Abstract
The present study investigates the spectrum and incidence of mitochondrial DNA (mtDNA) mutations associated with Leber's hereditary optic neuropathy (LHON) in a Han population using a multi-gene panel with 46 LHON-associated mutations among 13 mitochondrial genes. A total of 23 mutations were observed in a cohort of 275 patients and 281 control subjects using multi-gene panel analysis. The causative mutations associated with LHON were identified to be m.11778G>A, m.14484T>C, m.3460 G>A, m.3635G>A, m.3866T>C and m.3733G>A, responsible for 70.55% cases in the patient cohort. The secondary mutations in the Chinese LHON population were m.12811T>C, m.11696 G>A, m.3316G>A, m.3394T>C, m.14502T>C, m.3497C>T, m.3571C>T, m.12338T>C, m.14693A>G, m.4216T>C and m.15951A>G, with incidences of 5.09, 4.36, 4.00, 4.00, 4.00, 2.55, 1.82, 1.82, 1.45, 1.09 and 1.09%, respectively. Besides three hotspot genes, MT-ND1, MT-ND4 and MT-ND6, MT-ND5 also had a high incidence of secondary mutations. Those mutations reported as rare causative mutations in a European LHON population, m.3376G>A, m.3700G>A and m.4171C>A, m.10663T>C, m.13051G>A, m.14482C>G/A, m.14495A>G and m.14568C>T were undetected in the present study. The primary and secondary mutations associated with LHON in the present multi-gene panel will advance the current understanding of the clinical phenotype of LHON, and provide useful information for early diagnosis.
Keywords: Leber's hereditary optic neuropathy, mitochondrial DNA, causative mutation, multi-gene panel, gene-target sequencing
Introduction
Leber's hereditary optic neuropathy (LHON; OMIM 535000) is a classic mitochondrial disease, associated with a rapid, painless, acute or sub-acute bilateral visual loss in young adults, predominantly caused by the primary and secondary mutations in mitochondrial DNA (mtDNA). It has been reported that 1:8,500 individuals harbor a primary LHON-causing mutation and 1:31,000 experience visual loss as a result of LHON in the North East of England (1). Few significant improvements in visual acuity are reported following atrophy of the optic discs. LHON typically affects males more frequently than females, with the incomplete and variable penetrance estimated at ~50% in males and 10% in females (2–4). Additionally, certain LHON cases have additional clinical symptoms, such as movement disorders, dystonia, and multiple-sclerosis-like illness, which complicate the diagnosis in the clinical setting (5–7). Although the majority ofcases of LHON transmitted by maternal inheritance have a history of visual loss in families, up to 40% of cases are sporadic (5).
The genetic cause of LHON is mutations in the mitochondrial genome, which is a double-stranded 16,569-nucleotide pair, circular molecule, consisting of one D-Loop region and 37 genes. The three most causative mutations, m.11778G>A (MT-ND4), m.14484T>C (MT-ND6) and m.3460G>A (MT-ND1), have been reported to account for 90% of LHON patients in a Caucasian population, but for only 38.3 and 46.5% of cases in two large cohorts of Chinese Han subjects with LHON (7–10). Our previous studies have shown the spectrum of genes, MT-ND1, MT-ND4 and MT-ND6, and the frequency of the three primary mutations in a Chinese LHON population (8–10) using Sanger sequencing. In addition, secondary mutations that contributed to the high penetrance, including m.3394T>C (MT-ND1), m.11696G>A (MT-ND4), m.12338T>C (MT-ND5) and m.15951A>G (MT-TT) areusually synergized with m.11778G>A or m.14484T>C or m.3460G>A (11). According to Mitomap (http://www.mitomap.org/), >40 point mutations in mtDNA are associated with LHON, of which the incidence varies between different ethnic backgrounds.
To further understand the spectrum of mutations associated with LHON in a Chinese population, 46 LHON-associated mutations distributed among 13 mitochondrial genes were selected from Mitomap, and multi-gene target sequencing was performed in 275 cases of LHON as well as in 281 Chinese control subjects to distinguish the most frequent mtDNA mutations associated with LHON in the Han population.
Materials and methods
DNA samples, extraction, quantification and quality control
A total of 275 unrelated LHON samples and 281 Chinese control samples were enrolled from the ophthalmology clinics at Zhejiang University School of Medicine (Hangzhou, China) and Wenzhou Medical College (Wenzhou, China) between 2004 and 2015, as described previously (8–10,12), under protocols approved by Zhejiang University and Wenzhou Medical University Ethics Committees. DNA was extracted from 1 ml peripheral blood using a QIAamp DNA Blood Minikit (51106; Qiagen China Co., Ltd., Shanghai, China). The quality and quantity of DNA were assessed using Qubit 3.0 fluorometers (Thermo Fisher Scientific, Inc., Waltham, MA, USA). DNA samples with concentration >1.0 ng/µl were employed in the sequencing experiments.
