Abstract
Studied as a type of tumor suppressor, microRNA (miR) performs an important role in growth and apoptosis of various human carcinomas. However, the effects of miR-l44 on osteosarcoma growth and apoptosis, as well as possible underlying mechanisms, remain unclear. The present study investigated the expression of miR-144 in osteosarcoma MG-63 and U-2 OS cell lines compared with osteoblast cells. In order to elucidate the effects of miR-144 on osteosarcoma, miR-144 was upregulated in MG-63 and U-2 OS cells by transfecting chemically synthesized miR-144 mimics. Bioinformatics analysis of potential miR-144 target genes was performed using TargetScanHuman 7.0 and confirmed by luciferase assay. This analysis identified mammalian target of rapamycin (mTOR) as a target of miR-144. The present results indicated that the overexpression of miR-144 may significantly inhibit proliferation and promote apoptosis of MG-63 and U-2 cells compared with scramble control. Furthermore, the effects of miR-144 on osteosarcoma were associated with the mTOR signaling pathway via directly targeting the 3′ untranslated region of mTOR mRNA, resulting in a decrease in the level of mTOR protein. In summary, miR-144 was demonstrated to act as a tumor suppressor, which inhibits proliferation and promotes apoptosis of osteosarcoma cell lines. In addition, this effect was mediated by direct targeting on mTOR following inhibition of the mTOR signaling pathway. The present study suggested that miR-144 may be a candidate for the gene therapy of osteosarcoma.
Keywords: osteosarcoma, microRNA-144, mammalian target of rapamycin signaling pathway, gene therapy
Introduction
Osteosarcoma is a type of malignant tumor, which often occurs in children (1). Surgical methods in the treatment of osteosarcoma frequently involve wide resection or amputation, which is likely to result in a disability or deformity, with the consequence of poor quality of life (2). For this reason, early warning, diagnosis and treatment are exceptionally important. Previous studies revealed that microRNA (miRNA/miR) has a major role in the occurrence and development of numerous diseases such as ischemic cardiomyopathy, gastric cancer and hepatocellular carcinoma (3,4). As a non-coding single-chain micro-molecule RNA, miRNA is made up of 21–25 nucleotides (5,6). Through incomplete complementary sequence combination with the 3′ untranslated region (3′UTR) sequence of target mRNA, miRNA is able to inhibit mRNA translation or cause direct degradation to the extent that the miRNA level shows the effect of gene silencing following transcription (7). Studies have revealed that multiple miRNAs are able to serve as oncogenes or anti-oncogenes in various tumors, including osteosarcoma, while specific miRNA is involved in each stage of the occurrence and development of tumors (8,9).
Previous studies indicated that miR-144 is downregulated expression in colon cancer miR-144 is in close association with the development of colorectal cancer and can be used as a molecular diagnostic marker of colon cancer (10,11). A previous study demonstrated that downregulation of miR-144 is able to result in the proliferation of bladder cancer cells via EZH2 and Wnt signaling pathways (12). Another study investigating the function of miR-144 revealed that GATA4 may activate miR-144 precursors and induce their expression (13), and studies have also suggested that the overexpression of miR-144 may cause apoptosis of myocardial cells and inhibit their proliferation (14,15). Furthermore, miR-144 is considered to be a potential target for the treatment of ischemic heart disease (13,15). However, whether miR-144 can affect the biological function of osteosarcoma cells is yet to be reported, and its mechanism remains unknown. Therefore, the current study focused on investigating the effect of miR-144 on the proliferation and apoptosis of osteosarcoma cell lines (MG-63 and U-2) and investigating its mechanism of action.
Materials and methods
Cell lines and cell culture
The human osteosarcoma cell lines, MG-63 and U-2 OS, were obtained from the American Type Culture Collection (Manassas, VA, USA). MG-63 was maintained in minimum essential medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), and U-2 OS was cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% horse serum (Invitrogen; Thermo Fisher Scientific, Inc.) and 5% FBS. The two mediums were supplemented with 100 U/ml penicillin and 100 U/ml streptomycin sulfate (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and the cells were cultured in a humidified 5% CO2 incubator at 37°C.
Evaluation of miR-144 expression in osteosarcoma cells
For RNA analysis, each cell line was seeded at 1×105 cells/well in a volume of 2 ml on 6-well flat-bottomed plates. Total RNA from these cell lines was isolated using the mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol following 48 h incubation at 37°C. Then, cDNA was synthesized from 10 ng of total RNA using TaqMan MicroRNA hsa-miR-144 specific primers (Applied Biosystems; Thermo Fisher Scientific, Inc.) and a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to measure miR-144 expression, and the primers were as follows: miR-144 forward, 5′GCT GGG ATA TCA TCA TAT ACT G3′ and reverse, 5′CGG ACT AGT ACA TCA TCT ATA CTG3′; and U6 forward, 5′CTC GCT TCG GCA GCA CA3′ and reverse, 5′AAC GCT TCA CGA ATT TGC GT3′. The following temperature profile was used: Initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 10 sec and extension at 65°C for 10 sec. PCR was performed in a LightCyclerTM 480 system (Roche Applied Science, Penzburg, Germany) using the LightCyclerTM 480 Probes Master kit (Roche Applied Science). Expression levels of miR-144 were normalized to that of the small nuclear RNA U6 (Applied Biosystems; Thermo Fisher Scientific, Inc.) transcript. Relative quantification of miRNA expression was calculated using the 2−ΔΔCq method (16).
