Abstract
The dynamin-related GTPase atlastin (ATL) catalyzes membrane fusion of the endoplasmic reticulum and thus establishes a network of branched membrane tubules. When ATL function is compromised, the morphology of the endoplasmic reticulum deteriorates, and these defects can result in neurological disorders such as hereditary spastic paraplegia and hereditary sensory neuropathy. ATLs harness the energy of GTP hydrolysis to initiate a series of conformational changes that enable homodimerization and subsequent membrane fusion. Disease-associated amino acid substitutions cluster in regions adjacent to ATL's catalytic site, but the consequences for the GTPase's molecular mechanism are often poorly understood. Here, we elucidate structural and functional defects of an atypical hereditary spastic paraplegia mutant, ATL1-F151S, that is impaired in its nucleotide-hydrolysis cycle but can still adopt a high-affinity homodimer when bound to a transition-state analog. Crystal structures of mutant proteins yielded models of the monomeric pre- and post-hydrolysis states of ATL. Together, these findings define a mechanism for allosteric coupling in which Phe151 is the central residue in a hydrophobic interaction network connecting the active site to an interdomain interface responsible for nucleotide loading.
Keywords: dynamin-related proteins, GTPase, membrane fusion, allosteric regulation, enzyme structure, axonopathy, hereditary spastic paraplegia
Introduction
Hereditary spastic paraplegia (HSP)4 comprises a group of genetic neurological disorders that manifest as spasticity and weakness in the distal extremities. One of the predominantly mutated loci contributing to HSP (subtype SPG3A) is the atlastin-1 (atl1) gene, which accounts for ∼10% of autosomal dominant and the majority of early onset cases (1). Similarly, mutations in atl1 and atl3 were identified recently as a cause of hereditary sensory neuropathy (HSN) type I (2, 3), emphasizing the crucial role of atlastins (ATLs) in axonopathies and for normal cellular physiology (4, 5).
The atl genes encode for a subclass of the dynamin-related protein (DRP) superfamily (6). DRPs use the energy from GTP hydrolysis to control a multitude of cellular events, including vesicle scission, fusion and fission of organelle membranes, cytokinesis, and antiviral activity (7). DRPs often contain a GTPase (G) domain, helical middle domain, motifs for membrane association, and a GTPase effector domain (GED) (7, 8). The GED forms part of the protein's stalk that is important for oligomerization and assembly-dependent enzyme stimulation. Similarly, ATLs consist of G and middle domains and a transmembrane anchor but lack the canonical GED and consequentially appear not to form regular higher-order complexes akin to those observed with dynamin and MxA (9–12). Instead, ATLs contain a C-terminal, cytoplasm-facing amphipathic helix that is crucial for lipid bilayer thinning and membrane fusion, the main function described for this class of DRPs (13–15).
ATLs localize to the endoplasmic reticulum (ER), where they catalyze the fusion of membrane tubules to form three-way junctions, establishing the highly reticular morphology of the smooth ER found at the periphery of the cell body (16–18). The branched structure of the peripheral ER is suppressed upon the expression of dominant-negative ATL alleles, including disease mutants, gene deletions, and transient siRNA knockdowns (16, 18–20). In Xenopus egg extracts or reconstituted systems using an invertebrate Drosophila melanogaster ortholog, catalytically active ATL is required to generate and maintain a branched, tubular network; Drosophila ATL is also sufficient to create a stable tubular membrane network from synthetic proteoliposomes (19, 21, 22). Effective fusion of ER membranes both in vitro and in the cell relies on ATLs traversing multiple cycles of conformational changes and oligomerization events that are driven by both GTP binding and hydrolysis (23–26).
Genomes of higher eukaryotes typically encode three main ATL isoforms (ATL1–3), with ATL1 being enriched in neural tissues and ATL2 and ATL3 showing more ubiquitous expression patterns (16). On a molecular level, ATL isoforms vary in GTPase activity and their apparent distribution across the ER (27, 28). ATL1 is the most active isoform in vitro and decorates the tubular ER network evenly in cells, whereas ATL3 is a less efficient enzyme and localizes preferentially to junction points in the ER network that appear to be associated with membrane fusion events (27–29). A closer structural comparison of two ATL isoforms (ATL1 and ATL3) indicates that despite differences in GTP-hydrolysis kinetics, the overall sequence of molecular events leading to ATL dimerization (and presumably membrane fusion) is conserved (26). In both cases, monomeric ATL binds GTP, undergoes an intramolecular conformational change, and hydrolyzes GTP, which enables the sequential dimerization of the G and middle domains. Dimerization precedes phosphate release. As indicated by a structural study, an intramolecular arginine residue displaces the catalytic magnesium, allowing the cycle to be reset (26). Compromising the overall fidelity of this system is a hallmark in mutant variants that are associated with neurological disorders (e.g. HSP and HSN) (2, 16, 23, 30, 31).
There is an emerging theme that neurological disorders are caused or affected by defects in organelle structure and membrane trafficking, exemplified by HSP, HSN, and related neurodegenerative diseases (32). However, the underlying structural and functional principles often remain poorly understood. In the case of ATLs, part of the knowledge gap can be attributed to the lack of reoccurring mutations that would define residue-specific hot spots within the proteins. Overall, mutations appear family-specific and distribute across ATL's primary sequence (1, 30). When mapped onto the three-dimensional structures of ATL, mutants predominantly cluster in or near the protein's active site or a region connecting the active site to an allosteric interface between the G and middle domains responsible for nucleotide loading (23, 30, 33). The majority of reported HSP/HSN mutations elicit dominant-negative phenotypes, yet the mutations are buried within the folded protein (16, 31, 34–36), supporting the notion that the mutations' primary defects are intrinsic to ATLs. However, there is no unifying mechanistic model for how mutations affect ATL function, as the disease variants impair nucleotide hydrolysis, protein oligomerization, or both (30, 31, 33). Unfortunately, characteristic defects often cannot be gleaned from their location in the protein's structure, and this lack of predictive power necessitates studies on individual mutants to elucidate pathogenic mechanisms. Here, we biochemically characterize ATL1 harboring the HSP mutation F151S (34, 35) and show that despite being able to bind nucleotide, the mutant enzyme is deficient in forming higher-order oligomers in the presence of GTP or non-hydrolyzable GTP analogs. However, this HSP mutant can dimerize through G and middle domains when bound to a transition-state analog, GDP·AlFx. Phe151 clusters with other HSP mutations but displays a unique structure-function profile. We propose that Phe151 serves as a pivotal residue that couples ATL's nucleotide hydrolysis and conformational switching, with the F151S mutation breaking this allosteric path. Additional structural information from mutant proteins and the isolated G domain, all with varying capacities to dimerize, adds to our general understanding of ATL's switching between pre- and post-hydrolysis states.
Results
The non-canonical HSP variant ATL1-F151S fails to hydrolyze GTP but retains nucleotide-binding ability
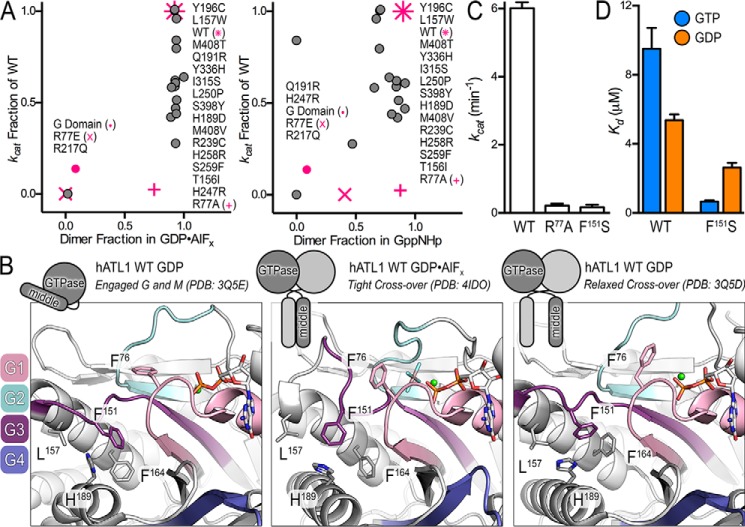
The molecular defects of ATL1 disease variants that drive HSP (and HSN) remain poorly defined. A re-analysis of available data for GTPase activity and dimerization propensities of HSP and engineered mutations highlights a lack of unified mechanism and motivated our current study (23, 30) (Fig. 1A). In this context, we focused on mutations introduced into the soluble catalytic core fragment comprising ATL1's G and middle domains but also a construct of the isolated G domain. Correlating catalytic activities and oligomerization in different nucleotide analogs of these variants establishes that the majority of mutations retain basic functionalities to various degrees and often exhibit only a modest reduction in GTPase activity with limited impairment of dimerization at equilibrium, consistent with previous reports (30, 31, 33). More precisely, GTPase activity and dimerization for HSP mutations illustrate that nearly all mutants can efficiently dimerize when bound to transition-state and non-hydrolyzable GTP analogs, GDP·AlFx and Gpp(NH)p, respectively (Fig. 1A) (30). The propensity to dimerize in the transition state is strong, as indicated by mutant dimer fractions ranging between 89 and 100% (Fig. 1A, left), and the non-hydrolyzable GTP analog shows similar dimerization potential ranging from 47 to 100% (Fig. 1A, right). At the same time, mutations have a broader effect on GTPase activity ranging between 28 and 100% of wild-type activity (Fig. 1A). Two exceptions are Q191R, which retains GTPase activity but cannot dimerize when bound to Gpp(NH)p, and R217Q, which cannot hydrolyze GTP or dimerize in the presence of either GDP·AlFx or Gpp(NH)p (30) (Fig. 1A). Effects of Q191R on ATL1's molecular mechanism have not been examined in greater detail, and hence it is not clear how the apparent defect in oligomerization propensities manifests. In contrast, the cause of R217Q's negative effect on ATL function is explicable. The guanidinium group of Arg217 directly contributes to the binding of the nucleotide by coordinating the guanine base; when mutated to glutamine, the affinity for nucleotide is lost (30). Similar to the R217Q mutant, engineered ATL variants such as R77E, a charge reversal mutation of the catalytic arginine, and a truncated construct expressing the isolated G domain both have impaired GTPase activity and cannot dimerize in GDP·AlFx (30, 33) (Fig. 1A, pink x and circles). The loss of function for both R77E and the isolated G domain can be attributed to a decreased affinity for nucleotide (23, 33). A more subtle mutation at the catalytic arginine finger, R77A, renders the protein's GTPase inactive despite preserving GDP and GTP binding with affinities comparable with wild-type enzyme; nevertheless, if left to equilibrate for extended periods of time, the enzyme begins to exhibit the propensity to dimerize in Gpp(NH)p and GDP·AlFx (23) (Fig. 1A, pink plus symbol). Overall, these mutational analyses show that even in cases where GTPase activity and nucleotide binding are compromised, ATLs are still able to oligomerize at equilibrium. At the same time, equilibrium oligomerization does not equate to proper function, as cellular ATL function depends on coordinated, GTPase-driven conformational and quaternary structure changes for membrane fusion (22, 25).