Multi-genepanel design
Multi-gene target sequencing was performed using the VariantPro™ Capture Technology by LC Sciences (Hangzhou, China) as described previously (13). The 46 LHON-associated mutations were selected from Mitomap and previous studies (8–10). As presented in Table I, they were distributed in the following 13 genes: MT-ND1, MT-ND2, MT-ATP6, MT-CO3, MT-ND3, MT-ND4L, MT-ND4, MT-ND5, MT-ND6, MT-CYB, MT-TM, MT-TT and MT-TE. Twenty-seven amplicons that covered all 46 mutations were designed by LC Sciences, as described previously (13). All amplicons were pooled into two polymerase chain reaction (PCR) tubes (tube 1 and tube 2) as VariantPro™ PCR mastermix with an average length of 184 nt (range, 167–203 nt).
Table I.
Mutations in the multi-gene panel (n=46).
| Index | Gene name | Var start | Var end | Ref allele | Var allele | Amino acid change |
|---|---|---|---|---|---|---|
| 1 | MT-ND1 | 3316 | 3316 | G | A | A-T |
| 2 | MT-ND1 | 3376 | 3376 | G | A | E-K |
| 3 | MT-ND1 | 3394 | 3394 | T | C | Y-H |
| 4 | MT-ND1 | 3460 | 3460 | G | A | A-T |
| 5 | MT-ND1 | 3497 | 3497 | C | T | A-V |
| 6 | MT-ND1 | 3571 | 3571 | C | T | L-F |
| 7 | MT-ND1 | 3635 | 3635 | G | A | S-N |
| 8 | MT-ND1 | 3700 | 3700 | G | A | A-T |
| 9 | MT-ND1 | 3733 | 3733 | G | A | E-K |
| 10 | MT-ND1 | 3866 | 3866 | T | C | I-T |
| 11 | MT-ND1 | 4025 | 4025 | C | T | T-M |
| 12 | MT-ND1 | 4171 | 4171 | C | A | L-M |
| 13 | MT-ND1 | 4216 | 4216 | T | C | Y-H |
| 14 | MT-ND2 | 4640 | 4640 | C | A | I-M |
| 15 | MT-ND2 | 5244 | 5244 | G | A | G-S |
| 16 | MT-ATP6 | 9101 | 9101 | T | C | I-T |
| 17 | MT-CO3 | 9804 | 9804 | G | A | A-T |
| 18 | MT-ND3 | 10237 | 10237 | T | C | I-T |
| 19 | MT-ND4L | 10663 | 10663 | T | C | V-A |
| 20 | MT-ND4L | 10680 | 10680 | G | A | A-T |
| 21 | MT-ND4 | 11253 | 11253 | T | C | I-T |
| 22 | MT-ND4 | 11696 | 11696 | G | A | V–I |
| 23 | MT-ND4 | 11778 | 11778 | G | A | R-H |
| 24 | MT-ND5 | 12338 | 12338 | T | C | M-T |
| 25 | MT-ND5 | 12811 | 12811 | T | C | Y-H |
| 26 | MT-ND5 | 12848 | 12848 | C | T | A-V |
| 27 | MT-ND5 | 13051 | 13051 | G | A | G-S |
| 28 | MT-ND5 | 13528 | 13528 | A | G | T-A |
| 29 | MT-ND5 | 13637 | 13637 | A | G | Q-R |
| 30 | MT-ND5 | 13730 | 13730 | G | A | G-E |
| 31 | MT-ND6 | 14279 | 14279 | G | A | S-L |
| 32 | MT-ND6 | 14325 | 14325 | T | C | N-D |
| 33 | MT-ND6 | 14482 | 14482 | C | A | M-I |
| 34 | MT-ND6 | 14482 | 14482 | C | G | M-I |
| 35 | MT-ND6 | 14484 | 14484 | T | C | M-V |
| 36 | MT-ND6 | 14495 | 14495 | A | G | L-S |
| 37 | MT-ND6 | 14498 | 14498 | T | C | Y-C |
| 38 | MT-ND6 | 14502 | 14502 | T | C | I–V |
| 39 | MT-ND6 | 14568 | 14568 | C | T | G-S |
| 40 | MT-ND6 | 14596 | 14596 | A | T | I-M |
| 41 | MT-CYB | 14831 | 14831 | G | A | A-T |
| 42 | MT-CYB | 15812 | 15812 | G | A | V-M |
| 43 | MT-TM | 4435 | 4435 | A | G | tRNAMet |
| 44 | MT-TT | 15951 | 15951 | A | G | tRNAThr |
| 45 | MT-TE | 14693 | 14693 | A | G | tRNAGlu |
| 46 | MT-TE | 14727 | 14727 | T | C | tRNAGlu |
Var start, start site of variant; var end, end site of variant; ref allele, referenced allele; Var allele, variant allele.
Library preparation and sequencing
Library generation was performed according to the manufacturer's protocol (LC Sciences). Briefly, 5 ng DNA per pool was amplified in 25 cycles of PCR using probe sequences of 27 library amplicons (Table II) and a VariantPro™ PCR mastermix as described previously (13). PCR runs of 18, 20, 24 and 26 cycles were performed to evaluate the influence of the PCR cycles on the experiment. The correlation coefficient, R2, ranged from 0.90–1.00 (average, 0.95), implicating that cycles between 18 and 26 had no influence on the experiment outcome. The amplified products were purified using Agencourt AMPure XP beads [Beckman Coulter (UK) Ltd., High Wycombe, UK]. Each library was diluted to 20 pM and sequenced on an Illumina Miseq with a minimum of 2X 150-bp paired-end reads.
