Abstract
The generation of clinical good manufacturing practices (GMP)-grade adeno-associated virus (AAV) vectors requires purification strategies that support the generation of vectors of high purity, and that exhibit a good safety and efficacy profile. To date, most reported purification schemas are serotype dependent, requiring method development for each AAV gene therapy product. Here, we describe a platform purification process that is compatible with the purification of multiple AAV serotypes. The method generates vector preparations of high purity that are enriched for capsids with full vector genomes, and that minimizes the fractional content of empty capsids. The two-column purification method, a combination of affinity and ion exchange chromatographies, is compatible with a range of AAV serotypes generated by either the transient triple transfection method or the more scalable producer cell line platform. In summary, the adaptable purification method described can be used for the production of a variety of high-quality AAV vectors suitable for preclinical testing in animal models of diseases.
Keywords: AAV purification, epitope swapping, analytical ultracentrifugation
Introduction
Adeno-associated virus (AAV) vector-based gene therapy has now reported many successes. In 2012, Europe approved the first gene therapy product, Glybera, an AAV1-LDL vector for the treatment of lipoprotein lipase deficiency.1 Additionally, positive results have been reported from AAV vector-based clinical trials for the treatment of an early childhood blindness, Leber’s congenital amaurosis (LCA2),2, 3, 4, 5 and hemophilia B.6 AAV-based gene delivery vectors comprise an AAV capsid harboring the therapeutic transgene, with capsid selection based on tropism for the target tissue. For example, the LCA2 trial evaluated AAV2 because of its predilection for retinal pigmented epithelial (RPE) cells. AAV8 transduces human hepatocytes and was the choice for the hemophilia B trial, while Glybera is an AAV1-based therapeutic that targets muscle;1, 2, 3, 4, 5, 6 consequently, large-scale good manufacturing practices (GMPs) that can support the production and purification of a range of AAV serotypes are essential, especially as the repertoire of new and more diverse AAV serotypes expands.7, 8 In support of this concept, scalable AAV production methods, compatible with a range of AAV serotypes, have been described;9 however, purification methods are less generic and are traditionally based on the unique properties of each AAV capsid, necessitating the optimization of capsid-specific purification methods.10, 11, 12
AAV purification methods based on affinity chromatography and, more specifically, AVB Sepharose, have increased in popularity.13, 14 AVB Sepharose is an affinity resin based on single-domain antibody fragments from the family Camelidae. These antibodies have high physical and thermal stability and excellent binding characteristics.14 The ligand used in AVB Sepharose was isolated from llamas naturally exposed to wild-type AAV; consequently, many AAV serotypes bind to this resin.15 Recently, an AVB-binding epitope, residing in a surface-exposed region of the AAV capsid, was identified. AAV capsids harboring this canonical epitope bound with high affinity to the AVB resin, while the substitution of this epitope into AAV capsids with poor affinity for AVB converted the capsids into AVB “binders.”15
A major disadvantage of affinity chromatography is the indiscriminate purification of both empty and vector-containing particles, an expected result based on their identical amino acid composition. Empty particles are considered a product-related impurity and are produced at a significant level during the biosynthesis of AAV vectors. Their presence in preclinical and clinical AAV vector stocks is problematic, because they contribute to an increased level of AAV antigen and unnecessary immune responses in animals and humans.16 Chromatographic separation methods based on charge differences can be useful in the separation of empty from vector genome-containing AAV capsids, suggesting that a subtle difference in charge between the two populations exists, facilitating their separation by traditional ion exchange chromatography (IEX).17
Herein, a universal purification method that combines affinity and IEX is described. The method is compatible with a range of AAV serotypes and production platforms, including the triple transfection method and the more scalable producer cell line method. The utility of the AVB Sepharose affinity resin for the purification of multiple AAV serotypes is confirmed; however, these studies suggest that vector recovery is improved when serotype-specific affinity resins are employed. Moreover, serotype-specific resins are useful for the purification of related AAV serotypes with conserved epitopes. Additionally, AAV capsids that fail to bind to AVB or serotype-specific resins, such as AAVDJ, can be converted to AVB binders by epitope swapping. Finally, the chromatographic-based purification strategy described here generates AAV vectors of high quality with a reduced fractional content of empty capsids and robust in vivo potency, as demonstrated by widespread retinal transduction in the context of an AAV5 EGFP vector.
Results
Purification of Various AAV Serotypes over AVB Resin
The AVB affinity resin was evaluated as the initial resin in the purification process, and a range of AAV serotypes was assessed for their binding kinetics under similar flow rates, ionic conditions, and elution parameters. The AVB resin was compatible with the purification of multiple AAV serotypes, including AAV1, AAV2, AAV5, AAV6, and AAVrh10. Tables 1, 2, and 3 include representative recoveries following AVB chromatography for these serotypes, with AAV5 showing the best performance in terms of recovery (>90%). Notably, the recoveries of serotypes, such as AAV8 and AAVrh8R, trended lower with AVB chromatography, while others, such as AAV9 or AAVDJ, failed to bind to the AVB resin at any appreciable level (data not shown). The AVB ligand was selected from a library created from llamas naturally exposed to AAV; consequently, it is not specific for any one serotype.13 Improved recoveries were achieved using affinity resins harboring an affinity ligand that was generated specifically against that serotype, e.g., POROS AAV9 and AAV8 CaptureSelect Affinity matrices. The yield of AAV9 following chromatography with POROS AAV9 CaptureSelect Affinity matrices was approximately 73%; similarly, the purification of an AAV8 vector using a POROS AAV8 CaptureSelect Affinity matrix yielded improved recoveries of approximately 80%, as shown in Tables 1, 2, and 3. Interestingly, AAVrh8R, which shares sequence homology with AAV9, failed to bind to the POROS AAV9 CaptureSelect yet was successfully purified to high yield (56% recovery) with a POROS AAV8 CaptureSelect affinity matrix (Tables 1, 2, and 3). Despite having significant homology to AAV8, no improvement in vector yield was achieved by purifying AAVDJ using the POROS AAV8 CaptureSelect affinity matrix, giving only a 1% yield (Tables 1, 2, and 3).8 The purification of the various AAV serotypes using the affinity resins generated vectors of high purity for all serotypes evaluated. Figure 1A represents the SDS-PAGE analysis of affinity-purified vectors, and the predominant bands were VP1, VP2, and VP3, with few other contaminants.
Table 1.
AVB Sepharose High Performance
| Serotype | Volume (mL) | vg/mL | Total vg | % Recovery |
|---|---|---|---|---|
| AAV1 | ||||
| Load | 1,625 | 1.43E11 | 2.32E14 | – |
| FT | 1,635 | 1.01E9 | 1.65E12 | <1 |
| Wash | 107.5 | 5.9E8 | 6.34E10 | <1 |
| Elution | 8.37 | 1.4E13 | 1.17E14 | 50 |
| AAV2 | ||||
| Load | 500 | 4.09E11 | 2.04E14 | – |
| FT | 500 | 3.90E7 | 1.95E10 | <1 |
| Wash | 20 | 3.83E7 | 7.70E8 | <1 |
| Elution | 15 | 1.08E13 | 1.62E14 | 79 |
| AAV5 | ||||
| Load | 513 | 3.93E11 | 2.0E14 | – |
| FT | 513 | 9.39E9 | 4.81E12 | 2.4 |
| Wash | 144 | 1.38E9 | 1.99E11 | <1 |
| Elution | 24 | 7.64E12 | 1.83E14 | 91.5 |
| AAV6 | ||||
| Load | 433 | 1.81E11 | 7.84E13 | – |
| FT | 433 | 1.35E10 | 5.85E12 | 7.5 |
| Wash | 75 | 5.51E9 | 4.13E11 | <1 |
| Elution | 6.6 | 9.76E12 | 6.44E13 | 82 |
| rh10 | ||||
| Load | 138 | 1.15E11 | 1.59E13 | – |
| FT | 138 | 1.5E10 | 2.07E12 | 13 |
| Wash | 22 | 6.12E9 | 1.35E11 | <1 |
| Elution | 6.6 | 1.33E12 | 8.78E12 | 55 |
FT, flow-through; vg, vector genomes.
Table 2.
POROS CaptureSelect AAV8 Affinity Matrix
| Serotype | Volume (mL) | vg/mL | Total vg | % Recovery |
|---|---|---|---|---|
| AAV8 | ||||
| Load | 150 | 1.15e12 | 1.73e14 | – |
| FT | 150 | 4.07e10 | 7.05e12 | 4.1 |
| Wash | 31 | 2.16e10 | 6.7e11 | <1 |
| Elution | 6.6 | 2.11e13 | 1.39e14 | 80 |
| AAVrh8R | ||||
| Load | 2.6 | 1.43E12 | 3.7E12 | – |
| FT | 2.6 | LOD | – | – |
| Wash | 5 | 2.19E9 | 1E10 | – |
| Elution | 1.5 | 1.38E12 | 2.1E12 | 56 |
| AAVDJ | ||||
| Load | 43 | 3.85E11 | 1.65E13 | – |
| FT | 43 | 3.15E11 | 1.35E12 | 82 |
| Wash | 23 | 4.46E10 | 1E12 | 6 |
| Elution | 4 | 3.98E10 | 1.59E11 | 1 |
FT, flow-through; vg, vector genomes.
Table 3.
POROS CaptureSelect AAV9 Affinity Matrix
| Serotype | Volume (mL) | vg/mL | Total vg | % Recovery |
|---|---|---|---|---|
| AAV9 | ||||
| Load | 71 | 1.8E11 | 1.3E 13 | – |
| FT | 71 | 3.9E9 | 2.77E11 | 2 |
| Wash | 60 | 8.82E9 | 5.3E11 | 4 |
| Wlution | 8 | 1.19E12 | 9.5E12 | 73 |
FT, flow-through; vg, vector genomes.
