complex structure of the KANK2 ankyrin domain with the same KIF21A peptide (Table 1). Region 1146–1167 of KIF21A, which also adopts a helix-linker-helix conformation, was built in the KANK2-KIF21A complex (Fig. 2E and Fig. S2 (C and D)). Overall, the conformations of protein and peptide are quite similar in two complexes, with an RMSD of 0.92 Å over 244 pairs of aligned protein Cα atoms and an RMSD of 0.38 Å over 15 pairs of peptide Cα atoms (calculated by PyMOL) (Fig. S5). Superposition of the two complexes showed that the KANK2 ankyrin domain only slightly differs from that of KANK1 at the N terminus. KANK2 contains only one longer helix (α1) preceding its ANK0 repeat and does not contain α2, as existing in KANK1. Of note, α2 of KANK1 is not involved in the interaction with KIF21A (Fig. S5).
Most of the intermolecular interactions between KANK1 and KIF21A, including hydrogen bonds and hydrophobic interactions, are observed between corresponding residues of KANK2 and KIF21A (Fig. S2D), which prompted us to test whether the KANK2 ankyrin domain displays a binding property similar to that of the KANK1 ankyrin domain. We first tested the binding of KANK2 to the mutated peptide by ITC and found that R1154A bound to KANK2 very weakly (Kd > 200 μm), whereas L1164A does not display KANK2-binding affinity (Fig. 4C and Table S1). Because Tyr702, Asp803, and Asp805 of KANK2 correspond to Tyr1197, Asp1298, and Asp1300 of KANK1, respectively, we made several single mutants and one double mutant of KANK2 (Y702A, Y702L, D803A, D805A, and D803A/D805A) and tested their binding to KIF21A(1146–1167). Similar to their equivalent KANK1 mutants, Y702A, Y702L, D803A, and D805A decreased the KIF21A-binding affinity by 4–33-fold, whereas D803A/D805A decreased the KIF21A-binding affinity by >180-folds (Fig. 4D and Table S1).
Not all KANK proteins display KIF21A-binding properties
Sequence alignment of KANK1–4 indicates that all KANK proteins contain a highly homologous C-terminal ankyrin domain (47–55% sequence identity). By analyzing the KIF21A-binding residues, we found that they are conserved in KANK1–2, but not in KANK4. All P2 residues of KANK1 are conserved in both KANK3 and KANK4 (Fig. S1A); however, two P1 residues of KANK1, Glu1276 and Asp1300, are replaced by histidine and alanine in KANK4, respectively (Fig. S1A). We further constructed three KANK4-mimic mutants of KANK1, including E1276H, D1300A, and E1276H/D1300A, and tested their binding to KIF21A by ITC. As shown above, D1300A decreased the KIF21A-binding affinity by >10-fold, whereas E1276H and E1276H/D1300A abolished the binding to KIF21A (Fig. S2D and Table S1). In this way, our data suggest that the KANK proteins probably possess different KIF21A-binding affinities, and KANK4 is unlikely to bind KIF21A via its ankyrin domain.
Discussion
Coordinated recognition of the KIF21A peptide by the ankyrin domains of KANK1 and KANK2
Repeat modules, such as ankyrin (20), WD40 (29), pumilio (30), and so forth, have received increasing attention because they are actively involved in protein-protein interactions (31) to regulate certain signaling pathway in vivo (32). Ankyrin domain proteins are versatile because they differ not only in the numbers of ankyrin repeats, but also in the modes of ligand recognition (26).
The ankyrin domain of KANK1 has been found to physically associate with KIF21A and play a critical role in controlling MT growth. However, the molecular mechanism by which KANK1 interacts and recruits KIF21A is poorly understood. Here we characterize the interactions between KIF21A and the ankyrin domains of KANK1 and KANK2 by using structural biology and biochemical experiments. Detailed structural analysis shows that KANK1 and KANK2 share a common KIF21A recognition mode (Fig. S2, B and D). Specifically, the peptide adopts a helix-linker-helix conformation in both complexes (Fig. 2, D and E). The thermodynamic data determined from the ITC experiment, especially the unfavorable entropy, also imply that the KIF21A peptide might undergo conformational change upon binding KANK1 (Table S1). The binding-induced folding transition has been implicated to create a protein-ligand interface in specific protein-ligand recognition (33).
