Abstract
Deregulated microRNAs (miRs) and their roles in carcinogenesis have attracted great attention in recent years. Although miR-204 was reportedly dysregulated in various types of cancer, its function and mechanism in cervical cancer remain unknown. The present study focused on the expression and mechanisms of miR-204 in cervical cancer development. Expression of miR-204 in cervical cancer tissues and non-tumor tissues was measured using PCR analysis. The effect of ectopic expression of miR-204 on cell motility was evaluated using wound-healing and Transwell invasion assays. Luciferase activity and western blot assays were used to verify the regulatory effect of miR-204 on its target gene. It was demonstrated that miR-204 was significantly decreased in primary cervical cancer tissues, and that downregulated miR-204 was associated with lymph node metastasis and poor survival. In addition, it was revealed that ectopic expression of miR-204 significantly inhibited the migratory and invasive ability of cervical cancer cells in vitro. In addition, bioinformatic prediction and experimental validation demonstrated that transcription factor 12 (TCF12) was a direct target of miR-204. Overexpression of TCF12 attenuated the inhibitory effect of miR-204 on cell motility. Taken together, the present data indicated that miR-204 is a metastasis-associated gene and may contribute to the progression of cervical cancer by regulating TCF12, providing novel insights, including that miR-204/TCF12 may be an important mechanism for cervical cancer metastasis.
Keywords: microRNA-204, cervical cancer, transcription factor 12, migration, prognosis
Introduction
Cervical cancer is the third most prevalent gynecological malignancy and the second leading cause of cancer-associated mortality among females worldwide, with an estimated 530,000 mortalities per year (1). It is the result of a multistep process involving the transformation of normal cervical epithelium to pre-neoplastic cervical intraepithelial neoplasia, which is subsequently transformed to cervical cancer (2). High-risk human papillomavirus (HPV) infection performs an important role in cervical cancer progression, whereas HPV infection alone is insufficient for tumor development (3). The molecular mechanisms underlying the development of cervical cancer have not been fully elucidated. Therefore, it is necessary to further explore the molecular mechanisms underlying cervical carcinogenesis to facilitate the development of novel and effective therapeutic targets for cervical cancer treatment.
MicroRNAs (miRNAs/miRs) represent a class of small non-coding regulatory RNA molecules (19–25 nucleotides), which can downregulate gene expression by binding to a complementary sequence in the 3′untranslated region (3′UTR) region of target mRNAs (4,5). Functionally, miRNAs are involved in the regulation of numerous critical biological processes, including cell proliferation, differentiation, migration and invasion (6). Accumulating evidence has indicated that dysregulation of miRNAs occurs in various tumor types, and that they function as oncogenes or tumor suppressors according to the nature of the target (7). Additionally, many studies have demonstrated that the deregulation of miRNAs is associated with cancer initiation and progression (8). Clinically, miRNAs can be used as potential biomarkers for the diagnosis and prognosis assessment of patient with cancer, and possibly as therapeutic targets (9).
Currently, more than several hundred, unique mature human miRNAs are known, a number of which are involved in tumorigenesis. Among them, miR-204 was reported as a tumor suppressor and is frequently downregulated in gastric cancer, breast cancer, renal cell carcinoma, non-small cell lung cancer (NSCLC) and glioma (10–14). Overexpression of miR-204 in cancer cells inhibits migration and proliferation, and promotes apoptosis (15–18). However, to the best of our knowledge, there are no studies regarding the association between miR-204 and cervical cancer. Therefore, the present study was performed to determine whether miR-204 serves a role in cervical carcinogenesis.
In the present study, it was revealed that miR-204 is significantly decreased in primary tumor tissues, and that reduced miR-204 was associated with lymph node status and poor overall survival. In vitro analysis demonstrated that overexpression of miR-204 inhibits cell migration and invasion in cervical cancer cells. Furthermore, transcription factor 12 (TCF12) was identified as a direct target of miR-204, and to be involved in the miR-204 regulated cell motility alteration. Thus, results of the present study, for the first time, provided novel clues regarding the role of miR-204 as a metastasis-associated gene by regulating TCF12 in cervical cancer.