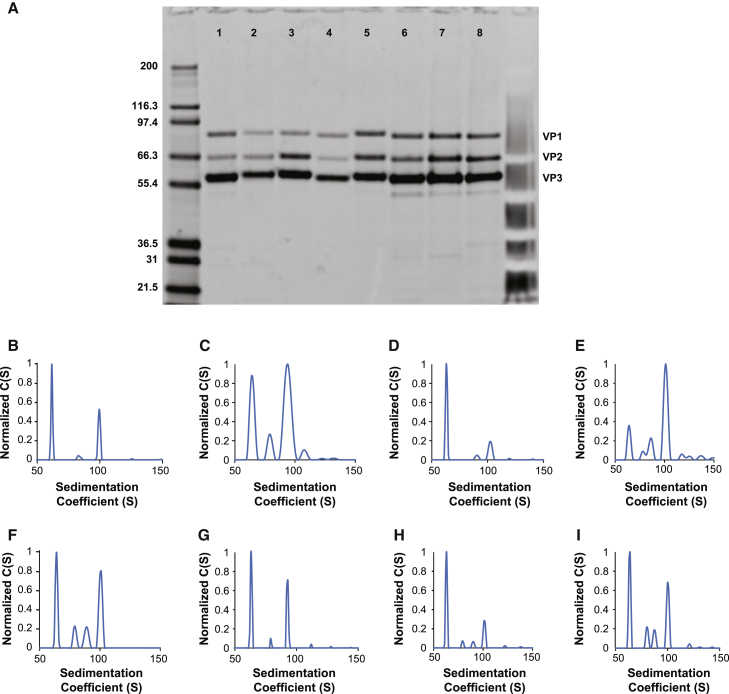
Figure 1.
Gel Purity and AUC Analysis of Various AAV Vector Serotypes following Affinity Chromatography
(A) SDS-PAGE gel analysis of various AAV vectors, followed by SYPRO Red staining; lanes 1–8 represent AAV1, AAV2, AAV5, AAV6, AAV8, AAV9, AAVrh8R, and AAVrh10; VP1, VP2, and VP3 AAV capsid proteins are labeled. (B–I) AUC analysis of eight AAV vector serotypes following affinity chromatography, including AAV1 (B), AAV2 (C), AAV5 (D), AAV6 (E), AAV8 (F), AAV9 (G), AAVrh8R (H), and AAVrh10 (I). (B) Sedimentation distribution plot for an AAV14400 vector harboring a 4,400-nucleotide single-stranded vector genome. The relative percentages of each capsid species are 90% (62S), 2% (84S), and 8% (100S), with the 100S species representing capsids harboring the full 4,400-nucleotide vector genome; the 62S species represents empty capsids, and the 84S represents capsids harboring a fragmented genome of approximately 2,000 nucleotides. (C) Sedimentation distribution plot for an AAV23400 vector harboring a 3,400-nucleotide vector genome. The relative percentages of each capsid species are 75% (64S), 7% (79S), and 18% (94S), with the 94S species representing capsids harboring the full 3,400-nucleotide vector genome, the 64S species representing empty capsids, and the 79S species representing capsids harboring an approximately 1,500-nucleotide fragmented genome. (D) Sedimentation distribution plot for an AAV54600 vector harboring a 4,600-nucleotide vector genome. The relative percentages of each capsid species are 95% (63S), 1% (91S), and 8% (102S), with the 102S species representing capsids harboring the full 4,600-nucleotide vector genome, the 63S species representing empty capsids, and the 91S species representing capsids harboring a fragmented genome of approximately 3,100 nucleotides. (E) Sedimentation distribution plot for an AAV64400 vector harboring a 4,400-nucleotide vector genome. The relative percentages of each capsid species are 61% (64S), 5% (78S), 9% (86S), and 25% (101S), with the 101S species representing capsids harboring the full 4,400-nucleotide vector genome, the 64S species representing empty capsids, and the 78S and 86S species representing capsids harboring fragmented genomes of approximately 1,400 and 2,200 nucleotides, respectively. (F) Sedimentation distribution plot for an AAV84600 vector harboring a 4,600-nucleotide vector genome. The relative percentages of each capsid species are 77% (64S), 7% (79S), 5% (89S), and 11% (101S), with the 101S species representing capsids harboring the full 4,600-nucleotide vector genome, the 64S species representing empty capsids, and the 79S and 89 species representing capsids harboring fragmented genomes of approximately 1,500 and 3,000 nucleotides, respectively. (G) Sedimentation distribution plot for an AAV93400 vector harboring a 3,400-nucleotide vector genome. The relative percentages of each capsid species are 88% (63S), 2% (79S), and 11% (93S), with the 93S species representing capsids harboring the full 3,400-nucleotide vector genome, the 63S species representing empty capsids, and the 79S species representing capsids harboring a fragmented genome of approximately 1,500 nucleotides. (H) Sedimentation distribution plot for an AAVrh8R4600 vector harboring a 4,600-nucleotide vector genome. The relative percentages of each capsid species are 92% (63S), 2% (79S), 1% (90S), and 4% (101S), with the 101S species representing capsids harboring the full 4,600-nucleotide vector genome, the 63S species representing empty capsids, and the 79S and 90S species representing capsids harboring fragmented genomes of approximately 1,500 and 3,000 nucleotides, respectively. (I) Sedimentation distribution plot for an AAVrh104400 vector harboring a 4,400-nucleotide vector genome. The relative percentages of each capsid species are 82% (64S), 6% (80S), 4% (87S), and 8% (100S), with the 100S species representing capsids harboring the full 4,400-nucleotide vector genome, the 64S species representing empty capsids, and the 80S and 87S species representing capsids harboring fragmented genomes of approximately 1,800 and 2,800 nucleotides, respectively.
Analytical ultracentrifugation (AUC), a highly accurate and reproducible way to resolve and quantify empty and genome-containing rAAV vectors,18 was applied to analyze the quality of a range of triple transfection produced rAAV vector preparations, following affinity purification. An AUC profile reveals the sedimentation coefficient, S value, of each constituent capsid species in a vector preparation. The S value is directly related to the size of the encapsidated vector genome, with empty capsids typically sedimenting at 63–65S.18 The fractional content of each capsid species was calculated using the molar extinction coefficient for each capsid species, as previously described.18 Figure 1 shows the AUC profiles of a series of affinity-purified AAV serotypes, including AAV1 (Figure 1B), AAV2 (Figure 1C), AAV5 (Figure 1D), AAV6 (Figure 1E), AAV8 (Figure 1F), AAV9 (Figure 1G), AAVrh8R (Figure 1H), and AAVrh10 (Figure 1I), and reveals the presence of both empty and genome-containing capsids. This result confirms that affinity chromatography does not selectively enrich for AAV capsids harboring DNA vector genomes. In agreement with previous observations, for the majority of AAV serotypes analyzed, greater than 80% of the AAV particles sedimented with an S value of 63–65S, the expected sedimentation coefficient for an AAV empty particle.18
Strategies to Improve the Affinity of AAV Vectors for AVB Sepharose
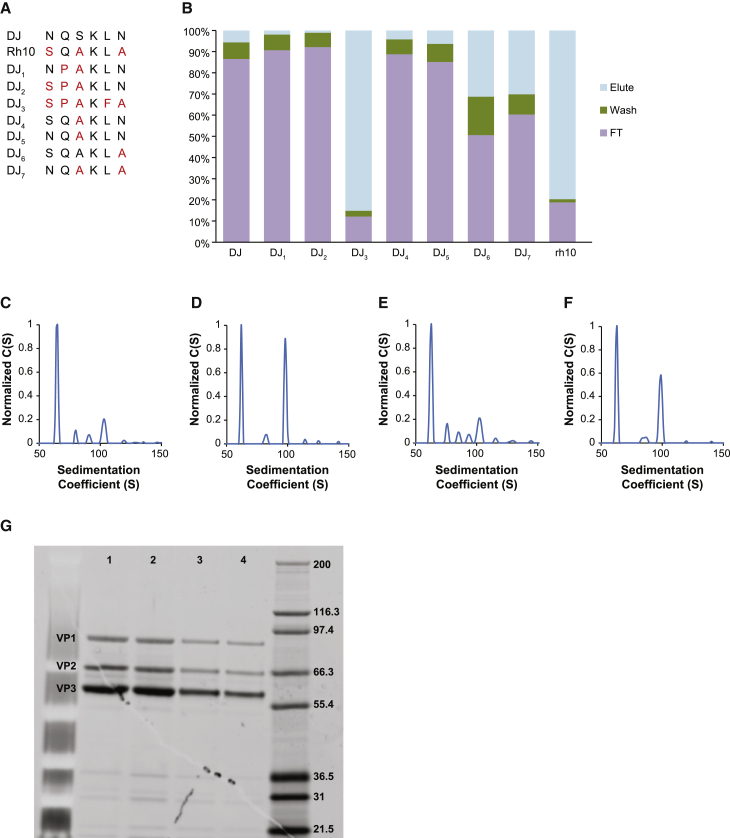
Recently, the amino acid epitope SPAKFA, at positions 663–668 in the AAV3B capsid, was identified as an epitope that correlated with high-affinity binding of an AAV capsid to the AVB resin.15 Moreover, it was determined that AAV capsids harboring elements of this canonical sequence emerged as AVB binders.15 Analysis of the AAVDJ sequence at amino acids 664–669 (NQSKLN) revealed close homology to AAVrh10 (SQAKLA), an AVB binder; thus, mutants were generated by sequentially changing single amino acids, converting AAVDJNQSKLN (DJ) to AAVDJSQAKLA (DJ6). Additionally, a mutant harboring the optimal AVB-binding motif SPAKFA was generated and designated AAVDJSPAKFA (DJ3) (Figure 2A). All mutants had equivalent or greater productivity (vector genomes [vg]/cell) as the parental AAVDJ capsid using the transient triple transfection method. Notably, AAVDJSPAKFA (DJ3) showed a trend toward an increased packaging efficiency compared to the parental AAVDJ (Table S1) range of values (AAVDJ: 1.5e4–1.6e4 vg/cell; AAVDJ3; 4e4–1e5 vg/cell). Additionally, differences were observed in vector homogeneity for AAVDJ3 and AAVDJ7, as assessed by AUC. In contrast to the parental AAVDJ vector, both mutant vector preparations trended toward a higher fractional content of full genome-containing particles (12% and 9%, respectively; Figures 2D and 2F). Figures 2C and 2E compare the parental AAVDJ and mutant AAVDJ6 vectors, which harbored approximately 4% full genome-containing capsids. Notably, AAVDJ6 contained a greater population of capsids harboring fragmented vector genomes (Figure 2E).
