Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) causes a persistent infection, presenting latent and lytic replication phases during its life cycle. KSHV-related diseases are associated with deregulated expression of inflammatory cytokines, including IL-6 and IL-10, but the mechanisms underlying this dysregulation are unclear. Herein, we report a molecular mechanism for KSHV-induced IL-10 gene expression. KSHV replication and transcription activator (K-RTA) is a molecular switch for the initiation of expression of viral lytic genes, and we describe, for the first time, that K-RTA significantly activates the promoter of the human IL-10 gene. Of note, mutations involving a basic region of K-RTA reduced the association of K-RTA with the IL-10 promoter. Moreover, the host-cell transcription factors, specificity proteins (SP) 1 and 3, play a pivotal cooperative role in K-RTA–mediated transactivation of the IL-10 promoter. K-RTA can interact with SP1 and SP3 directly in vitro, and electrophoresis mobility shift assays (EMSAs) revealed co-operative interaction involving K-RTA, SP1, and SP3 in binding to the IL-10 promoter. As DNase I footprinting assays indicated that K-RTA did not affect SP3 binding to the IL-10 promoter, SP3 can function to recruit K-RTA to the IL-10 promoter. These findings indicate that K-RTA can directly contribute to IL-10 up-regulation via a functional interplay with the cellular transcription factors SP1 and SP3.
Keywords: herpesvirus, interleukin, specificity protein 1 (Sp1), transcription, viral transcription, IL-10, SP3, transactivation
Introduction
Kaposi's sarcoma-associated herpesvirus (KSHV)2/human herpesvirus-8 (HHV-8) is a large DNA virus, belonging to the human gammaherpesvirus family (1, 2), and is associated with the pathogenesis of diseases such as Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman disease (MCD) (3, 4). Like other herpesviruses, KSHV presents two phases during its life cycle: latent infection and lytic replication. Biopsies from latently-infected KSHV carriers show lytic replication, albeit at low frequency, suggesting that the lytic phase plays a significant role in KSHV-associated malignancies (5).
Following stimulation to enter the lytic phase, transcription activation protein (K-RTA), an immediate-early gene product encoded by KSHV open reading frame 50 (Orf50), plays an essential role in the induction of lytic phase genes (6–8). Disruption of Orf50 causes a null phenotype both in viral DNA synthesis and virus production (9). Ectopic K-RTA expression can trigger lytic replication (10), and conversely, spontaneous reactivation is suppressed by K-RTA dominant-negative mutants (7). Therefore, K-RTA has a critical function as a molecular switch between latency and lytic reactivation in the viral life cycle.
K-RTA is a strong transactivator protein with an N-terminal DNA-binding domain and a C-terminal transactivation motif that regulates the expression of various lytic genes (11). K-RTA directly binds to the core element AAATGGGTGGCTAACCCCTACATAA in the viral polyadenylated nuclear (PAN) RNA promoter with high affinity (12, 13). The K-RTA N-terminal DNA-binding domain contains a region rich in basic residues, termed the basic domain, which is involved in direct DNA binding (14). In contrast, K-RTA has been shown to regulate a number of cellular genes (15), and an indirect mechanism is also involved in K-RTA–mediated gene expression (16). Various cellular proteins, such as RBP-Jκ (17, 18), CCATT/enhancer-binding protein C/EBPα (19), OCT-1 (20), STAT3 (21), K-RBP (22), and CBP (23) are demonstrated to interact with K-RTA at the sites of various DNA elements. These cellular proteins may function as recruiters of K-RTA to promoter sequences and are likely to affect transactivation by K-RTA in a promoter context-dependent manner.
IL-10 immunosuppressive activity is effective in various cell types, including Th1 cells and macrophages, and IL-10-mediated suppression of macrophage activity helps to maintain mycobacterial infections (24). IL-10 activates STAT3, indirectly inhibiting gene transcription or modulating post-transcriptional mRNA processing of specific genes (25, 26). In addition, IL-10 inhibits Ets2 expression and LPS-induced miR-155 expression during the inflammatory response (27). Similarly, the IL-10 pathway might enable the virus to evade host immunity (28). Interestingly, another gammaherpesvirus, EBV, and a betaherpesvirus, CMV, encode viral homologs of IL-10, which contribute to evasion from virus-specific immune responses and to the establishment of latent infections (29, 30). In KSHV-associated diseases, such as PEL and MCD in patients with HIV/AIDS, elevated levels of viral cytokine IL-6 and of cellular cytokines IL-6, IL-10, stromal cell-derived factor 1, and vascular endothelial growth factor are thought to contribute to viral persistence in and pathogenesis of these virus-associated lymphoproliferative disorders (31–34).
IL-10 is produced by a number of cells of the immune system, such as T-, B-, and dendritic cells and macrophages, and the IL-10 promoter is regulated by various transcriptional networks in different cell types (35). Recent interesting studies have demonstrated that infection by KSHV or expression of the viral miRNAs miR-K12-3 and miR-K12-7 induces IL-10 in macrophages through targeting the negative regulator of IL-10 transcription, C/EBPβ (36). STAT3 activation contributes to IL-10 production by dendritic cells exposed to UV-inactivated KSHV (37). These observations support the idea that lytic replication caused by intermittent minor reactivation during latency may be associated with IL-10 production by KSHV-infected cells.
In this study, we examined the effect of K-RTA on IL-10 up-regulation and explored the possible mechanism of K-RTA-mediated IL-10 promoter activation. We show, for the first time, that K-RTA significantly activates the human IL-10 promoter through association with SP1 and SP3 in KSHV-infected and non-infected human cells. We also show that specific residues such as Lys-152, Lys-154, and His-145 on K-RTA play a role in K-RTA–mediated IL-10 promoter activation. These data indicate that the functional activity of K-RTA can contribute to IL-10 induction in KSHV-associated diseases.
Results
K-RTA expression activates IL-10 gene promoter
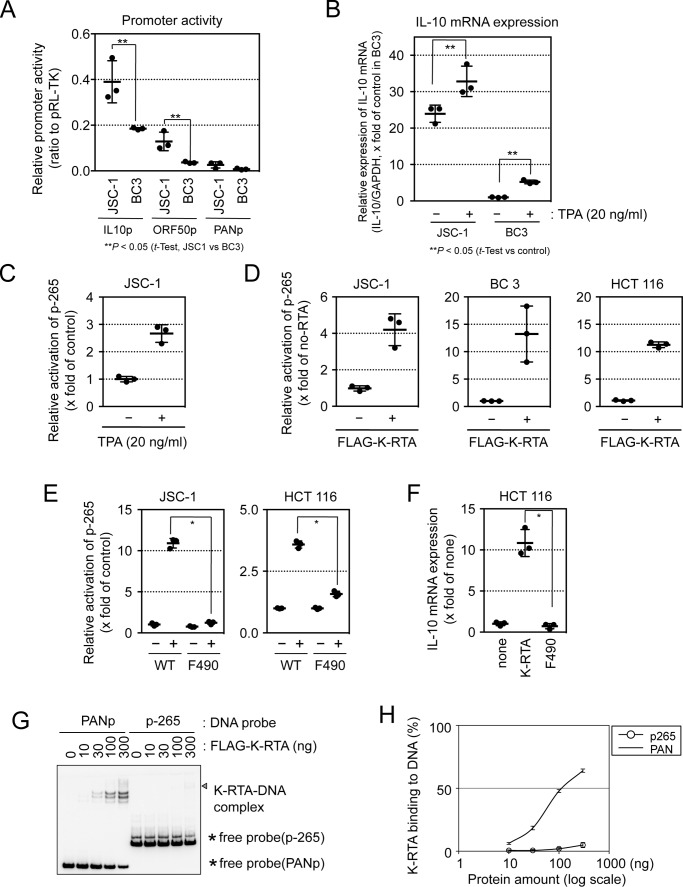
KSHV-infected PEL cells produce various cytokines, including IL-10 (38, 39), and some PEL cells induce lytic viral replication under latent conditions (34). The KSHV-infected PEL cell line JSC-1 is reported to show higher basal and induced expression of lytic phase genes and to produce higher titers of infectious viruses than KSHV-infected BC3 cells (40). In line with those results, luciferase reporter assays confirmed that promoter activities for the lytic phase initiator ORF50/K-RTA and human IL-10 genes were higher in the KSHV-infected PEL cell line JSC-1 than in BC3 cells (Fig. 1A). Quantitative real-time PCR showed that the relative expression of IL-10 mRNA was higher in JSC-1 cells than in BC3 cells and that 12-O-tetradecanoylphorbol-13-acetate (TPA), a chemical initiator of lytic viral replication (41, 42), caused an increase in IL-10 mRNA expression in both cell lines (Fig. 1B). These results suggest that lytic replication and IL-10 mRNA expression are correlated.