Materials and methods
Clinical samples and cell lines
Fresh cervical cancer and matched adjacent non-cancerous tissues were collected from 58 patients who underwent surgery between April 2009 and January 2011 at the Department of Obstetrics and Gynecology, Huzhou Central Hospital (Huzhou, China). No patients received preoperative radiotherapy, chemotherapy or other treatment history. The corresponding non-cancerous tissues were obtained 4 cm beyond the boundary of cervical cancer tissues. Clinicopathological variables, including age, tumor size, differentiation and International Federation of Gynecology and Obstetrics (FIGO) stage (19) were collected prospectively. All the histological diagnoses for tumor tissues and normal tissues were examined and confirmed by two independent pathologists who were blinded to clinical outcomes of the patients. The fresh tissue specimens were immediately frozen in liquid nitrogen until use. The study protocol was approved by the institutional review board of Huzhou Central Hospital, and the study was conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all patients.
Four human cervical cancer cell lines, HeLa, C33A, SiHa and CaSki, and an immortalized HPV-negative skin keratinocyte cell line, HaCaT, were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI 1640 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100 IU/ml of penicillin and 100 lg/ml of streptomycin. All cells were incubated in a humidified incubator at 37°C and 5% CO2.
miRNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total miRNA was extracted from cell lines and tissues using the miRNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. An RT-qPCR assay was conducted to detect the expression levels of miR-204 in cell lines and tissues. Briefly, 10 µg total RNA was subjected to RT. The cDNA was used for the amplification of mature miR-204, and the endogenous control U6, by PCR. The PCR conditions were as follows: Initial denaturation at 95°C for 3 min, followed by 45 cycles of 95°C for 15 sec, 62°C for 1 min and 72°C for 1 min. RT-qPCR was performed using SYBR® Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) on an ABI 7000 HT Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used were as follows: miR-204 forward, 5′-GTCCCTGTGTCATCCT-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGTAT-3′; U6 forward, 5′-CTCTCTGCGGCAGCACA-3′ and reverse, 5′-AACGCTGTACGAATGTGAGT-3′. The quantity of miR-204 was normalized to U6, and calculated using the 2−ΔΔCq method (20). All RT-qPCR reactions were performed in triplicate.
Plasmid construction, oligonucleotide synthesis and transfection
The miR-204 mimic (sequence: 5′-UUCCCUUUGUCAUCCUAUGCCU-3′) and the mimic negative control (mim-NC, sequence: 5′-UUUGUACUACACAAAAGUACUG-3′) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). HeLa and C33A cells were transfected with 400 µM mimic or 400 µM mim-NC using Lipofectamine® 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 24 h post-transfection, cells were used for in vitro migration and invasion assays. TCF12 cDNA without its 3′UTR was inserted into a pcDNA3.1(+) vector (Invitrogen; Thermo Fisher Scientific, Inc.) to generate the recombinant vector.
Migration assay
A wound-healing assay was conducted to detect cell migration. Cells (5×105 cells/well) were seeded onto 6-well plates and grown to 80% confluency in RPMI 1640 medium supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were then transfected with miR-204 mimics, and the miRNA mimic control (mim-NC) was used as the control group. After 24 h, a wound was created in the confluent cell monolayer using a 200-ml pipette tip. The cells were washed with RPMI 1640 medium three times and then incubated at 37°C for 24 h. The mean distance of the migrated cells was determined at 24 h.
Invasion assay
For the invasion assay, cells were serum-starved for 6 h in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) containing 0.1% FBS (Invitrogen; Thermo Fisher Scientific, Inc.). Serum-starved cells were trypsinized and resuspended in DMEM containing 0.1% FBS, following which cells (4×104) were added to the upper chambers of Transwell assay plate; each well was coated with 30 mg/cm2 Matrigel (Sigma-Aldrich; Merck KGaA). After 24 h at 37°C, cells remaining on the upper membrane surface were removed by careful wiping with a cotton swab, the migrated cells were fixed in 95% ethanol at room temperature for 30 min and then stained with 0.2% crystal violet solution (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature. Invasive cells adhering to the undersurface of the filter were then counted using an inverted microscope (×100).
Target prediction
Three publically available databases [including TargetScan, www.targetscan.org/vert_71 (21), PicTar, www.pictar.org (22) and miRanda, www.microrna.org (23)] were used to predict the candidate targets of miR-204.