Figure 2.
Strategies to Improve the Affinity Purification of AAVDJ
(A) Table describing the AAVDJ mutants with amino acid substitutions at positions 664–669 in the AAVDJ parental capsid. (B) Performance of AAVDJ mutants described in (A) using AVB affinity chromatography. AAVDJ and AAVDJ vector mutants, all harboring an EGFP transgene, were loaded onto an AVB resin under the same conditions of pH, ionic strength, and flow rate. The flow-through (FT), wash (Wash), and eluted fractions (Elution) were collected, analyzed by qPCR to quantitate their vector genomes, and are represented as percent genome copies of the total applied to the resin. The common qPCR target used for all vectors was the polyA sequence. (C–F) AUC sedimentation distribution plots for AAVDJ (C), AAVDJ3 (D), AAVDJ6 (E), and AAVDJ7 (F) following AVB affinity chromatography. (C) Sedimentation distribution plot for the AAVDJ parental vector, harboring a 4,400-nucleotide EGFP vector genome. The relative percentages of each capsid species are 93% (65S), 2% (80S), 1% (91S), and 4% (103S), with the 103S species representing capsids harboring the full 4,400-nucleotide vector and the 65S species representing empty capsids. The 80S and 91S capsid species represent capsids harboring fragmented vector genomes of approximately 1,700 and 3,100 nucleotides, respectively. (D) Sedimentation distribution plot for an AAVDJ3 vector harboring a 4,400-nucleotide EGFP vector genome. The relative percentages of each capsid species are 85% (62S), 3% (82S), and 12% (100S), with the 100S species representing capsids harboring the full 4,400-nucleotide vector genome, the 62S species representing empty capsids, and the 82S species representing capsids harboring an approximately 1,900-nucleotide fragmented genome. (E) Sedimentation distribution plot for the AAVDJ6 vector harboring a 4,400-nucleotide EGFP vector genome. The relative percentages of each capsid species are 89% (63S), 5% (76S), 2% (85S), 1% (94S), and 3% (101S) species, with the 101S species representing capsids harboring the full 4,400-nucleotide vector genome and the 76S, 85S, and 94S species representing capsids harboring fragmented genomes. (F) Sedimentation distribution plot for the AAVDJ7 vector harboring a 4,400-nucleotide EGFP vector genome. The relative percentages of each capsid species are 89% (65S), 2% (85S), and 9% (100S) species, with the 100S species representing capsids harboring the full 4400-nucleotide vector genome and the 85S species representing capsids harboring a fragmented genome of approximately 2,250 nucleotides. (G) SDS-PAGE gel analysis followed by SYPRO Red staining of AAVDJ, AAVDJ3, AAVDJ6, and AAVDJ7; lanes 1–4 represent AAVDJ, AAVDJ3, AAVDJ6, and AAVDJ7.
Mutants AAVDJ SQAKLA (DJ6) and AAVDJ NQAKLA (DJ7) demonstrated appreciable levels of binding to the AVB resin (approximately 30% recovery with both mutants), suggesting that substituting the AAVDJ capsid sequence NQSKLN with either the entire putative AVB-binding epitope of AAVrh10, SQAKLA, or the less divergent sequence NQAKLA converts AAVDJ into an AVB binder. In contrast, AAVDJNQAKLN, (DJ5), which differs from AAVDJNQAKLA by one amino acid (an asparagine in place of an alanine at the sixth position), failed to bind to the AVB resin. Similarly, AAVDJSQAKLN (DJ4), which deviates from the AVB binder AAVDJSQAKLA by the same asparagine/alanine, also failed to bind AVB. In aggregate, these data underscore the importance of the terminal alanine for AVB binding in the epitope SQAKLA. Converting the amino acid sequence from NQSKLN to SPAKFA, the optimal AVB-binding motif,15 generated an AAVDJ mutant capsid (AAVDJ3) that bound with even greater affinity to the AVB resin; vector recoveries were as high as 65% (Figure 2B). This result confirms the significance of the SPAKFA epitope for AAV capsid affinity to the AVB resin and further underscores the importance of the phenylalanine and alanine at positions five and six, respectively; mutant DJ2, AAVDJSPAKLN, had little affinity for AVB (Figure 2B).
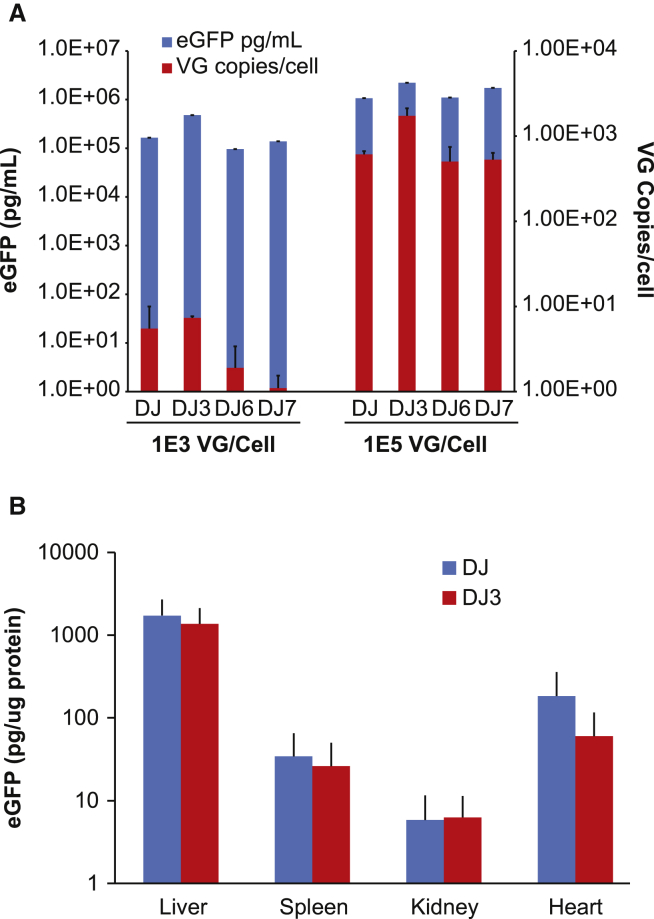
SDS-PAGE analysis of the AAVDJ and AAVDJ mutants following AVB chromatography, as shown in Figure 2G, revealed comparable purity and VP1:VP2:VP3 capsid protein ratio, suggesting that the amino acid changes had not adversely affected the capsid protein assembly. Finally, using an infectivity assay in HuH7 cells, we assessed the effect of epitope swapping on vector potency. Two vector concentrations (1e3 and 1e5 vg/cell) of the parental AAVDJ and mutants AAVDJ3, AAVDJ6, and AAVDJ7 were compared for potency, all using vectors encoding the EGFP transgene. The relative infectivity of AAVDJ, AAVDJ3, AAVDJ6, and AAVDJ7 was assessed by measuring EGFP protein levels (pg/mL) in infected cell lysates; additionally, the transduction efficiency of each vector was ranked by assessing the vector genomes per cell. As shown in Figure 3A, the infectivity of the AAVDJ mutants was comparable to the parental AAVDJ; notably, the infectivity of AAVDJ3 was increased approximately 2-fold; however, this was not statistically significant. Similarly, HuH7 cells infected with AAVDJ3 harbored more vector genomes per cell relative to cells infected with AAVDJ, AAVDJ6, or AAVDJ7, in agreement with the increased EGFP protein levels reported in Figure 3A. AAVDJ3 was further evaluated in vivo, using parental AAVDJ as a comparator. C57Bl6 mice were administered, systemically, 3e11 vg of either AAVDJ EGFP or AAVDJ3 EGFP vector. Four weeks after vector administration, tissues including liver, spleen, kidney, and heart were harvested and EGFP protein levels measured. Figure 3B reveals that the in vivo transduction properties of AAVDJ3 were comparable to AAVDJ; the EGFP protein levels, in all tissues analyzed, were equivalent between vectors.
Figure 3.
Assessment of In Vitro and In Vivo Transduction of AAVDJ and AAVDJ Mutants
(A) AAVDJ, AAVDJ3, AAVDJ6, and AAVDJ7 vectors harboring the identical EGFP expression cassette were used to infect HuH7 cells at two vector concentrations, 1e3 and 1e5 vg/cell. Forty-eight hours following infection, the cells were lysed, and cellular lysates were assayed for EGFP protein levels (ELISA) and vector genome copy numbers per cell (qPCR). (B) Assessment of in vivo performance of AAVDJEGFP and AAVDJ3EGFP. C57Bl6 mice were administered, systemically, 3e11 vg of either AAVDJEGFP or AAVDJ3EGFP. Spleen, liver, heart, and lung were assayed for EGFP protein levels, 4 weeks after vector administration. Error bars represent SD.