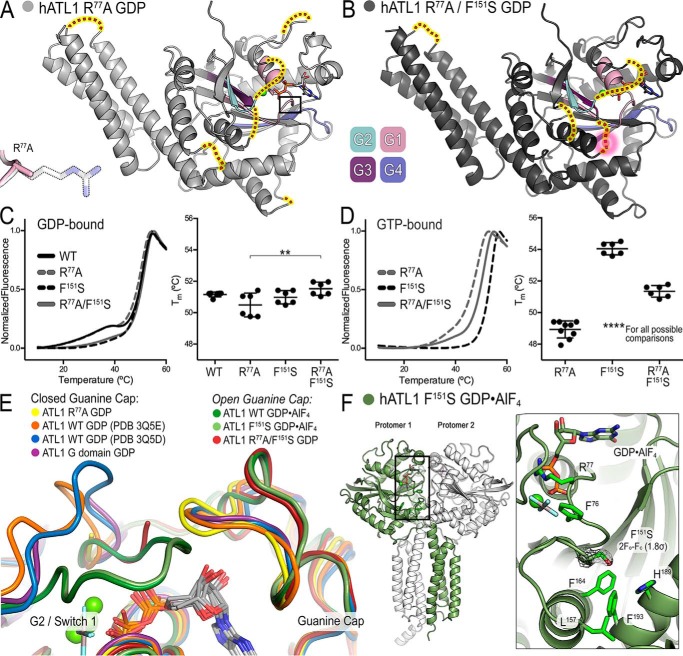
Figure 1.
ATL1 with the atypical HSP mutation F151S is catalytically deficient but retains ability to bind nucleotide. A, re-evaluation of the correlation between GTPase activity and dimerization propensities of HSP mutations found in ATLs. B, the atypical F151S mutation is located between the active site and intramolecular domain interface responsible for nucleotide loading. Motifs characteristic of G proteins including the switch regions are shown in color. G1/P-loop (pink), G2/switch 1 (teal), G3/switch 2 (dark purple), and G4 (blue/purple) are indicated along with HSP mutations proximal to Phe151. A schematic depicting the engaged, tight cross-over, and relaxed cross-over states is shown above the panels to indicate the crystallographically determined conformations. C, turnover number (kcat) of phosphate release for wild-type ATL1, catalytically deficient ATL1-R77A mutant, and ATL1-F151S. Kinetic experiments were conducted with the soluble catalytic core fragments, comprising G and middle domains. D, affinity of wild-type and F151S-containing ATLs for GTP and GDP were calculated using mant-nucleotides. Graphs showing means and S.D. (error bars) are plotted from two biological replicates with three technical repeats each.
Here we characterized the HSP mutant ATL1-F151S, assessing its GTPase activity, nucleotide affinity, and oligomerization propensity. Phe151 is located in G3/switch 2 (Fig. 1B, dark purple motif), positioned between the site of nucleotide hydrolysis and an intramolecular interface where the middle domain docks onto the G domain in a conformation that we previously identified as a requirement for GTP loading (23, 30) (Fig. 1B). Additional conserved elements responsible for phosphate and metal coordination, guanine-base specificity, and catalytic residues include G1/P-loop (pink), G2/switch 1 (teal), and G4 (blue/purple) (37); neighboring HSP mutations and select hydrophobic residues are also illustrated (Fig. 1B).
The biochemical properties of ATL1-F151S indicate a deviation from trends presented by previously analyzed HSP and engineered mutants (30, 31, 33) (Fig. 1A), although F151S does present the same dominant-negative effect on ER morphology as other HSP mutations (16, 38). In particular, ATL1-F151S was unable to hydrolyze GTP with a turnover number equivalent to the catalysis-deficient ATL1-R77A mutant, which is in agreement with previous reports (33) (Fig. 1C). Notably, F151S confers one of the lowest GTPase rates of any HSP mutation that is not in direct contact with the nucleotide. Because the mutation is removed from the active site, it was not surprising that ATL1-F151S retains the ability to bind both GTP and GDP, although it is curious that it does so at higher affinities than wild-type protein (Fig. 1D).
ATL1-F151S dimerizes only when bound to the transition-state analog at equilibrium
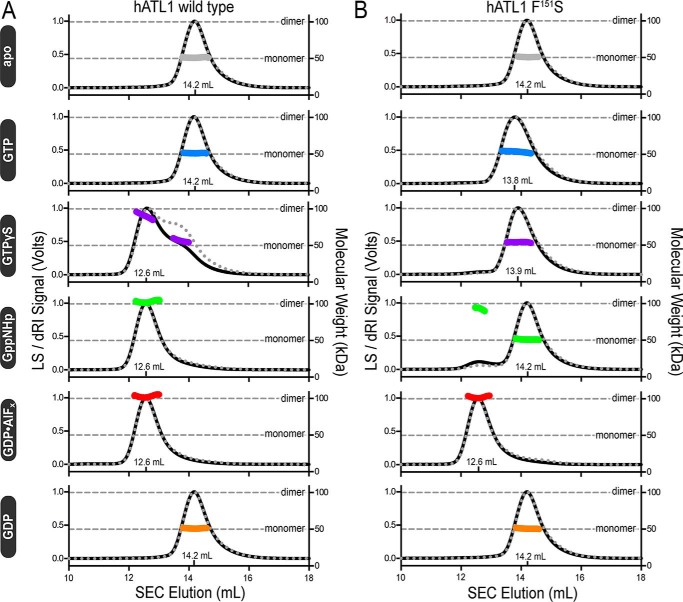
Considering that ATL1-F151S is catalytically inactive but retains the ability to bind nucleotide (Fig. 1, C and D), we would predict that its dimerization profile would be similar to that of the R77A mutant (23). The oligomerization capability of ATL1-F151S was assessed upon incubating the protein with various nucleotides and nucleotide analogs and measuring the molecular weights of the protein/nucleotide mixtures using size exclusion chromatography–coupled multiangle light scattering (SEC-MALS) (39). The wild-type ATL1 catalytic core exhibited nucleotide-dependent dimerization consistent with previous studies where nucleotide-free (apo) and GDP-bound proteins were monomeric with a molecular mass of ∼50 kDa, and Gpp(NH)p and GDP·AlFx were dimeric with a molecular mass of ∼100 kDa (26, 30) (Fig. 2A). GTPγS-bound, wild-type ATL1 comprises a mixture of dimeric and monomeric species (60% dimer, 40% monomer; cumulative weight analysis), which can be rationalized by either hydrolysis of GTPγS at high protein concentrations (40 μm) or weaker overall dimerization that is more affected by buffer exchange (GTPγS was not present in the mobile phase) (Fig. 2A). ATL1 equilibrated with GTP eluted as a monomer due to both substrate consumption and buffer exchange into the mobile phase void of GTP (as indicated above) (Fig. 2A).
Figure 2.
Molecular weight determination indicates that ATL-F151S only dimerizes when bound to the transition-state analog, GDP·AlFx. A, absolute molecular weights (colored data points across elution peaks are plotted on the right axis; theoretical monomer and dimer molecular weights are shown as horizontal gray lines) of wild-type ATL catalytic core fragment (injection: 40 μm ± 1 mm nucleotide) were determined using SEC-MALS (90° light scattering (black solid line) and refractive index signal (gray dotted line) are plotted on the left axis). Wild-type enzyme samples the canonical monomer/dimer states described previously (23, 26). B, SEC-MALS data for the corresponding ATL1-F151S construct using the same experimental conditions as in A. Representative data, collected on the same day, from at least two independent experiments are shown.