Figure 1.
K-RTA transactivates the human IL-10 promoter. A, KSHV-infected PEL cell line JSC-1 and BC3 cells were co-transfected with the reporter plasmids IL-10 p-1000, ORF50, or PANp and pRL-TK vector (internal control). Two days after transfection, a luciferase assay was performed. Results are presented as the mean ± S.D. (n = 3). Student's t test; **, p < 0.05. B, KSHV-infected PEL cell line JSC-1 and BC3 cells were treated with or without TPA (20 ng/ml) for 24 h, and human IL-10 mRNA expression was determined by RT-qPCR. GAPDH mRNA was used to normalize data. Results are presented as the mean ± S.D. (n = 3). Student's t test; **, p < 0.05. C, human IL-10 promoter reporter plasmid p-265 and internal control pRL-TK were transfected into JSC-1 cells. One day later, cells were treated or not with TPA (20 ng/ml) for 24 h. Luciferase activities were determined 2 days after transfection. Results are presented as the mean ± S.D. (n = 3). The activation was statistically significant by Student's t test; p < 0.01. D, three human cell lines, JSC-1, BC3, and HCT 116 cells, were co-transfected with FLAG–K-RTA, IL-10 promoter reporter plasmid p-265, and the internal control pRL-TK plasmid (ratio 1:1:1). Two days after transfection, luciferase activity was examined. Results are presented as the mean ± S.D. (n = 3). The activation was statistically significant by Student's t test; *, p < 0.01. E, human IL-10 promoter reporter plasmid p-265 and internal control pRL-TK were co-transfected into cells with either an empty plasmid (i.e. without K-RTA) or with a plasmid carrying the wild-type K-RTA (WT) or the transactivation domain-deleted K-RTA (F490). Luciferase activity was determined 2 days after transfection. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01. F, HCT 116 cells were transiently transfected with either K-RTA WT or K-RTA F490 expression plasmids. Two days after transfection, mRNA was prepared, and human IL-10 gene expression was measured by RT-qPCR. GAPDH mRNA was used to normalize data. Results are presented as the mean ± S.D. (n = 3). Student's t test. *, p < 0.01. G and H, binding efficiencies of K-RTA to PAN (PANp) or IL-10 (−265/−1 from p-265) promoter DNA probes were analyzed by EMSA. Five-fmol biotin-labeled promoter probes were incubated with increasing amounts of FLAG–K-RTA protein (aa 1–531). The amount of FLAG–K-RTA increased from 0 to 300 ng. A complex of FLAG–K-RTA–DNA probes was examined by EMSA, indicated by the triangle (G). The asterisk indicates free probe DNA. The binding curves were calculated (H).
Various transcription factors are involved in IL-10 mRNA induction in different cell types and conditions. We examined the influence of KSHV lytic replication on IL-10 gene promoter activity, by luciferase reporter assays, using reporter plasmids (p-265) containing IL-10 promoter regions corresponding to the 5′-upstream sequence of the human IL-10 gene open reading frame (position −265 to −1 from the first ATG codon). The activity of the IL-10 promoter was increased after TPA treatment (a stimulus for lytic viral replication) in KSHV-infected JSC-1 cells (Fig. 1C). Expression of FLAG–K-RTA activated IL-10 promoter activity from reporter plasmids in JSC-1, BC3, and HCT 116 cells (Fig. 1D).
The N-terminal portion of K-RTA (amino acids (aa) 1–490), lacking its C-terminal transactivation domain, can bind directly to the K-RTA-responsive element (43). Transactivation domain is also responsible for binding to other transcriptional co-factors (8, 11). FLAG–K-RTA (F490, containing aa 1–490) did not activate p-265 in KSHV-infected JSC-1 or non-infected HCT 116 cells (Fig. 1E). In addition, transcription of IL-10 mRNA was induced by full-length K-RTA but not by F490 in HCT 116 cells (Fig. 1F). Thus, the transactivation domain of K-RTA is required for K-RTA–mediated IL-10 mRNA induction. To examine direct binding of K-RTA to the IL-10 promoter DNA, electrophoresis mobility shift assay (EMSA) was performed. The results showed that direct binding of FLAG–K-RTA (1–531) with IL-10 promoter sequences (−265/−1) was not detected, although FLAG–K-RTA (1–531) bound to PAN promoter DNA in a dose-dependent manner (Fig. 1, G and H). These data suggested that K-RTA has the potential to transactivate the IL-10 promoter by co-operating with other factors.
Impaired DNA-binding ability of K-RTA leads to less activation of IL-10 promoter
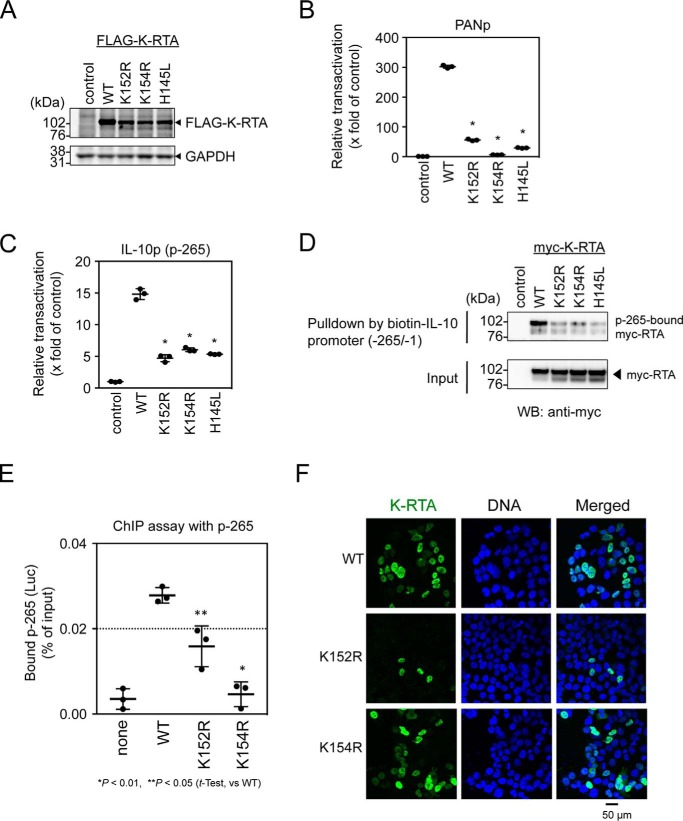
The basic region at the N terminus of K-RTA is proposed to have a critical role in the direct binding to DNA (16). We introduced single lysine-to-arginine substitutions in the basic region of K-RTA (K94R, K124R, K142R, K152R, and K154R), and we found that Lys-152 and Lys-154 are responsible for the transactivation of IL-10 and PAN promoters (Fig. S1). Next we compared the activities of the mutant K-RTAs K152R and K154R with that of an already published mutant, H145L (44). H145L K-RTA is shown to lose the capacity to promote degradation of IRF7 as well as self-ubiquitination (44). These mutant K-RTAs were expressed at comparable levels in cells (Fig. 2A), and luciferase reporter assays showed that the K152R, K154R, and H145L mutations caused a loss of transactivational activity of K-RTA in both PAN and IL-10 p-265 promoters (Fig. 2, B and C). Consistently, our in vitro EMSA experiments indicated that these three mutant K-RTAs lost direct DNA-binding ability to the PAN promoter DNA (Fig. S2).
Figure 2.
Effect of basic domain of K-RTA on IL-10 promoter activation. A, expression of basic domain mutated K-RTAs (K152R, K154R, and H145L) was analyzed in HCT 116 cells by Western blotting. FLAG-tagged K-RTAs were detected with anti-FLAG antibody. GAPDH is shown as loading control. B and C, HCT 116 cells were co-transfected with the mutant FLAG–K-RTA expression plasmids, promoter reporter plasmid, viral PANp (B) or IL-10 promoter p-265 (C), and internal control pRL-CMV (ratio 1:1:1). A luciferase assay for these mutant K-RTAs was performed. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01 (WT versus mutant). D, loading of mutant K-RTA on IL-10 promoter p-265 was examined by ABCD assay. An extract from cells expressing mutant K-RTAs was subjected to a pulldown assay, using p-265 DNA probe, and p-265-bound K-RTA was detected by Western blotting. Two independent experiments showed similar results. E, occupancy of IL-10 promoter by FLAG–K-RTA was examined by ChIP assay. HEK293 cells were transfected with FLAG–K-RTA-expressing plasmid and p-265 reporter plasmid (ratio 1:1) and 2 days later were fixed with 3.7% formalin. Nuclei were sonicated, and FLAG–K-RTA-bound DNA was immunoprecipitated with anti-FLAG antibody. Eluted DNA was quantified by qPCR using primers targeting p-265. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01, and **, p < 0.05 (WT versus mutant). F, subcellular locations of mutant K-RTAs (K152R and K154R) were examined by indirect immunofluorescent microscopic analysis. FLAG–K-RTAs (green) were detected by anti-FLAG antibody, and nuclei were stained with DAPI (blue). Representative images are shown. Scale bar, 50 μm.