Chromatographic Enrichment of Full Capsids
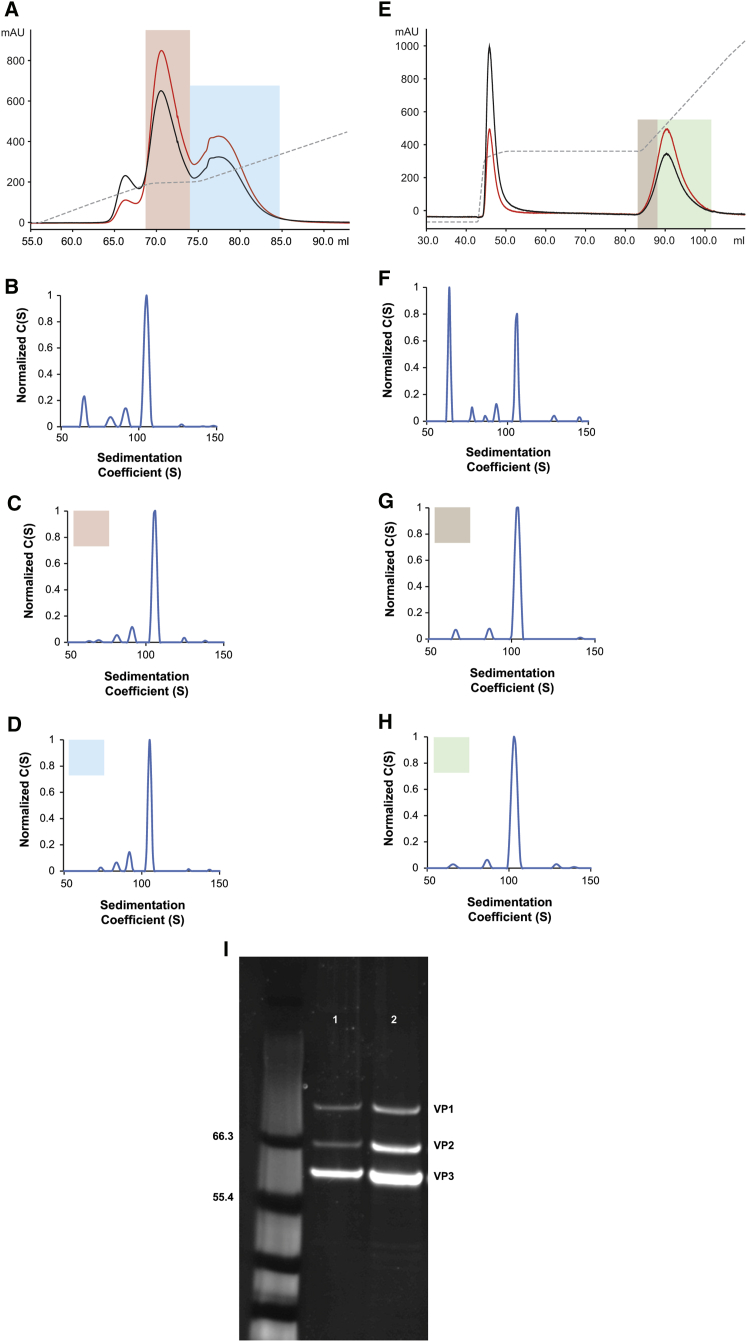
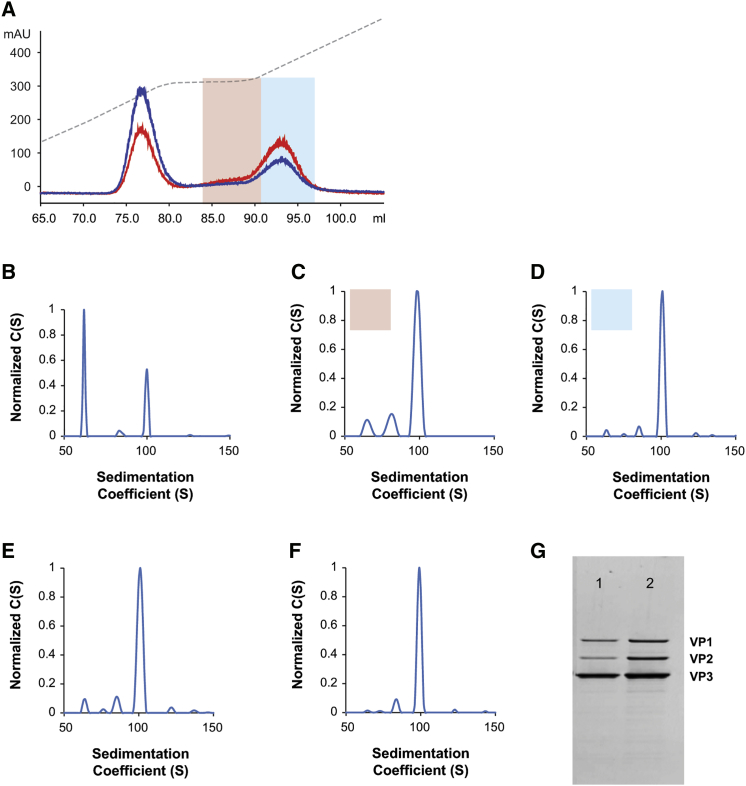
AUC analysis of AAV vector preparations following affinity chromatography revealed the presence of both empty and full particles, irrespective of serotype (Figures 1B–1I). A reduction in the fractional content of empty particles in AAV vector preparations was achieved using IEX. An AAV5 vector harboring a GUCY2D gene (retinal guanylate cyclase 1) was produced by either triple transfection or the producer cell line method and purified via AVB chromatography. AUC analysis of the AVB eluate for both vector preparations revealed that the fractional content of empty particles was higher in the triple transfection-generated vector (Figures 4B and 4F). In the context of the AAV5GUCY2DTTX vector preparation, the fractional content of capsids harboring a full vector genome was only 10% (Figure 4F). In contrast, a similar AAV5GUCY2DPCL vector, generated via a producer cell line, yielded an AAV vector preparation that harbored 32% genome-containing capsids (Figure 4B). IEX of an AVB-purified AAV5GUCY2DPCL vector resulted in a significant reduction in the fractional content of empty particles (Figures 4C and 4D). The elution of the bound AAV5GUCY2DPCL vector from the ion exchange resin resulted in three distinct peaks (Figure 4A). Peak I represents empty capsids, and peaks II and III represent genome-containing capsids. The AUC analysis of representative fractions from peaks II and III revealed that the fractional content of the empty capsids had been significantly reduced (Figures 4C and 4D); empty capsids typically sediment at 64S.18 The AUC analysis of the vector from the second elution peak (peak II) revealed the fractional content of empty capsids to be as low as 14% (Figure 4C), while 67% of the vector preparation contained capsids with a full vector genome, represented by the 103S species. Similarly, the AUC analysis of the vector from elution peak III revealed a vector preparation harboring 6% empty capsids and 69% capsids with a full vector genome (Figure 4D).
Figure 4.
IEX of AAV5GUCY2D Vectors: Full Vector Capsid Enrichment
(A–H) Affinity-purified AAV5GUCY2D vector, generated via either the producer cell line production method (A–D) or the triple transfection method (E–H), was subjected to IEX to remove empty capsids. (A) Elution profile of affinity-purified AAV5GUCY2DPCL from the ion exchange resin showing both A260 (red line) and A280 (black line) profiles for the three resolved peaks. Conductivity is represented by the dashed line. The first elution peak contained empty capsids; the A280 (black line) reading was higher than the A260 (red line) reading. Encapsidated DNA contributes to the A260 reading, and absorbance at this wavelength monitors the full- or vector-genome-containing capsids. Peaks II (pink shading) and III (blue shading) represent fractions enriched for full capsids, as the absorbance at 260 nm (red line) is greater than absorbance at 280 nm (black line). (B–D) AUC analysis of fractions from peaks II (pink shading) (C) and III (blue shading) (D), confirming that the fractional content of full capsids is higher than that of the starting affinity-purified material, AAV5GUCY2DPCL (B). Sedimentation distribution plot for affinity-purified AAV5GUCY2DPCL. (B) The relative percentages of each capsid species are 55% (65S), 6% (82S), 7% (91S), and 32% (105S), with the 105S species representing capsids harboring the full GUCY2D vector genome and the 65S species representing empty capsids. The 91S and 82S species represent capsids harboring fragmented genomes. (C) Sedimentation distribution plot for peak II (pink shading) eluted from the ion exchange resin. The relative percentages of each capsid species are 14% (67S), 8% (81S), 10% (91S), and 67% (105S), with the 105S species representing capsids harboring the full GUCY2D vector genome and the 67S species representing empty capsids. The 91S and 81S species represent capsids harboring fragmented genomes. (D) Sedimentation distribution plot for peak III (blue shading) eluted from the ion exchange resin. The relative percentages of each capsid species are 6% (74S), 10% (84S), 15% (92S), and 69% (105S) species, with the 105S species representing capsids harboring the full GUCY2D vector genome. The 84S and 92S species represent capsids harboring fragmented genomes of approximately 2,200 and 3,200 nucleotides, respectively. (E) Elution profile of affinity-purified AAV5GUCY2DTTX from the ion exchange resin, revealing both A260 (red line) and A280 (black line) profiles for the two resolved peaks. Conductivity is represented by the dashed line. The first elution peak contained empty capsids, as the A280 (black line) reading was higher than the A260 (red line). Encapsidated DNA contributes to the A260 (red line), and absorbance at this wavelength monitors the full- and vector-genome-containing capsids. The leading (brown shading) and trailing fractions (green shading) from peak II represent vectors enriched with full capsids, as the absorbance at 260 nm (red line) is greater than the absorbance at 280 nm (black line). (F–H) AUC analysis of fractions from peak II (G, brown shading, and H, green shading) confirms that the fractional content of full capsids was higher than that of the starting, affinity-purified AAV5GUCY2DTTX vector (F). Sedimentation distribution plot for affinity-purified AAV5GUCY2DTTX. (F) The relative percentages of each capsid species are 86% (64S), 3% (78S), 1% (86S), 2% (93S), and 10% (105S), with the 105S species representing capsids harboring the full GUCY2D vector genome and the 64S species representing empty capsids. The 78S, 86S, and 93S species represent capsids harboring fragmented genomes of 1,400, 2,500, and 3,300 nucleotides, respectively. (G) Sedimentation distribution plot for the leading edge of elution peak II (brown shading) from the ion exchange resin. The relative percentages of each capsid species are 29% (67S), 6% (87S), and 65% (103S), with the 103S species representing capsids harboring the complete GUCY2Dvector genome and the 67S species representing empty capsids. The 87S species represents capsids harboring a fragmented genome of approximately 2,600 nucleotides. (H) Sedimentation distribution plot for the trailing edge of peak II (green shading) eluted from the ion exchange resin. The relative percentages of each capsid species are 18% (66S), 6% (87S), and 76% (103S), with the 103S species representing capsids harboring the full GUCY2D vector genome and the 66S species representing empty capsids. The 87S species represents capsids harboring a fragmented genome of approximately 2,600 nucleotides. (I) SDS-PAGE/SYPRO Red stain of AVB-IEX-purified AAV5GUCY2DPCL (lane 1) and AAV5GUCY2DTTX vectors (lane 2).
