Abstract
Accumulating evidence demonstrated that Hox antisense intergenic RNA (HOTAIR) serves essential roles in the development and metastasis of several types of cancer. In hepatocellular carcinoma (HCC), high expression of HOTAIR is associated with poor prognosis, and HOTAIR regulates cell migration and proliferation. However, the downstream molecular targets of HOTAIR depend on the cancer cell types, and little is known about the precise molecular mechanisms of HOTAIR involved in cancer development. The present study investigated the role of HOTAIR in HCC cell lines. Notably, the overexpression of HOTAIR in HCC cell lines did not affect cell migration and proliferation capability. In the microarray analysis, C-C motif chemokine ligand (CCL)2 was identified to be differentially expressed in HOTAIR-overexpressing cells, and it was confirmed that HOTAIR promotes the secretion of CCL2. Furthermore, it was revealed that the proportion of macrophages and myeloid-derived suppressor cells (MDSCs) were increased when peripheral blood mononuclear cells were co-cultured with HOTAIR-overexpressing cells. Collectively, these data suggest that HOTAIR regulates CCL2 expression, which may be involved in the recruitment of macrophages and MDSCs to the tumor microenvironment.
Keywords: hepatocellular carcinoma, HOTAIR, CCL2, tumor-associated macrophages, PBMC
Introduction
Hox antisense intergenic RNA (HOTAIR), a lncRNA that acts as an oncogenic molecule in various types of cancer, is localized to the HOXC gene cluster. HOTAIR interacts with PRC2 (polycomb repressive complex 2) to enhance H3K27 trimethylation, and thereby decreases the expression of a large number of genes. Several groups, including our laboratory, have reported that high HOTAIR expression is correlated with a poor prognosis in several types of cancer, including breast (1), colorectal (2), non-small lung cell (3), and gastric cancer (4). Interestingly, recent report suggested that effects of HOTAIR are strongly tissue-dependent and can even differ within the same type of cancer (5). Thus, the underlying mechanism by which HOTAIR is involved in malignant progression remains uncertain.
Hepatocellular carcinoma (HCC) is the fifth leading cause of cancer and second leading cause of cancer-related mortality worldwide. More than 600,000 deaths are associated with HCC every year worldwide (6). Previous report suggested that the expression of HOTAIR is associated with tumor recurrence and poor prognosis in hepatocellular carcinoma (7,8). Several in vitro analyses were reported using HCC cell line, HepG2; HOTAIR is a FOXC1-activated driver of malignancy, which acts in part through the repression of miR-1 in HepG2 cells (9). HOTAIR silence activates P16Ink4a and P14ARF signaling by enhancing miR-218 expression and suppressing Bmi-1 expression, resulting in the suppression of tumorigenesis in HepG2 cells (10). Introduction of human HOTAIR into HepG2 cells revealed that HOTAIR promoted more rapid proliferation (7). Although different intracellular signaling are expected among multiple HCC cell lines, the research of HOTAIR using HCC cell lines except for HepG2 is not fully performed.
The tumor microenvironment is known to play important roles in cancer development and behavior (11). Macrophages are a component of the microenvironment in tumors, which called tumor-associated macrophages (TAMs). Four decades ago, TAMs from malignant metastatic cancers were reported to promote tumor growth and metastasis (12). Recently, TAMs can also promote initiation of tumor cells, inhibit antitumor immune responses mediated by T cells, and stimulate tumor angiogenesis and subsequently tumor progression (13).
Myeloid-derived suppressor cells (MDSCs) are another critical component in the microenvironment (14). MDSCs are a heterogeneous group of immature myeloid cells and expanded in response to a variety of tumor factors. An increased presence of MDSCs is associated with tumor progression and poorer outcomes. In HCC patients, MDSCs defined as CD14+HLA-DR−/low cells exert their immunosuppressive function through the induction of CD4+CD25+Foxp3+ regulatory T cells (15).