Table II.
PCR amplicons.
| Index | Gene name | Tgt start | Tgt end | Prb strand | Prb start | Prb end | Var start | Var end | Prb length (bp) | Amp length (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MT-ND1 | 3,316 | 3,866 | − | 3,269 | 3,439 | 3,288 | 3,425 | 171 | 303 |
| 2 | MT-ND1 | 3,316 | 3,866 | + | 3,402 | 3,585 | 3,415 | 3,571 | 184 | 316 |
| 3 | MT-ND1 | 3,316 | 3,866 | − | 3,526 | 3,699 | 3,543 | 3,684 | 174 | 306 |
| 4 | MT-ND1 | 3,316 | 3,866 | + | 3,643 | 3,812 | 3,661 | 3,794 | 170 | 302 |
| 5 | MT-ND1 | 3,316 | 3,866 | − | 3,745 | 3,936 | 3,767 | 3,918 | 192 | 324 |
| 6 | MT-ND1 | 4,025 | 4,025 | − | 3,952 | 4,140 | 3,962 | 4,126 | 189 | 321 |
| 7 | MT-ND1 | 4,171 | 4,216 | + | 4,132 | 4,334 | 4,145 | 4,315 | 203 | 335 |
| 8 | MT-TM | 4,435 | 4,435 | − | 4,363 | 4,529 | 4,380 | 4,514 | 167 | 299 |
| 9 | MT-ND2 | 4,640 | 4,640 | − | 4,482 | 4,667 | 4,494 | 4,655 | 186 | 318 |
| 10 | MT-ND2 | 5,244 | 5,244 | + | 5,140 | 5,317 | 5,154 | 5,302 | 178 | 310 |
| 11 | MT-ATP6 | 9,101 | 9,101 | − | 9,026 | 9,223 | 9,041 | 9,208 | 198 | 330 |
| 12 | MT-CO3 | 9,804 | 9,804 | − | 9,639 | 9,839 | 9,653 | 9,824 | 201 | 333 |
| 13 | MT-ND3 | 10,237 | 10,237 | − | 10,161 | 10,331 | 10,178 | 10,310 | 171 | 303 |
| 14 | MT-ND4L | 10,663 | 10,680 | − | 10,541 | 10,710 | 10,560 | 10,692 | 170 | 302 |
| 15 | MT-ND4 | 11,253 | 11,253 | + | 11,226 | 11,404 | 11,239 | 11,386 | 179 | 311 |
| 16 | MT-ND4 | 11,696 | 11,778 | + | 11,642 | 11,833 | 11,660 | 11,813 | 192 | 324 |
| 17 | MT-ND5 | 12,338 | 12,338 | − | 12,213 | 12,399 | 12,230 | 12,385 | 187 | 319 |
| 18 | MT-ND5 | 12,811 | 12,848 | + | 12,764 | 12,941 | 12,776 | 12,924 | 178 | 310 |
| 19 | MT-ND5 | 13,051 | 13,051 | − | 12,967 | 13,146 | 12,984 | 13,128 | 180 | 312 |
| 20 | MT-ND5 | 13,528 | 13,730 | + | 13,488 | 13,682 | 13,503 | 13,665 | 195 | 327 |
| 21 | MT-ND5 | 13,528 | 13,730 | + | 13,577 | 13,774 | 13,590 | 13,760 | 198 | 330 |
| 22 | MT-ND6 | 14,279 | 14,325 | − | 14,251 | 14,421 | 14,267 | 14,408 | 171 | 303 |
| 23 | MT-TE; MT-ND6; MT-CYB | 14,482 | 14,831 | + | 14,444 | 14,645 | 14,460 | 14,628 | 202 | 334 |
| 24 | MT-TE; MT-ND6; MT-CYB | 14,482 | 14,831 | − | 14,587 | 14,762 | 14,610 | 14,748 | 176 | 308 |
| 25 | MT-TE; MT-ND6; MT-CYB | 14,482 | 14,831 | + | 14,699 | 14,866 | 14,720 | 14,857 | 168 | 300 |
| 26 | MT-CYB; MT-TT | 15,812 | 15,951 | + | 15,780 | 15,966 | 15,801 | 15,945 | 187 | 319 |
| 27 | MT-CYB; MT-TT | 15,812 | 15,951 | − | 15,792 | 15,989 | 15,809 | 15,974 | 198 | 330 |
PCR, polymerase chain reaction; Tgt, target; prb, probe; var, variation; +, forward primer; -, antisense primer.
Data analysis
Low quality reads (reads containing sequencing adaptors or nucleotides with quality scores <20) were removed before alignment. Cleaned, paired-end sequence reads in paired FASTQ files were aligned using Burrows-Wheeler Alignment version 0.1.19 (14). Variant calling was generated using the Genome Analysis Toolkit version 3.3.0 and its Unified Genotyper module (https://www.broadinstitute.org/gatk/guide/tagged?tag=unifiedgenotyper). A Gaussian mixture model was used to evaluate the confidence score for each putative mutation call and novel potential variants. Sequence reads were aligned to the human mtDNA sequence data relative to the revised Cambridge Reference Sequence (GenBank accession no. NC_012920) (15).