N-terminal and C-terminal ends of the KIF21A peptide are recognized by the acid patch and the hydrophobic groove of the ankyrin domains of KANK1/2, respectively (Fig. 3). By aligning the KIF21A peptide sequence across the species, we found that several residues are absolutely conserved from C. elegans to humans, including Lys1152, Arg1154, Arg1155, Thr1156, Leu1163, and Leu1164 (Fig. 3). The first three positively charged residues of KIF21A are recognized by P1 of KANK1 via hydrogen bonds, whereas the two leucines are accommodated into P2 of KANK1 via hydrophobic interactions (Fig. 3). Mutating Arg1154 or Leu1164 to Ala would severely diminish or disrupt the binding to KANK1/2 (Fig. 4, A and C). Mutation of residues in either P1 or P2 of KANK1/2 also impairs or disrupts the binding to KIF21A, indicating that both pockets of KANK1/2 are required for binding to KIF21A (Fig. 4, B and D).
In addition, we found that the KIF21A-binding residues of KANK1 are also conserved in KANK1 orthologs from C. elegans to humans (Fig. S1A). Together, our results indicated that the recognition mode of KIF21A by KANK1 are conserved in higher eukaryotes.
Comparison of KANK1-KIF21A with other ankyrin-peptide complexes
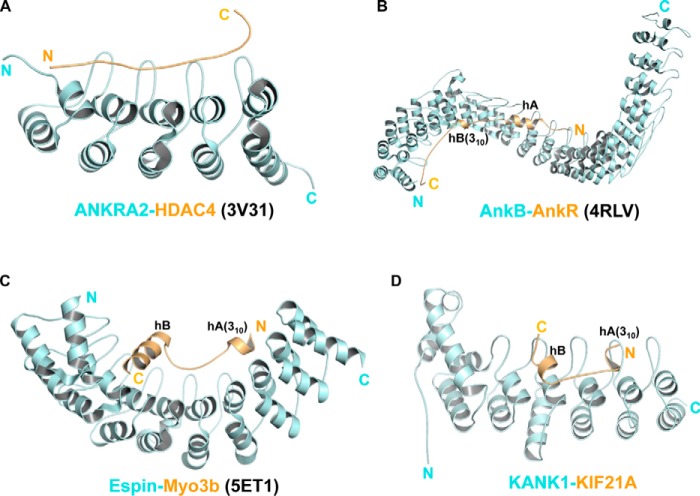
In addition to KANK1/2-KIF21A complexes, we and other groups have reported several structures of peptide-bound ankyrin domains, including ANKRA2-HDAC4 (22), AnkB-AnkR (26), and Espin-Myo3b (27). In the ANKRA2-HDAC4 complex, the HDAC4 peptide does not adopt any secondary structure (Fig. 5A), whereas the peptides adopt helical structures in the other two complexes (Fig. 5, B and C), which prompts us to compare the latter two with the KANK1-KIF21A complex (Fig. 5D). However, the recognition mode of KANK1-KIF21A differs from that observed in the AnkB-AnkR and Espin-Myo3b complex in two aspects. First, hA and hB of the KIF21A peptide are spaced in a side-to-side fashion (Fig. 5D), whereas the two helices of the other two peptides are both arranged in a head-to-tail orientation (Fig. 5, B and C). Second, the KIF21A peptide interacts with KANK1 via N-terminal and C-terminal sequences, with the linker region making fewer contacts with KANK1 (Fig. 2). In the other two complexes, the linker regions of the peptides are much longer and interact extensively with the inner grooves of the ankyrin domains (Fig. 5, C and D). In addition, the fact that Thr1157-Thr1159 could be putatively phosphorylated by receptor tyrosine kinases (28, 34) suggests that the interaction between KANK1 and KIF21A might also be mediated by post-translational modifications, as observed between ANKRA2 and HDAC4 (22).
Figure 5.