Chemoattractants including chemokines such as CCL2 and CCL5, and cytokines (for example, CSF-1 and members of the VEGF family) are important mediators of the recruitment and functional polarization of TAMs. CCL2 is highly upregulated in HCC patients (16), and inhibition of CCL2 could be an effective therapeutic approach against hepatocellular carcinoma (17). The CCL2 is required for recruitment of monocytes/macrophages and is implicated in various aspects of liver pathology, including HCC (16). Furthermore, recent study suggested that microenvironment-derived CCL2 results in the accumulation of MDSCs in glioma (18). Thus, CCL2 is critical for immunosuppression to promote cancer metastasis. It is well-known that tumor cells as well as stromal cells were thought to be the source of CCL2 in established tumors (19). However, it is not still elucidated how HCC cells regulate CCL2 production.
In this study, we examined whether HOTAIR-expressing cancer cells exert the malignant phenotypes. We investigate the effect of HOTAIR against both cancer cell itself and peripheral blood monocyte cells (PBMCs) as immune cells in tumor microenvironment.
Materials and methods
Ethics statement
The present study was conducted according to the principles expressed in the Declaration of Helsinki and was approved (MCC-AE-2016-7) by the Ethics Committees at the Miyagi Cancer Center Research Institute (Natori, Japan). Experimental protocols involving animals were approved by the Miyagi Cancer Center Animal Care and Use Committee.
Cell lines and cell culture
Hepatoma cell line Li-7 and Hep3B was obtained from RIKEN BioResource Center (Tsukuba, Japan) and Cell Resource Center for Biomedical Research Cell Bank, Institute of Development, Aging, and Cancer, Tohoku University (Sendai, Japan), respectively. These cells were maintained in Dulbeccos modified Eagles medium (DMEM; Gibco Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Euro-Clone, Milano, Italy) and 1% penicillin-streptomycin (Gibco Life Technologies). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
Retroviral transfection
Human HOTAIR cDNA (obtained from Addgene, Cambridge, MA, USA) was amplified by PCR and inserted into the pBabe-hygro vector (pBabe-HOTAIR). The recombinant retrovirus was produced with the Platinum-A packaging-cell line (Plat-A, kindly provided by Prof. Kitamura, Tokyo University) as described previously (3). Briefly, Plat-A cells were transfected with pBabe-HOTAIR or a pBabe-hygro empty vector (Empty). FugeneHD (Roche Applied Science, Mannheim, Germany) and Opti-MEM I (Gibco Life Technologies) were added following the manufacturers protocol. The retrovirus-containing supernatant was collected 48 h after transfection and passed through a 0.45-µm filter. Li7 and Hep3B cells were infected with the recombinant retroviruses and then selected with hygromycin.
Isolation of peripheral blood mononuclear cells (PBMCs)
The heparinized blood was collected from healthy donors, and isolated by density gradient centrifugation using Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) following the manufacturers protocol.
Wound-healing assay
Cells were seeded in 24-well plates in normal cell-growth medium, incubated until confluent, and a yellow pipette tip was used to make a straight scratch, simulating a wound. After a 24 h incubation, the area occupied by cells that had migrated into the scratch area was measured using NIS-Elements software (Nikon, Tokyo, Japan).
Co-incubation assay
A trans-membrane of 0.4 µm pore size (Corning Life Science, Pittsburgh, PA, USA) was used for the analysis of soluble factors secreted by HOTAIR overexpressing hepatoma cell lines or HOTAIR-overexpressing hepatoma cell lines. The upper chamber included PBMCs (1×106 cells/500 µl) from healthy donors, and the lower chamber included HOTAIR-overexpressing hepatoma cell lines (1.0×105 cells/500 µl). After 72 h co-incubation at 37°C under an atmosphere of 5% CO2, multicolor FACS analysis was performed.