Analogous to wild-type protein, apo- and GDP-bound species of ALT1-F151S eluted as monomers (Fig. 2B). The light-scattering chromatogram from GTP-bound ATL1-F151S displays two interesting signatures; the GTP-bound species eluted ∼0.4 ml earlier and was band-broadened by ∼0.3 ml, as determined from the difference between initial and final elution volumes at half-maximal intensities when compared with other monomer SEC profiles from both wild-type ATL1 and ATL1-F151S (Fig. 2, A and B). Because the molecular weight is consistently monomeric across the peak, this observation suggests that ATL1-F151S is able to undergo a specific conformational change when bound to GTP that has not been observed in other protein-nucleotide complexes. We expected ATL1-F151S to dimerize in the presence of GTPγS and Gpp(NH)p due to the similar nucleotide-binding abilities shared with R77A (23); however, ATL1-F151S remained monomeric when bound to GTPγS, and cumulative weight fraction analysis indicated that only ∼5% of the population was dimeric when bound to Gpp(NH)p (Fig. 2B). In contrast to the GTPγS and Gpp(NH)p results, ATL1-F151S can fully dimerize when bound to GDP·AlFx. This nucleotide-dependent dimerization profile makes ATL1-F151S unique and the only HSP mutant protein characterized to date that is deficient in catalytic cycling, unable to dimerize when bound to non-hydrolyzable GTP analogs, but capable of dimerizing with a transition-state analog.
ATL1-F151S retains its ability to dimerize through both G and middle domains when bound to GDP·AlFx
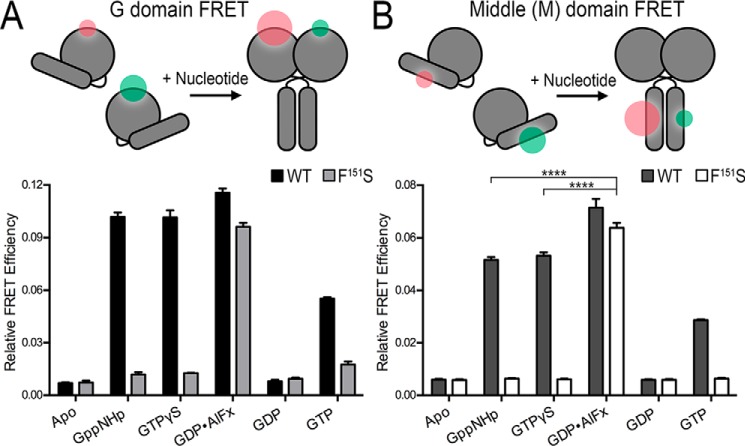
SEC-MALS analysis reports on the absolute molecular weight of the species in solution but cannot discriminate between the relative orientation of the G and middle domains in dimeric assemblies. Using FRET-based approaches, we have previously shown that ATLs proceed through a series of conformational changes where dimerization of the G and middle domains occurs in close succession (23, 26). To determine the conformations that ATL1-F151S is able to adopt, we used the same FRET sensors to differentiate between G and middle domain dimerization (Fig. 3). In short, engineered surface-exposed cysteine residues were incorporated into the G or middle domains and labeled with small organic donor or acceptor fluorophores (23, 26) (Fig. 3, A and B, top). Equilibrium FRET efficiencies in the presence of various nucleotides were calculated for ATL1 wild type and the corresponding F151S variant; the duration required to reach equilibrium was determined via kinetic time courses (Fig. S1). As in previous studies (23, 26), the G and middle domains of ATL1 wild type formed homodimers in the presence of Gpp(NH)p, GTPγS, and GDP·AlFx, whereas the enzyme remained monomeric when bound to GDP or in the absence of nucleotide (Fig. 3, A and B). In agreement with the SEC-MALS molecular weight determination, the ATL1-F151S catalytic core fragment was unable to dimerize with non-hydrolyzable nucleotides but formed G and middle domain homodimers when bound to GDP·AlFx. Because the middle-domain FRET efficiency for ATL1-F151S bound to GDP·AlFx was greater than wild type bound to non-hydrolyzable analogs (****, p ≤ 0.0001), we can conclude that the mutant is competent in forming the high-affinity transition-state dimer (25, 26) (Fig. 3B). A non-equilibrium measurement was conducted at steady state in the presence of GTP. The wild-type G and middle domain FRET exhibit intermediate efficiencies, which is in agreement with a cycling system that samples both monomeric and dimeric states. In contrast, F151S lacks pronounced G and middle domain FRET under the same conditions (Fig. 3, A and B).
Figure 3.
ATL1-F151S utilizes both the G and middle domains in the transition-state dimer. A, equilibrium and steady-state G domain FRET dimerization. Both wild-type and ATL1-F151S proteins were labeled site-specifically (K295C) with donor (Alexa Fluor 488) and acceptor (Alexa Fluor 647) FRET fluorophores, and measurements including 1 μm protein and 500 μm nucleotide were taken once equilibrium (GTPγS, Gpp(NH)p, GDP, or GDP·AlFx) or steady-state (GTP) conditions were fulfilled. B, equilibrium and steady-state middle domain FRET dimerization. Analogous FRET measurements were conducted with wild-type and ATL1-F151S proteins labeled site-specifically at their middle domains (K400C). ATL-F151S has a statistically higher FRET efficiency than wild-type bound to non-hydrolyzable analogs (****, p ≤ 0.0001). Graphs showing means and S.D. (error bars) are plotted from two biological replicates with three technical repeats each. Sizes of the green and red halos in the schematic diagrams (top panels) illustrate the fluorophore intensities in FRET and non-FRET states.
Structural characterization of ATL1 variants with diverse dimerization capacities
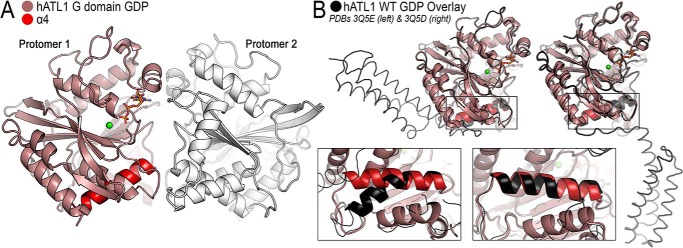
GTPase-driven dimerization is crucial for ATL function (19, 21, 22, 24). Previous studies shed light on the dimer conformations that ATLs sample (23, 26, 30, 33, 40, 41), but the conformations of the monomeric states are less well-defined. Here, we determined crystal structures of ATL1 mutants R77A, R77A/F151S, and F151S, all of which display distinct oligomerization propensities. We also crystallized the isolated G domain in the presence of GDP·Mg2+ as another reference model for a catalytic and dimerization-incompetent state. The G domain fails to dimerize in any nucleotide because it cannot adopt the GTP loading–competent state without the middle domain (23).
A structure for the isolated G domain of ATL1 (residues 1–339) was solved in space group F23, containing four molecules per asymmetric unit, at a maximum resolution of 3.0 Å (Table 1). The four GDP·Mg2+-bound protomers assemble into two identical crystallographic dimers via a G domain dimer interface seen in previously published, GDP·Mg2+-bound ATL structures despite the protein's inability to oligomerize in solution (23, 26, 30, 33, 42) (Fig. 4A). One key feature of this structure is the configuration of the fourth α-helix (α4) of the G domain fold, which is straight. Based on previous ATL1 structures, this helix can adopt two principal configurations, straight and bent (26, 30, 33, 42) (Fig. 4B). When bent, G and middle domains are engaged, and this interaction enables GTP loading (23). GTP binding is followed by an intramolecular conformational change that is thought to represent the release of the middle domain from the G domain (26). The straightening of helix α4 is one potential mechanism for the kick-off of the middle domain. In conclusion, the structure of the isolated G domain represents a GTPase cycling–incompetent state and provides further structural evidence that helix α4 bending requires middle domain docking, which is associated with GTP binding. Of note, Phe151 is resolved in the isolated G domain structure. The residue shares the same conformation and environment as seen in a crystallographic state of ATLs bound to GDP·Mg2+ (see Fig. 1B (right) for reference), where the middle domain is dislodged from the G domain, and G and middle domains form weak homodimers with a adjacent protomer in the crystal lattice, which is thought to represent a post-hydrolysis dimer (30, 33).
Table 1.