Sanger validation
Thirteen LHON cases associated with m.11778G>A (11) or m.14484T>C (2) mutations and four healthy control samples (available upon request) were selected as the positive and negative controls, respectively, for runs of the panel following validation by Sanger sequencing. Furthermore, 100% correlation was derived from the panel assay and the Sanger sequencing for the positive and negative controls.
Results
Summary of sequencing data
A total of 118 milion reads were obtained, on average 89% of which were mapped to the amplicon targets and resultedin a mean of 7,001 reads to each sample per amplicon (Table III). The mean reads over the 27 amplicons were distrubed with an average uniformity of coverage of 98.0% (amplicon mean coverage, 20%) and an average read-depth of 1,000 X (Fig. 1). A total of 363 variants were distributed in 46 LHON-associated mutations of all samples.
Table III.
Summary of sequencing data in the panel for 556 samples.
| Variable | Outcome |
|---|---|
| Total no. of reads | 118,156,518 |
| Reads mapped to the amplicons (forward primer) | 110,440,487 |
| Reads mapped to the amplicons (reverse primer) | 108,812,285 |
| Reads mapped to the amplicon targets | 105,107,193 |
| Reads mapped to each amplicon (average) | 3,892,859 |
| Reads mapped to each sample per amplicon (mean) | 7,001.5 |
| Reads enrichment to the targets,% (average) | 89 |
| Uniformity of coverage, % (20% mean) | 98 |
| Total no. of variants among 46 point mutations | 363 |
Figure 1.
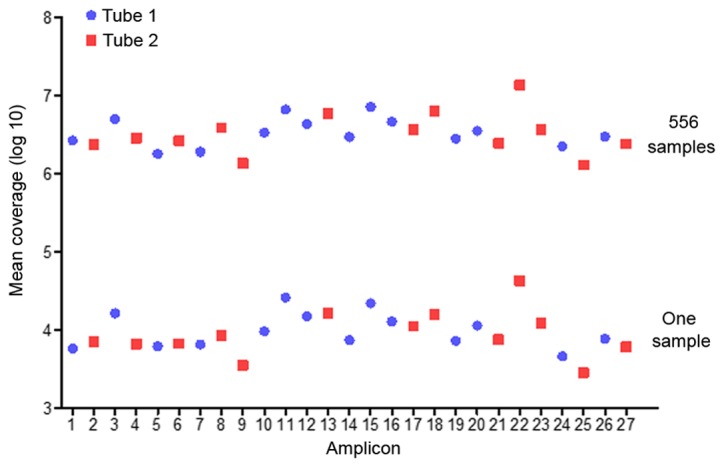
Mean coverage of 27 amplicons. Upper: Average read coverages (log 10) for each amplicon (556 samples). Lower: Read coverages (log10) for each amplicon from one sample.
Mutations analysis
A total of 363 variants were identified in the cohort of all 556 samples; 285 variants were detected in LHON cases as an average incidence of 104%, whereas only 78 variants were identified in 281 controls with a mean incidence of 28% (Table IV).
Table IV.
Summary information of 46-point mutations in the cohort.
| Primer mutations | Incidence (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Indexa | Gene | SNP | Total variants | Patients (n=275) | 11778 | 14484 | 3460 | Controls (n=281) | Patients | Controls |
| 1 | MT-ND1 | 3316G>A | 15 | 11 | 7 | 1 | 0 | 4 | 4.00 | 1.42 |
| 2 | 3376G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 3 | 3394T>C | 19 | 11 | 7 | 0 | 0 | 8 | 4.00 | 2.85 | |
| 4 | 3460G>A | 2 | 2 | 0 | 0 | 2 | 0 | 0.73 | 0.00 | |
| 5 | 3497C>T | 14 | 7 | 4 | 0 | 0 | 7 | 2.55 | 2.50 | |
| 6 | 3571C>T | 11 | 5 | 3 | 0 | 0 | 6 | 1.82 | 2.14 | |
| 7 | 3635G>A | 1 | 1 | 0 | 0 | 0 | 0 | 0.36 | 0.00 | |
| 8 | 3700G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 9 | 3733G>A | 1 | 1 | 0 | 0 | 0 | 0 | 0.36 | 0.00 | |
| 10 | 3866T>C | 4 | 4 | 1 | 0 | 0 | 0 | 1.45 | 0.00 | |
| 11 | 4025C>T | 1 | 1 | 0 | 0 | 0 | 0 | 0.36 | 0.00 | |
| 12 | 4171C>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 13 | 4216T>C | 11 | 3 | 0 | 0 | 0 | 8 | 1.09 | 2.85 | |
| 14 | MT-ND2 | 4640C>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 15 | 5244G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 16 | MT-ATP6 | 9101T>C | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 17 | MT-CO3 | 9804G>A | 2 | 1 | 1 | 0 | 0 | 1 | 0.36 | 0.36 |
| 18 | MT-ND3 | 10237T>C | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 19 | MT-ND4L | 10663T>C | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 20 | 10680G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 21 | MT-ND4 | 11253T>C | 1 | 1 | 1 | 0 | 0 | 0 | 0.36 | 0.00 |
| 22 | 11696G>A | 18 | 12 | 10 | 1 | 0 | 6 | 4.36 | 2.14 | |
| 23 | 11778G>A | 162 | 162 | 162 | 0 | 0 | 0 | 58.90 | 0.00 | |
| 24 | MT-ND5 | 12338T>C | 19 | 5 | 2 | 0 | 0 | 14 | 1.82 | 4.98 |
| 25 | 12811T>C | 21 | 14 | 12 | 0 | 0 | 7 | 5.09 | 2.49 | |
| 26 | 12848C>T | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 27 | 13051G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 28 | 13528A>G | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 29 | 13637A>G | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 30 | 13730G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 31 | MT-ND6 | 14279G>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 32 | 14325T>C | 2 | 0 | 0 | 0 | 0 | 2 | 0.00 | 0.71 | |