Flow cytometry analysis
To determine the frequency of proportions of macrophages and MDSCs, the cells were stained with specific antibodies for 30 min at 4°C, washed twice, and subsequently analyzed using a FACSCanto II (Becton Dickinson, CA). The antibodies used: anti-human APC-CD14 (clone: HCD14), FITC-CD68 (clone: Y1/82A), Pacific Blue-HLA-DR (clone: L243), PE-CD33 (clone: P67.6), APC/Cy7-CD11b (clone: ICRF44), and 7-AAD for dead cell removal. All monoclonal antibodies used in this study were purchased from BioLegend, Inc., (San Diego, CA, USA).
ELISA assay
Enzyme-linked immunosorbent assay (ELISA) kits for CCL2 were purchased (R&D Systems, Inc., Minneapolis, MN, USA), and ELISA was performed according to manufacturers instruction.
Microarray analysis
A microarray analysis was performed to search for genes regulated by HOTAIR. We used the SurePrint G3 Human GE 8×60K Microarray (Agilent Technologies, Santa Clara, CA, USA) to profile the gene expression in control and HOTAIR-overexpressing Li-7 cells. The obtained gene expression data were expressed in logarithmic scale, and heat maps were generated using the weighted average difference (WAD) algorithm (20). The analyses were performed using R software.
RNA preparation, cDNA synthesis, and quantitative real-time RT-PCR
Total RNA was extracted from HCC cell lines by RNeasy mini kit (Qiagen Inc., Valencia, CA, USA) according to the manufacturers protocol. cDNAs were synthesized from 1.0 µg of total RNA with the PrimeScript 1st Strand cDNA Synthesis kit (TaKaRa Bio, Shiga, Japan) following the manufacturers protocol.
HOTAIR expression was quantified as previously described (3). The primer sequences used: for HOTAIR, F 5′-ggt aga aaa agc aac cac gaa gc-3′ and R 5′-aca taa acc tct gtc tgt gag tgc c-3′. HOTAIR expression was normalized to RNA polymerase II (RPII) expression in each sample, and calculated by ddCt methods (21) (LighCycler480; Roche, Basel, Switzerland). All statistics were performed at the ∆Ct stage in order to exclude potential bias due to averaging of data transformed through the equation 2−∆∆Ct. Statistical analysis was undertaken using Student t-test.
Statistical analysis
All statistical analyses were performed using JMP 11 (SAS Institute, Cary, NC, USA). P<0.05 indicated that the difference was statistically significant using Students t-test. Graphs show average and standard deviation unless otherwise specified.
Results
Overexpression of HOTAIR in Li-7 cells did not alter cell proliferation and migration
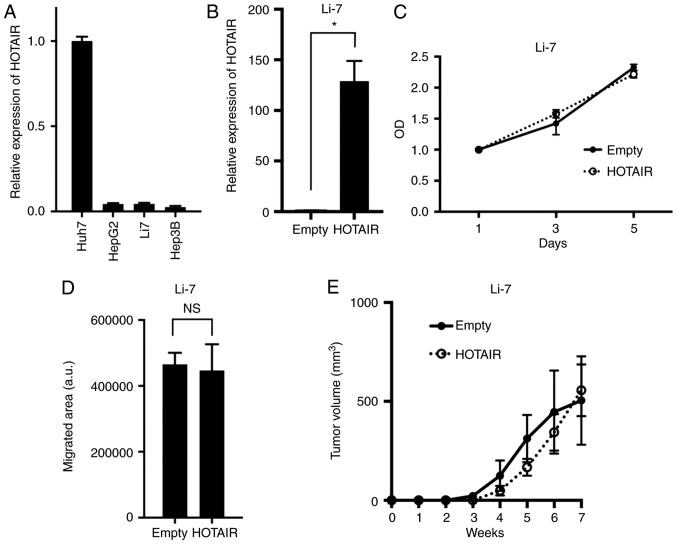
To evaluate the role of HOTAIR in HCC, we used human HCC cell line Li-7 which expressed relatively lower HOTAIR (Fig. 1A) to generate cells that stably overexpressed HOTAIR (Fig. 1B). These HOTAIR-transduced cells overexpressed HOTAIR at levels 128-fold higher (Li7-HOTAIR) than did empty vector-transduced cells. We assayed the effect of HOTAIR on cell proliferation using a MTT and found that HOTAIR overexpression in Li-7 cells did not alter their proliferation compared to control cells (Fig. 1C). We evaluated HOTAIRs role in migratory capability with a wound-healing assay, and found that the HOTAIR overexpression did not alter their migration compared to control cells (Fig. 1D). We further tested tumorigenicity of HOTAIR-overexpressing cells using NOD/Shi-scid-IL2Rγnull (NOG) mice. Because NOG mice are severely immunodificient, tumorigenicity assay using NOG mice can reveal the proliferation and colonization capability of cancer cell, and not evaluate the tumor immunity. No significant difference was found between HOTAIR-overexpression and control cells (Fig. 1E). These data indicated that HOTAIR did not alter malignant phenotypes in Li-7 cells.