To compare the loading of mutant K-RTA on IL-10 promoter DNA, extracts of cells expressing wild type or mutant K-RTAs were subjected to DNA affinity precipitation, by an avidin–biotin-coupled DNA (ABCD) assay. We found that affinity precipitation of mutant K-RTAs with p-265 DNA was clearly reduced (Fig. 2D). In addition, loading of K152R and K154R mutants on IL-10 promoter p-265 reporter plasmid was analyzed by ChIP assay. The results showed that the targeting of the K152R and K154R mutant K-RTAs to p-265 plasmid was clearly diminished (Fig. 2E). Furthermore, we compared the affinity of mutant K-RTAs for chromatin by a biochemical cell fractionation method (Fig. S3). In contrast to the case of wild-type K-RTA, significant amounts of mutant K-RTAs (K152R, K154R, and H145L) were readily extracted by low-salt buffer, and there was little mutant K-RTA in nuclear extracts (Fig. S3, B and C). Because microscopic analysis showed nuclear localization of both K152R and K154R mutant K-RTAs (Fig. 2F), the DNA-binding ability of the basic domain plays a pivotal role in chromatin retention and IL-10 promoter targeting of K-RTA, despite the fact that, in vitro, direct binding of K-RTA to the IL-10 promoter DNA was not detected (Fig. 1G). Collectively, these data suggested that K-RTA might require other co-factors for IL-10 promoter activation.
SP1 and SP3 are involved in K-RTA–mediated IL-10 promoter activation
Transcription of the IL-10 gene is regulated in a context- and cell type-specific manner. Although the IL-10 promoter region present in the p-265 plasmid does not contain the canonical RBP-Jκ-binding element 5′-GTGGGAA-3′, the K-RTA-binding CANT repeat-like sequence (45) is present (sequence from −236 to −229, 5′-CAATCATT-3′). Additionally, various transcription factors, including the AP-1 complex, NFY, c-musculoaponeurotic fibrosarcoma factor (c-MAF), interferon regulatory factors (IRF), SP1, SP3, STAT3, nuclear factor κB (NF-κB), PBX1, and C/EBP, have been reported to bind to this region (46). We constructed mutant IL-10 promoter reporter p-265 plasmids harboring mutations on these transcriptional factor-targeting elements to test their contribution to K-RTA-mediated IL-10 promoter transactivation (Fig. 3A). The results of luciferase reporter assays showed that mutations involving SP1-, SP3-, PBX1-, or NF-κB-targeting elements clearly reduced K-RTA–mediated p-265 reporter activation (mSP, mPBX, and mNF-κB in Fig. 3, A and B).
Figure 3.
Mutation analysis of IL-10 promoter. A, schematic representation of putative transcription factor-binding sites in the fragment from nucleotides −265 to −1 of the human IL-10 promoter. The binding elements for transcription factors are shown in dotted boxes, and mutation sites are shown in capital letters. B, HCT 116 cells were co-transfected with the FLAG–K-RTA–expressing plasmid, internal control pRL-CMV, and mutant p-265 plasmid (as illustrated in A) (ratio 1:1:1), and luciferase assays were performed 2 days later. Basal activity of wild-type p-265 reporter plasmid without K-RTA (in empty box) was used as relative control 1. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01 (WT versus mutant p-265). C, SP-binding site-dependent K-RTA loading onto the IL-10 promoter was determined by ABCD assay. Nuclear extracts (100 μg) from myc-K-RTA–expressing HEK293 cells were incubated with biotin-labeled IL-10 promoter probe (from either wild type or mutated mSP p-265). Myc-K-RTA co-precipitating with biotinylated DNA probes was detected by Western blotting (upper panel). Signal intensity was quantified from triplicate Western blotting experiments and calculated as p-265-bound K-RTA/input K-RTA ratio (lower plot). Student's t test; **, p < 0.05 (WT versus mSP). D, WT or mutant IL-10 promoter occupancy by K-RTA was analyzed by ChIP assay. HCT 116 cells were co-transfected with FLAG–K-RTA–expressing plasmid and p-265 (either WT or mSP) reporter plasmid (ratio 1:1) and 2 days later were fixed with 3.7% formalin. Nuclei were sonicated, and FLAG–K-RTA-bound DNA was immunoprecipitated with anti-FLAG antibody. Eluted DNA was quantified by qPCR, using primers targeting p-265. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01.
SP1 has been shown to interact with various cellular proteins (47). PBX1 also forms a complex with SP1 and activates expression of a subset of promoters containing SP1-binding sites (48). Thus, we examined the effect of the SP1/3-targeting site on K-RTA–mediated transactivation of the IL-10 promoter. The loading of K-RTA on IL-10 promoter DNA was analyzed by ABCD assay using biotin-labeled wild type or mSP IL-10 promoter DNA (fragment from −265 to −1). The results showed that co-precipitation of K-RTA was significantly decreased by mutation at the SP1/3-targeting element (from AGGAGG to GAATCC, positions −177 to −172 in mSP) (Fig. 3C). ChIP assay confirmed that the occupancy of IL-10 promoter p-265 by K-RTA was reduced by a mutation at the SP1/3-targeting element (mSP), suggesting that the SP1/3-binding site is involved in the loading of K-RTA onto the IL-10 promoter (Fig. 3D). In addition, luciferase reporter assays showed that co-expression of SP1-HA or SP3-HA with K-RTA elevated K-RTA–mediated IL-10 promoter activation (Fig. 4A), whereas siRNA-mediated knockdown of SP1 or SP3 decreased K-RTA–mediated IL-10 promoter activation (Fig. 4, B and C). Collectively, these results indicated that SP1/3 participates in K-RTA–mediated IL-10 promoter activation.
Figure 4.
K-RTA–mediated IL-10 promoter activation involves SP1 and SP3. A, dose-dependent increase of K-RTA–mediated IL-10 promoter activation by SP1 and by SP3. HEK293 cells (1.5 × 105 cells/well) were co-transfected with increasing amounts of SP1 or SP3 expression plasmid (0, 25, 50, 100, 200, and 400 ng), 100 ng of K-RTA–expressing plasmid, 100 ng of p-265 reporter plasmid, and 30 ng of pRL-CMV vector (internal control). Luciferase activity was measured 2 days later. Results are presented as the mean ± S.D. (n = 3). Student's t test; *, p < 0.01 (K-RTA only versus +SP1 or +SP3). Western blotting data for the expression of SP1-HA, SP3-HA, and FLAG–K-RTA are shown in the lower panels. B, knockdown of SP1 and SP3 by siRNA. HeLa cells were transfected with 5 nm SP1- or SP3-specific siRNA or with a scrambled siRNA. Knockdown of SP1 and SP3 proteins was confirmed by Western blotting at 72 h post-transfection. GAPDH was used as loading control. C, effect of SP1 and SP3 knockdown on K-RTA–mediated IL-10 promoter activation. HeLa cells were transfected with siRNA as described in C. Twenty four hours later, HeLa cells were co-transfected with the K-RTA–expressing plasmid, the reporter plasmid p-265, and the internal control pRL-CMV (ratio 1:1:1). Luciferase activity was measured after 2 days. Results are presented as the mean ± S.D. (n = 3). Student's t test. *, p < 0.01 (scramble versus SP1 or SP3).
The ABCD assay using biotin-labeled wild type or mSP IL-10 promoter DNA (fragment from −265 to −1) showed that K-RTA and SP3 co-precipitated with wild-type IL-10 promoter DNA in a dose-dependent manner, but this did not occur with mSP DNA probe (Fig. 5A). In this ABCD assay, binding of SP1 to p-265 probe appeared relatively weaker than that of SP3. EMSA using immunopurified SP1-HA and SP3-HA proteins (Fig. 5B) with an IL-10 promoter DNA probe showed that SP3, rather than SP1, preferentially binds to wild-type IL-10 promoter DNA (Fig. 5, C and D), and the formation of a specific complex of DNA probe with SP3 was decreased for mSP probe (Fig. 5, E and F). Thus, SP3, rather than SP1, binds directly to the IL-10 promoter (sequence from −265 to −1).