X-ray data collection and refinement statistics
| ATL1-R77A; Mg2+, GDP 1–446 | ATL1-R77A/F151S; Mg2+, GDP 1–446 | ATL1-F151S; Mg2+, GDP·AlFx 1–446 | ATL1 G domain; Mg2+, GDP 1–339 | |
|---|---|---|---|---|
| Data collection | ||||
| X-ray source | CHESS | CHESS | CHESS | CHESS |
| Wavelength | 0.9759 | 0.6314 | 0.977 | 0.9759 |
| Space group | P1 | P1 | P212121 | F23 |
| Unit cell | ||||
| a, b, c (Å) | 51.1, 68.4, 75.9 | 52.1, 68.1, 76.0 | 49.6, 115.7, 184.3 | 308, 308, 308 |
| α, β, γ (degrees) | 117.0, 89.8, 99.0 | 116.9, 90.8, 98.5 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 42.36–1.95 (2.06 – 1.95)a | 42.75–1.99 (2.10–1.99) | 49.0–1.90 (2.00–1.90) | 48.70–3.00 (3.16–3.00) |
| No. of reflections | ||||
| Total | 153,067 (22,418) | 117,856 (15,186) | 571,474 (55,219) | 1,229,123 |
| Unique | 63,315 (9210) | 60,236 (7994) | 83,775 (11,190) | 48,244 (7006) |
| Completeness (%) | 96.4 (96.2) | 95.7 (86.9) | 98.8 (92.6) | 99.9 (99.8) |
| Multiplicity | 2.4 (2.4) | 2.0 (1.9) | 6.8 (4.9) | 25.5 (25.3) |
| I/σ(I) | 7.1 (2.1) | 9.9 (1.5) | 10.7 (1.3) | 22.8 (3.0) |
| Rmeas (%) | 10.3 (56.0) | 8.6 (85.7) | 12.9 (137.3) | 20.1 (145.5) |
| Rmerge (%) | 7.9 (43.0) | 6.4 (63.4) | 11.9 (123.5) | 19.7 (142.6) |
| CC½ | 0.991 (0.784) | 0.996 (0.598) | 0.997 (0.419) | 0.999 (0.847) |
| Refinement | ||||
| Rwork/Rfree (%) | 20.5/23.9 | 20.3/24.4 | 17.8/22.2 | 17.0/20.5 |
| RMSD | ||||
| Bond length (Å) | 0.007 | 0.009 | 0.007 | 0.002 |
| Bond angles (degrees) | 0.929 | 0.973 | 0.865 | 0.534 |
| No. of atoms | ||||
| Protein | 5872 | 6088 | 6660 | 9730 |
| Ligands | 94 | 63 | 116 | 116 |
| Water | 281 | 198 | 557 | 92 |
| Average B-factors (Å2) | ||||
| Protein (total) | 43.3 | 44.3 | 35.3 | 68.0 |
| Ligands | 40.8 | 54.8 | 33.2 | 49.7 |
| Water | 44.0 | 38.8 | 42.4 | 52.9 |
| Ramachandran (%) | ||||
| Favored | 97.7 | 98.0 | 98.2 | 96.4 |
| Outliers | 0 | 0 | 0 | 0 |
| PDB code | 6B9D | 6B9E | 6B9F | 6B9G |
a Values in parentheses are for the highest-resolution bin.
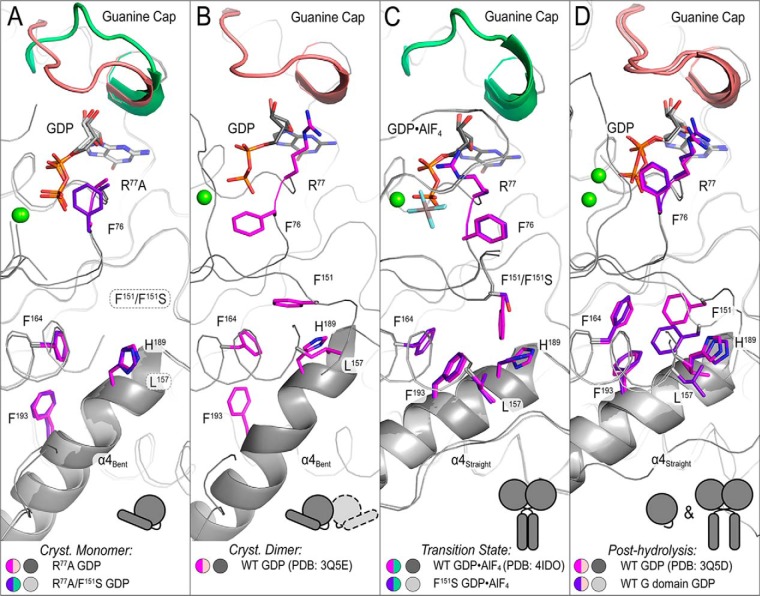
Figure 4.
Crystal structure of the isolated G domain of ATL1 bound to GDP·Mg2+. A, the G domain structure displays weak crystal packing interactions that involve switch regions. The G domain (colored salmon), helix α4 (red), and a second protomer (white) are illustrated. B, comparison of the isolated G domain with either the engaged (PDB code 3Q5E; left) or relaxed cross-over (PDB code 3Q5D; right) structures (both black).
One caveat with previous structural studies of ATL, including the isolated G domain above, stems from a preferred crystal packing interaction where the G domains of two molecules form a loose dimer even when the protein is bound to GDP·Mg2+ (23, 26, 33, 42). This conformation is reminiscent of the tight dimer observed in the transition state and involves the switch regions (G1–G4) of the G domains but has a smaller interfacial area than the transition state (23). On the other hand, in solution, GDP·Mg2+ does not support protein dimerization (23, 26, 43) (Figs. 2 and 3), and hence the corresponding structures may not portray the ensemble of solution conformations accurately. Here, we developed a strategy to capture a structure of GDP·Mg2+-bound ATL1 with monomeric G domains by mutating Arg77, a residue that forms a salt bridge with Glu224 from an adjacent G domain in all GDP-bound crystal structures determined to date. Using crystals that diffracted X-rays to 1.95 Å, we were able to determine the structure of ATL1-R77A (residues 1–446) bound to GDP·Mg2+. The structure was solved by molecular replacement in space group P1 with two molecules in the asymmetric unit (Table 1). The R77A mutation led to novel crystal packing contacts within the asymmetric unit and between unit cells that differ from those described previously (Fig. S2A). Within the asymmetric unit (Fig. S2B, molecules 1a and 1b), protomers have an average interface area of 491 Å2; between unit cells, the G domains have an interface area of 969 Å2 (molecules 1b and 2a), and middle domains (molecules 3a and 1b) have an interface area of 576 Å2 (Fig. S2C). However, none of these interfaces have been predicted to be biologically relevant (44), and they do not involve the G domain's switch regions or active site. As a result of these new packing interactions, the switch regions are free to move and are consequently disordered (Fig. 5A), consistent with the conformations observed for other G proteins bound to GDP (45). Notably, the loop containing the R77A mutation is resolved, and the alanine residue occupies the same conformation and orientation as the catalytic arginine residue in the wild-type structure (PDB entry 3Q5E) (Fig. 5A, inset). Based on the structural changes, we propose that this structure represents the prehydrolysis, monomeric form of ATL1.
Figure 5.
Structures of ATL1 mutants have diverse dimerization capacities and guanine cap configuration. A, ATL1-R77A bound to GDP·Mg2+ (light gray) exists in a monomeric state with G and middle domains engaged. Switch regions (G1 (pink), G2 (teal), G3 (dark purple), and G4 (blue/purple)) are largely disordered, and their hypothetical locations are indicated (red dots highlighted yellow). Inset, α-carbon backbone of loop containing R77A and the β-carbon of residue 77 are not perturbed. B, ATL1-R77A/F151S bound to GDP·Mg2+ (dark gray) also exists in a monomeric state with G and middle domains engaged. Switch regions, colored as in A, are more ordered than in the isomorphic ATL1-R77A structure, and their hypothetical locations are indicated (red dots highlighted yellow). The F151S mutation is part of the G3 switch region and is disordered in this structure; hypothetical location of the mutation is outlined by a pink halo. C, thermal melting data for wild-type ATL1, ATL1-R77A, ATL1-F151S, and ATL1-R77A/F151S bound to GDP. The Tm associated with the major unfolding phase is significantly different for ATL1-R77A and ATL1-R77A/F151S (**, p ≤ 0.01). D, thermal melting data for ATL1-R77A, ATL1-F151S, and ATL1-R77A/F151S bound to GTP. A single unfolding phase exhibits very different Tm values for the three mutant proteins (****, p ≤ 0.0001 for all possible comparisons). E, the indicated crystal structures were aligned using the G domain as a reference to observe the conformation of the guanine cap. The ATL1-R77A structure (yellow) had this loop closed onto the guanine base like other GDP·Mg2+-bound structures (orange, blue, and purple). However, both the ATL1-R77A/F151S GDP-bound (red) and the ATL1 -F151S GDP·AlF4−-bound (light green) structures mirror the wild-type transition-state structure (dark green) and have a retracted guanine cap. F, ATL1-F151S bound to GDP·AlF4− (protomer 1 (green) and protomer 2 (white)) is fully capable of equilibrating into the high-affinity transition state despite the F151S mutation being buried within the core of the enzyme. 2Fo − Fc map for the F151S mutation contoured to 1.8σ is depicted in black mesh. In C and D, melting curves showing means and Tm values showing means ± S.D. (error bars) are plotted from two biological replicates with a minimum of three technical repeats each.