In parallel, the AAV5GUCY2D vector was produced by the triple transfection production method and purified by AVB. The AUC analysis of the AVB-purified AAV5GUCY2DTTX vector revealed the population of empty capsids to be 86% (Figure 4F). IEX was performed on the AVB-purified AAV5GUCY2DTTX vector preparation to reduce the fractional content of empty capsids; however, due to the high empty capsid burden, typical of TTX produced vectors,18 an adjusted wash step was required prior to the gradient elution to achieve an acceptable level of full capsid enrichment (Figure 4E). This modified method resulted in two elution peaks: the first peak contained empty particles, while the second peak contained capsids harboring the vector genome. Fractions were collected from the second elution peak, and AUC analysis was performed (Figures 4G and 4H). The leading edge of the elution peak contained capsids that were 65% full (Figure 4G), while the fractional content of genome-containing capsids in vectors isolated from the trailing edge of the peak was higher, at 78% (Figure 4H). Finally, purity analysis of both AAV5GUCY2DTTX and AAV5GUCY2DPCL following full capsid enrichment revealed both vector preparations to be highly purified, containing only capsid proteins VP1, VP2, and VP3 with no contaminants (Figure 4I). The recovery of AAV5GUCY2DTTX and AAV5GUCY2DPCL vectors over IEX was comparable at 54% and 74%, respectively (Table S2).
The versatility of this purification method was further demonstrated using another AAV serotype, AAV1 (Figure 5). The AAV1EGFP vector was made by either triple transfection or producer cell line (PCL) production methods and purified using the two-column purification method (AVB-IEX). Comparable to what was achieved with AAV5GUCY2D vectors (Figure 4), theAAV1EGFPTTX and AAV1EGFPPCL vectors, following AVB-IEX chromatography, were highly purified (Figure 5G). Moreover, the fractional content of empty capsids in the AAV1EGFPPCL vector preparation, as assessed by AUC, was reduced from 35% to 7% following IEX chromatography (Figures 5E and 5F). In the context of the AVB-IEX-purified AAV1EGFPTTX vector, the reduction in the fractional content of empty capsids from 90% to 19% was even more impressive (Figures 5B and 5D). The performance of the AAV1EGFPTTX vector on IEX was similar to that of AAV5GUCY2DTTX, in that a step wash was necessary prior to a gradient elution (Figure 5A). The AAV1EGFPTTX vector that eluted later in the gradient harbored more full genome-containing particles, 72% (Figure 5D), than the AAV1EGFP vector that eluted earlier (Figure 5C). AUC analysis of vectors from the fractions eluted earlier in the gradient revealed the presence of both empty and genome-containing capsids at equivalent levels (Figure 5C). Finally, the recoveries of AAV1EGFPTTX and AAV1EGFPPCL following IEX were 67% and 52%, respectively (Table S2), in agreement with recoveries achieved for the AAV5GUCY2D vectors following IEX chromatography.
Figure 5.
IEX of AAV1EGFP Vectors for Full Vector Capsid Enrichment
(A–F) Affinity-purified AAV1EGFP vector generated via either the triple transfection production method (A–D) or the producer cell line method (E and F) was subjected to IEX to remove empty capsids. (A) Elution profile of affinity-purified AAV1EGFPTTX from the ion exchange resin, showing A260 (red line) and A280 (black line) profiles for the two resolved peaks. Conductivity is represented by the dashed line. The first elution peak contained empty capsids; the A280 (black line) reading was higher than the A260 (red line) reading. Encapsidated DNA contributes to the A260 reading, and the absorbance at this wavelength monitors the full- or vector-genome-containing capsids. Peak II (pink and blue shading) represents fractions enriched for full capsids, and the absorbance at A260 (red line) is greater than absorbance at A280 (black line). AUC analysis of fractions from the leading edge (C, pink shading) and trailing edge (D, blue shading) of peak II confirmed that the fractional content of full capsids is higher than that of the starting, affinity-purified AAV1EGFPTTX vector. (B) Sedimentation distribution plot for affinity-purified AAV1EGFPTTX (B). The relative percentages of each capsid species are 90% (62S), 2% (84S), and 8% (100S), with the 100S species representing capsids harboring the complete EGFP vector genome and the 62S species representing empty capsids. The 84S species represents capsids harboring a fragmented genome of approximately 2,200 nucleotides. (C) Sedimentation distribution plot for the trailing edge of peak II (pink shading) eluted from the ion exchange resin. The relative percentages of each capsid species are 40% (65S), 15% (81S), and 45% (98S), with the 98S species representing capsids harboring the complete EGFP vector genome and the 65S species representing empty capsids. The 81S species represents capsids harboring a fragmented genome of approximately 1,700 nucleotides. (D) Sedimentation distribution plot for the leading edge of elution peak II (blue shading). The relative percentages of each capsid species are 19% (63S), 2% (75S), 7% (85S), and 72% (100S), with the 100S species representing capsids harboring the complete EGFP vector genome and the 63S species representing empty capsids. The 75S, 85S, and 92S species represent capsids harboring fragmented genomes of approximately 1,100, 2,400, and 3,200 nucleotides, respectively. (E and F) AUC analysis of the affinity purified AAV1EGFPPCL vector before (E) and after (F) IEX. A sedimentation distribution plot for affinity-purified AAV1EGFPPCL (E). The relative percentages of each capsid species were 35% (64S), 3% (76S), 2% (85S), and 60% (101S), with the 101S species representing capsids harboring the complete EGFP vector genome and the 64S species representing empty capsids. The 76S and 85S species represent capsids harboring fragmented genomes of approximately 1,400 and 2,200 nucleotides, respectively. (F) Sedimentation distribution plot for affinity-purified AAV1EGFPPCL following IEX. The relative percentages of each capsid species are 7% (64S), 2% (73S), 12% (84S), and 78% (99S), with the 99S species representing capsids harboring the complete EGFP vector genome and the 64S species representing empty capsids. The 73S and 84S species represent capsids harboring fragmented genomes of approximately 1,000 and 2,200 nucleotides, respectively. (G) SDS-PAGE/SYPRO Red stain of AVB-IEX-purified AAV1EGFPPCL (lane 1) and AAV1EGFPTTX (lane 2).