Luciferase reporter assay
To construct the pMIR-TCF12-3′UTR plasmid that contained the potential binding sites of TCF12 3′UTR downstream of the firefly luciferase gene, a fragment was amplified and inserted into the pMIR-REPORT Luciferase Vector (Ambion; Thermo Fisher Scientific, Inc.). Amplification conditions were as follows: 94°C for 5 min, 40 cycles of 60°C for 20 sec and 72°C for 20 sec, and Taq DNA Polymerase (Thermo Fisher Scientific, Inc.) was used in this procedure. The primers were used as follows: forward, 5′-AAGCAACTGGTCAACACTTCC-3′, and reverse, 5′-CGGTGAGTGGAGAGATCGAG-3′. HeLa and C33A cells were used to measure luciferase activity. Cells were co-transfected with 100 ng luciferase plasmid and 50 ng Renilla plasmid (Ambion; Thermo Fisher Scientific, Inc.), along with 600 ng miR-204 mimic or NC-mim using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Mutations in the miR-204-binding site of TCF12 3′-UTR were introduced by the QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA) according to manufacturer's protocol. Luciferase activity was measured 48 h post-transfection using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA). Firefly luciferase activity was then normalized to the corresponding Renilla luciferase activity. Data are presented as the mean value obtain from triplicate experiments.
Western blot analysis
Proteins were extracted from the cells using the protein extraction reagent radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with protease inhibitors (cat no. 8553; Cell Signaling Technology, Inc., Danvers, MA, USA), and quantified by a bicinchoninic acid protein assay (Beyotime Institute of Biotechnology). Subsequently, 50 µg protein extractions were separated via 12% SDS-PAGE, transferred to nitrocellulose membranes (Whatman® Ptotran®; GE Healthcare, Chicago, IL, USA) and incubated with specific primary antibodies, as follows: TCF12 Rabbit mAb (cat no. 11825; dilution, 1:1,000; Cell Signaling Technology, Inc.) and GAPDH antibody (cat no. sc-47724; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. The membranes were then washed and probed with horseradish peroxidase (HRP)-conjugated secondary antibodies for GAPDH (mouse IgG binding protein-HRP; cat no. sc-516102; 1:5,000; Santa Cruz Biotechnology, Inc.) and for TCF12 (Mouse Anti-rabbit IgG; cat no. 5127; 1:5,000; Cell Signaling Technology, Inc.) at room temperature for 30 min. The signals were detected using the Chemiluminescent ECL Detection system (Merck KGaA), and analysed using ImageJ 2× version 2.1.4.7 software (Rawak Software, Inc., Dresden, Germany).
Statistical analysis
Statistical analysis was performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA) and Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The expression of miR-204 in tissues were presented as the mean ± the standard deviation. The difference in expression levels between paired samples was analyzed by a paired t-test. Comparisons between groups were performed using the Mann-Whitney U test for continuous variables. The Kaplan-Meier method was used for survival analysis, and differences in survival curves were compared using the log-rank test. All tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Results
miR-204 is downregulated in cervical cancer tissues
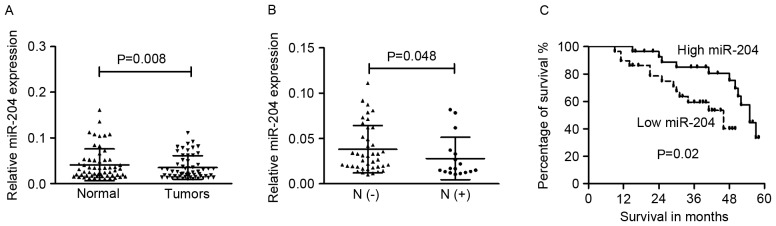
To determine the expression level of miR-204 in cervical cancer cells, RT-qPCR was performed in four cancer cells (HeLa, C33A, SiHa and CaSki), and HaCaT cells as the normal control. The cervical cancer cells exhibited significantly lower expression of miR-204 compared with HaCaT (data not shown). The level of miR-204 in tissue specimens from patients with cervical cancer was then examined. The expression of miR-204 in the primary tumor tissues (0.035±0.025) was significantly lower than that in the adjacent non-cancerous tissues (0.041±0.034; Fig. 1A; P=0.008).