Figure 1.
HOTAIR-overexpressing cells did not alter malignant phenotypes. (A) HOTAIR expression in four human hepatocellular carcinoma (HCC) cell lines. HOTAIR expression was determined by real-time PCR, and normalized to the Huh7 cell line. (B) HOTAIR expression in Li-7 cells transduced with a HOTAIR-overexpressing (HOTAIR) or empty vector (Empty), determined by real-time PCR. *P=0.0004, n=3. (C) Cell-proliferation assays in Li-7 cell line. The optical density was normalized at day 1. HOTAIR expression was normalized to the Empty cells. (D) Wound-healing assays in Li-7 cell line. N.S, not significant. (E) Tumorigenicity was assayed by injecting HOTAIR-overexpressing or control cells subcutaneously into NOG mice; graph shows average tumor volume and standard error.
CCL2 secretion was induced by HOTAIR
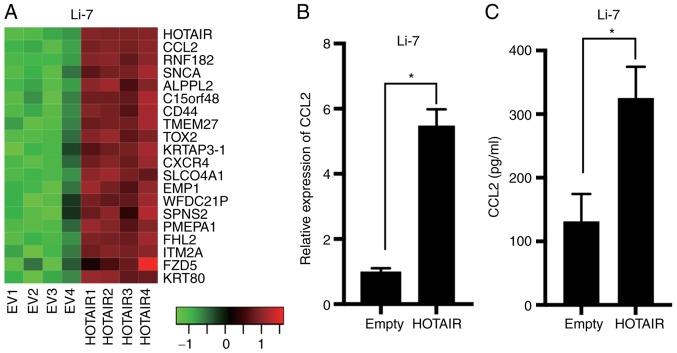
Since our results showed that HOTAIR did not regulate cancer phenotypes, which is different from previous studies using HepG2 cells, we screen the effect of HOTAIR in HCC cell lines to identify the different role of HOTAIR in HCC cells. We compared gene expression profiles between HOTAIR-overexpression and control Li-7 cells by microarray analysis using a weighted average difference (WAD) algorithm (Fig. 2A). Analysis of the microarray data showed that CCL2, also referred as monocyte chemoattractant protein 1, was differentially expressed in the overexpressing and control Li-7 cells. Real-time PCR and ELISA results confirmed that CCL2 was upregulated in the Li-7-HOTAIR cells (Fig. 2B and C). These results suggested that CCL2 acts downstream of HOTAIR in HCC.
Figure 2.
The secretion of CCL2 was increased in HOTAIR-overexpressing cells. (A) Heatmap showing the expression profiles of top 20 genes upregulated in HOTAIR-overexpressing (HOTAIR1-4) cells compared to control (EV1-4) Li-7 cells as determined by microarray analysis using the weighted average difference (WAD) algorithm. Red and green indicate higher and lower expression, respectively. (B) CCL2 expression determined by real-time PCR in Li-7 cells. *P<0.0001, n=4. CCL2 expression was normalized to the Empty cells. (C) CCL2 expression of culture supernatant determined by ELISA in Li-7 cells. *P=0.0002, n=5.