| 33 | 14482C>A | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 34 | 14482C>G | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 35 | 14484T>C | 25 | 25 | 0 | 25 | 0 | 0 | 9.10 | 0.00 | |
| 36 | 14495A>G | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 37 | 14498T>C | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 38 | 14502T>C | 18 | 11 | 8 | 3 | 0 | 7 | 4.00 | 2.49 | |
| 39 | 14568C>T | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 40 | 14596A>T | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| 41 | MT-CYB | 14831G>A | 5 | 1 | 0 | 0 | 0 | 4 | 0.36 | 1.42 |
| 42 | 15812G>A | 1 | 0 | 0 | 0 | 0 | 1 | 0.00 | 0.36 | |
| 43 | MT-TM | 4435A>G | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| 44 | MT-TT | 15951A>G | 5 | 3 | 1 | 2 | 0 | 2 | 1.09 | 0.71 |
| 45 | MT-TE | 14693A>G | 5 | 4 | 3 | 1 | 0 | 1 | 1.45 | 0.36 |
| 46 | 14727T>C | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 | |
| Total | 162b | 25c | ||||||||
| variants | 363 | 285 | (60) | (8) | 2 | 78 | 103.64 | 27.76 | ||
The index number is according to Table I
162 subjects carrying the m.11778G>A mutation, of which 60 subjects were carrying the m.11778G>A and one of the secondary variants
25 subjects carrying the m.14484T>C mutation, of which 8 subjects were carrying the m.14484T>C and a secondary variant. The three common mutations for LHON are emboldened. SNP, single nucleotide polymorphism.
As 46 mutations of 13 mitochondrial genes were selected from all populations of the world, variants from LHON cases in the current study were deposited in 8 mitochondrial genes, MT-ND1, MT-CO3, MT-ND4, MT-ND5, MT-ND6, MT-CYB, MT-TT and MT-TE, with frequencies of 16.14, 0.35, 61.40, 6.67, 12.63, 0.35, 1.05 and 1.40%, respectively. Consistent with our previous reports (8–10), MT-ND1, MT-ND4 and MT-ND6 were the hotspots associated with LHON, and almost cover 90% of variants in the present study.
Twenty-three out of the 46 LHON-associated mutations were detected in all subjects. These were as follows: 3316G>A, 3394T>C, 3460G>A, 3497C>T, 3571C>T, 3635G>A, 3733G>A, 3866T>C, 4025C>T and 4216T>C mutations in MT-ND1, 9804G>A in MT-CO3, 11253T>C, 11696G>A and 11778G>A in MT-ND4, m.12338T>C and 12811T>C in MT-ND5, 14325T>C, 14484T>C and 14502T>C in MT-ND6, 14831G>A and 15812G>A in MT-CYB, 15951A>G in MT-TT and 14693A>G in MT-TE. The incidence of these three common mutations m.11778G>A, m.14484T>C and m.3460G>A in this Chinese cohort were 58.90, 9.10 and 0.73%, respectively. In addition, two causative mutations were detected; m.3635G>A in 1 patient and m.3866T>C in 4 patients (one case carrying both m.11778G>A and m.3866T>C mutations), as reported previously (16). Notably, three mutations, m.3733G>A, m.4025C>T and m.11253T>C, were observed in one LHON case each, but absent in the control cohort. Whereas, m.4025C>T and m.11253T>C were observed in the control population in our recent studies (8,9).
Thirteen secondary mutations were identified in 30.55% patients. The incidence of these known secondary mutations, m.12811T>C, m.11696G>A, m.3316G>A, m.3394T>C, m.14502T>C, m.3497C>T, m.3571C>T, m.12338T>C, m.14693A>G, m.4216T>C, m.15951A>G, m.14831G>A and m.9804G>A were 5.09, 4.36, 4.00, 4.00, 4.00, 2.55, 1.82, 1.82, 1.45, 1.09, 1.09, 0.36 and 0.36%, respectively. A total of 88 variations from these 13 mutations were observed in patients. Among these, 67 variations were concurrent with either m.11778G>A (59 variations, except 1 from the m.3866T>C mutation) or m.14484T>C (8 variations). Besides the hotspots of MT-ND1, MT-ND4 and MT-ND6, the MT-ND5 gene was frequently accumulated in the distribution of secondary mutations with an incidence of 6.91% in the patients. In addition, the incidence of MT-ND1, MT-ND4 and MT-ND6 for secondary mutations, were 13.45, 4.36 and 4%, respectively. The secondary mutations were predominantly present in the patient and control populations. Of those, mutations m.12338T>C m.4216T>C, m.3571C>T and m.14831 G>A had higher incidences in the control cohort than in the patients. Two LHON-associated mutations (m.14325T>C and m.15812G>A) only arose in the control population in the present study.