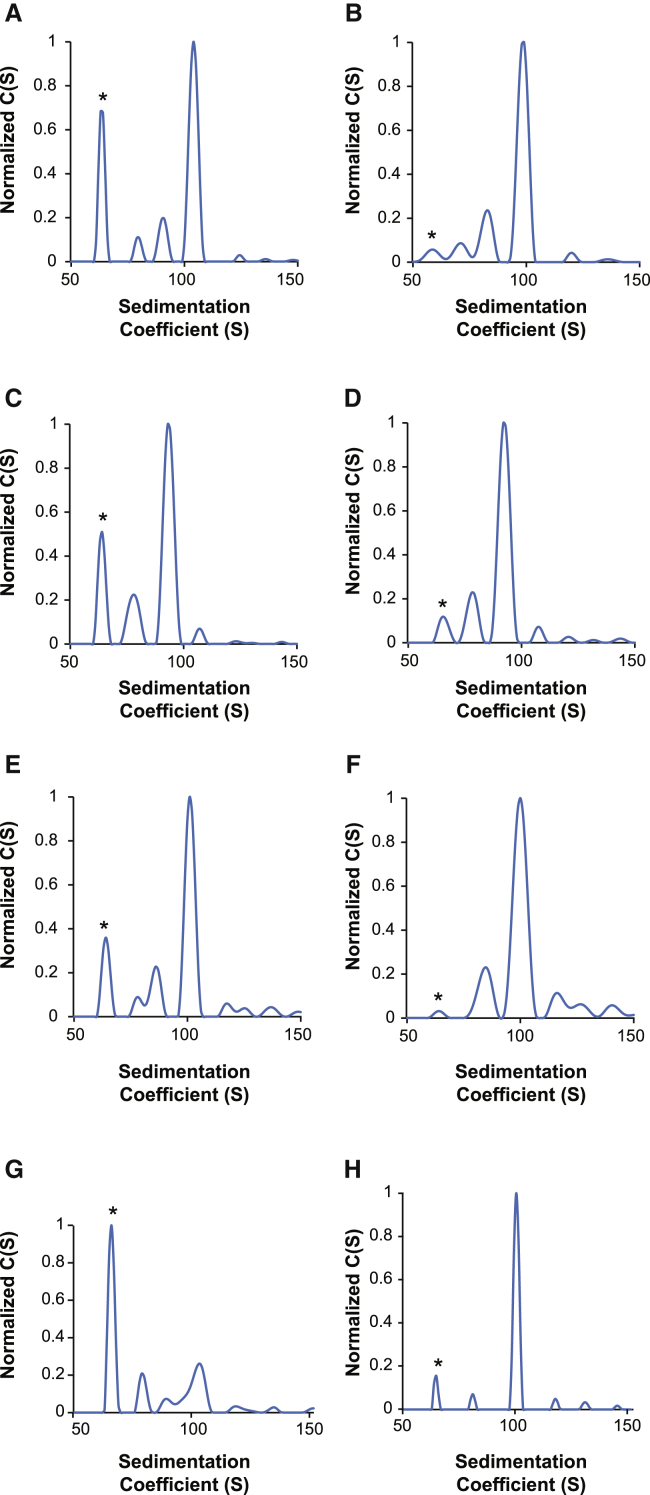
Figure 6 shows the AUC profiles for a range of AAV serotypes following AVB-IEX chromatography, demonstrating the versatility of this two-column purification method for additional serotypes, including AAV2 (Figures 6A and 6B), AAVrh8R (Figures 6C and 6D), AAV6 (Figures 6E and 6F), and AAVDJ (Figures 6G and 6H). For all serotypes evaluated, IEX chromatography resulted in a significant decrease in the fractional content of empty capsids. Additionally, IEX chromatography was useful for the removal of empty capsids from affinity-purified AAV8 and AAVDJ8 (data not shown), further underscoring the versatility of this method.
Figure 6.
AUC Analysis of Various AAV Vectors following AVB-IEX Chromatography
(A–H) AUC analysis of four AAV vector serotypes following affinity chromatography (A, C, E, and G) and following a combination of AVB-IEX chromatography to enrich for full capsids (B, D, F, and H). The serotypes analyzed were AAV2 (A and B), AAVrh8R (C and D), AAV6 (E and F), and AAVDJ (G and H). All vector preparations were generated using the transient triple transfection production method. (A and B) Sedimentation distribution plots for an AAV24600 vector harboring a 4,600-nucleotide vector genome. The relative percentages of each capsid in the affinity-purified AAV24400 vector prep are 75% (64S), 4% (80S), 5% (90S), and 17% (103S), with the 103S species representing capsids harboring the complete 4,600-nucleotide vector genome and the 64S species representing empty capsids. (B) Sedimentation distribution plot for the same AAV24600 vector shown in (A) following IEX to remove the empty capsids. The relative percentages of each capsid in the enriched AAV24600 vector preparation were 0% (64S), 14% (80S), 19% (90S), and 67% (103S), with the 103S species representing capsids harboring the complete vector genome and the 64S species representing empty capsids. (C) Sedimentation distribution plot for an AAVrh8R3200 vector harboring a 3,200-nucleotide vector genome. The relative percentages of each capsid in the affinity-purified AAVrh8R3200 vector prep are 54% (63S), 12% (77S), and 30% (92S), with the 92S species representing capsids harboring the complete 4,400-nucleotide vector genome and the 63S species representing empty capsids. (D) Sedimentation distribution plot for the same AAVrh8R3200 vector shown in (C), following IEX to remove empty capsids. The relative percentages of each capsid in the enriched AAVrh8R3200 vector prep were 21% (64S), 26% (77S), and 55% (92S), with the 92S species representing capsids harboring the complete 3,200-nucleotide vector genome and the 64S species representing empty capsids. (E) Sedimentation distribution plot for an AAV64400 vector harboring a 4,400-nucleotide vector genome. The relative percentages of each capsid in the affinity-purified AAV64400 vector prep are 61% (64S), 5% (78S), 9% (86S), and 25% (101S), with the 101S species representing capsids harboring the complete 4,400-nucleotide vector genome and the 64S species representing empty capsids. (F) Sedimentation distribution plot for the same AAV64400 vector shown in (E) following IEX to remove empty capsids. The relative percentages of each capsid in the enriched AAV64400 vector prep are 11% (64S), 26% (84S), and 63% (100S), with the 100S species representing capsids harboring the complete 4,400-nucleotide vector genome and the 64S species representing empty capsids. (G) Sedimentation distribution plot for an AAVDJ4400 vector harboring a 4,400-nucleotide vector genome. The relative percentages of each capsid in the affinity-purified AAVDJ4400 vector preparations are 86% (65S), 6% (78S), 2% (89S), and 6% (101S), with the 101S species representing capsids harboring the complete 4,400-nucleotide vector genome and the 65S species representing empty capsids. (H) Sedimentation distribution plot for the same AAVDJ4400 vector shown in (G) following IEX to remove empty capsids. The relative percentages of each capsid in the enriched AAVDJ4400 vector prep are 46% (65S), 5% (81S), and 49% (100S), with the 100S species representing capsids harboring the complete 4,400-nucleotide vector genome and the 65S species representing empty capsids. The asterisk (*) in each AUC profile highlights the peak that corresponds to empty particles.
In Vivo Assessment of Vector Potency
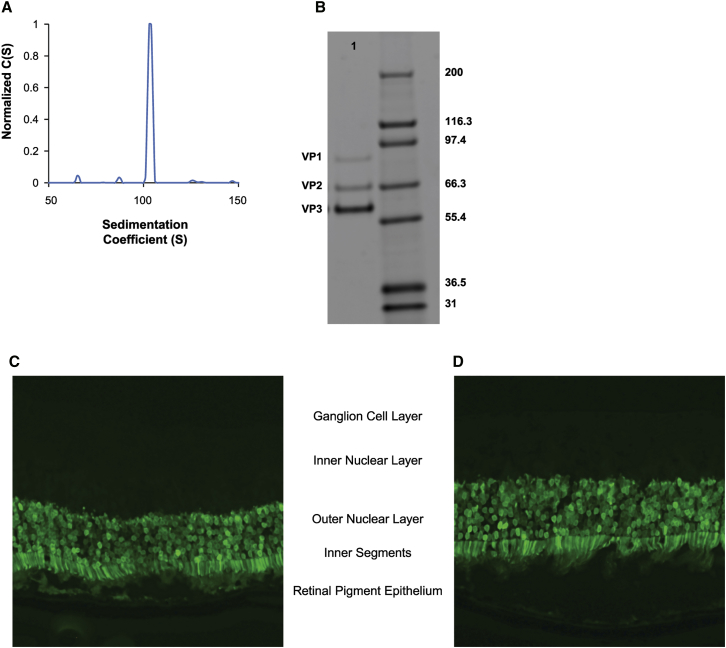
Finally, the in vivo performance of the AAV5EGFP vector following AVB-IEX chromatography was assessed (Figure 7). Wild-type mice were injected subretinally with 1e9 vg of AAV5-EGFPTTx, a vector in which the hGRK1 photoreceptor-specific human rhodopsin kinase promoter19 drives EGFP expression. This vector was purified by AVB-IEX, and AUC analysis revealed that the vector preparation was 80% full and of high purity (Figures 7A and 7B). Analysis of retinal cross sections by epifluorescence 28 days post-injection (Figures 7C and 7D) revealed strong expression in the outer nuclear layer and inner segments, consistent with robust transduction of both rod and cone photoreceptor cells. No EGFP expression was detected in any additional retinal layers, confirming the fidelity of the hGRK1 promoter for restricting gene expression to the photoreceptors.19 Additionally, the AAV5EGFP exhibited extensive lateral spread following subretinal delivery, as evidenced by robust EGFP staining throughout the entire retina (data not shown).
Figure 7.
In Vivo Assessment of AVB-IEX-Purified AAV5EGFP
(A) Sedimentation profile for the AAV5EGFP vector purified using a combination of AVB-IEX chromatography, revealing that greater than 80% of the vector harbors the complete EGFP vector genome (101S species). (B) SDS-PAGE/SYPRO purity assessment of the AAV5EGFP vector preparation lane 1. (C and D) Representative fluorescence of mouse eyes treated subretinally with 1e9 vg of AAV5EGFP. The images represent EGFP raw fluorescence and show the restriction of robust EGFP expression to photoreceptor cells, both outer nuclear layer and inner segments, confirming the fidelity of hGRK1, the photoreceptor-specific promoter.
Discussion
The development of a generic protocol that supports the purification of a range of AAV serotypes provides considerable flexibility in a gene therapy manufacturing paradigm. Several groups13, 14, 15 have reported on the versatility of the AVB affinity resin for the purification of a range of AAV serotypes, an observation confirmed in this study. However, here, the concept of affinity chromatography was further extended to the use of affinity resins harboring antibodies generated specifically to AAV capsids, notably AAV9 and AAV8. Although AAV8 can be captured by the AVB resin, the affinity is low, precluding its use for process scale-up. Improved recoveries were achieved by using an AAV8-specific resin. Similarly, AAV9, a weak AVB binder, showed improved binding to an AAV9-specific resin. Notably, an AAV8-selective affinity resin was useful for the purification of a related serotype, AAVrh8R, while the performance of this serotype on an AAV9 selective resin was unsatisfactory. This result suggests that despite considerable sequence homology between AAV9 and AAVrh8R,20 the epitope responsible for binding to the AAV9 resin is not shared; however, a common, as yet unidentified epitope facilitates binding of AAVrh8R to an AAV8-specific resin. These data are important, because they suggest that existing off-the-shelf serotype-specific affinity resins may be useful for the purification of disparate AAV capsids with conserved epitopes.