Comparison of different ankyrin-peptide complex structures. A, the structure of the ANKRA2 ankyrin repeats (cyan) in complex with the HDAC4 peptide (yellow) (PDB code 3V31). B, the complex structure of the AnkB ankyrin domain (cyan) and the AnkR peptide (yellow) (PDB code 4RLV). C, the structure of the Espin ankyrin domain (cyan) with the Myo3b peptide (yellow) (PDB code 5ET1). D, the structure of the KANK1 ankyrin domain (cyan) with the KIF21 peptide (yellow). All ankyrin repeats are arranged from N to C (left to right). The 310-helices of AnkR, Myo3b, and KIF21A are also indicated.
Functional implication of the differential interactions between KANK proteins and KIF21A
Considering the high similarities between the ankyrin domains of KANK proteins, it is unexpected to find that KIF21A interaction residues are conserved in KANK1–2, but not in KANK4 (Fig. S1A). All three KANK4-mimic mutants weaken or disrupt the binding to KIF21A, suggesting that KANK4 may not be a KIF21A binding partner (Fig. S1D and Table S1).
Although all four KANK members exhibit high expression levels in kidney, the functions of two of them, KANK3 and KANK4, remain largely unknown. We could not exclude the possibility that the KANK4 ankyrin domain might also bind another unknown ligand with high affinity.
Compared with the other three KANK members, KANK1 has received more attention as a potential tumor suppressor (8). At the cell cortex, KANK1 interacts with LL5b, ELKS, and liprin-α1/β1 and activates talin via the KN motif before recruiting KIF21A. One advantage of recruiting KIF21A is the ability to restrict the MT growth at the cell periphery, thereby maintaining the stability of the MT array, at least to some extent (7). KANK1 has been reported to join the talin-based molecular clutch to mediate the force transmission (35). The high binding affinity between the KANK1 ankyrin domain and the KIF21A peptide also suggests that KANK1 contains distinct function modules and acts as the scaffold molecule. Therefore, our determined KANK1/2-KIF21A complexes not only unveil the molecular mechanism of the KANK1/2-KIF21A binding mode, but also would advance our understanding of the role of KANK1 as the scaffold protein in clustering cortical complexes to control MT growth precisely.
Experimental procedures
Cloning, expression, and purification of the ankryin domains of KANK1 and KANK2, the GST-KIF21A
Genes encoding ankyrin domains of KANK1 (residues 1080–1329) and KANK2 (residues 578–832) were synthesized by Sangon Biotech (Shanghai) and cloned into a modified pET28-MHL (GenBankTM accession number EF456735) (36), respectively. Genes encoding 1137–1167 and 1158–1187 of KIF21A were synthesized by Sangon Biotech (Shanghai) and cloned into pGEX-4T-1 (GE Healthcare), respectively. All expression plasmids were transformed into E. coli BL21 (DE3), and proteins were overexpressed at 16 °C for 18 h in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside.
Recombinant ankyrin domains of KANK1 and KANK2 were first purified by a fast flow nickel-nitrilotriacetic acid column (GE Healthcare). N-terminal His6 tags of recombinant proteins were removed by tobacco etch virus protease. Then the cleaved recombinant proteins were further purified by Superdex 75 gel filtration and mono Q ion exchange (GE Healthcare). The purified protein was concentrated to ∼15 mg/mol and stored at −80 °C. The mutants of the KANK1/2 ankyrin domains were constructed by conventional PCR using the MutanBEST kit (TaKaRa) and further verified by DNA sequencing. The mutants are expressed and purified in the same way as the wild-type proteins.
Crystallization, data collection, and structure determination
All crystals were grown using the sitting-drop vapor diffusion method at 18 °C. The apo-form of the KANK1 ankyrin domain (residues 1080–1329) was crystallized by mixing an equal volume of 15 mg/ml protein with crystallization buffer (0.1 m HEPES, pH 7.5, 0.2 m lithium sulfate monohydrate, and 25% PEG 3350). For complex crystallization, KANK1(1080–1329) and KANK2(578–832) were preincubated with the KIF21A peptide (residues 1146–1167) at a molar ratio of 1:3 and then mixed with different crystallization buffers (0.05 m magnesium formate and 21% PEG 3350 for KANK1-KIF21A complex; 0.1 m HEPES, pH 7.5, 0.15 m ammonium acetate, and 31% PEG 3350 for KANK2-KIF21A complex), respectively. Before flash-freezing crystals in liquid nitrogen, all crystals were soaked in a cryoprotectant consisting of 90% reservoir solution plus 10% glycerol. The diffraction data were collected on BL17U1 at the Shanghai Synchrotron Facility (37). Data sets were collected at 0.9796 Å and processed by using the HKL2000 program (38). The initial models of the KANK1 complex, the KANK2 complex, and the KANK1 ankyrin domain in its apo-form were all solved by molecular replacement in PHASER (39) with our previous structure of the KANK2 ankyrin domain (PDB code 4HBD) as the search model. Then all of the models were refined manually and built with Coot (40). The final structures were refined by PHENIX (41). The statistics for data collection and structural refinement are summarized in Table 1.