Figure 5.
Superior SP3 binding to IL-10 promoter in vitro. A, ABCD assay was employed to detect in vitro IL-10 promoter association of K-RTA. Increasing amounts of nuclear extracts (NE) from myc-K-RTA–expressing HEK293 cells were incubated with biotin-labeled DNA probes from either wild type (WT) or SP-binding site-mutated (mSP) IL-10 promoter (fragment from nucleotides −265 to −1). Myc-K-RTA and endogenous SP1 and SP3 co-precipitating with biotin-DNA probes were detected by Western blotting. Myc-K-RTA and SP3 co-precipitated with p-265 DNA probe in a nuclear extract dose-dependent manner. Two independent experiments showed similar results. B, SP1-HA and SP3-HA were expressed in HEK293 cells and immunopurified using anti-HA-agarose. Purified SP1-HA and SP3-HA were visualized by silver staining after SDS-PAGE. An eluted sample from an empty plasmid-transfected lysate was used as control; the molecular mass marker is shown (left lane, Marker). C and D, binding affinities of SP1-HA and SP3-HA to the IL-10 promoter were analyzed by EMSA. The biotin-labeled IL-10 promoter probes (fragment from nucleotides −265 to −1) were incubated with increasing amounts of SP1-HA (left lanes) and SP3-HA (right lanes). The amount of SP1-HA and SP3-HA increased from 0 to 15 ng. The triangle indicates SP3–DNA complex, and the asterisk indicates free DNA probe (C). EMSA experiments were performed three times, and the binding curves and the Kd values for SP3-HA on the IL-10 promoter probe were determined as in Fig. 1H (D). E and F, binding affinities of SP3-HA to wild type (WT p-265; left lanes) or SP3-binding site-mutated (mSP; right lanes) IL-10 promoter were analyzed by EMSA. The biotin-labeled IL-10 promoter probes (fragment from nucleotides −265 to −1) were incubated with increasing amounts of SP3-HA. The amount of SP3-HA increased from 0 to 15 ng. The triangle indicates SP3–DNA complex, and the asterisk indicates free DNA probe (E). EMSA experiments were performed three times, and the binding curve of WT is identical to that of SP3 in D (F).
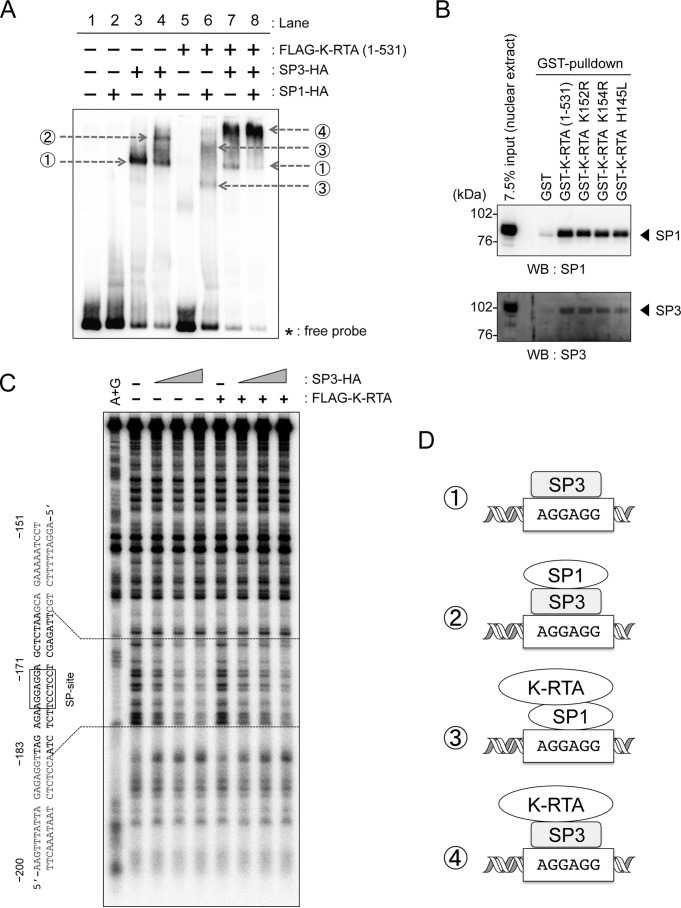
To study possible combinational effects of SP1, SP3, and K-RTA, EMSA was performed using immunopurified FLAG–K-RTA (aa 1–531), SP1-HA, and SP3-HA. The results showed that a specific complex of SP3 and wild-type IL-10 promoter DNA (Fig. 6A, lane 3, arrow 1) was supershifted by a combination with SP1 (Fig. 6A, lane 4, arrow 2) or K-RTA (Fig. 6A, lane 7, arrow 4). Although SP1 alone (Fig. 6A, lane 2) or K-RTA alone (Fig. 6A, lane 5) did not show a specific signal indicating formation of a DNA–protein complex, the SP1 and K-RTA combination apparently induced a DNA–protein complex signal (Fig. 6A, lane 6, arrows 3). A triple combination of SP1, SP3, and K-RTA also showed a specific signal (Fig. 6A, lane 8, arrow 4). As unique signals by combinations of K-RTA with SP1/3 in EMSA disappeared when mSP DNA probes were used (Fig. S4), specific binding of the K-RTA–SP1/3 complex to the IL-10 promoter requires the SP-binding element (AGGAGG, position at −177 to −172). Next, we examined the existence of a possible interaction between K-RTA, SP1, and SP3 proteins. A GST-pulldown assay using nuclear extracts showed that both SP1 and SP3 were co-precipitated with GST-K-RTA (aa 1–531) and mutations involving the basic region did not affect such interactions (Fig. 6B). Collectively, these data suggested that SP3 binding to the element AGGAGG in the IL-10 promoter (sequence from −177 to −172) is involved in the recruitment of K-RTA to the IL-10 promoter, and both SP1 and SP3 could mediate loading of K-RTA onto the IL-10 promoter (possible models are shown in Fig. 6D).
Figure 6.
Co-operative occupancy of IL-10 promoter by K-RTA, SP1, and SP3. A, immunopurified FLAG–K-RTA (aa 1–531), SP1-HA, and SP3-HA proteins from HEK293 cells were incubated with biotin-labeled IL-10 promoter probe (fragment from nucleotides −265 to −1) in the indicated combinations, and specific protein–DNA complexes were analyzed by EMSA. Gray arrows indicate protein–DNA complexes. Specific protein–DNA complexes are numbered as 1–4. Two independent experiments showed similar results. B, GST-pulldown experiments. Wild-type or mutant GST-K-RTA (aa 1–531) proteins were incubated with nuclear extracts, and the co-precipitated endogenous SP1 or SP3 was subsequently detected by Western blotting (WB). Two independent experiments showed similar results. C, DNase I footprinting assay with the IL-10 promoter DNA probe (−206/−72) labeled with [32P]dATP at the 3′ terminus. The 32P-labeled DNA probe was incubated with either immunopurified FLAG–K-RTA (0 or 2000 ng) and increasing amounts of SP3-HA (0, 55, 110, and 220 ng) An autoradiogram produced after gel electrophoresis of DNase I-digested samples is shown. A+G is DNA marker ladder by the Maxam-Gilbert A+G reaction. Boxes around predicted SP-binding site (−177/−172) and SP3-bound footprint are indicated. D, possible models of K-RTA recruitment to the IL-10 promoter. The protein–DNA complex of K-RTA, SP1, and SP3, as shown in A, may regulate IL-10 promoter transactivation. Specific protein–DNA complexes numbered as 1–4 correspond to those in A.
Next, we investigated target sites for K-RTA and SP3 complex formation with DNase I footprinting experiments. The results showed that SP3 alone bound to the predicted site (around −177/−172 in p-265) (Fig. S5). Intriguingly, K-RTA alone did not show any apparent binding site, and specific SP3 binding appeared to be independent of the addition of K-RTA (Fig. 6C and Fig. S6). Thus, we concluded that SP3 binding to the SP site (−177/−172) on the IL-10 promoter was not affected by the presence of K-RTA, and SP3 can function as a guide for K-RTA at the IL-10 promoter. Overall, our data suggested a mechanism that SP3 can recruit K-RTA to the IL-10 promoter (Fig. 6D, number 4), but transactivational activity of K-RTA would be required for IL-10 promoter activation.