The generation of an affinity resin specific for any one AAV serotype is both laborious and expensive, requiring the generation of large amounts of AAV antigen for injection into llamas and the lengthy screening of antibody libraries to identify a final candidate. Thus, a capsid engineering approach that could potentially convert an AAV capsid with low affinity for AVB into one that has high affinity has obvious merit. Recently, Wang et al.15 identified the canonical epitope SPAKFA that mediates high-affinity AAV3B binding to the AVB resin; additionally, it was determined that AAV capsids harboring elements of this canonical sequence have emerged as AVB binders,15 e.g., AAVrh10. In this study, using AAVDJ as a prototype, the practice of epitope swapping was evaluated. The NQSKLN sequence at amino acid positions 664–669 in the AAVDJ capsid sequence was converted into the canonical epitope, SPAKFA; additionally, a mutant with the less divergent sequence SQAKLA, the epitope found in AAVrh10, was generated. An important consideration is the effect of epitope substitutions on yield and tropism of the vector; thus, single amino acid changes were initially made to convert AAVDJNQSKLN into AAVDJSQAKLA with the goal of determining the minimum number of amino acid changes needed to convert AAVDJ into an AVB binder. In contrast to the parental AAVDJ mutants, AAVDJSQAKLA and AAVDJNQAKLA showed appreciable levels of binding to the AVB resin, suggesting that substituting the AAVDJ capsid sequence NQSKLN with either the entire putative AVB-binding epitope of AAVrh10 (SQAKLA) or the less divergent sequence NQAKLA converts AAVDJ into an AVB binder. Notably, the performance of these mutants on AVB, although significantly improved over the parental AAVDJ, did not approximate that of the parental AAVrh10, suggesting that in the context of AAVrh10, an additional, as yet to be determined epitope(s) contributes to high-affinity AVB binding. In contrast, AAVDJSPAKFA demonstrated high-affinity AVB binding, while a closely related mutant, AAVDJSPAKLN, was a poor AVB binder, underscoring the importance of the terminal phenylalanine and alanine in the canonical epitope SPAKFA. Moreover, the significance of the terminal alanine was further highlighted by mutants AAVDJNQAKLN and AAVDJSQAKLN, which, in contrast to AAVDJNQAKLA or AAVDJSQAKLA, failed to bind to the AVB resin (Figure 2B).
Mutant AAVDJSPAKFA showed greater productivity (vg/cell) than the parental AAVDJ capsid and improved transduction efficiencies in a human Huh7 liver cell line. In contrast, in vivo, AAVDJSPAKFA transduced mouse liver with a similar efficiency to parental AAVDJ, underscoring discordance between in vitro and in vivo performance of AAV vectors. However, the influence of species differences on vector transduction efficiency needs also to be considered. In summary both the in vitro and in vivo performance of AAVDJSPAKFA compared to parental AAVDJ suggest that the amino acid changes had not adversely affected packaging efficiency, transduction, or, by extension, tropism. Interestingly, the amino acid changes were confined to a variable region within the HI loop of the AAV capsid, a region important for capsid assembly and viral genome packaging,21 suggesting that the SPAKFA epitope may confer some advantage in these processes. Interestingly, the length of the HI loop in relation to the underlying subunit is crucial for proper capsid assembly, while HI loop amino acid interactions with the underlying capsid subunits dictate genome packaging efficiency.21 Further data to support improvements in mutant AAVDJSPAKFA vector quality were revealed by AUC analysis, which demonstrated a genome-containing capsid content that was higher than that of the parental AAVDJ. The versatility of epitope swapping was confirmed in the context of another AAV capsid, AAVDJ8. Similar to AAVDJ, grafting of the SPAKFA epitope onto the AAVDJ8 capsid rendered the capsid an AVB binder. Recoveries following AVB chromatography increased from <5% with AAVDJ8, to >70% with the AAVDJ8SPAKFA variant (data not shown).
During the course of these studies, Thermo Fisher released a new AAV affinity resin, AAVX, purported by the manufacturer to be useful for the purification of multiple AAV serotypes. However, the performance of the AAVX resin remains to be evaluated by the larger gene therapy community. Additionally, one potential caveat to its use is the recommendation, by the manufacturer, to include glycine during vector elution. In the context of GMP manufacturing, the presence of glycine raises obvious concerns, as its effective removal from clinical vector lots would need to be confirmed prior to human use.
Affinity chromatography, although highly selective for the AAV capsid, has one disadvantage: the affinity ligand cannot discriminate between a full-genome-containing and an empty capsid. Both populations are indistinguishable at the amino acid level and, by extension, share epitopes. Empty capsids are considered a product-related impurity; however, to circumvent the possible deleterious effects of this population on vector efficacy and safety, a scalable chromatographic purification method to separate empty capsids from vector particles was evaluated.17 This separation method harnesses the subtle differences in surface charge between an empty and a genome-containing particle, postulated to be created following a capsid conformational change concomitant with DNA insertion into the pre-formed AAV capsid.22 Importantly, this subtle difference in surface charge allows the effective separation of these two populations using traditional charge-based separation methods, such as IEX.17 Traditionally, empty capsids are separated from vector particles by gradient density centrifugation using either cesium chloride or iodixanol gradients.23 However, these methods do not meet the challenge of manufacturing gene therapy vectors consistently and at the quality necessary for routine preclinical or clinical use. Additionally, density gradient purification strategies are not amenable to scaling up and are challenging to validate in a GMP setting.
In conclusion, we describe a generic purification method useful for a range of AAV serotypes. In this study, the concept of epitope swapping to facilitate the use of AVB affinity chromatography for the purification of the AAVDJ serotype was validated. However, additional data reported here suggest that the use of serotype-specific affinity resins have the potential to purify related AAV serotypes, provide additional tools for the purification of novel AAV serotypes, and obviate the need for capsid engineering. The purification strategy described herein is compatible with common, as well as novel, AAV serotypes and purifies AAV with a high yield (approximately 35% accumulative yield) and quality, with excellent in vivo potency. Notably, purified AAV vector preparations have a reduced fractional content of empty capsids, which is an important quality attribute, considering that capsid proteins can elicit immune responses against the vector and the host cell transduced by the vector.15 Importantly, the purification strategy advances the field of AAV product manufacturing toward the ideal of a universal “plug and play” platform, capable of supporting the demand for large-scale AAV vector production of increasingly diverse AAV serotypes.
Materials and Methods
Vector Production and Purification
AAV vectors were produced either via transient transfection24 or the producer cell line method,25, 26 as previously described. For the production of AAV vectors by transfection, HEK293 cells were transfected using polyethyleneimine, polyethylenimine (PEI), and a 1:1:1 ratio of the three plasmids (inverted terminal repeat [ITR] vector, AAV rep/cap, and Ad helper plasmid). The pAd helper used was pHelper (Stratagene/Agilent Technologies, Santa Clara, CA, USA). The generation of AAV vectors by the producer cell line method was performed as previously described.25, 26 In brief, a HeLa-based producer cell line was created following transfection of HeLaS3 cells (ATCC CCL-2.2) with a single plasmid containing the following elements: AAV2 rep genes and cap gene of the desired serotype, the vector genome flanked by AAV2 ITRs, and a puromycin resistance gene. Transfected cells were grown in the presence of puromycin to isolate stable integrants, which were subsequently screened for AAV productivity following infection with wtAd5 virus.25, 26 Purification of AAV from both production platforms was achieved using a two-column purification method. For vectors generated using the triple transfection method, cell pellets were harvested following centrifugation (1,500× rpm) and resuspended in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 10 mM MgCl2) prior to freeze/thawing (3×). Following the addition of Benzonase and 0.1%Triton X-100, the lysate was incubated at 37°C and centrifuged at 3,400 rpm before sequential filtrations using 0.8- and 0.45-μm filters. For producer cell line-generated vectors, cells were maintained in shaker flask suspension cultures in EX-CELL HeLa medium (Sigma-Aldrich) with 6 mM L-glutamine at 37°C in 10% CO2. For virus production, cells were switched into production media and infected with wtAd5 (100 vg/cell) for 3 days. The cells were lysed with 0.1% Triton X-100; then 2 mM MgCl2 and 50 U/mL Benzonase was added and incubated at 37°C for 2 hr; 500 mM NaCl was then added and incubated for 10 min, followed by centrifugation. The supernatant was collected and filtered before tangential flow filtration (TFF). Post-lysis, an additional incubation at 52°C for 20 min was performed to inactivate the adenovirus; this step was followed by filtration using a 0.22-μm filter.
Chromatography
The clarified cell lysate containing the rAAV vector was loaded onto an AVB Sepharose HP (GE Healthcare) Tricorn 10 × 100 column (8 mL, column volume [CV]). In addition, serotype-specific resins AAV8 and AAV9 Capture Select were evaluated for the purification of AAV8, AAV9, AAVrh8R, and AAVDJ. Following chromatography, the bound AAV was eluted with low-pH buffer. The eluted virus solution was neutralized by adding 1 M Tris-HCl (pH 8.7) at 1/10 of the fraction volume directly into the fraction collection tube prior to elution.