To attain a monomeric form of ATL1-F151S, we paired the HSP mutation with R77A and determined the structure of ATL1-R77A/F151S bound to GDP·Mg2+ in space group P1 with two molecules in the asymmetric unit (Table 1). Globally, ATL1-R77A/F151S adopts the same crystal packing and contacts as found in GDP·Mg2+-bound ATL1-R77A (Fig. S3). Unfortunately, the F151S mutation was located on a flexible loop of the G3/switch 2, which could not be resolved in either the ATL1-R77A or -R77A/F151S structures; the presumed location of the mutation is indicated (Fig. 5B, pink halo). Despite the high degree of similarity between the two structures (RMSD = 0.25 Å), the ATL1-R77A/F151S structure has fewer disordered loops than ATL1-R77A, which is most likely due to the addition of the F151S mutation (Fig. 5B).
We used an established fluorescence-based thermal shift assay to determine whether the F151S mutation confers temperature-dependent stability in solution. When bound to GDP, wild-type and R77A proteins exhibit two phases of unfolding, with a minor unfolding transition occurring at 40 °C (accounting for 20% of the total fluorescence signal) and a major, second transition occurring at 56 °C. Upon the addition of the F151S mutation into both wild-type and R77A constructs, the initial unfolding phase at 40 °C was not observed (Fig. 5C, left). Tm values for the GDP-bound proteins were calculated for the predominant unfolding phase, and they were significantly different between ATL1-R77A and -R77A/F151S with a difference of 1.0 °C (**, p ≤ 0.01) (Fig. 5C, right). We also tested the thermal stability of ATL1-R77A, -F151S, and -R77A/F151S bound to GTP and determined that the F151S mutations alone or in combination with R77A confers a 5.2 °C and 2.8 °C stabilization, respectively (Fig. 5D). The stabilization exhibited by F151S·GTP is additional evidence for the conformational change that resulted in the shifted elution and peak broadening determined via SEC-MALS (Fig. 2B).
To further delineate the effect that the addition of R77A contributes to the double mutant, we conducted SEC-MALS–based molecular weight determination for both ATL1-R77A and -R77A/F151S in the presence of various nucleotides (Fig. S4). The data indicate that ATL1-R77A/F151S does not dimerize with GTP, non-hydrolyzable analogs, or the transition state, unlike the variant with the R77A mutation alone. Arg77 is also required for the apparent band broadening and peak shift observed with ATL1-F151S (Fig. 2B), as elution peak volume and peak width changes are affected in the ATL1-R77A/F151S double mutant compared with ATL1-F151S (Fig. S4B and Fig. 2B). This result suggests a conformation where Arg77 assumes its nucleotide-coordinating conformation in ATL1-F151S·GTP, and, based on the SEC-MALS data above, this occurs in a monomer.
Additional structural differences pertain to the guanine cap, a motif that is important for guanine-base binding and specificity (37). In the structure of GDP-bound ATL1-R77A/F151S, this motif adopts an open conformation (Fig. 5E (dark red trace) and Fig. S5). Based on an overlay with all other crystal forms (i.e. G/M-engaged, transition state, and relaxed crossover (23, 30)), this guanine cap conformation and the resolved portion of G2 mirrors the transition-state form of ATL1 (Fig. 5E, green traces). In contrast, ATL1-R77A adopts a conformation more similar to all other GDP-bound structures, with the guanine cap adopting a closed configuration (Fig. 5E).
We were also able to attain the transition-state structure of ATL1-F151S with crystals diffracting X-rays to 1.9 Å. Phasing by molecular replacement and subsequent refinement in space group P212121 yielded the final model with two molecules per asymmetric unit (Table 1 and Fig. 5F). Despite including a non-conservative mutation in the buried core of the G domain, the structure is well-resolved and displays the anticipated change in electron density at position 151 (Fig. 5F, inset). ATL1-F151S bound to GDP·AlF4− is virtually identical to the synonymous wild-type transition-state structure (PDB code 4IDO) with an RMSD of 0.22 Å (23). This structure indicates that the protein with the F151S mutation is fully capable of equilibrating into the high-affinity transition state but is unable to reach this energetic state with the endogenous substrate GTP.
Discussion
Here we have characterized the atypical HSP mutation ATL1-F151S that is deficient in GTP-hydrolysis and cannot dimerize when bound to GTP or non-hydrolyzable analogs but retains the ability to adopt the dimeric transition state when bound to GDP·AlFx. Mechanistically, this implicates Phe151 in reducing the activation energy required for GTP hydrolysis and/or interdomain allostery linking nucleotide hydrolysis and middle domain engagement. Structural comparisons suggest a model where Phe151 may represent the central residue that couples the enzyme's active site with the functionally important G/middle domain interface (Fig. 6). Specifically, crystal structures of various states reveal a hydrophobic tract that links the active site to helix α4, a motif that transitions from bent to straight during GTP hydrolysis and the intramolecular release of G and middle domains. The proposed hydrophobic tract begins with Phe76 of the G1/P-loop, a residue that is adjacent to the catalytic arginine (Arg77) and thought to sense the γ-phosphate moiety. In the engaged, monomeric ATL1-R77A structure, the side chain of Phe76 is disordered but is resolved in the isomorphic ATL1-R77A/F151S structure, where it points toward the bound GDP (Fig. 6A). These monomeric ATL structures may represent the hitherto elusive nucleotide-loading state, as they are the first structures where the switch regions and guanine cap are not stabilized through crystallographic contacts (Fig. 6A). Weak crystal packing interactions that involve the G domain and its switch regions (PDB code 3Q5E) allow Phe76 to rotate ∼180° toward switch 1 (Fig. 6B). In the presence of a nucleotide with γ-phosphate or a transition-state analog, Phe76 moves outward in concert with major reorientations of the switch regions that stabilize the phosphate moieties and align Arg77 for nucleophilic attack (Fig. 6C) (23). In the post-hydrolysis states, Phe76 may return to the configuration seen in the engaged monomeric structures, which may aid phosphate release and resetting of the enzymatic cycle (Fig. 6D).
Figure 6.
Structure-based model for a hydrophobic interaction network that establishes interdomain allostery. Hydrophobic residues connect the active site (GDP (white/black) and guanine cap (green/salmon)) to helix α4 (schematic helix). A, structures depicting crystallographic monomers (ATL1-R77A (pink/dark gray) and ATL1-R77A/F151S (purple/light gray)) are flexible with Phe76 (in ATL1-R77A structure), F151S, and Leu157 being disordered and helix α4 bent. B, when switch regions are stabilized via homotypic crystallographic dimer contacts, the G/middle domain-engaged structure (PDB code 3Q5E, pink/dark gray) shows Phe76 rotating away from the nucleotide and Phe151 being resolved. C, both wild-type (PDB code 4IDO, pink/dark gray) and ATL1-F151S (purple/light gray) transition-state structures exhibit a switch reorganization that is accompanied with a downward rotation of Phe151 that pushes against His189 and Leu157. The helix α4 becomes straight, and Phe193, which is located at the helix's bend and may contribute energetically to helix straightening, rotates to form hydrophobic interactions with other residues labeled in this panel. D, structures resembling the post-hydrolysis state (PDB code 3Q5D (pink/dark gray) and isolated G domain (purple/light gray)) exhibit a configuration that is en route to resetting the cycle. The helix α4 remains straight, and upstream residues (Phe76 and Phe193) begin to take on conformations seen in A.
Phe76 is proximal to Phe151, the site of the HSP disease mutation investigated above. Phe151 adopts a number of conformations when comparing the various crystallographic states and is not resolved in the monomeric, engaged structures where the switch regions are largely disordered (Fig. 6A). During the large reorganization of the switch regions leading to the conformation depicted in the transition-state crystal structures, Phe151 transitions from interacting with a single hydrophobic residue (Phe164) to an environment that is highly hydrophobic. As Phe151 rotates away from the active site, it pushes against Leu157 and His189 located on helix α4 (Fig. 6, B and C). Pushing against these residues coincides with a rotation of helix α4, which results in its straightening. The straightening of helix α4 also involves Phe193, which is located at helix α4's bend; this residue rotates to interact with the newly formed cluster of hydrophobic residues (Phe151, Leu157, Phe164, and His189) and may provide energetics for the aforementioned molecular rearrangement (Fig. 6C). It should also be noted that this structural mechanism is not limited to an active site-outward relay for the GTPase-driven release of the middle domain from the G domain, but is most likely bidirectional based on observations that the G/middle domain interface is required for nucleotide loading (23, 26). Finally, the described network of hydrophobic interactions is probably a fundamental feature of this DRP subfamily, considering the high conservation of these residues in ATL orthologs, including isoforms from Drosophila, silkworm, zebrafish, and higher eukaryotes.