GST pull-down and Western blotting
Recombinant GST-fused proteins were incubated with glutathione-Sepharose (GE Healthcare), washed with precooled PBS containing 1% glycerol, and used for a GST pull-down assay. All GST pull-down samples were loaded onto an SDS-polyacrylamide gel and then transferred to nitrocellulose membrane (Thermo Fisher Scientific) for Western blotting. The nitrocellulose membrane was blocked with PBS supplemented with 0.1% Nonidet P-40 and 5% milk and then was incubated with primary anti-His antibody (Santa Cruz Biotechnology, Inc.), followed by incubation of anti-mouse HRP-conjugated antibody (Santa Cruz Biotechnology). ECL chemiluminescent substrate (Pierce) was used to detect His-tagged KANK1 and KANK2, and images were obtained by ImageQuant LAS 4000 (GE Healthcare).
ITC
Peptides KIF21A(1138–1160), KIF21A(1150–1167), KIF21A (1142–1167), KIF21A(1146–1167), KIF21A(1146–1167)-L1164A, and KIF21A(1146–1167)-R1154A were synthesized by GL Biochem (Shanghai) Ltd. Peptides were dissolved in water as a stock, and pH of stock solution was adjusted to pH 7.4. Peptides and concentrated proteins were diluted with ITC buffer (20 mm Tris, 150 mm NaCl, 1% glycerol, 1 mm EDTA, pH 7.4). ITC experiments were performed by titrating 2 μl of peptide (0.6–0.8 mm) into a cell containing 30–50 μm proteins on ITC200 (Malvern) at 25 °C, with a spacing time of 120 s and a reference power of 5 μcal/s. Control experiments were performed by injection of peptides in buffer. 16 successive injections were monitored, and 15 sets of injection data (all except for the first one) were used for data analyses. Control was subtracted from data, and data were analyzed using the single-site binding model with the Origin software package. The representative ITC curves of ITC binding measurements are shown in Fig. S6.
CD
All CD experiments were performed on a ChirascanTM qCD spectrophotometer (Applied Photophysics Ltd.) over wavelengths ranging from 190 to 260 nm at 20 °C. All peptides were diluted in CD buffer (20 mm NaH2PO4, 20 mm Na2HPO4, pH 7.4) to a concentration of 0.4 mg/ml. Measurements were taken in a 1-mm path length quartz cuvette at a data pitch of 1 nm. The CD spectrum of the buffer was measured as control and subtracted. Three successive scans were recorded, and results were smoothed and averaged.
Accession numbers
Coordinates and structure factors for the structures of the KANK1 ankyrin domain alone and in complex with the KIF21A peptide, as well as the structure of the KANK2 ankyrin domain with KIF21A, have been deposited into the PDB under accession codes 5YBJ, 5YBU, and 5YBV.
Author contributions
G. Q. and S. L. performed experiments and data analysis with assistance from Z. Z., Y. L., and F. L. S. L. and C. X. conceived and supervised the project. C. X. wrote the manuscript. All authors contributed to editing the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Jinrong Min for critical reading of the manuscript. We are grateful to the staff members at beamline BL17U1 at Shanghai Synchrotron Radiation Facility for assistance with data collection.
This work was supported by National Natural Science Foundation of China Grants 31570737 and 31770806 (to C. X.) and 31500601 (to S. L.) and the “Thousand Young Talent program” (to C. X.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 5YBJ, 5YBU, and 5YBV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MT
- microtubule
- aa
- amino acid(s)
- ITC
- isothermal titration calorimetry
- RMSD
- root mean square deviation
- PDB
- Protein Data Bank.