Figure 1.
Expression of miR-204 was downregulated in cervical cancer. (A) Expression levels of miR-204 in paired cervical cancer tissues and their adjacent non-tumor tissues. (B) Downregulation of miR-204 was associated with lymph node metastasis. (C) Low level of miR-204 predicts poor prognosis in patients with cancer. miR-204, microRNA-204.
Low expression of miR-204 is associated with poor prognosis in patients with cervical cancer
The associations between miR-204 expression levels and different clinicopathological characteristics were tested. It was revealed that miR-204 expression was significantly decreased in the tumor tissues of patients with lymph node metastasis (0.028±0.023) compared with those without lymph node metastasis (0.038±0.026; P=0.048; Fig. 1B). The downregulated miR-204 had no association with other clinicopathological characteristics of cervical cancer cases. It was speculated that downregulated miR-204 may be involved in the regulation of metastasis in the development of cervical cancer.
The median level of miR-204 was used as a cut-off value to divide 58 cases into two groups (low- or high- group). The association between miR-204 and the overall survival of cervical cancer patients was determined via Kaplan-Meier analysis and the log-rank test. As shown in Fig. 1C, patients with cervical cancer and low miR-204 expression had poorer overall survival rates compared with those with high miR-204 expression (P=0.02).
miR-204 inhibits cell migration and invasion in vitro
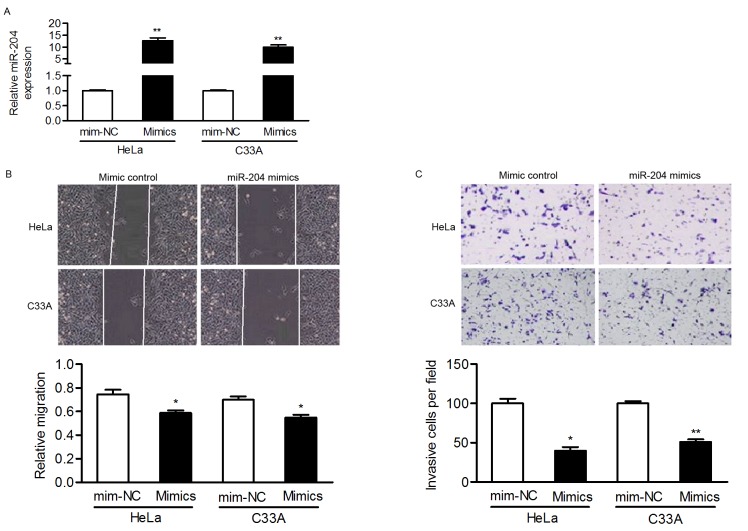
Considering the association between miR-204 levels and lymph node metastasis, the present study then focused on the effect of miR-204 on cell motility. HeLa and C33A cells, selected as they express relatively low levels of miR-204, were transfected with miR-204 mimics and it was confirmed that the expression of miR-204 was upregulated in transfected cells (Fig. 2A).
Figure 2.
miR-204 inhibits the migration and invasion of cervical cancer cells in vitro. (A) HeLa and C33A cells were infected with miR-204 mimics or the mimic control, and the expression of miR-204 was analyzed by reverse transcription-quantitative polymerase chain reaction. Endogenous miR-204 levels in HeLa and C33A cells were increased with miR-204 mimics, and the effects of miR-204 on cell migration and invasion were determined using (B) wound healing and (C) Transwell assays, using crystal violet. Magnification, ×100. *P<0.05; **P<0.01. miR-204, microRNA-204; mim-NC, mimic negative control.