The two F151S-containing crystal structures establish that the mutant proteins in principle can sample the mechanistically relevant nucleotide-loading and transition states, but cannot adopt the latter under physiological conditions due to lack of GTP hydrolysis (Fig. 6, A and C). In the transition-state structure, Arg77 adopts its catalytically competent conformation by coordinating the phosphate moieties, identical to the conformation of the wild-type protein in this state (Fig. 6C) (23). Arg77 is also involved in the GTP-induced changes in hydrodynamic properties of monomeric ATL1-F151S (Fig. 2B and Fig. S4). Together, these results suggest that Arg77 not only serves as the catalytic arginine finger but may also contribute to the allosteric network by communicating with Phe76 and Phe151, and this communication is broken in the F151S mutant. Curiously, the protein with the F151S mutation has an even higher affinity for GTP than the corresponding wild-type ATL1, which can be rationalized based on the crystal structure of ATL1-R77A/F151S (G/middle-engaged) (Fig. 5E and Fig. S5). With F151S, the monomeric protein exhibits an open conformation of the guanine cap that was previously reserved for structures adopting the tight cross-over dimer (e.g. the transition-state structure) (23). The guanine cap is not stabilized through crystal packing in either ATL1-R77A or ATL1-R77A/F151S structures, indicating the side chain identity at position 151 as the intrinsic reason for the apparent differences in ATL conformation and/or dynamics observed in the crystal structures.
Notably, two residues that Phe151 interacts with (Leu157 and His189) are both HSP disease mutants (46). Biochemical characterization of the corresponding disease variants, ATL1-L157W and ATL1-H189D, established that they are both catalytically active and able to fully dimerize when bound to either Gpp(NH)p or GDP·AlFx (Fig. 1A) (30). This difference in mutant protein switching and activity argues that Phe151 is a central residue responsible for the allosteric coupling between the active site and intramolecular interface where the middle domain docks onto the G domains and regulates nucleotide binding, hydrolysis, and conformational changes.
This study was centered on understanding the mechanistic underpinnings of a disease-causing mutation in ATL1 and, in turn, revealed a pivotal intrinsic allosteric network that couples the enzyme's active site to the interface between G and middle domains. Perturbation at position Phe151 affects regulation across this allosteric pathway, and because other disease mutations often localize to the region between the active site and helix α4, we speculate that mutations are similarly impacting allostery, just less severely. The varying degrees of severity in HSP mutations are unique to their biochemistry, but the resulting cellular phenotypes invariably manifest as an unbranched ER (16, 31). Rationales for the potent dominant-negative ER phenotype seen with HSP mutations are based on ATL's higher-order oligomerization in cells and its involvement in larger, functional complexes with other protein, presumably involving the transmembrane segments of ATLs (14, 47). A functionally compromised copy of ATL is likely to poison these homo- and heterocomplexes, leading to futile membrane fusion attempts and unbranched ER tubules, even if wild-type proteins are present. Our observations indicate that comprehensive structural and biochemical analysis of disease mutations is an advantageous route to verify the specifics of aberrant ATL regulation and potentially elucidate novel regulatory mechanisms (exemplified by studying F151S), many of which could be indispensable for proper function and disease pathogenesis.
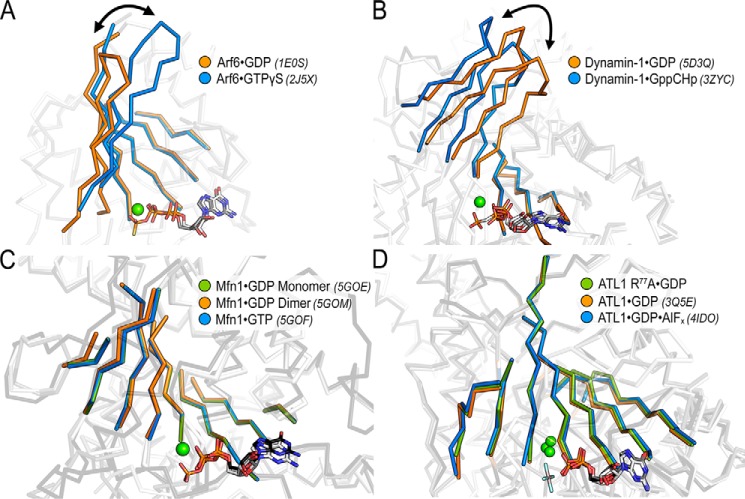
Similar to the straightening of helix α4 in ATL, both small and large G proteins have established means of linking nucleotide sensing to functional outputs. Small Arf-like G proteins couple GTP binding to the release of an N-terminal helix for membrane association (termed “front-to-back communication”), which is achieved through a two-residue registry shift in the β-sheet connecting switches 1 and 2 (48–50) (Fig. 7A). This positional shift of β-sheets has also been described for dynamin, where GTP binding initiates a tilting of the core β-sheet away from the active site enabling the release of the bundle signaling element (a domain analogous to ATL's middle domain) (51–53) (Fig. 7B). To determine whether this feature is conserved among other DRPs, we analyzed structures of mitofusin bound to GDP and GTP. Based on the available structures, mitofusin lacked both the positional change in the core β-sheet and the straightening of an α-helix akin to helix α4 in ATL (54) (Fig. 7C). The β-sheet modulation was also absent in ATL; however, structural information of the monomeric, GTP-bound state remains elusive for ATL. Thus, we cannot rule out a mode of intradomain allostery described for Arf and dynamin until corresponding structural data are available. However, comparing the various modes of allosteric coupling based on current knowledge illustrates the unique differences in regulation that have been adopted by various DRPs.
Figure 7.
Evidence for nucleotide-sensing via β-sheet modulation is elusive for Mfn or ATL. A, Arf-like G proteins exhibit a nucleotide dependent registry shift in a β-sheet connecting switches 1 and 2 (Arf6·GDP (orange), PDB code 1E0S; Arf6·GTP (blue), PDB code 2J5X), enabling the release of an N-terminal helix (46, 47). B, the core β-sheet of dynamin tilts in response to nucleotide (dynamin-1·GDP (orange), PDB code 5D3Q; dynamin-1·GppCHp (blue), PDB code 3ZYC) and has been proposed to release the bundle signal element (49–51). C, the core β-sheet of mitofusin does not change in response to GDP (Mfn1·GDP monomer (green), PDB code 5GOE; Mtfn-1·GDP dimer (orange), PDB code 5GOM) or GTP (Mtf1·GTP (blue), PDB code 5GOF) (52). D, the core β-sheet of ATL also remains unperturbed between crystallographic states and various bound nucleotides (ATL1-R77A·GDP, green; ATL1·GDP, orange, PDB code 3Q5E; ATL1·GDP·AlF4−, blue, PDB code 4IDO).
Experimental procedures
Phosphate release kinetics
The release of free phosphate was detected using the Enzchek phosphate assay kit and the manufacturer's protocol (Molecular Probes, Inc., Eugene, OR) with the exception of the addition of 100 mm NaCl to the reaction buffer. Reactions consisted of a 100-μl volume, 1 μm enzyme, and 500 μm GTP (Thermo Fisher Scientific, Waltham, MA), incubated in half-area, 96-well plates (Corning, Inc., Corning, NY) at 25 °C. Data were recorded at an absorbance of 366 nm using a BioTek Synergy II plate reader, and the initial rate before 10% substrate conversion was fit to a linear regression and converted to units of μm/min using a standard curve. Turnover number is defined as kcat = V/[enzyme].
Determination of dissociation constants with N-methylanthraniloyl (mant)-nucleotides
On and off rates were determined from mant-nucleotide binding experiments (23, 55) and used to calculate dissociation constants for mant-GDP and mant-GTP (Thermo Fisher Scientific, Waltham MA). A KinTek SF-2004 stopped flow was used to rapidly mix mant-nucleotides and ATL to a final concentration of 2.5 μm nucleotide and increasing amounts of ATL (5–20 μm) at 25 °C. Samples were excited at 366 nm, and signal from emission filter HQ460/40 M (Chroma Technology Corp., Rockingham, VT) was collected. Data were fit to exponential association equations, and the rate constants were plotted versus ATL concentration; the resulting slope of the linear fit corresponds to kon. koff was directly measured by preloading ATL with mant-nucleotide and chasing with a high concentration of nucleotide (final concentrations of 2.5 μm ATL·mant-nucleotide and 2.5 mm unlabeled nucleotide). The resulting data were fit to a single-exponential decay curve where the observed rate constant corresponds to koff. Dissociation constants (Kd) were calculated as koff/kon.
Size-exclusion chromatography coupled to multiangle light scattering
Protein/nucleotide mixtures (40 μm ATL and 1 mm nucleotide) were injected onto a Superdex 200 Increase 10/300 column (GE Healthcare), equilibrated with 25 mm HEPES, pH 7.5, 100 mm NaCl, 4 mm MgCl2, and 2 mm EGTA. The GDP·AlFx sample was supplemented with 2 mm aluminum chloride and 20 mm sodium fluoride. Before injection, protein/nucleotide mixtures were incubated for a minimum of 1 h. The gel filtration column was coupled to a static 18-angle light scattering detector (DAWN HELEOS-II) and a refractive index detector (Optilab T-rEX) (Wyatt Technology, Goleta, CA). Data were collected every second at a flow rate of 0.7 ml/min. Data analysis was carried out using ASTRA VI, yielding the molar mass and mass distribution (polydispersity) of the sample. The monomeric fraction of BSA (Sigma; 5 mg/ml) was used to normalize the light scattering detectors and used as a positive quality control.