HOTAIR-overexpression induced macrophage and MDSC proliferation in Li-7 and Hep3B cells
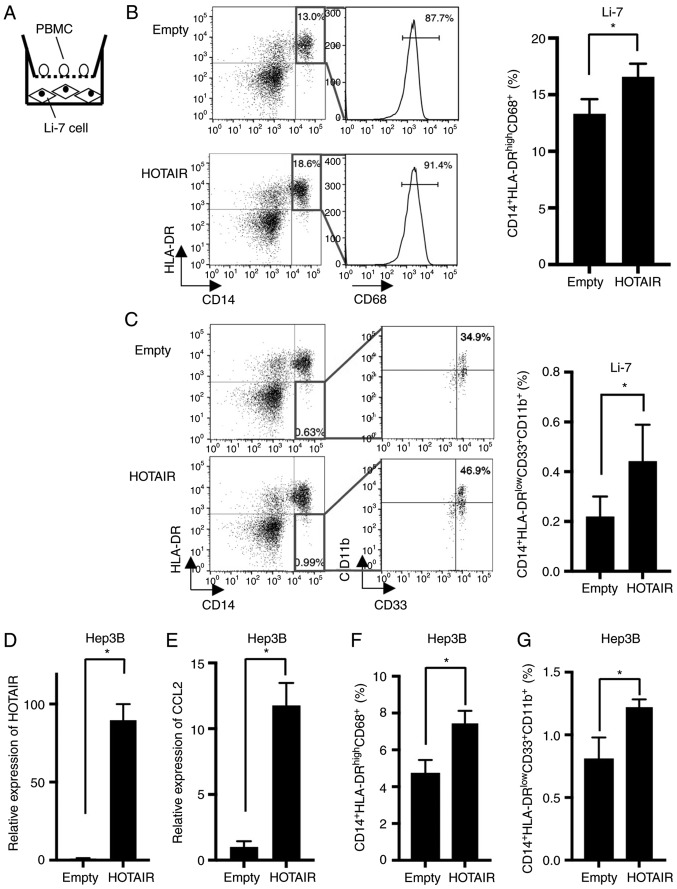
Because CCL2 is known to recruit TAMs and MDSCs, we examined the effect of HOTAIR-overexpressing Li-7 cells against PBMCs (Fig. 3A). The proportion of CD14+HLA-DR+CD68+ macrophages were increased in PBMCs co-cultured with HOTAIR-overexpressing cells (Fig. 3B). Furthermore, CD14+HLA-DR−CD33+CD11b+ MDSCs were also increased in co-cultured PBMCs (Fig. 3C).
Figure 3.
HOTAIR overexpressing Li-7 increased the proportion of macrophage. (A) A scheme of the co-culture assay. (B) (Left) Representative dot plots of PBMC co-cultured with Li-7 cells after 72 h. Macrophages were found in CD14+CD68+HLA-DR+ gate. (Right) Quantification of macrophages of PBMC co-cultured with HOTAIR-overexpressing and control Li-7 cells. *P=0.0031, n=5. (C) (Left) Representative dot plots of PBMC co-cultured with Li-7 cells after 72 h. MDSCs were found in CD14+ HLA-DRlow/−CD33+CD11b+ gate. (Right) Quantification of MDSCs of PBMC co-cultured with HOTAIR-overexpressing and control cells. *P=0.018, n=5. (D) HOTAIR expression in Hep3B cells transduced with a HOTAIR-overexpressing (HOTAIR) or empty vector (Emtpy), determined by real-time PCR. HOTAIR expression was normalized to Empty cells. *P=0.0001, n=3. (E) CCL2 expression determined by real-time PCR in Hep3B cells. HOTAIR expression was normalized to Empty cells. *P<0.0001, n=4. (F) Quantification of macrophages of PBMC co-cultured with HOTAIR-overexpressing and control Hep3B cells. *P=0.0007, n=4. (G) Quantification of MDSCs of PBMC co-cultured with HOTAIR-overexpressing and control Hep3B cells. *P=0.0014, n=4.