Discussion
We demonstrated that K-RTA can transactivate the human IL-10 gene promoter. The N-terminal basic domain and C-terminal transactivational domain of K-RTA were required for K-RTA–mediated IL-10 promoter activation. Both SP1 and SP3 were critical regulators for K-RTA–mediated IL-10 promoter activation, and we also uncovered the functional interplay between K-RTA, SP1, and SP3. K-RTA alone did not bind to the IL-10 promoter DNA in vitro. However, ChIP and ABCD assays and EMSAs testing the combinational effect of K-RTA with SP1 and SP3 indicated that SP1 and SP3 mediate the loading of K-RTA onto the IL-10 promoter. Collectively, K-RTA can activate the IL-10 promoter via SP1 and SP3.
The IL-10 promoter contains various cellular transcription factor-targeting sites such as those for STAT3, NF-κB, C/EBPα, AP1, SP1, and SP3 (35), and previous studies suggested that K-RTA may interact with those proteins (15, 19, 21, 42). Although we did not observe functional co-operation between K-RTA and STAT1/3 in K-RTA–mediated IL-10 promoter activation,3 herein, we show that both SP1 and SP3 have a co-operative effect on K-RTA–mediated IL-10 promoter activation. IL-10 transcription in macrophages is positively regulated by both SP1 and SP3 (49), and two previous studies also support functional interplay between K-RTA and SP1. KSHV thymidine kinase TATAA-less promoter is co-operatively regulated by K-RTA and SP1 (50), and K-RTA–mediated transcription of KSHV viral interferon regulatory factor (vIRF) gene involves SP1-binding sites in the vIRF promoter (51). Unfortunately, however, the precise molecular mechanism of the interplay between K-RTA, SP1, and SP3 was unresolved. We failed to confirm the existence of a protein complex composed of K-RTA, SP1, and SP3 by co-immunoprecipitation assays3; ChIP and ABCD assays and EMSAs revealed its presence on DNA (Fig. 6D). Although K-RTA alone did not form a specific complex with IL-10 promoter DNA, an interaction with SP1/3 enables K-RTA to target the IL-10 promoter.
In this study, we showed that basic domain mutations K152R, K154R, and H145L of K-RTA disrupted its DNA-binding ability to viral PAN promoter, and we also found that the K-RTA N-terminal mutants K152R and K154R caused a reduction of IL-10 promoter transactivation. Although we did not observe direct K-RTA binding to the IL-10 promoter in vitro, results from the ABCD assay, EMSA, and biochemical cell fractionation experiments suggested that the basic region of K-RTA can contribute to the loading of K-RTA onto the IL-10 promoter. As our data did not support the possible role of the basic region in the physical interaction between K-RTA, SP1, and SP3 (Fig. 6), it may suggest another partner protein in K-RTA–mediated IL-10 promoter activation. It should be mentioned that SP1 and SP3 are not the only transcription factors involved in K-RTA–mediated IL-10 activation. In fact, we observed that mutations at PBX1- or NF-κB-targeting elements, located near the SP1/3-targeting element, resulted in a clear reduction of K-RTA–mediated IL-10 promoter activation. Intriguingly, PBX1 has been shown to interact with SP1 and SP3 competitively (52) or cooperatively (48). Thus, such PBX1–SP1/3 interplay might be the mechanism affecting K-RTA–mediated IL-10 promoter activation, and there may be possible interactions between basic domains of K-RTA with other transcription factors. It is also of some concern that most of the results were obtained from transient expression of proteins combined with reporter constructs and from in vitro experiments. More precise investigations surrounding the authentic regulation of the IL-10 promoter in its cellular context are required. The functional interaction between K-RTA and other regulators involving the IL-10 promoter should be analyzed in greater detail in the future.
Collectively, this study demonstrated a novel functional interaction between K-RTA, SP1, and SP3 in K-RTA–mediated IL-10 promoter activation. Although there is the possibility of as-yet-unknown molecules that interact with SP1, SP3, and K-RTA, overall, our observations and those in other studies suggest that SP1/3-K-RTA complex-mediated gene transcription is not restricted to IL-10 promoter activation but might be widely associated with various KSHV-associated diseases. Further studies should uncover the intricate mechanisms of functional associations between SP1, SP3, and K-RTA.
Experimental procedures
Cells and reagents
HEK293, HCT 116, HeLa, BC3, and JSC-1 cell lines (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's or RPMI 1640 medium (Sigma) supplemented with 7% fetal bovine serum (FBS) and kanamycin (50 μg/ml), at 37 °C, in 5% CO2. TPA was purchased from Sigma.
Quantitative RT-PCR
Expression of IL-10 mRNA was analyzed by the TaqMan probe-based real-time, reverse-transcriptase PCR (RT-qPCR) method. Total cellular RNA was extracted using an RNeasy kit (Qiagen Sciences, Germantown, MD), according to the manufacturer's protocol. RT-qPCR was performed using the QuantiTect® probe RT-PCR kit (Qiagen), with TaqMan probes (IL-10, Hs00961622_m1; GAPDH, Hs03929097_g1; purchased from Applied Biosystems, Waltham, MA) on the Applied Biosystems 7300 real-time PCR system. The PCR conditions were as follows: 1 cycle at 50 °C for 30 min and 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s and 60 °C for 1 min. Relative gene expression was calculated from triplicates by the ΔΔCt method, using GAPDH as a reference gene.
Plasmids
K-RTA/ORF50 cDNA was amplified by conventional PCR from genomic DNA of BCBL-1 cells. Sequence-verified DNA was inserted into either pFLAG-CMV2 (Sigma), to express N-terminal FLAG-tagged K-RTA, or pD3myc vector (Myc tag was inserted at the multiple cloning site of pcDNA3), to express N-terminal Myc-tagged K-RTA. To express F490 mutant K-RTA, a PCR fragment encoding K-RTA aa 1–490 was inserted into the pCMV2-FLAG plasmid. Mutant K-RTA–expressing plasmids were generated using a QuickChangeTM lightning multisite-directed mutagenesis kit (Agilent Technologies, Stratagene, La Jolla, CA). KSHV PAN promoter region (−671 to +15, corresponding to 27,996–28,681 in HHV-8 type-M; GenBankTM U75698.1) and the ORF50 promoter region (−867 to +79, corresponding to 70646–71592 in HHV-8 type-M; GenBankTM U75698.1) were inserted into pLuc-MCS plasmid (Agilent Technologies) to produce firefly luciferase-expressing reporter plasmid pPANp-Luc and pORF50p-Luc, respectively. The human IL-10 gene promoter region (−1000 to −1 and −265 to −1 upstream from the first ATG codon, corresponding to 3047–4046 and 3781–4046; GenBankTM AF295024.1) was inserted into pGL4.10 vector (Promega Corp., Madison, WI) to produce the reporter plasmid p-1000 and p-265. SP1-encoding cDNA was generated by conventional PCR and inserted into pD3HA plasmid (53) to express C-terminal HA-tagged SP1. SP3-expressing plasmid pMCS-Sp3-HA was a generous gift from Dr. Guntram Suske (Institute of Molecular Biology and Tumor Research, Philipps-University Marburg, Germany) (54). The K-RTA WT-, K152R-, K154R-, and H145L-)-531STOP (aa 1–531) cDNAs were PCR-amplified from each FLAG–K-RTA–expressing plasmid and subcloned into pCMV2–FLAG, pD3myc, or pGEX-6P-3 (EcoRI restriction site; GE Healthcare). PCR primers are described in Table S1, and DNA sequences of PCR-generated constructs were confirmed.
Luciferase reporter assay
Cells were seeded on 24-well plates (1.5 × 105 cells/well for HCT 116 and HEK293 and 5 × 105 cells/well for JSC-1 and BC3 cells) 1 day before transfection. FuGENE® HD transfection reagents (Roche Diagnostics GmbH, Mannheim, Germany) were used for transfection of HEK293 and HCT 116 cells, and LipofectamineTM 2000 (Life Technologies, Inc., Invitrogen) was used for JSC-1 and BC3 cells, according to the manufacturers' protocols. Cells were collected 48 h post-transfection, and the Dual-GloTM luciferase assay system (Promega) was used to measure luciferase activity. As an internal control to normalize transfection efficiency, pRL-TK or pRL-CMV plasmids (Promega) were co-transfected. Renilla luciferase activities from the pRL plasmid were not affected by co-transfection with K-RTA. For all the transfections, the amount of DNA in each well was made constant by adding empty plasmid DNA pCMV2. Triplicate transfection was performed in each reporter assay. The ratio between firefly and Renilla luciferase activities from the same sample was calculated to obtain normalized firefly activity. Average and standard deviations (S.D.) were calculated (n = 3), and reproducibility was confirmed.