References
- 1. Vale R. D. (1987) Intracellular transport using microtubule-based motors. Annu. Rev. Cell Biol. 3, 347–378 10.1146/annurev.cb.03.110187.002023 [DOI] [PubMed] [Google Scholar]
- 2. Kapitein L. C., and Hoogenraad C. C. (2015) Building the neuronal microtubule cytoskeleton. Neuron 87, 492–506 10.1016/j.neuron.2015.05.046 [DOI] [PubMed] [Google Scholar]
- 3. Akhmanova A., Stehbens S. J., and Yap A. S. (2009) Touch, grasp, deliver and control: functional cross-talk between microtubules and cell adhesions. Traffic 10, 268–274 10.1111/j.1600-0854.2008.00869.x [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez O. C., Schaefer A. W., Mandato C. A., Forscher P., Bement W. M., and Waterman-Storer C. M. (2003) Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5, 599–609 10.1038/ncb0703-599 [DOI] [PubMed] [Google Scholar]
- 5. Stehbens S. J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T. (2014) CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siegrist S. E., and Doe C. Q. (2007) Microtubule-induced cortical cell polarity. Genes Dev. 21, 483–496 10.1101/gad.1511207 [DOI] [PubMed] [Google Scholar]
- 7. van der Vaart B., van Riel W. E., Doodhi H., Kevenaar J. T., Katrukha E. A., Gumy L., Bouchet B. P., Grigoriev I., Spangler S. A., Yu K. L., Wulf P. S., Wu J., Lansbergen G., van Battum E. Y., Pasterkamp R. J., et al. (2013) CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell 27, 145–160 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 8. Kakinuma N., Zhu Y., Wang Y., Roy B. C., and Kiyama R. (2009) Kank proteins: structure, functions and diseases. Cell Mol. Life Sci. 66, 2651–2659 10.1007/s00018-009-0038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu Y., Kakinuma N., Wang Y., and Kiyama R. (2008) Kank proteins: a new family of ankyrin-repeat domain-containing proteins. Biochim. Biophys. Acta 1780, 128–133 10.1016/j.bbagen.2007.09.017 [DOI] [PubMed] [Google Scholar]
- 10. Sarkar S., Roy B. C., Hatano N., Aoyagi T., Gohji K., and Kiyama R. (2002) A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J. Biol. Chem. 277, 36585–36591 10.1074/jbc.M204244200 [DOI] [PubMed] [Google Scholar]
- 11. Allison S. J. (2015) Nephrotic syndrome: the KANK family in podocyte function. Nat. Rev. Nephrol. 11, 387 10.1038/nrneph.2015.84 [DOI] [PubMed] [Google Scholar]
- 12. Gee H. Y., Zhang F., Ashraf S., Kohl S., Sadowski C. E., Vega-Warner V., Zhou W., Lovric S., Fang H., Nettleton M., Zhu J. Y., Hoefele J., Weber L. T., Podracka L., Boor A., et al. (2015) KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J. Clin. Invest. 125, 2375–2384 10.1172/JCI79504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouchet B. P., Gough R. E., Ammon Y. C., van de Willige D., Post H., Jacquemet G., Altelaar A. M., Heck A. J., Goult B. T., and Akhmanova A. (2016) Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife 5, e18124 10.7554/eLife.18124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Z., Guo S. S., and Fässler R. (2016) Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 10.1083/jcb.201609037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Z., Tseng H. Y., Tan S., Senger F., Kurzawa L., Dedden D., Mizuno N., Wasik A. A., Thery M., Dunn A. R., and Fässler R. (2016) Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 18, 941–953 10.1038/ncb3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ihara S., Hagedorn E. J., Morrissey M. A., Chi Q., Motegi F., Kramer J. M., and Sherwood D. R. (2011) Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat. Cell Biol. 13, 641–651 10.1038/ncb2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lerer I., Sagi M., Meiner V., Cohen T., Zlotogora J., and Abeliovich D. (2005) Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum. Mol. Genet. 14, 3911–3920 10.1093/hmg/ddi415 [DOI] [PubMed] [Google Scholar]
- 18. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- 19. Yamada K., Andrews C., Chan W. M., McKeown C. A., Magli A., de Berardinis T., Loewenstein A., Lazar M., O'Keefe M., Letson R., London A., Ruttum M., Matsumoto N., Saito N., Morris L., et al. (2003) Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1). Nat. Genet. 35, 318–321 10.1038/ng1261 [DOI] [PubMed] [Google Scholar]