To confirm the effect of HOTAIR against macrophages and MDSCs, we established HOTAIR-overexpressing cells using another HCC cell line, Hep3B. HOTAIR was expressed 89-fold higher in HOTAIR-overexpressing Hep3B cells than in control cells (Fig. 3D). We measured the expression of CCL2, and found that CCL2 was upregulated in the Hep3B-HOTAIR cells (Fig. 3E). Then we investigated the effect of HOTAIR-overexpressing Hep3B cells against PBMCs using co-culture method, and found that the proportion of macrophages (Fig. 3F) and MDSCs (Fig. 3G) were increased by co-cultured HOTAIR-overexpressing Hep3B cells, which is compatible with the data using Li-7 cells.
Collectively, these data suggested that HOTAIR in HCC cell lines plays critical roles in the promotion of macrophage and MDSC, through the secretion of cytokines and/or chemokines by HCC cells.
Discussion
In this study, we demonstrated that HOTAIR-overexpressing cells secreted higher CCL2 and promotes macrophage/MDSC proliferation. Previous report indicated that CCL2 is required for recruitment of macrophage and accumulation of MDSCs in the tumor microenvironment (16,18), suggesting that high expression of HOTAIR in cancer cells recruit TAMs/MDSCs through the secretion of CCL2, resulting in promoting tumor growth and metastasis. To the best of our knowledge, this is the first report that HOTAIR can affect the surrounding immune cells through humoral factor such as CCL2. CCL2 is known to be highly expressed in HCC, and believed to be a good target for immunotherapy (16). To test whether HOTAIR could be a specific target for immunotherapy against CCL2, we analyzed the correlation between CCL2 and HOTAIR expression in Gene Expression Omnibus (GEO) data set of HCC (GSE2109, n=40). We found that the group consist of patients with high expression level of both CCL2 and HOTAIR tended to associated with advanced clinical stages (P=0.078, data not shown). This data could be compatible with our findings in this study, although statistical significant was not reached due to the small scale of cases. Further study will be required to elucidate the role of HOTAIR in human tissue.
Several reports indicated the regulatory mechanisms of CCL2 production: the transcription of the CCL2 gene is shown to increase in response to stimuli, such as TNF or LPS, by the binding of NF-kB dimers to two distal NF-kB binding sites in murine fibroblasts (22). SP1 also regulates the basal level transcription of the CCL2 gene by binding to a GC-box located in the proximal region of the 5′-UTR (23). In this study, we demonstrated that HOTAIR can regulate CCL2 production in a HCC cell line. Recent study suggested that SP1 is a target of miR-326, and HOTAIR can modulate the pathway of miR-326/SP1 pathway (24). Thus, we speculate that HOTAIR regulate CCL2 through the transcriptional factors such as SP1.
Previous study suggested that introduction of human HOTAIR into liver cancer cells revealed that HOTAIR promoted more rapid proliferation compared to control HepG2 cells (7). Another report demonstrated that HOTAIR knockdown dramatically inhibited cell viability and induced G1-phase arrest in vitro and suppressed tumorigenicity in vivo by promoting miR-218 expression in HepG2 and Bel7404 cell lines. In this study, using Li-7 cell line, no significant difference was found between HOTAIR-overexpressing and control cells in cell proliferation, migration, and in vivo tumorigenicity assay. Because the function of HOTAIR are strongly cell type-dependent (5), we speculate that the context of intracellular signaling is different between HepG2 and other HCC cell lines. Further study will be required to elucidate the cell type-dependent molecular function of HOTAIR in types of HCC cell lines, and identify the critical molecule(s) which cooperate with HOTAIR.
In conclusion, we newly identified CCL2 as a downstream molecule of HOTAIR, which may be involved in the recruitment of macrophages/MDSC in tumor microenvironment.