Discussion
The present study evaluated the distribution of mitochondrial genes and mutations among 275 Chinese LHON patients, in parallel with a control cohort of 281 subjects, using a multi-gene panel. Mutations of the multi-gene panel were designed from the Mitomap database, previous reports and our previous study in a Chinese Han population (8–10). Twenty-seven amplicons, the specific primers for the point mutations, covered those of 46 mutations, as well as other substitutions in the amplicons, such as pathogenic mutations, m.3697G>A, m.10197G>A and m.14459G>A in the fragments of amplicon 4, 13 and 23, respectively. None of these three rare causative mutations were recorded among subjects in the test. Of the 46 mutations, the m.14727T>C variant in the MT-TE gene reported in encephalomyopathy patients (17), was set as a negative control variant in the panel and was absent in all of the subjects. The panel with high-throughput sequencing makes it possible to screen multi-genes or multi-single nucleotide polymorphisms (SNPs) for subjects in a run, and provide more information than traditional Sanger sequencing. Certainly, sequencing the whole mtDNA genome is an optional selection using next generation sequencing. The information from the primary and secondary mutations may be indicative regarding the incomplete penetrance and other clinical symptoms.
It is generally accepted that LHON-associated mutations and their incidence are varied in populations with different ethnic backgrounds (18). Consistent results were confirmed in our multi-gene panel screening. Twenty-three mutations in the panels were absent in the patients and control cohort in the current study. Those mutations reported as rare causative mutations in a European LHON population, m.3376G>A, m.3700G>A and m.4171C>A in the MT-ND1 gene (19–21), m.10663T>C in the MT-ND4L (20), m.13051G>A in the MT-ND5 gene (22), m.14482C>G/A, m.14495A>G and m.14568C>T in the MT-ND6 gene (8,20,23), were undetected in the present study. However, 6 causative mutations, m.11778G>A, m.14484T>C, m.3460G>A, m.3635G>A, m.3866T>C and m.3733G>A were observed in 194 LHON cases with their contribution of 83.51, 12.89, 1.04, 0.52, 2.06 and 0.52. Of these, the incidence of m.3460G>A was markedly lower in this cohort than that in a Caucasian population reported by Mackey et al (24). Additionally, the spectrum of secondary mutations associated with LHON was also dependent on their ethnic background, and were distinct between the Chinese and Caucasian cohorts. This panel screening demonstrated that the secondary mutations, m.12811T>C, m.11696G>A, m.3394T>C, m.3316G>A, m.14502T>C and m.12338T>C had higher frequencies in the patient cohort, as these mutations were assigned to Asian mtDNA lineage, including the macro-haplo group of M and N. Certainly, m.12811T>C is considered to be a polymorphic variant in sub-haplo groups of M7. While, mutations, m.3394T>C and m.11696G>A are categorized as haplo group-specific variants of M9a and D4j, respectively (25,26). Congruent results were obtained in our previous reports (27).
Usually, secondary mutations, proposed to increase the penetrance of LHON (25–27), are observed in LHON cases associated with m.11778G>A or m.14484T>C mutations. In the present study, 77.27% of variations of secondary mutations were coexistent with one of the primary mutations, m.11778G>A and m.14484T>C. Their detailed distribution was illustrated in Table V. Secondary mutations m.12811T>C, m.11696G>A, m.14502T>C, m.3394T>C and m.3316G>A exhibited the most co-occurrence with m.11778G>A. Meanwhile, three LHON cases carried m.14484T>C and m.14502T>C together. Notably, the m.14502T>C mutation was evidenced as a modifier in the phenotypic manifestation of LHON (28), although it was reported as a causative mutation elsewhere (https://www.mitomap.org/foswiki). Furthermore, more than one secondary mutation co-occurred with m.11778G>A, but not with m.14484T>C in this panel. m.3497C>T and m.3571C>T, which belong to the haplo group variants of B4c1, arose in three cases associated with m.11778G>A. In addition, m.9804G>A and m.14831G>A, reported as LHON-associated mutations in Caucasian cases, were common in the present control cohort according to the panel analysis. It was confirmed that the spectrum of mutations varied between ethnic backgrounds and indicated that the selected SNPs of the panel would be optimized for a Han population in the future.
Table V.
