Abstract
Brevibacterium spp. are aerobic, nonbranched, asporogenous, gram-positive, rod-shaped bacteria which may exhibit a rod-coccus cycle when cells get older and can be found in various environments. Several Brevibacterium species have industrial importance and are capable of biotransformation of various contaminants. Here we describe the draft genome sequence of Brevibacterium epidermidis EZ-K02 isolated from nitrocellulose-contaminated wastewater environments. The genome comprises 3,885,924 bp, with a G + C content of 64.2%. This whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PDHL00000000.
Keywords: Brevibacterium epidermidis, Draft genome, Wastewater
Specifications Table
| Subject area | Biology |
| More specific subject area | Genome analysis |
| Type of data | Table, figures |
| How data was acquired | Illumina Miseq |
| Data format | Analyzed |
| Experimental factors | Genomic DNA from pure culture |
| Experimental features | Isolation of bacteria, genome sequencing, draft genome assembly and annotation |
| Data source location | Nitrocellulose-contaminated wastewater environments, Kazan, Russia |
| Data accessibility | Data are in public repository. This whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PDHL00000000 (https://www.ncbi.nlm.nih.gov/nuccore/PDHL00000000). The 16S rRNA gene sequence has been deposited at GenBank under the accession number MG050737 (https://www.ncbi.nlm.nih.gov/nuccore/MG050737). |
Value of the data
-
•
Several Brevibacterium species have industrial importance and are capable of biotransformation of various contaminants; therefore, more investigations at the genomic level are necessary to improve our understanding of their ecology, genetics, as well as potential biotechnological applications.
-
•
Data shown here can be useful for other groups working or studying in the field of application of brevibacteria in bioremediation processes.
-
•
Data demonstrated here can be used by other researchers working or studying in the field of genome analysis.
1. Data
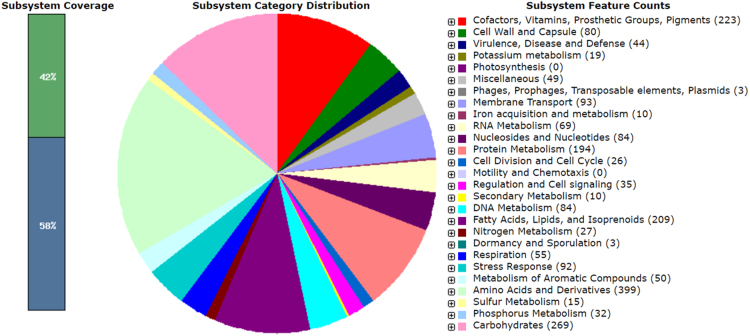
The draft genome sequence of B. epidermidis strain EZ-K02 constituted a total of 65 contigs (> 500 bp) with 3,885,924 bp, and a G + C content of 64.2% (Table 1). The RAST server predicted 3443 coding sequences where 1436 coding sequences (42%) were annotated as seed subsystem features and 2007 coding sequences (58%) annotated as outside of the seed subsystem. In total 2457 and 986 coding sequences were assigned as non-hypothetical and hypothetical, respectively. The genome was shown to encode at least 3 rRNAs and 47 tRNAs. The strain EZ-K02 possesses a substantial number of genes which are responsible for nitrate/nitrite ammonification (for example, in case with nitrate released during nitrocellulose denitration) as well as for metabolism of aromatic compounds, including genes involved in benzoate, p-hydroxybenzoate, acetophenone, catechol, gentisate and several other compounds biodegradation. Numerous genes responsible for resistance to toxic compounds, including arsenic, cobalt and cadmium, were additionally detected. Hence, B. epidermidis EZ-K02 may have high importance in the field of development of several effective environmental biotechnologies, such as environmental bioremediation and wastewater treatment.
Table 1.
Comparison of the genomic feature of Brevibacterium epidermidis EZ-K02 strain with various Brevibacterium strains. The information regarding the reference genomes was received from the EzBioCloud database [11].