Wound-healing and Matrigel Transwell assays were performed to verify whether miR-204 inhibited the migration and invasion of cervical cancer cells. In HeLa and C33A cells, overexpressing miR-204 inhibited the migratory ability of cells in comparison to cells transfected with the mim-NC (Fig. 2B). Furthermore, the invasive capacity of cervical cancer cells was markedly suppressed in miR-204-transfected cells (Fig. 2C). These results indicated that miR-204 may be a metastasis-associated gene and involved in cervical cancer pathogenesis.
miR-204 directly inhibits TCF12 expression by targeting its 3′UTR
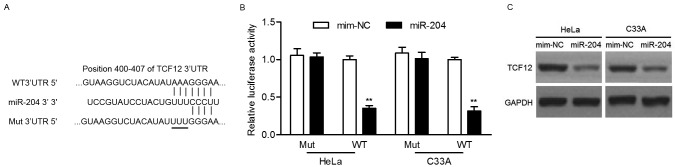
To explore the molecular mechanism by which miR-204 inhibits cervical cancer cells, potential targets of miR-204 were predicted using the PicTar, TargetScan and miRanda tools. Among the predicted candidate targets, the present study focused on TCF12 and considered the involvement of TCF12 in tumor metastasis (24,25). As presented in Fig. 3A, the 3′-UTR of TCF12 possesses a potential binding site of miR-204. To determine if miR-204 could directly target the 3′UTR of TCF12, luciferase activity assay was performed. Luciferase reporter assays revealed that the upregulation of miR-204 significantly decreased the relative luciferase activity of the 3′UTR of TCF12 in HeLa or C33A cells, but had no effect on the mutant 3′UTR of TCF12 (Fig. 3B). In addition, western blot analysis revealed that overexpression of miR-204 noticeably suppressed the protein expression of TCF12 (Fig. 3C). Taken together, these results demonstrated that miR-204 suppresses TCF12 expression by directly targeting its 3′UTR.
Figure 3.
miR-204 directly inhibits TCF12 expression by targeting its 3′UTR. (A) Predicted binding sites of miR-204 in the 3′UTR of TCF12. Mutant-type was generated in the seed region of the 3′UTR of TCF12 by mutating 3 nucleotides that is recognized by miR-204. (B) Luciferase activity assay was performed in HeLa and C33A cells co-transfected with miR-204 mimics and a luciferase reporter containing the WT Mut 3′UTR of TCF12. (C) The expression of TCF12 in HeLa and C33A cells after 48 h transfection with miR-204 mimics and NC-mim was measured by western blot analysis. **P<0.01. TCF12, transcription factor 12; miR-204, microRNA-204′ 3′UTR, 3′untranslated region; mim-NC, mimic negative control; Mut, mutant; WT, wild-type.
miR-204 inhibits the malignant phenotypes of cervical cancer cells by regulating TCF12
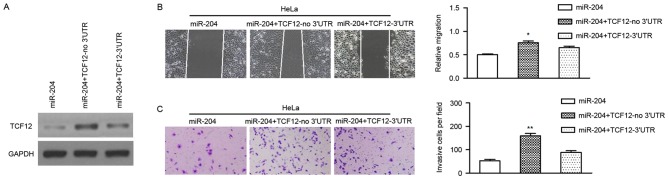
As miR-204 regulates the expression of TCF12 post-transcriptionally, and miR-204 inhibits the migration and invasion of cervical cancer cells, it was postulated that these effects were attributed to TCF12. To verify this hypothesis, a vector was constructed using cDNA that lacked the 3′UTR of TCF12, and the vector was transiently co-transfected with miR-204 mimics into cervical cancer cells. It was revealed that the transfection of TCF12 (no 3′UTR) could effectively reverse the expression of TCF12 in HeLa and C33A cells (Fig. 4A). Subsequently, it was revealed that the restoration of TCF12 could significantly attenuate the inhibitory role of miR-204 in the migration and invasion of cervical cancer cells (Fig. 4B and C).
Figure 4.
TCF12 is involved in miR-204 modulation of cervical cancer cell migration. HeLa and C33A cells were transfected with miR-204 mimics or co-transfected with miR-204 mimics and TCF12 lacking the 3′UTR (TCF12-no 3′UTR). (A) TCF12 expression was confirmed in HeLa and C33A cells by western blot analysis. (B) Analysis of cell migration using the wound healing assay. (C) Measurement of cell invasion ability by Transwell assay using 0.2% crystal violet. Magnification, ×100. *P<0.05; **P<0.01. TCF12, transcription factor 12; miR-204, microRNA-204; 3′UTR, 3′untranslated region.