Distribution of secondary mutations with m.11778 G>A and m.14484 T>C.
| Secondary mutation (cases harboring successive secondary mutations, n) | ||||
|---|---|---|---|---|
| Primary mutation | 1 | 2 | 3 | Samples, n |
| 12811T>C | − | − | 12 | |
| 11696G>A | 3394 T>C (1) | − | 10 | |
| 14502T>C | 14693 A>G (1) | 9804 G>A (1) | 8 | |
| 3866 T>C (1) | − | |||
| 3316G>A | − | − | 7 | |
| 11778 G>A | 3394T>C | 11696 G>A (1) | − | 7 |
| 3497C>T | 3571 C>T (3) | − | 4 | |
| 14693A>G | 14502 T>C (1) | − | 3 | |
| 3571C>T | 3497 C>T (3) | − | 3 | |
| 12338T>C | − | − | 2 | |
| 9804G>A | 14502 T>C (1) | 14693 A>G (1) | 1 | |
| 11253T>C | − | − | 1 | |
| 15951A>G | − | − | 1 | |
| 14484 T>C | 14502T>C | − | − | 3 |
| 15951A>G | − | − | 2 | |
| 11696G>A | − | − | 1 | |
| 3316G>A | − | − | 1 | |
| 14693A>G | − | − | 1 | |
In conclusion, the current data indicates that the spectrum and incidence of mtDNA mutation-associated LHON cases in the Han population are different to those in a Caucasian population. Here, the causative mutations associated with LHON, m.11778G>A, m.14484T>C, m.3460G>A, m.3635G>A, m.3866T>C and m.3733G>A, were observed in 70.55% of the patient cohort. The common secondary mutations in the Chinese LHON population were m.12811T>C, m.11696G>A, m.3394T>C, m.3316G>A, m.14502T>C and m.12338T>C. Furthermore, besides the three hotspots genes MT-ND1, MT-ND4 and MT-ND6, MT-ND5 also had a high incidence of secondary mutations including m.12811T>C and m.12338T>C; this finding was comparable with a previous study (29). The primary and secondary mutation-associated LHON cases in the present multi-gene panel will advance current understanding of the clinical phenotype of LHON, and offer valuable information for the early diagnosis and subsequent options of intervention for mitigating risk of additional vision loss in LHON patients.
Acknowledgements
The present study was supported by the National Technologies R&D Program (grant no. 2012BAI09B03) and a grant from the National Natural Science Foundation of China (grant no. 31671303).
Glossary
Abbreviations
- LHON
Leber's hereditary optic neuropathy
- mtDNA
mitochondrial DNA
References
- 1.Y-W-Man P, Griffiths PG, Brown DT, Howell N, Turnbull DM, Chinnery PF. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am J Hum Genet. 2003;72:333–339. doi: 10.1086/346066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saracchi E, Difrancesco JC, Brighina L, Marzorati L, Curtò NA, Lamperti C, Carrara F, Zeviani M, Ferrarese C. A case of Leber hereditary optic neuropathy plus dystonia caused by G14459A mitochondrial mutation. Neurol Sci. 2013;34:407–408. doi: 10.1007/s10072-012-1013-1. [DOI] [PubMed] [Google Scholar]
- 3.Howell N, Mackey DA. Low-penetrance branches in matrilineal pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet. 1998;63:1220–1224. doi: 10.1086/302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser JA, Biousse V, Newman NJ. The neuro-ophthalmology of mitochondrial disease. Surv Ophthalmol. 2010;55:299–334. doi: 10.1016/j.survophthal.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies – disease mechanisms and therapeutic strategies. Prog Retin Eye Res. 2011;30:81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SH, Kim JS. Leber's ‘Plus’ in a Korean patient with 14484/ND6 mutation. J Clin Neurol. 2016;12:512–514. doi: 10.3988/jcn.2016.12.4.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia X, Li S, Xiao X, Guo X, Zhang Q. Molecular epidemiology of mtDNA mutations in 903 Chinese families suspected with Leber hereditary optic neuropathy. J Hum Genet. 2006;51:851–856. doi: 10.1007/s10038-006-0032-2. [DOI] [PubMed] [Google Scholar]
- 8.Ji Y, Liang M, Zhang J, Zhu L, Zhang Z, Fu R, Liu X, Zhang M, Fu Q, Zhao F, et al. Mitochondrial ND1 variants in 1281 Chinese subjects with Leber's hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2016;57:2377–2389. doi: 10.1167/iovs.16-19243. [DOI] [PubMed] [Google Scholar]
- 9.Jiang P, Liang M, Zhang J, Gao Y, He Z, Yu H, Zhao F, Ji Y, Liu X, Zhang M, et al. Prevalence of mitochondrial ND4 mutations in 1281 Han Chinese subjects with Leber's hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2015;56:4778–4788. doi: 10.1167/iovs.14-16158. [DOI] [PubMed] [Google Scholar]
- 10.Liang M, Jiang P, Li F, Zhang J, Ji Y, He Y, Xu M, Zhu J, Meng X, Zhao F, et al. Frequency and spectrum of mitochondrial ND6 mutations in 1218 Han Chinese subjects with Leber's hereditary optic neuropathy. Invest Ophthalmol Vis Sci. 2014;55:1321–1331. doi: 10.1167/iovs.13-13011. [DOI] [PubMed] [Google Scholar]
- 11.Yu-Wai-Man P, Chinnery PF. Leber hereditary optic neuropathy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, editors. GeneReviews® [Internet] University of Washington; Seattle, WA: pp. 1993–2016. [Google Scholar]