Western blotting and antibodies
Proteins were resolved by SDS-PAGE and electrotransferred to Immobilon-P membrane (EMD Millipore, Billerica, MA), and membranes were blocked and incubated with antibodies. Western blotting signals were developed with the SuperSignal® West Dura Extended Duration Substrate (Thermo Fisher Scientific, Pierce) and recorded with an ImageQuant LAS4000 mini image analyzer (GE Healthcare). The following primary antibodies were used: anti-FLAG M2 (Sigma); anti-HA (3F10; Roche Applied Science, Upper Bavaria, Germany); anti-Myc (9E10; Roche Applied Science); anti-Sp3 (H-225; Santa Cruz Biotechnology, Santa Cruz, CA); anti-Sp1 (D4C3; Cell Signaling Technology, Inc., Danvers, MA); anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH, 6C5; Millipore Corp., Chemicon, Billerica, MA); anti-α-tubulin (10G10; Wako Pure Chemical Industries, Ltd., Osaka, Japan); and anti-TopoIIβ (40/TopoIIβ; BD Biosciences, Tokyo, Japan). Horseradish peroxidase-conjugated donkey anti-rabbit IgG, horseradish peroxidase-conjugated sheep anti-mouse IgG (both from Amersham Biosciences), and horseradish peroxidase-conjugated goat anti-rat IgG (Santa Cruz Biotechnology) were used as secondary antibodies.
Immunofluorescence confocal microscopy
Cells were seeded on ScientificTM NuncTM Lab-TekTM II CC2TM chamber slides (5–10 × 104 cells/0.5 ml/well; Thermo Fisher Scientific) and cultured overnight. Cells were transfected with the plasmid expressing FLAG–K-RTA, using FuGENE® HD transfection reagent (Promega) and, on the following day, were fixed with 3.7% formalin/phosphate-buffered saline (PBS) for 10 min at room temperature. Incubation with the antibodies was performed as described previously (55). Briefly, cells were permeabilized, blocked, and incubated with primary and secondary antibodies. Anti-FLAG M2 antibody was used at a 1:4000 dilution and the Alexa-Fluor-488-conjugated anti-mouse antibody (Invitrogen, Molecular Probes) at 1:2000. Cells were washed three times with PBS and mounted with Prolong® gold antifade reagent with DAPI (Invitrogen). Acquisition of images (640 × 640 pixels) was performed using an FV1000-D IX81 confocal microscope equipped with an UPLFLN 40 × NA1.3 oil objective lens (Olympus Corp., Tokyo), with identical parameter settings. Confocal two-dimensional TIFF images were merged with Adobe® Photoshop CS4 extended software (Adobe Systems Inc., San Jose, CA).
Cell fractionation
Cells were fractionated into cytosolic, nuclear, and pellet fractions as described elsewhere (56), with modifications. Briefly, cells were washed with PBS and suspended at 2 × 106 cells/ml in hypotonic buffer (HB) (10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 1 mm dithiothreitol, 0.1% Nonidet P-40) plus a supplement. Cells were incubated for 5 min on ice, and after a brief centrifugation, the low-salt fraction and the nuclei were recovered. Nuclei were resuspended in nuclear extract buffer (20 mm HEPES (pH 7.9), 0.42 m KCl, 1.5 mm MgCl2, 1 mm dithiothreitol, 25% glycerol) plus the supplement, to recover the nuclear fraction. The remaining pellet was resuspended in 1× SDS buffer and sonicated.
Immunopurification
For immunopurification of SP1-HA, SP3-HA, FLAG–K-RTA (aa 1–531), and myc-K-RTA (aa 1–531), HEK293 cells were transfected with the respective plasmids using Lipofectamine 2000 (Thermo Fisher Scientific), according to the manufacturer's protocol. Alternatively, HEK293 cells were transfected by the calcium phosphate method. Two days after transfection, nuclear extracts or NEB350 extracts of transfected HEK293 cells were prepared as described (57) and incubated with 10 μl of anti-HA or anti-FLAG M2 affinity gel (EZviewTM red, Sigma) for 2 h, at 4 °C, with constant rotation. Affinity gel was recovered by gentle centrifugation and washed six times with binding buffer. Tagged proteins were eluted by competitive peptide-containing binding buffer (100 μg/ml HA peptide or 400 μg/ml FLAG peptide). Competitive peptides were purchased from Sigma.
Expression and purification of GST-fusion proteins
The K-RTA-531STOP fragment was cloned into pGEX-6P-3 to express a GST-fusion protein. Recombinant K-RTA protein was induced in Escherichia coli Rosetta 2(DE3) pLysS (Novagen®, Merck Millipore, Darmstadt, Germany) with 0.1 mm isopropyl d-thiogalactopyranoside or 2 h at 30 °C. The cells was harvested and resuspended in lysis buffer (20 mm Tris-HCl (pH 8.0), 5 mm EDTA, 500 mm NaCl, 0.5% Nonidet P-40, 0.01% SDS, and protease inhibitor mixture from Roche Applied Science) and sonicated. The lysate was incubated with glutathione-Sepharose 4B (GE Healthcare Japan Corp.) for 2 h at 4 °C, with continual rotation. Sepharose was recovered by gentle centrifugation and washed five times with lysis buffer. The GST-fusion protein was eluted using elution buffer (20 mm GSH in lysis buffer).
EMSA
The biotin-labeled PANp probe (sequence from nucleotide −122 to −1) was generated by conventional PCR using the primers biotin-5′-AGGGTCAGCTTGAAGGATG-3′ and 5′-GCAGTCCCAGTGCTAAAC-3′ and the plasmid pPANp-Luc as template. The biotin-labeled IL-10 promoter probe (sequence from nucleotide −265 to −1) was similarly generated, using the primers 5′-CTCGAGATAGCTGTAATGCAGAAGTTCATG-3′ and biotin-5′-AGATCTGCCTTCTTTTGCAAGTCTGTCTTG-3′ and p-265, either wild type or mSP, as template. Amplified probes were purified by ethanol precipitation and diluted to 10 fmol/μl in ultrapure water. The binding mixtures contained purified proteins, 5 fmol of biotin probe, 1 μg of poly(dI-dC), 10 mm HEPES (pH 7.45), 50 mm NaCl, 5% glycerol, 5.7 mm MgCl2, 50 mm β-mercaptoethanol, and 0.38 mm dithiothreitol, in a volume of 20 μl. After incubation at room temperature, for 30 min, samples were electrophoresed on 4% non-denaturing polyacrylamide gels in 0.5× TBE (44.5 mm Tris, 44.5 mm boric acid, 1 mm EDTA (pH 8.3)), for 60 min at 10 mA. Probes were then transferred to nylon membranes (Hybond N+, Amersham Biosciences), at 380 mA, for 30 min, in 0.5× TBE. Transferred DNA was cross-linked by UVC irradiation at 120 mJ/cm2, and biotin-labeled probes were detected by LightShiftTM chemiluminescent EMSA kit (Thermo Fisher Scientific).
Knockdown experiments
For SP1 and SP3 knockdown by siRNA transfection, control (AllStars negative control siRNA from Qiagen) and SP1 and SP3 siRNAs (siGENOME SP1 and SP3; D-026959 and D-023096, respectively; GE Healthcare) were used. In siRNA transfection experiments, HiPerfect reagent (Qiagen) was used for transfection of HeLa cells, in accordance with the manufacturer's instructions.
Chromatin–immunoprecipitation assay
HEK293 or HCT 116 cells were co-transfected with FLAG–K-RTA and IL-10 promoter reporter plasmid p-265, and 2 days post-transfection, cells were harvested, and nuclei were collected and cross-linked with 1% formalin for 5 min. DNA bound to FLAG–K-RTA was immunoprecipitated from sonicated nuclear extracts using anti-FLAG M2 antibody, as described earlier (56). Collected DNA was subjected to qPCR, performed with the QuantiTect SYBR® Green PCR kit (Qiagen) and primers for the IL-10 promoter fragment from nucleotides −265 to −1 (primer −265, 5′-CTCGAGATAGCTGTAATGCAGAAGTTCATG-3′, and primer −1, 5′-GCCTTCTTTTGCAAGTCTGTCTTGTGG-3′). A portion of pre-cleared lysate was separated as “input standard” and subjected to the same qPCR experiments to obtain a standard curve. The ratio of K-RTA-bound IL-10 promoter was calculated from this standard curve.