Discussion
Exploring the underlying molecular mechanisms of carcinogenesis is essential for early diagnosis and effective therapy. Furthermore, one miRNA is able to regulate the expression of multiple genes, since it can bind to its mRNA targets either as an imperfect or a perfect complement. It is, therefore, not surprising that miRNAs are involved in the regulation of all major tumor-associated biological processes, including differentiation, proliferation, migration and apoptosis. The present study demonstrated that miR-204 expression was downregulated in cervical cancer tissues. The results of Kaplan-Meier analysis revealed that the downregulation of miR-204 was associated with a poor prognosis. It was demonstrated that the overexpression of miR-204 suppressed migration and invasion in cervical cancer cells. Furthermore, TCF12 was also identified as a novel and direct target of miR-204.
miR-204, located on chromosome 9q21.12, has attracted attention due to its involvement in numerous tumor types. Previous studies have documented that miR-204 expression was frequently downregulated in gastric cancer, breast cancer, renal cell carcinoma, glioma, prostate cancer and NSCLC (10–14). In accordance with these studies, the present data demonstrated the decreased expression of miR-204 in primary cervical cancer tissues compared with adjacent normal tissues, indicating the potential tumor suppressor role of miR-204 in cervical cancer progression.
In addition, the prognostic value of the expression of miR-204 in patients with cervical cancer was analyzed. It was revealed that low expression of miR-204 was associated with lymph node metastasis and poor overall survival. Consistent with the present data, Bao et al (26) reported that lower miR-204-5p expression was associated with advanced FIGO stages, and possibly a lower rate of survival in patients with endometrial carcinoma. A low level of miR-204 was significantly associated with a more aggressive and poor prognostic phenotype of nasopharyngeal carcinoma (27). Low miR-204 expression was also significantly associated with higher tumor-node-metastasis stage and poorer overall survival in breast cancer cases (11). These results indicated that miR-204 expression may be a promising prognostic marker for patients with cancer.
Furthermore, miR-204 expression was associated with chemotherapeutic resistance of patients with breast cancer (11). Enhanced miR-204 expression significantly increased sensitivity to cisplatin and etoposide in vitro (28). Therefore, additional studies regarding the association between expression of miR-204 and anti-carcinogen, such as cisplatin and etoposide, may provide new insights into chemotherapy of cervical cancer.
miR-204 acts as a tumor suppressor in various types of tumors by regulating its target genes and is involved in several tumor-associated processes. Wu et al (17) indicated that miR-204 could regulate the proliferation and invasion of retinoblastoma cells by directly targeting the cyclin D2 and matrix metalloproteinase-9 genes. Downregulation of miR-204 and the consequent overexpression of SRY sex-determining region-Y box 4 promoted epithelial-mesenchymal transition (EMT) and enhanced the invasion and proliferation of gastric cancer cells in vitro (29). Loss of miR-204 promotes to cancer cell migration and invasion, while restoration of miR-204 inhibited EMT by targeting the EMT inducer, including slug and sirtuin 1 (30,31). Shi et al (13) indicated that miR-204 inhibited NSCLC tumor migration and invasion by directly targeting and downregulating NUAK family kinase 1 expression. By contrast, overexpression of miR-204 suppressed papillary thyroid carcinoma cell proliferation, induced cell cycle arrest and apoptosis and inhibited insulin-like growth factor binding protein 5 mRNA (32). In the present study, it was observed that the overexpression of miR-204 suppressed cell migration and invasion ability. The present study expanded the knowledge of miR-204 in cervical cancer and demonstrated that miR-204 acts as a metastasis-associated gene in cervical cancer. More importantly, it was revealed that miR-204 directly regulates TCF12 expression by targeting its 3′UTR. Previous studies reported that TCF12 functions as a transcriptional repressor of E-cadherin and that its overexpression was significantly associated with the occurrence of colorectal cancer metastasis (24,25). Thus, it was proposed that miR-204/TCF12 axis may serve important roles in the migration and invasion of cervical cancer.