- 20. Sedgwick S. G., and Smerdon S. J. (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 10.1016/S0968-0004(99)01426-7 [DOI] [PubMed] [Google Scholar]
- 21. Li J., Mahajan A., and Tsai M. D. (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178 10.1021/bi062188q [DOI] [PubMed] [Google Scholar]
- 22. Xu C., Jin J., Bian C., Lam R., Tian R., Weist R., You L., Nie J., Bochkarev A., Tempel W., Tan C. S., Wasney G. A., Vedadi M., Gish G. D., Arrowsmith C. H., Pawson T., Yang X. J., and Min J. (2012) Sequence-specific recognition of a PxLPxI/L motif by an ankyrin repeat tumbler lock. Sci. Signal. 5, ra39 [DOI] [PubMed] [Google Scholar]
- 23. Collins R. E., Northrop J. P., Horton J. R., Lee D. Y., Zhang X., Stallcup M. R., and Cheng X. (2008) The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 15, 245–250 10.1038/nsmb.1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gallagher D., Voronova A., Zander M. A., Cancino G. I., Bramall A., Krause M. P., Abad C., Tekin M., Neilsen P. M., Callen D. F., Scherer S. W., Keller G. M., Kaplan D. R., Walz K., and Miller F. D. (2015) Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell 32, 31–42 10.1016/j.devcel.2014.11.031 [DOI] [PubMed] [Google Scholar]
- 25. Nie J., Xu C., Jin J., Aka J. A., Tempel W., Nguyen V., You L., Weist R., Min J., Pawson T., and Yang X. J. (2015) Ankyrin repeats of ANKRA2 recognize a PxLPxL motif on the 3M syndrome protein CCDC8. Structure 23, 700–712 10.1016/j.str.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 26. Wang C., Wei Z., Chen K., Ye F., Yu C., Bennett V., and Zhang M. (2014) Structural basis of diverse membrane target recognitions by ankyrins. Elife 3 10.7554/eLife.04353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu H., Li J., Raval M. H., Yao N., Deng X., Lu Q., Nie S., Feng W., Wan J., Yengo C. M., Liu W., and Zhang M. (2016) Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin. Elife 5, e12856 10.7554/eLife.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu C., and Min J. (2011) Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 10.1007/s13238-011-1018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quenault T., Lithgow T., and Traven A. (2011) PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21, 104–112 10.1016/j.tcb.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 31. Xu C., Ishikawa H., Izumikawa K., Li L., He H., Nobe Y., Yamauchi Y., Shahjee H. M., Wu X. H., Yu Y. T., Isobe T., Takahashi N., and Min J. (2016) Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 30, 2376–2390 10.1101/gad.288340.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mosavi L. K., Cammett T. J., Desrosiers D. C., and Peng Z. Y. (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 10.1110/ps.03554604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spolar R. S., and Record M. T. Jr. (1994) Coupling of local folding to site-specific binding of proteins to DNA. Science 263, 777–784 10.1126/science.8303294 [DOI] [PubMed] [Google Scholar]
- 34. Moritz A., Li Y., Guo A., Villén J., Wang Y., MacNeill J., Kornhauser J., Sprott K., Zhou J., Possemato A., Ren J. M., Hornbeck P., Cantley L. C., Gygi S. P., Rush J., and Comb M. J. (2010) Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 3, ra64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elosegui-Artola A., Oria R., Chen Y., Kosmalska A., Pérez-González C., Castro N., Zhu C., Trepat X., and Roca-Cusachs P. (2016) Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 10.1038/ncb3336 [DOI] [PubMed] [Google Scholar]
- 36. Xu C., Wang X., Liu K., Roundtree I. A., Tempel W., Li Y., Lu Z., He C., and Min J. (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- 37. Wang Q. S., Yu F., Huang S., Sun B., Zhang K. H., Liu K., Wang Z. J., Xu C. Y., Wang S. S., Yang L. F., Pan Q. Y., Li L., Zhou H., Cui Y., Xu Q., et al. (2015) The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 26, 12–17 10.13538/j.1001-8042/nst.26.010102 [DOI] [Google Scholar]
- 38. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 39. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.