Acknowledgements
This work was supported by JSPS KAKENHI (grant nos. 15K09055 and 16K07132), Japan Agency for Medical Research and Development, and Novartis Pharma Research Grants. This work was partly supported by the Biomedical Research Core of Tohoku University School of Medicine.
Glossary
Abbreviations
- CCL2
chemokine (C-C motif) ligand 2
- cDNA
complementary DNA
- DMEM
Dulbeccos modified Eagles medium
- ELISA
enzyme linked immunosorbent assay
- FBS
fetal bovine serum
- HOTAIR
HOX antisense intergenic RNA
- HCC
hepatocellular carcinoma
- lincRNA
large intervening non-coding RNA
- MDSC
myeloid-derived suppressor cell
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate buffered saline
- PRC2
polycomb repressive complex 2
- RPII
RNA polymerase II
- TAM
tumor associated macrophage
References
- 1.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 4.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heubach J, Monsior J, Deenen R, Niegisch G, Szarvas T, Niedworok C, Schulz WA, Hoffmann MJ. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol Cancer. 2015;14:108. doi: 10.1186/s12943-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee R, Mitra A. An overview of effective therapies and recent advances in biomarkers for chronic liver diseases and associated liver cancer. Int Immunopharmacol. 2015;24:335–345. doi: 10.1016/j.intimp.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 8.Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, Liang WC, Wang SS, Ko CH, Waye MM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A. Effects on in vitro tumor growth of murine macrophages isolated from sarcoma lines differing in immunogenicity and metastasizing capacity. Int J Cancer. 1978;22:741–746. doi: 10.1002/ijc.2910220617. [DOI] [PubMed] [Google Scholar]
- 13.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pyzer AR, Cole L, Rosenblatt J, Avigan DE. Myeloid-derived suppressor cells as effectors of immune suppression in cancer. Int J Cancer. 2016;139:1915–1926. doi: 10.1002/ijc.30232. [DOI] [PubMed] [Google Scholar]
- 15.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Krüger C, Manns MP, Greten TF, Korangy F. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology. 2008;135:234–243. doi: 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X, Wang H. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–167. doi: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 17.Teng KY, Han J, Zhang X, Hsu SH, He S, Wani NA, Barajas JM, Snyder LA, Frankel WL, Caligiuri MA, et al. Blocking the CCL2-CCR2 Axis Using CCL2-Neutralizing Antibody Is an Effective Therapy for Hepatocellular Cancer in a Mouse Model. Mol Cancer Ther. 2017;16:312–322. doi: 10.1158/1535-7163.MCT-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, Kanojia D, Pituch KC, Qiao J, Pytel P, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76:5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimura T. The production of monocyte chemoattractant protein-1 (MCP-1)/CCL2 in tumor microenvironments. Cytokine. 2017;pii:S1043–4666. doi: 10.1016/j.cyto.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Kadota K, Nakai Y, Shimizu K. A weighted average difference method for detecting differentially expressed genes from microarray data. Algorithms Mol Biol. 2008;3:8. doi: 10.1186/1748-7188-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Ping D, Boekhoudt GH, Rogers EM, Boss JM. Nuclear factor-kappa B p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J Immunol. 1999;162:727–734. [PubMed] [Google Scholar]
- 23.Ueda A, Okuda K, Ohno S, Shirai A, Igarashi T, Matsunaga K, Fukushima J, Kawamoto S, Ishigatsubo Y, Okubo T. NF-kappa B and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J Immunol. 1994;153:2052–2063. [PubMed] [Google Scholar]
- 24.Li J, Li S, Chen Z, Wang J, Chen Y, Xu Z, Jin M, Yu W. miR-326 reverses chemoresistance in human lung adenocarcinoma cells by targeting specificity protein 1. Tumour Biol. 2016;37:13287–13294. doi: 10.1007/s13277-016-5244-2. [DOI] [PubMed] [Google Scholar]