ABCD assay
HEK293 cells were transfected with myc-K-RTA expression vectors by the calcium phosphate method. Two days after transfection, nuclear extracts or NEB350 extracts of HEK293 cells were prepared as described (56). Extracts were diluted 1:3 in buffer D (20 mm HEPES (pH 7.9), 20% (v/v) glycerol, 0.5 mm PMSF, and 0.5 mm DTT) and incubated with 1 μg of biotin-labeled IL-10 promoter probe (fragment from nucleotides −265 to −1), in a binding buffer consisting of 20 mm HEPES (pH 7.9), 140 mm KCl, 20% glycerol, 1.5 mm MgCl2, 2.5 μm ZnSO4, 1 mm DTT, 2.5 μm aprotinin, 1 mm AEBSF, and a phosphatase inhibitor mixture (Sigma). Streptavidin affinity gel (EZviewTM Red, Sigma) was then added, and the reaction mixtures were incubated at 4 °C for 20 h. Affinity gel was washed six times with binding buffer, and the proteins were dissolved in SDS sample buffer and subjected to Western blotting.
GST-pulldown assay
Nuclear extracts of HCT 116 cells (350 μg) were incubated with 1.2 pmol of GST-fused mutant or K-RTA proteins bound to glutathione-Sepharose, in a binding buffer consisting of 20 mm HEPES (pH 7.9), 105 mm KCl, 20% glycerol, 0.4 mm MgCl2, 1 mm dithiothreitol, 2.5 μm aprotinin, and 1 mm AEBSF. The mixtures were incubated at 4 °C for 16 h. Beads were washed six times in binding buffer and eluted with 20 mm GSH in binding buffer. Proteins were analyzed by Western blotting.
DNase I footprinting analysis
A DNA probe containing the IL-10 promoter region (sequence from nucleotides −206 to −72) was amplified by PCR using the primers (5′-CAAGAATTCAAAACTAAGTTTATTAGAGAGGTTAG-3′ and 5′-CTCTGTCCCCCTTTTATATTGTAAGCTCAGGGAGG-3′) with plasmid p-265 as template. The amplified probe was digested by EcoRI and purified. The probe was labeled at the 3′ terminus with [α-32P]dATP using DNA polymerase I, large (Klenow) fragment (Life Technologies, Inc., Invitrogen), and purified. The 32P-labeled probes (10,000 cpm/sample) were incubated with immunopurified proteins in 1.75 μg of poly(dI-dC), 10 mm HEPES (pH 7.45), 50 mm NaCl, 5% glycerol, 5.7 mm MgCl2, 50 mm β-mercaptoethanol, and 0.38 mm dithiothreitol. After incubation at room temperature for 30 min, samples were treated with DNase I (Roche Applied Science) at room temperature for 15 min. Reactions were halted by adding 20 mm EDTA and subjected to ethanol precipitation. The purified DNA samples were electrophoresed on 6% denaturing polyacrylamide gel containing 8.3 m urea in 1× TBE (89 mm Tris, 89 mm boric acid, 2 mm EDTA (pH 8.3)) for 120 min at 1000 V. The dried gels were analyzed using an FLA 7000 bio-image analyzer (Fujifilm, Tokyo, Japan).
Statistical analysis
In vitro experiments were repeated independently at least twice, and consistent results were observed. Quantitative results are presented as the mean ± S.D. (n = 3). GraphPad Prism7 software (La Jolla, CA) was applied to statistical analyses and to generate scatter plots. Error bars represent S.D. Two-tailed Student's t test was used to evaluate the significance of differences between two groups of data with similar variances. Values of p < 0.05 were considered to indicate statistically significant differences.
Author contributions
K. N. conceived the idea of this study. M. M., K. N., and M. K. conducted the experiments. S. Y. contributed to plasmid construction. M. M., K. N., K. K., and Y. S. analyzed the results and wrote the paper. All authors reviewed the results and have approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Guntram Suske (Institute of Molecular Biology and Tumor Research, Philipps-University Marburg, Germany) for providing SP3-expressing plasmids. We also thank Hiroki Umetsu and Koki Hashimoto for their help with initial experiments.
This work was supported in part by Grants-in-aid from the Japan Society for the Promotion of Science KAKENHI S1411004, 15K14409, and 26460076. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Table S1.
M. Miyazawa, K. Noguchi, M. Kujirai, K. Katayama, S. Yamagoe, and Y. Sugimoto, unpublished observations.
- KSHV
- Kaposi's sarcoma-associated herpesvirus
- HHV-8
- human herpesvirus-8
- K-RTA
- KSHV replication and transcription activator
- SP
- specificity protein
- PEL
- primary effusion lymphoma
- MCD
- multicentric Castleman disease
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- C/EBP
- CCAAT/enhancer-binding protein
- v
- viral
- IRF
- interferon regulatory factor
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride
- aa
- amino acid
- PAN
- polyadenylated nuclear
- ABCD
- avidin–biotin-coupled DNA
- qPCR
- quantitative PCR.
References
- 1. Chang Y., Cesarman E., Pessin M. S., Lee F., Culpepper J., Knowles D. M., and Moore P. S. (1994) Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266, 1865–1869 10.1126/science.7997879 [DOI] [PubMed] [Google Scholar]
- 2. Mesri E. A., Cesarman E., and Boshoff C. (2010) Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10, 707–719 10.1038/nrc2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boshoff C., and Weiss R. A. (1998) Kaposi's sarcoma-associated herpesvirus. Adv. Cancer Res. 75, 57–86 10.1016/S0065-230X(08)60739-3 [DOI] [PubMed] [Google Scholar]
- 4. Boshoff C., and Chang Y. (2001) Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52, 453–470 10.1146/annurev.med.52.1.453 [DOI] [PubMed] [Google Scholar]
- 5. Deng H., Liang Y., and Sun R. (2007) Regulation of KSHV lytic gene expression. Curr. Top. Microbiol. Immunol. 312, 157–183 [DOI] [PubMed] [Google Scholar]
- 6. Sun R., Lin S. F., Gradoville L., Yuan Y., Zhu F., and Miller G. (1998) A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U.S.A. 95, 10866–10871 10.1073/pnas.95.18.10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukac D. M., Kirshner J. R., and Ganem D. (1999) Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73, 9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Staudt M. R., and Dittmer D. P. (2007) The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr. Top. Microbiol. Immunol. 312, 71–100 [DOI] [PubMed] [Google Scholar]
- 9. Xu Y., AuCoin D. P., Huete A. R., Cei S. A., Hanson L. J., and Pari G. S. (2005) A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79, 3479–3487 10.1128/JVI.79.6.3479-3487.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gradoville L., Gerlach J., Grogan E., Shedd D., Nikiforow S., Metroka C., and Miller G. (2000) Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74, 6207–6212 10.1128/JVI.74.13.6207-6212.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. West J. T., and Wood C. (2003) The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22, 5150–5163 10.1038/sj.onc.1206555 [DOI] [PubMed] [Google Scholar]
- 12. Song M. J., Li X., Brown H. J., and Sun R. (2002) Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76, 5000–5013 10.1128/JVI.76.10.5000-5013.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang P. J., Shedd D., Gradoville L., Cho M. S., Chen L. W., Chang J., and Miller G. (2002) Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76, 3168–3178 10.1128/JVI.76.7.3168-3178.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang P. J., Shedd D., and Miller G. (2005) Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79, 8750–8763 10.1128/JVI.79.14.8750-8763.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roan F., Inoue N., and Offermann M. K. (2002) Activation of cellular and heterologous promoters by the human herpesvirus 8 replication and transcription activator. Virology 301, 293–304 10.1006/viro.2002.1582 [DOI] [PubMed] [Google Scholar]
- 16. Guito J., and Lukac D. M. (2012) KSHV Rta promoter specification and viral reactivation. Front. Microbiol. 3, 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang Y., Chang J., Lynch S. J., Lukac D. M., and Ganem D. (2002) The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 16, 1977–1989 10.1101/gad.996502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Persson L. M., and Wilson A. C. (2010) Wide-scale use of Notch signaling factor CSL/RBP-Jκ in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes. J. Virol. 84, 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S. E., Wu F. Y., Fujimuro M., Zong J., Hayward S. D., and Hayward G. S. (2003) Role of CCAAT/enhancer-binding protein α (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77, 600–623 10.1128/JVI.77.1.600-623.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carroll K. D., Khadim F., Spadavecchia S., Palmeri D., and Lukac D. M. (2007) Direct interactions of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation. J. Virol. 81, 8451–8467 10.1128/JVI.00265-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gwack Y., Hwang S., Lim C., Won Y. S., Lee C. H., and Choe J. (2002) Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277, 6438–6442 10.1074/jbc.M108289200 [DOI] [PubMed] [Google Scholar]