- 12.Qu J, Li R, Zhou X, Tong Y, Lu F, Qian Y, Hu Y, Mo JQ, West CE, Guan MX. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophthalmol Vis Sci. 2006;47:475–483. doi: 10.1167/iovs.05-0665. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Liu L, Xi Q, Zhao X, Fang M, Ma J, Zhu Z, Wang X, Shi C, Wang J, et al. Short-term serum deprivation causes no significant mitochondrial DNA mutation in vascular smooth muscle cells revealed by a new next generation sequencing technology. Acta Biochim Biophys Sin (Shanghai) 2016;48:862–864. doi: 10.1093/abbs/gmw059. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Qian Y, Zhang J, Tong Y, Jiang P, Liang M, Dai X, Zhou H, Zhao F, Ji Y, et al. Leber's hereditary optic neuropathy is associated with the T3866C mutation in mitochondrial ND1 gene in three Han Chinese families. Invest Ophthalmol Vis Sci. 2012;53:4586–4594. doi: 10.1167/iovs.11-9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truong HT, Nguyen VA, Nguyen LV, Pham VA, Phan TN. Screening of common point-mutations and discovery of new T14727C change in mitochondrial genome of Vietnamese encephalomyopathy patients. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:441–448. doi: 10.3109/19401736.2014.900665. [DOI] [PubMed] [Google Scholar]
- 18.Wallace DC, Lott MT. Leber hereditary optic neuropathy: Exemplar of an mtDNA disease. Handb Exp Pharmacol. 2017;240:339–376. doi: 10.1007/164_2017_2. [DOI] [PubMed] [Google Scholar]
- 19.Blakely EL, de Silva R, King A, Schwarzer V, Harrower T, Dawidek G, Turnbull DM, Taylor RW. LHON/MELAS overlap syndrome associated with a mitochondrial MTND1 gene mutation. Eur J Hum Genet. 2005;13:623–627. doi: 10.1038/sj.ejhg.5201363. [DOI] [PubMed] [Google Scholar]
- 20.Achilli A, Iommarini L, Olivieri A, Pala M, Kashani B Hooshiar, Reynier P, La Morgia C, Valentino ML, Liguori R, Pizza F, et al. Rare primary mitochondrial DNA mutations and probable synergistic variants in Leber's hereditary optic neuropathy. PLoS One. 2012;7:e42242. doi: 10.1371/journal.pone.0042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JY, Hwang JM, Park SS. Mitochondrial DNA C4171A/ND1 is a novel primary causative mutation of Leber's hereditary optic neuropathy with a good prognosis. Ann Neurol. 2002;51:630–634. doi: 10.1002/ana.10177. [DOI] [PubMed] [Google Scholar]
- 22.Fauser S, Leo-Kottler B, Besch D, Luberichs J. Confirmation of the 14568 mutation in the mitochondrial ND6 gene as causative in Leber's hereditary optic neuropathy. Ophthalmic Genet. 2002;23:191–197. doi: 10.1076/opge.23.3.191.7881. [DOI] [PubMed] [Google Scholar]
- 23.Chinnery PF, Brown DT, Andrews RM, Singh-Kler R, Riordan-Eva P, Lindley J, Applegarth DA, Turnbull DM, Howell N. The mitochondrial ND6 gene is a hot spot for mutations that cause Leber's hereditary optic neuropathy. Brain. 2001;124:209–218. doi: 10.1093/brain/124.1.209. [DOI] [PubMed] [Google Scholar]
- 24.Mackey DA, Oostra RJ, Rosenberg T, Nikoskelainen E, Bronte-Stewart J, Poulton J, Harding AE, Govan G, Bolhuis PA, Norby S. Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet. 1996;59:481–485. [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Zhou X, Li C, Zhao F, Zhang J, Yuan M, Sun YH, Wang J, Tong Y, Liang M, et al. Mitochondrial haplogroup M9a specific variant ND1 T3394C may have a modifying role in the phenotypic expression of the LHON-associated ND4 G11778A mutation. Mol Genet Metab. 2010;101:192–199. doi: 10.1016/j.ymgme.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Qu J, Li R, Zhou X, Tong Y, Yang L, Chen J, Zhao F, Lu C, Qian Y, Lu F, Guan MX. Cosegregation of the ND4 G11696A mutation with the LHON-associated ND4 G11778A mutation in a four generation Chinese family. Mitochondrion. 2007;7:140–146. doi: 10.1016/j.mito.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudoyo H, Suryadi H, Lertrit P, Pramoonjago P, Lyrawati D, Marzuki S. Asian-specific mtDNA backgrounds associated with the primary G11778A mutation of Leber's hereditary optic neuropathy. J Hum Genet. 2002;47:594–604. doi: 10.1007/s100380200091. [DOI] [PubMed] [Google Scholar]
- 28.Jiang P, Liang M, Zhang C, Zhao X, He Q, Cui L, Liu X, Sun YH, Fu Q, Ji Y, et al. Biochemical evidence for a mitochondrial genetic modifier in the phenotypic manifestation of Leber's hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum Mol Genet. 2016;25:3613–3625. doi: 10.1093/hmg/ddv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seong MW, Choi J, Park SS, Kim JY, Hwang JM. Novel MT-ND5 gene mutation identified in Leber's hereditary optic neuropathy patient using mitochondrial genome sequencing. J Neurol Sci. 2017;375:301–303. doi: 10.1016/j.jns.2017.01.064. [DOI] [PubMed] [Google Scholar]