Dye labeling and FRET measurements
Site-specific dye labeling was achieved by replacing non-conserved surface amino acids with cysteine residues within the G or middle domains (23, 26). To obtain singly labeled protein, a surface-exposed cysteine residue was replaced with an alanine residue (C375A). A cysteine residue for site-specific labeling was introduced at a surface-exposed position on either the G (K295C) or middle (K400C) domain. Labeling was conducted at 100 μm protein, 150 μm Alexa Fluor 488 or Alexa Fluor 647 (Molecular Probes) in a buffer containing 25 mm HEPES, pH 7, 100 mm NaCl. This reaction was incubated on ice for 30 min, and excess dye was removed with a NAP-5 column (GE Healthcare) pre-equilibrated with buffer containing 25 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm MgCl2. Data for FRET equilibrium and time-course measurements were collected with a Gemini EM microplate reader (Molecular Devices, Sunnyvale, CA) with sensor excitation at 473 nm and fluorescence recorded at 515 and 665 nm for donor and acceptor channels, respectively. All experimental reactions consist of 1 μm protein and 500 μm nucleotide in a buffer containing 25 mm HEPES, pH 7.5, 100 mm NaCl, and 2 mm MgCl2. Raw FRET donor and acceptor signals were used to calculate a FRET efficiency, where FRET efficiency = Iacceptor/(Idonor + Iacceptor).
ATL mutant and isolated G domain crystallization and structure determination
Proteins were expressed and purified as reported previously (23, 26, 30). ATL crystals were obtained with the catalytic core fragment for mutants (residues 1–446) and the isolated G domain (residues 1–339) through sitting drop vapor diffusion. Protein (10–30 mg/ml) was incubated with 2 mm nucleotide and 4 mm MgCl2 for 1 h at 25 °C, subsequently mixed with an equal volume of reservoir solution, and stored at 20 °C. Initial crystal hits were optimized via hanging drop vapor diffusion at a protein concentration of 30 mg/ml. Final optimized conditions are as follows: ATL1-R77A, 0.2 m sodium malonate, pH 6, 20% PEG 3350, 1 mm GDP, 2 mm MgCl2; ATL1-R77A/F151S, 0.1 m BisTris, pH 5.5, 0.2 m ammonium sulfate, 22% PEG 3350, 1 mm GDP, 2 mm MgCl2, 10% xylitol (cryoprotectant); ATL1-F151S, 0.1 m ammonium citrate tribasic, pH 7, 0.1 m imidazole, pH 7, 20% PEG monomethyl ether 2000, 1 mm GDP, 2 mm MgCl2, 2 mm AlCl2, 20 mm NaF; ATL1 G domain, 0.2 m dl-malic acid, pH 7, 20% PEG 3350, 1 mm GDP, 2 mm MgCl2. Single crystals appeared between 24 and 48 h at 20 °C, and if crystals were not grown in a cryoprotectant (see above), they were supplemented with 25% glycerol for 5 min and frozen in liquid nitrogen.
X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS), and data processing and scaling were carried out with X-ray Detector Software (XDS) and CCP4 software suite (56, 57). Phases were attained through molecular replacement methods using the PHENIX software package (58) with the coordinates of ATL1 (3Q5E, GDP-bound structures; 4IDO, GDP·AlFx-bound structure) as search models. Because space groups varied between crystallographic states, a new test set was chosen for each structure before refinement. To reduce model bias, we included simulated-annealing steps early in the refinement. In addition, composite-omit maps were used for each structure refinement. Refinements were carried out in PHENIX (58) and COOT (59) to produce the final models. Data collection and model statistics are summarized in Table 1. Resolution cutoffs were selected based on a CC½ > 0.4 (60). Figures were made in PyMOL (version 1.8.4; Schrodinger, LLC). The aforementioned software packages were accessed through SBGrid (www.sbgrid.org (61)).5
Thermal shift assay
A fluorescence-based thermal shift assay using SPYRO Orange (Invitrogen) was used to generate protein melting curves and calculate Tm values (62). The reaction contains 2 μm ATL, 1 mm nucleotide, 2 mm MgCl2, and 5× SYPRO Orange in 25 mm HEPES, pH 7.5, 100 mm NaCl. A 20-μl reaction was melted in a MicroAmp 384-well plate (Applied Biosystems, Foster City, CA) using a ViiA 7 real-time PCR system (Applied Biosystems, Foster City, CA). The protocol was initiated with a 5-min incubation at 10 °C followed by increasing the temperature to 95 °C at the rate of 0.03 °C/s; excitation and emission wavelengths were 470 ± 15 and 586 ± 10 nm, respectively. Tm values are defined by calculating the temperature where the second derivative of the thermal unfolding curve is equal to zero using GraphPad Prism (GraphPad, La Jolla, CA).
Author contributions
J.P.O., L.J.B., and H.S. conceptualization; J.P.O., L.J.B., R.B.C., and H.S. formal analysis; J. P. O., L. J. B., and H. S. funding acquisition; J.P.O. and H.S. validation; J.P.O., L.J.B., and H.S. investigation; J.P.O. and H.S. visualization; J.P.O. and H.S. writing-original draft; L.J.B. and R.B.C. writing-review and editing; H.S. supervision; H.S. project administration.
Supplementary Material
Acknowledgments
CHESS is supported by National Science Foundation Grant DMR1332208. The macromolecular diffraction at the CHESS facility is supported by National Institutes of Health Grant GM103485.
This work was supported by National Institutes of Health Grants T32 GM008500 (to J. P. O.) and T32 GM008267/F31 NS077650 (to L. J. B.) and the Spastic Paraplegia Foundation (to H. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs S1–S5.
The atomic coordinates and structure factors (codes 6B9D, 6B9E, 6B9F, and 6B9G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HSP
- hereditary spastic paraplegia
- HSN
- hereditary sensory neuropathy
- ATL
- atlastin
- DRP
- dynamin-related protein
- G domain
- GTPase domain
- GED
- GTPase effector domain
- ER
- endoplasmic reticulum
- SEC-MALS
- size exclusion chromatography–coupled multiangle light scattering
- RMSD
- root mean square deviation
- mant
- N-methylanthraniloyl
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank
- CHESS
- Cornell High Energy Synchrotron Source
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- AlFx
- aluminum fluoride.
References
- 1. Fink J. K. (2013) Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 126, 307–328 10.1007/s00401-013-1115-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guelly C., Zhu P.-P., Leonardis L., Papić L., Zidar J., Schabhüttl M., Strohmaier H., Weis J., Strom T. M., Baets J., Willems J., De Jonghe P., Reilly M. M., Fröhlich E., Hatz M., et al. (2011) Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am. J. Hum. Genet. 88, 99–105 10.1016/j.ajhg.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fischer D., Schabhüttl M., Wieland T., Windhager R., Strom T. M., and Auer-Grumbach M. (2014) A novel missense mutation confirms ATL3 as a gene for hereditary sensory neuropathy type 1. Brain 137, e286–e286 10.1093/brain/awu091 [DOI] [PubMed] [Google Scholar]
- 4. Hübner C. A., and Kurth I. (2014) Membrane-shaping disorders: a common pathway in axon degeneration. Brain 137, 3109–3121 10.1093/brain/awu287 [DOI] [PubMed] [Google Scholar]
- 5. Li J., Yan B., Si H., Peng X., Zhang S. L., and Hu J. (2017) Atlastin regulates store-operated calcium entry for nerve growth factor-induced neurite outgrowth. Sci. Rep. 7, 43490 10.1038/srep43490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferguson S. M., and De Camilli P. (2012) Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13, 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Praefcke G. J. K., and McMahon H. T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- 8. Daumke O., and Praefcke G. J. K. (2016) Invited review: mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers 105, 580–593 10.1002/bip.22855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chappie J. S., Acharya S., Leonard M., Schmid S. L., and Dyda F. (2010) G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature 465, 435–440 10.1038/nature09032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faelber K., Posor Y., Gao S., Held M., Roske Y., Schulze D., Haucke V., Noé F., and Daumke O. (2011) Crystal structure of nucleotide-free dynamin. Nature 477, 556–560 10.1038/nature10369 [DOI] [PubMed] [Google Scholar]
- 11. Ford M. G. J., Jenni S., and Nunnari J. (2011) The crystal structure of dynamin. Nature 477, 561–566 10.1038/nature10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haller O., Gao S., Malsburg von der A. Daumke O., and Kochs G. (2010) Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J. Biol. Chem. 285, 28419–28424 10.1074/jbc.R110.145839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moss T. J., Andreazza C., Verma A., Daga A., and McNew J. A. (2011) Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. U.S.A. 108, 11133–11138 10.1073/pnas.1105056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu T. Y., Bian X., Sun S., Hu X., Klemm R. W., Prinz W. A., Rapoport T. A., and Hu J. (2012) Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. U.S.A. 109, E2146–E2154 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faust J. E., Desai T., Verma A., Ulengin I., Sun T.-L., Moss T. J., Betancourt-Solis M. A., Huang H. W., Lee T., and McNew J. A. (2015) The atlastin C-terminal tail is an amphipathic helix that perturbs the bilayer structure during endoplasmic reticulum homotypic fusion. J. Biol. Chem. 290, 4772–4783 10.1074/jbc.M114.601823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rismanchi N., Soderblom C., Stadler J., Zhu P. P., and Blackstone C. (2008) Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17, 1591–1604 10.1093/hmg/ddn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Orso G., Pendin D., Liu S., Tosetto J., Moss T. J., Faust J. E., Micaroni M., Egorova A., Martinuzzi A., McNew J. A., and Daga A. (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- 18. Hu J., Shibata Y., Zhu P.-P., Voss C., Rismanchi N., Prinz W. A., Rapoport T. A., and Blackstone C. (2009) A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang S., Tukachinsky H., Romano F. B., and Rapoport T. A. (2016) Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife 5, e18605 10.7554/eLife.18605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao G., Zhu P.-P., Renvoisé B., Maldonado-Báez L., Park S. H., and Blackstone C. (2016) Mammalian knock out cells reveal prominent roles for atlastin GTPases in ER network morphology. Exp. Cell Res. 349, 32–44 10.1016/j.yexcr.2016.09.015 [DOI] [PubMed] [Google Scholar]