Full capsid enrichment was performed as previously described by Qu et al.17 In brief, following AVB affinity purification, the AAV vector was further purified using anion exchange chromatography as described in section 2.4 of Qu et al.17 The bound AAV capsids, both empty and genome-containing, were sequentially eluted with increasing conductivity in the presence of a 10-mM to 300-mM Tris-acetate gradient (pH 8).17 Both populations were subsequently analyzed by TaqMan analysis and AUC.18
Sample Preparation for AUC Analysis
The purified vector, at a concentration of 2e12 to 5e12 vg/mL, was buffer exchanged into PBS (pH 7.2) using a 10K MWCO Slide-a-Lyzer (Thermo Scientific, Waltham, MA, USA). The AAV vector absorbance signal was determined by optical density measurement at 260 nm (OD260) using spectrophotometric methods. For consistency, the samples were adjusted to a target concentration (OD260 of between 0.2 and 0.8) either by direct dilution with PBS or further concentrated using an Amicon Ultra-0.5/30K MWCO Centrifugal Filter Device (Merck, Darmstadt, Germany).
Sedimentation Velocity AUC Data Acquisition
Sedimentation velocity AUC (SV-AUC) analysis was performed using a Proteome Lab XL-I (Beckman Coulter, Indianapolis, IN, USA). A 400-μL volume of sample was loaded into the sample sector of a two-sector velocity cell, and 410 μL of PBS was loaded into the corresponding reference sector. The sample was placed in the four-hole rotor and allowed to equilibrate in the instrument until a temperature of 20°C and full vacuum were maintained for 1 hr. Sedimentation velocity centrifugation was performed at 20,000 rpm and 20°C. Absorbance (260 nm) optics was used to record the radial concentration as a function of time until the lightest sedimenting component had cleared the optical window (1.2 hr). AUC data were analyzed as previously described.18
qPCR Analyses
The AAV vector was quantified using a real-time qPCR assay (7500 Real-Time PCR System; Applied Biosystems, Foster City, CA, USA) with primers specific for the polyadenylation signal. Vector levels are expressed as vector genomes per milliliter.
In Vivo Subretinal Administration and Histological Tissue Processing
C57BL/6J mice were anesthetized with 3%–3.5% isoflurane over oxygen. The bevel of a 30-gauge needle was used to make a pilot incision into the cornea of the eye along the margin of the pupil. A 35-gauge blunt needle attached to a UMPII Microinjection Pump (World Precision Instruments, Sarasota, FL, USA) was threaded through the pilot incision between the lens and the iris, and extended through the posterior segment into the retina. A retinal detachment was created by administering 0.5 μL of viral vector (1 × 109 vg) at a rate of 300 nL/s under the retina, between the RPE and the photoreceptors. The eyes were enucleated 28 days post-injection and placed in 10% neutral-buffered formalin for 48 hr. Eyes were then transferred to 15% sucrose for 24 hr before being frozen in Optimal Cutting Temperature compound (Sakura Finetek USA, Torrance, CA, USA). Seven-micron sections were collected on a CryoStar NX70 cryostat (Thermo Fisher Scientific, Waltham, MA, USA). Raw GFP signaling was visualized and imaged using epifluorescence. Animal care and use complied with Sanofi’s Institutional Animal Care and Use Committee (IACUC).
In Vitro and In Vivo Transduction Assessment of AAVDJ and AAVDJ Mutants
HuH7 cells were seeded at 2e5 cells/well and infected 24 hr later, in triplicate, with AAV at an MOI of either 1e3 or 1e5 vg/cell in a 500-μL volume. The media were replaced 24 hr post-infection with 1 mL of complete DMEM containing 10% fetal bovine serum (FBS), penicillin/streptomycin (pen/strep), and L-glutamine. After 72 hr, cells were lysed and assayed for vector genome copy number by TaqMan PCR assay (BGH target) and EGFP protein levels using an EGFP ELISA kit from Abcam (ab 171581). Additionally, C57BL6 mice, 8–10 weeks, were administered 3e11 vg of either AAVDJ or AAVDJ3, by tail vein injection. Approximately 4 weeks after vector administration, liver, spleen, kidney, and heart were harvested and analyzed for EGFP expression, using an EGFP ELISA kit from Abcam (ab 171581).
Author Contributions
Conceptualization, S.A.N. and C.R.O.; Methodology and Investigation, S.A.N., M.A.M., D.A.W., J.A.A., S.E.O., and A.M.F.; Writing – Original Draft, C.R.O.; Writing – Review and Editing, C.R.O., S.A.N., A.S., and S.H.C.
Conflicts of Interest
All authors are Sanofi employees.
Acknowledgments
Authors acknowledge technical support from the Gene Therapy Development group at Sanofi and graphics support from Alison Schroeer, Sanofi.
Footnotes
Supplemental Information includes two tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2017.12.004.
Supplemental Information
References
- 1.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauswirth W.W., Aleman T.S., Kaushal S., Cideciyan A.V., Schwartz S.B., Wang L., Conlon T.J., Boye S.L., Flotte T.R., Byrne B.J., Jacobson S.G. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum. Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maguire A.M., High K.A., Auricchio A., Wright J.F., Pierce E.A., Testa F., Mingozzi F., Bennicelli J.L., Ying G.S., Rossi S. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simonelli F., Maguire A.M., Testa F., Pierce E.A., Mingozzi F., Bennicelli J.L., Rossi S., Marshall K., Banfi S., Surace E.M. Gene therapy for Leber’s congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther. 2010;18:643–650. doi: 10.1038/mt.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury S.R., Fitzpatrick Z., Harris A.F., Maitland S.A., Ferreira J.S., Zhang Y., Ma S., Sharma R.B., Gray-Edwards H.L., Johnson J.A. In vivo selection yields AAV-B1 capsid for central nervous system and muscle gene therapy. Mol. Ther. 2016;24:1247–1257. doi: 10.1038/mt.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayuso E., Mingozzi F., Bosch F. Production, purification and characterization of adeno-associated vectors. Curr. Gene Ther. 2010;10:423–436. doi: 10.2174/156652310793797685. [DOI] [PubMed] [Google Scholar]
- 10.O’Riordan C.R., Lachapelle A.L., Vincent K.A., Wadsworth S.C. Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV) J. Gene Med. 2000;2:444–454. doi: 10.1002/1521-2254(200011/12)2:6<444::AID-JGM132>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Yang X., Wright J.F., High K.A., Couto L., Qu G. PEG-modulated column chromatography for purification of recombinant adeno-associated virus serotype 9. J. Virol. Methods. 2011;173:99–107. doi: 10.1016/j.jviromet.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Davidoff A.M., Ng C.Y., Sleep S., Gray J., Azam S., Zhao Y., McIntosh J.H., Karimipoor M., Nathwani A.C. Purification of recombinant adeno-associated virus type 8 vectors by ion exchange chromatography generates clinical grade vector stock. J. Virol. Methods. 2004;121:209–215. doi: 10.1016/j.jviromet.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Mietzsch M., Grasse S., Zurawski C., Weger S., Bennett A., Agbandje-McKenna M., Muzyczka N., Zolotukhin S., Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum. Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oranje P.P.A., Verheesen P., Verbart D., Mijnsbergen Y., Haard H.J.W., Hermans P. Isolation of an adeno-associated virus (AAV)-specific camelid-derived single chain antibody fragment: a novel tool for purification of AAV vectors of different serotypes. Mol. Ther. 2004;9:S162. [Google Scholar]
- 15.Wang Q., Lock M., Prongay A.J., Alvira M.R., Petkov B., Wilson J.M. Identification of an adeno-associated virus binding epitope for AVB sepharose affinity resin. Mol. Ther. Methods Clin. Dev. 2015;2:15040. doi: 10.1038/mtm.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 17.Qu G., Bahr-Davidson J., Prado J., Tai A., Cataniag F., McDonnell J., Zhou J., Hauck B., Luna J., Sommer J.M. Separation of adeno-associated virus type 2 empty particles from genome containing vectors by anion-exchange column chromatography. J. Virol. Methods. 2007;140:183–192. doi: 10.1016/j.jviromet.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Burnham B., Nass S., Kong E., Mattingly M., Woodcock D., Song A., Wadsworth S., Cheng S.H., Scaria A., O’Riordan C.R. Analytical ultracentrifugation as an approach to characterize recombinant adeno-associated viral vectors. Hum. Gene Ther. Methods. 2015;26:228–242. doi: 10.1089/hgtb.2015.048. [DOI] [PubMed] [Google Scholar]
- 19.Khani S.C., Pawlyk B.S., Bulgakov O.V., Kasperek E., Young J.E., Adamian M., Sun X., Smith A.J., Ali R.R., Li T. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest. Ophthalmol. Vis. Sci. 2007;48:3954–3961. doi: 10.1167/iovs.07-0257. [DOI] [PubMed] [Google Scholar]
- 20.Halder S., Van Vliet K., Smith J.K., Duong T.T., McKenna R., Wilson J.M., Agbandje-McKenna M. Structure of neurotropic adeno-associated virus AAVrh.8. J. Struct. Biol. 2015;192:21–36. doi: 10.1016/j.jsb.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiPrimio N., Asokan A., Govindasamy L., Agbandje-McKenna M., Samulski R.J. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 2008;82:5178–5189. doi: 10.1128/JVI.02721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King J.A., Dubielzig R., Grimm D., Kleinschmidt J.A. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 24.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorne B.A., Takeya R.K., Peluso R.W. Manufacturing recombinant adeno-associated viral vectors from producer cell clones. Hum. Gene Ther. 2009;20:707–714. doi: 10.1089/hum.2009.070. [DOI] [PubMed] [Google Scholar]
- 26.Martin J., Frederick A., Luo Y., Jackson R., Joubert M., Sol B., Poulin F., Pastor E., Armentano D., Wadsworth S., Vincent K. Generation and characterization of adeno-associated virus producer cell lines for research and preclinical vector production. Hum. Gene Ther. Methods. 2013;24:253–269. doi: 10.1089/hgtb.2013.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.