In conclusion, miR-204 was significantly downregulated in cervical cancer tissues and ectopic miR-204 inhibited cell motility. TCF12 was a direct target of miR-204 and upregulated TCF12 impaired the inhibitory effect of miR-204 on cell migration, indicating that miR-204 functions as a metastasis-associated gene, possibly by targeting TCF12 in cervical cancer. Thus, these results demonstrated that miR-204 performs important roles in the migration and invasion of cervical cancer by targeting TCF12.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Waggoner SE. Cervical cancer. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- 3.Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: A review. Virus Res. 2002;89:229–240. doi: 10.1016/S0168-1702(02)00191-0. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated posttranscriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 7.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/S1044-579X(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 8.Davidson B, Tropé CG, Reich R. The clinical and diagnostic role of microRNAs in ovarian carcinoma. Gynecol Oncol. 2014;133:640–646. doi: 10.1016/j.ygyno.2014.03.575. [DOI] [PubMed] [Google Scholar]
- 9.Seven M, Karatas OF, Duz MB, Ozen M. The role of miRNAs in cancer: From pathogenesis to therapeutic implications. Future Oncol. 2014;10:1027–1048. doi: 10.2217/fon.13.259. [DOI] [PubMed] [Google Scholar]
- 10.Zhang B, Yin Y, Hu Y, Zhang J, Bian Z, Song M, Hua D, Huang Z. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32:331. doi: 10.1007/s12032-014-0331-y. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Jin X, Zhang Q, Zhang G, Deng X, Ma L. Decreased expression of miR-204 is associated with poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2014;7:3287–3292. [PMC free article] [PubMed] [Google Scholar]
- 12.Munari E, Marchionni L, Chitre A, Hayashi M, Martignoni G, Brunelli M, Gobbo S, Argani P, Allaf M, Hoque MO, Netto GJ. Clear cell papillary renal cell carcinoma: micro-RNA expression profiling and comparison with clear cell renal cell carcinoma and papillary renal cell carcinoma. Hum Pathol. 2014;45:1130–1138. doi: 10.1016/j.humpath.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y, Li H, Wang L, Wang X, Zhao C. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316–2327. doi: 10.1038/bjc.2014.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, Chen HI, Chang YF, Panneerdoss S, Zoghi B, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7:e52397. doi: 10.1371/journal.pone.0052397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, Song M, Bian Z, Wang Q, Ni S, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–6199. doi: 10.1158/1078-0432.CCR-14-1030. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Zeng Y, Wu S, Zhong J, Wang Y, Xu J. MiR-204, down-regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting CyclinD2 and MMP-9. FEBS Lett. 2015;589:645–650. doi: 10.1016/j.febslet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Role of miR-204 in the regulation of apoptosis, endoplasmic reticulum stress response, and inflammation in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:2999–3007. doi: 10.1167/iovs.10-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–104. doi: 10.1016/j.ijgo.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CC, Chen WS, Chen CC, Chen LL, Lin YS, Fan CS, Huang TS. TCF12 protein functions as transcriptional repressor of E-cadherin, and its overexpression is correlated with metastasis of colorectal cancer. J Biol Chem. 2012;287:2798–2809. doi: 10.1074/jbc.M111.258947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen WS, Chen CC, Chen LL, Lee CC, Huang TS. Secreted heat shock protein 90α (HSP90α) induces nuclear factor-κB-mediated TCF12 protein expression to down-regulate E-cadherin and to enhance colorectal cancer cell migration and invasion. J Biol Chem. 2013;288:9001–9010. doi: 10.1074/jbc.M112.437897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, Zhang HJ, Wang LH, Wan XP. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155. doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Deng X, Wu M, Zhang G, Huang J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014;588:1562–1570. doi: 10.1016/j.febslet.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 28.Ryan J, Tivnan A, Fay J, Bryan K, Meehan M, Creevey L, Lynch J, Bray IM, O'Meara A, Tracey L, et al. MicroRNA-204 increases sensitivity of neuroblastoma cells to cisplatin and is associated with a favourable clinical outcome. Br J Cancer. 2012;107:967–976. doi: 10.1038/bjc.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS One. 2014;9:e101457. doi: 10.1371/journal.pone.0101457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Qiu YH, Wei YP, Shen NJ, Wang ZC, Kan T, Yu WL, Yi B, Zhang YJ. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem. 2013;32:1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290. doi: 10.1186/1471-2407-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Wang J, Li X, Ma J, Shi C, Zhu H, Xi Q, Zhang J, Zhao X, Gu M. miR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457:621–626. doi: 10.1016/j.bbrc.2015.01.037. [DOI] [PubMed] [Google Scholar]