- 22. Wang S., Liu S., Wu M. H., Geng Y., and Wood C. (2001) Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75, 11961–11973 10.1128/JVI.75.24.11961-11973.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gwack Y., Byun H., Hwang S., Lim C., and Choe J. (2001) CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75, 1909–1917 10.1128/JVI.75.4.1909-1917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murray P. J., Wang L., Onufryk C., Tepper R. I., and Young R. A. (1997) T cell-derived IL-10 antagonizes macrophage function in mycobacterial infection. J. Immunol. 158, 315–321 [PubMed] [Google Scholar]
- 25. Murray P. J. (2005) The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc. Natl. Acad. Sci. U.S.A. 102, 8686–8691 10.1073/pnas.0500419102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murray P. J. (2006) Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6, 379–386 10.1016/j.coph.2006.01.010 [DOI] [PubMed] [Google Scholar]
- 27. McCoy C. E., Sheedy F. J., Qualls J. E., Doyle S. L., Quinn S. R., Murray P. J., and O'Neill L. A. (2010) IL-10 inhibits miR-155 induction by toll-like receptors. J. Biol. Chem. 285, 20492–20498 10.1074/jbc.M110.102111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brooks D. G., Trifilo M. J., Edelmann K. H., Teyton L., McGavern D. B., and Oldstone M. B. (2006) Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 12, 1301–1309 10.1038/nm1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jochum S., Moosmann A., Lang S., Hammerschmidt W., and Zeidler R. (2012) The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog. 8, e1002704 10.1371/journal.ppat.1002704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spencer J. V., Lockridge K. M., Barry P. A., Lin G., Tsang M., Penfold M. E., and Schall T. J. (2002) Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76, 1285–1292 10.1128/JVI.76.3.1285-1292.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones K. D., Aoki Y., Chang Y., Moore P. S., Yarchoan R., and Tosato G. (1999) Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi's sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood 94, 2871–2879 [PubMed] [Google Scholar]
- 32. Oksenhendler E., Carcelain G., Aoki Y., Boulanger E., Maillard A., Clauvel J. P., and Agbalika F. (2000) High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman disease in HIV-infected patients. Blood 96, 2069–2073 [PubMed] [Google Scholar]
- 33. Polizzotto M. N., Uldrick T. S., Hu D., and Yarchoan R. (2012) Clinical manifestations of Kaposi sarcoma herpesvirus lytic activation: multicentric castleman disease (KSHV-MCD) and the KSHV inflammatory cytokine syndrome. Front. Microbiol. 3, 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gasperini P., Sakakibara S., and Tosato G. (2008) Contribution of viral and cellular cytokines to Kaposi's sarcoma-associated herpesvirus pathogenesis. J. Leukocyte Biol. 84, 994–1000 10.1189/jlb.1107777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saraiva M., and O'Garra A. (2010) The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 10.1038/nri2711 [DOI] [PubMed] [Google Scholar]
- 36. Qin Z., Kearney P., Plaisance K., and Parsons C. H. (2010) Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J. Leukocyte Biol. 87, 25–34 10.1189/jlb.0409251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santarelli R., Gonnella R., Di Giovenale G., Cuomo L., Capobianchi A., Granato M., Gentile G., Faggioni A., and Cirone M. (2014) STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci. Rep. 4, 4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asou H., Said J. W., Yang R., Munker R., Park D. J., Kamada N., and Koeffler H. P. (1998) Mechanisms of growth control of Kaposi's sarcoma-associated herpes virus-associated primary effusion lymphoma cells. Blood 91, 2475–2481 [PubMed] [Google Scholar]
- 39. Aoki Y., Yarchoan R., Braun J., Iwamoto A., and Tosato G. (2000) Viral and cellular cytokines in AIDS-related malignant lymphomatous effusions. Blood 96, 1599–1601 [PubMed] [Google Scholar]
- 40. Cannon J. S., Ciufo D., Hawkins A. L., Griffin C. A., Borowitz M. J., Hayward G. S., and Ambinder R. F. (2000) A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74, 10187–10193 10.1128/JVI.74.21.10187-10193.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Renne R., Zhong W., Herndier B., McGrath M., Abbey N., Kedes D., and Ganem D. (1996) Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2, 342–346 10.1038/nm0396-342 [DOI] [PubMed] [Google Scholar]
- 42. Wang S. E., Wu F. Y., Chen H., Shamay M., Zheng Q., and Hayward G. S. (2004) Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78, 4248–4267 10.1128/JVI.78.8.4248-4267.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chang P. J., and Miller G. (2004) Autoregulation of DNA binding and protein stability of Kaposi's sarcoma-associated herpesvirus ORF50 protein. J. Virol. 78, 10657–10673 10.1128/JVI.78.19.10657-10673.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu Y., Wang S. E., and Hayward G. S. (2005) The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22, 59–70 10.1016/j.immuni.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 45. Palmeri D., Carroll K. D., Gonzalez-Lopez O., and Lukac D. M. (2011) Kaposi's sarcoma-associated herpesvirus Rta tetramers make high-affinity interactions with repetitive DNA elements in the Mta promoter to stimulate DNA binding of RBP-Jk/CSL. J. Virol. 85, 11901–11915 10.1128/JVI.05479-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer S. S., and Cheng G. (2012) Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 32, 23–63 10.1615/CritRevImmunol.v32.i1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beishline K., and Azizkhan-Clifford J. (2015) Sp1 and the 'hallmarks of cancer'. FEBS J. 282, 224–258 10.1111/febs.13148 [DOI] [PubMed] [Google Scholar]
- 48. Bjerke G. A., Hyman-Walsh C., and Wotton D. (2011) Cooperative transcriptional activation by Klf4, Meis2, and Pbx1. Mol. Cell. Biol. 31, 3723–3733 10.1128/MCB.01456-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tone M., Powell M. J., Tone Y., Thompson S. A., and Waldmann H. (2000) IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 165, 286–291 10.4049/jimmunol.165.1.286 [DOI] [PubMed] [Google Scholar]
- 50. Zhang L., Chiu J., and Lin J. C. (1998) Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17, 735–742 10.1089/dna.1998.17.735 [DOI] [PubMed] [Google Scholar]
- 51. Chen J., Ueda K., Sakakibara S., Okuno T., and Yamanishi K. (2000) Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74, 8623–8634 10.1128/JVI.74.18.8623-8634.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jackson B., Brown S. J., Avilion A. A., O'Shaughnessy R. F., Sully K., Akinduro O., Murphy M., Cleary M. L., and Byrne C. (2011) TALE homeodomain proteins regulate site-specific terminal differentiation, LCE genes and epidermal barrier. J. Cell Sci. 124, 1681–1690 10.1242/jcs.077552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noguchi K., Fukazawa H., Murakami Y., and Uehara Y. (2004) Nucleolar Nek11 is a novel target of Nek2A in G1/S-arrested cells. J. Biol. Chem. 279, 32716–32727 10.1074/jbc.M404104200 [DOI] [PubMed] [Google Scholar]
- 54. Sapetschnig A., Koch F., Rischitor G., Mennenga T., and Suske G. (2004) Complexity of translationally controlled transcription factor Sp3 isoform expression. J. Biol. Chem. 279, 42095–42105 10.1074/jbc.M404989200 [DOI] [PubMed] [Google Scholar]
- 55. Noguchi K., Hongama K., Hariki S., Nonomiya Y., Katayama K., and Sugimoto Y. (2017) Functional effects of AKT3 on aurora kinase inhibitor-induced aneuploidy. J. Biol. Chem. 292, 1910–1924 10.1074/jbc.M116.747048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noguchi K., Vassilev A., Ghosh S., Yates J. L., and DePamphilis M. L. (2006) The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 25, 5372–5382 10.1038/sj.emboj.7601396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schreiber E., Matthias P., Müller M. M., and Schaffner W. (1989) Rapid detection of octamer binding proteins with mini-extracts′, prepared from a small number of cells. Nucleic Acids Res. 17, 6419 10.1093/nar/17.15.6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.