| Organism | DB accession number | Isolation source | Contigs | Genome size (bp) | G + C (%) | CDSs | r + tRNA genes |
|---|---|---|---|---|---|---|---|
| B. epidermidis EZ-K02 | GCA_002573745.1 | Wastewater | 65 | 3,885,924 | 64.2 | 3443 | 3+47 |
| B. album DSM 18261 | GCA_000426445.1 | Saline alkali soil | 15 | 4,094,970 | 70.9 | 3559 | 9+59 |
| B. casei S18 | GCA_000314575.1 | Human skin | 43 | 3,664,641 | 68.1 | 3233 | 6+46 |
| B. epidermidis NBRC 14811 | GCA_001570805.1 | Human skin | 25 | 3,703,261 | 64.3 | 3261 | 3+48 |
| B. linens SMQ-1335 | GCA_001729525.1 | Cheese | 1 | 4,209,935 | 62.6 | 3863 | 12+49 |
| B. ravenspurgense CCUG 56047 | GCA_001584615.1 | Human specimens | 14 | 2,297,397 | 62.4 | 2022 | 6+43 |
| B. senegalense JC43 | GCA_000285835.2 | Human stool | 17 | 3,427,329 | 69.4 | 3058 | 3+46 |
2. Experimental design, materials and methods
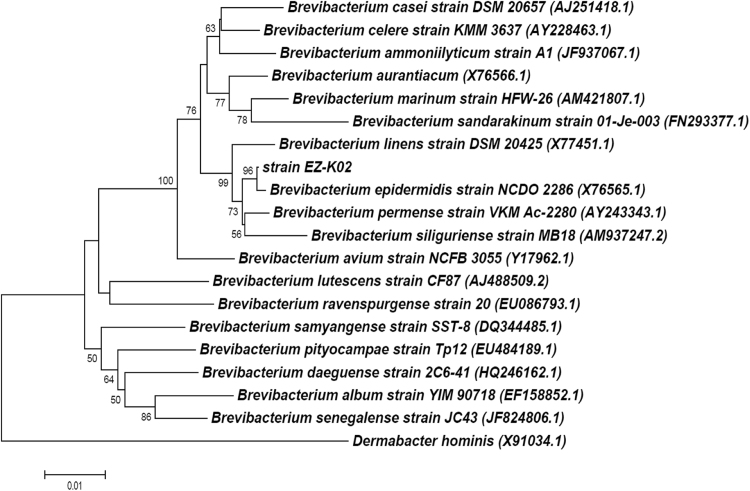
B. epidermidis EZ-K02 was isolated from nitrocellulose-contaminated wastewater environments, Kazan, the Republic of Tatarstan, Russia. Such industrial wastes are represented by large amounts of wastewaters contaminated with different dissolved chemical compounds and nitrocellulose powder particles. Also, these microbes are of high importance for the development of effective bioremediation strategies of various polluted environments [1], [2], [3]. The bacterium optimally grown on LB agar at +30 °C had been cultivated for 24–48 hours. Genomic DNA of the bacterial strain was then extracted and purified with a FastDNA spin kit (MP Biomedicals) and a FastPrep-24 homogenizer (MP Biomedicals) according to the manufacturer's protocol. Concentration and purity (A260/A280) of the extracted genomic DNA were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and stored at –20 °C until further processing. The bacterial strain EZ-K02 was morphologically identified and confirmed by PCR amplification using the primers UniBac27f, Bakt_805R and Univ1492r, followed by sequencing using an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems) and phylogenetic analysis (16S rRNA gene sequence; 1413 bp; Fig. 1). In order to perform whole genome analysis, DNA was fragmented using a Q800R2 Sonicator (Qsonica), and DNA library was then prepared with a NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturers’ protocols. Both efficiency of DNA fragmentation and DNA library preparation were monitored using a 2100 Bioanalyzer (Agilent) and a High Sensitivity DNA kit (Agilent). Further sequencing was performed with a high-throughput Illumina MiSeq platform (Illumina) at Joint KFU-Riken Laboratory, Kazan Federal University (Kazan, Russia) by a MiSeq Reagent Kit v2PE 500 cycles (Illumina). Briefly, sequence read quality was assessed using PRINSEQ lite version 0.20.4 [4], the genome was assembled using Velvet version 1.2.10 [5], and the ordering of contigs was achieved using Mauve version 2.4.0 [6]. The whole genome sequence of B. epidermidis was annotated using the Rapid Annotation System Technology (RAST) server [7]. The pie chart demonstrated the counts for each subsystem feature and the subsystem coverage (Fig. 2). The rRNA and tRNA genes were identified using RNAmmer 1.2 [8] and tRNA scan-SE 1.23 [9], respectively.
Fig. 1.
A phylogenetic tree based on 16S rRNA gene sequences demonstrating the relationship between Brevibacterium epidermidis EZ-K02 (NCBI accession number of 16S rRNA gene: MG050737) and the type strains from the LPSN site (www.bacterio.net). Analysis was conducted in MEGA7 [10] using the neighbor-joining method based on Jukes-Cantor evolutionary distances. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Dermabacter hominis was used as outgroup.
Fig. 2.
An overview of the subsystem categories assigned to the genome of Brevibacterium epidermidis EZ-K02. The whole genome sequence of the strain EZ-K02 was annotated using the Rapid Annotation System Technology (RAST) server [7]. The pie chart demonstrates the count of each subsystem feature and the subsystem coverage.
Acknowledgements
The work was supported by the grant of the President of the Russian Federation for the young scientists [Grant no. МД-100.2017.4].
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2017.12.053.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Ziganshina E.E., Ibragimov E.M., Ilinskaya O.N., Ziganshin A.M. Bacterial communities inhabiting toxic industrial wastewater generated during nitrocellulose production. Biologia. 2016;71:70–78. [Google Scholar]
- 2.Ziganshin A.M., Gerlach R., Naumenko E.A., Naumova R.P. Aerobic degradation of 2,4,6-trinitrotoluene by the yeast strain Geotrichum candidum AN-Z4. Microbiology. 2010;79:178–183. [Google Scholar]
- 3.Ziganshin A.M., Ziganshina E.E., Byrne J., Gerlach R., Struve E., Biktagirov T., Rodionov A., Kappler A. Fe(III) mineral reduction followed by partial dissolution and reactive oxygen species generation during 2,4,6-trinitrotoluene transformation by the aerobic yeast Yarrowia lipolytica. AMB Express. 2015;5:1–12. doi: 10.1186/s13568-014-0094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerbino D.R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinforma. 2010;11.5:1–13. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rissman A.I., Mau B., Biehl B.S., Darling A.E., Glasner J.D., Perna N.T. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe T.M., Eddy S.R. tRNA scan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material