- 21. Wang S., Romano F. B., Field C. M., Mitchison T. J., and Rapoport T. A. (2013) Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J. Cell Biol. 203, 801–814 10.1083/jcb.201308001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Powers R. E., Wang S., Liu T. Y., and Rapoport T. A. (2017) Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257–260 10.1038/nature21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Byrnes L. J., Singh A., Szeto K., Benvin N. M., O'Donnell J. P., Zipfel W. R., and Sondermann H. (2013) Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 32, 369–384 10.1038/emboj.2012.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pendin D., Tosetto J., Moss T. J., Andreazza C., Moro S., McNew J. A., and Daga A. (2011) GTP-dependent packing of a three-helix bundle is required for atlastin-mediated fusion. Proc. Natl. Acad. Sci. U.S.A. 108, 16283–16288 10.1073/pnas.1106421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu T. Y., Bian X., Romano F. B., Shemesh T., Rapoport T. A., and Hu J. (2015) Cis and trans interactions between atlastin molecules during membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 112, E1851–E1860 10.1073/pnas.1504368112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Donnell J. P., Cooley R. B., Kelly C. M., Miller K., Andersen O. S., Rusinova R., and Sondermann H. (2017) Timing and reset mechanism of GTP hydrolysis-driven conformational changes of atlastin. Structure 25, 997–1010.e4 10.1016/j.str.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan L., Sun S., Wang W., Shi J., Hu X., Wang S., Su D., Rao Z., Hu J., and Lou Z. (2015) Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J. Cell Biol. 210, 961–972 10.1083/jcb.201502078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu X., Wu F., Sun S., Yu W., and Hu J. (2015) Human atlastin GTPases mediate differentiated fusion of endoplasmic reticulum membranes. Protein Cell 6, 307–311 10.1007/s13238-015-0139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nixon-Abell J., Obara C. J., Weigel A. V., Li D., Legant W. R., Xu C. S., Pasolli H. A., Harvey K., Hess H. F., Betzig E., Blackstone C., and Lippincott-Schwartz J. (2016) Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, aaf3928–aaf3928 10.1126/science.aaf3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Byrnes L. J., and Sondermann H. (2011) Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. U.S.A. 108, 2216–2221 10.1073/pnas.1012792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ulengin I., Park J. J., and Lee T. H. (2015) ER network formation and membrane fusion by atlastin1/SPG3A disease variants. Mol. Biol. Cell 26, 1616–1628 10.1091/mbc.E14-10-1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blackstone C., O'Kane C. J., and Reid E. (2011) Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat. Rev. Neurosci. 12, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bian X., Klemm R. W., Liu T. Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T. A., and Hu J. (2011) Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 3976–3981 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ivanova N., Claeys K. G., Deconinck T., Litvinenko I., Jordanova A., Auer-Grumbach M., Haberlova J., Löfgren A., Smeyers G., Nelis E., Mercelis R., Plecko B., Priller J., Zámecník J., Ceulemans B., et al. (2007) Hereditary spastic paraplegia 3A associated with axonal neuropathy. Arch. Neurol. 64, 706–713 10.1001/archneur.64.5.706 [DOI] [PubMed] [Google Scholar]
- 35. Namekawa M., Muriel M.-P., Janer A., Latouche M., Dauphin A., Debeir T., Martin E., Duyckaerts C., Prigent A., Depienne C., Sittler A., Brice A., and Ruberg M. (2007) Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol. Cell. Neurosci. 35, 1–13 10.1016/j.mcn.2007.01.012 [DOI] [PubMed] [Google Scholar]
- 36. Khan T. N., Klar J., Tariq M., Anjum Baig S., Malik N. A., Yousaf R., Baig S. M., and Dahl N. (2014) Evidence for autosomal recessive inheritance in SPG3A caused by homozygosity for a novel ATL1 missense mutation. Eur. J. Hum. Genet. 22, 1180–1184 10.1038/ejhg.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wittinghofer A., and Vetter I. R. (2011) Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 80, 943–971 10.1146/annurev-biochem-062708-134043 [DOI] [PubMed] [Google Scholar]
- 38. Zhao X., Alvarado D., Rainier S., Lemons R., Hedera P., Weber C. H., Tukel T., Apak M., Heiman-Patterson T., Ming L., Bui M., and Fink J. K. (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat. Genet. 29, 326–331 10.1038/ng758 [DOI] [PubMed] [Google Scholar]
- 39. De N., Navarro M. V. A. S., Wang Q., Krasteva P. V., and Sondermann H. (2010) Biophysical assays for protein interactions in the Wsp sensory system and biofilm formation. Methods Enzymol. 471, 161–184 10.1016/S0076-6879(10)71010-7 [DOI] [PubMed] [Google Scholar]
- 40. Saini S. G., Liu C., and Lee T. H. (2014) Membrane tethering by the atlastin GTPase depends on GTP hydrolysis but not on forming the cross-over configuration. Mol. Biol. Cell. 25, 3942–3953 10.1091/mbc.E14-08-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winsor J., Hackney D. D., and Lee T. H. (2017) The crossover conformational shift of the GTPase atlastin provides the energy driving ER fusion. J. Cell Biol. 216, 1321–1335 10.1083/jcb.201609071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu F., Hu X., Bian X., Liu X., and Hu J. (2015) Comparison of human and Drosophila atlastin GTPases. Protein Cell. 6, 139–146 10.1007/s13238-014-0118-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morin-Leisk J., Saini S. G., Meng X., Makhov A. M., Zhang P., and Lee T. H. (2011) An intramolecular salt bridge drives the soluble domain of GTP-bound atlastin into the postfusion conformation. J. Cell Biol. 195, 605–615 10.1083/jcb.201105006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 45. Vetter I. R. (2014) The structure of the G domain of the Ras superfamily. in Ras Superfamily Small G Proteins: Biology and Mechanisms 1: General Features, Signaling (Wittinghofer A., ed) pp. 25–50, Springer, Vienna [Google Scholar]
- 46. Smith B. N., Bevan S., Vance C., Renwick P., Wilkinson P., Proukakis C., Squitieri F., Berardelli A., Warner T. T., Reid E., and Shaw C. E. (2009) Four novel SPG3A/atlastin mutations identified in autosomal dominant hereditary spastic paraplegia kindreds with intra-familial variability in age of onset and complex phenotype. Clin. Genet. 75, 485–489 10.1111/j.1399-0004.2009.01184.x [DOI] [PubMed] [Google Scholar]
- 47. Blackstone C. (2012) Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 35, 25–47 10.1146/annurev-neuro-062111-150400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ménétrey J., Macia E., Pasqualato S., Franco M., and Cherfils J. (2000) Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat. Struct. Biol. 7, 466–469 10.1038/75863 [DOI] [PubMed] [Google Scholar]
- 49. Pasqualato S., Ménétrey J., Franco M., and Cherfils J. (2001) The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2, 234–238 10.1093/embo-reports/kve043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pasqualato S., Renault L., and Cherfils J. (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Rep. 3, 1035–1041 10.1093/embo-reports/kvf221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chappie J. S., Mears J. A., Fang S., Leonard M., Schmid S. L., Milligan R. A., Hinshaw J. E., and Dyda F. (2011) A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell 147, 209–222 10.1016/j.cell.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chappie J. S., and Dyda F. (2013) Building a fission machine: structural insights into dynamin assembly and activation. J. Cell Sci. 126, 2773–2784 10.1242/jcs.108845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anand R., Eschenburg S., and Reubold T. F. (2016) Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state. Biochem. Biophys. Res. Commun. 469, 76–80 10.1016/j.bbrc.2015.11.074 [DOI] [PubMed] [Google Scholar]
- 54. Cao Y.-L., Meng S., Chen Y., Feng J.-X., Gu D.-D., Yu B., Li Y.-J., Yang J.-Y., Liao S., Chan D. C., and Gao S. (2017) MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542, 372–376 10.1038/nature21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kunzelmann S., Praefcke G. J. K., and Herrmann C. (2005) Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1). Methods Enzymol. 404, 512–527 10.1016/S0076-6879(05)04045-0 [DOI] [PubMed] [Google Scholar]
- 56. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 60. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. eLife 2, e01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lo M.-C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., and Ellestad G. (2004) Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332, 153–159 10.1016/j.ab.2004.04.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.