Transfection of miR-144 mimics and transfection efficiency evaluation
Designed and provided by RiboBio (Guangzhou RiboBio Co., Ltd., Guangzhou, China), human miR-144 duplex mimics and scramble control (with the final concentration of 10 nM) were transfected into cells using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 24 h post-transfection at 37°C, cells transfected with miRNA mimics or scramble were used for subsequent experiments, including proliferation and apoptosis assays.
Cell proliferation and viability assays
MG-63 or U-2 OS cells in the logarithmic phase were transfected with miR-144 mimics or scramble control and seeded at 4.0×103 cells/well in 96-well flat-bottomed microtiter plates in a final volume of 100 µl culture medium per well. The cells were incubated in a humidified atmosphere at 37°C and 5% CO2 for 24, 48, 72 and 96 h. MTT assays were used to measure cell viability during each time period. Dimethyl sulfoxide was added into each well to dissolve the purple formazan. Subsequent to the formazan crystals being dissolved, the absorbance was determined spectrophotometrically at 570 nm using a reference wavelength of 630 nm on an ELX800 UV universal microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Proliferation and apoptosis assays
Proliferation status of the cells was determined by measuring the incorporation of 5-bromo-2′-deoxyuridine (BrdU). The cells were incubated with 100 µM BrdU (Sigma-Aldrich; Merck KGaA) for 1–2 h at 37°C, and BrdU labeling was detected by confocal laser scanning microscopy or flow cytometry following incubation for 30 min at 37°C with a fluorescein isothiocyanate- or allophycocyanin-conjugated anti-BrdU antibody (cat. no. 558599; dilution, 1:200; BD Biosciences, Franklin Lakes, NJ, USA). The images were captured on a BD Pathway™ 435 confocal imager with an objective magnification, ×20, and merged using BD AttoVision™ software v1.6 (BD Biosciences). Staining of samples without the addition of BrdU was used as a negative control. The cells were subsequently analyzed and quantified by flow cytometry. In brief, the cells were stained for 30 min at 37°C with the following antibody: Anti-BrdU (cat. no. 558599; dilution, 1:200; BD Biosciences), then the labeled cells were thoroughly washed with 2X PBS and analyzed on BD FACSCantoII flow cytometer (BD Biosciences). Data were analyzed by BD FACSDiva Software v6.0 (BD Biosciences).
Dexamethasone was used to induce cell apoptosis
Duration the culturing of cells, dexamethasone with a concentration of 0.1 µM was added into culture medium. In situ Cell Death Detection kit (Roche Diagnostics GmbH, Mannheim, Germany) was used for TUNEL staining to determine the apoptotic status of the cells, according to the manufacturer's protocol. Zeiss Axio Imager 2 fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany) was used to observe and capture images at a magnification, ×100. A total of 3 fields of view in each group were obtained.
Protein expression analysis
Western blot analysis was used to confirm mammalian target of rapamycin (mTOR) or proteins associated with proliferation or apoptosis expression in miR-144-mimic-transfected cells and control cells. In brief, cell proteins were collected with radioimmunoprecipitation assay lysis buffer (Auragene Bioscience, Changsha, China) to obtain cellular protein and the protein concentrations were measured using bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 30 µg of cellular proteins were separated on 10 or 12% SDS-PAGE gels, and were subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). Then, the membranes were blocked in 5% milk for 1 h at room temperature and incubated with primary antibodies overnight at 4°C, and subsequently incubated with horseradish peroxidase-conjugated anti-rabbit IgG (cat. no. 7074; dilution, 1:2,000; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature for 1 h. The following primary antibodies and dilutions were used: mTOR rabbit monoclonal antibody (cat. no. 2983; dilution, 1:1,000; Cell Signaling Technology, Inc.) and GAPDH rabbit monoclonal antibody (cat. no. 5014; dilution, 1:1,000; Cell Signaling Technology, Inc.), p53 rabbit monoclonal antibody (cat. no. 2527; dilution, 1:1,000; Cell Signaling Technology, Inc.), PCNA rabbit monoclonal antibody (cat. no. 13110; dilution, 1:1,000; Cell Signaling Technology, Inc.), Bcl-2 rabbit monoclonal antibody (cat. no. 3498; dilution, 1:1,000; Cell Signaling Technology, Inc.), Bcl-xL rabbit monoclonal antibody (cat. no. 2764; dilution, 1:1,000; Cell Signaling Technology, Inc.), Bax rabbit monoclonal antibody (cat. no. 5023; dilution, 1:1,000; Cell Signaling Technology, Inc.), caspase 3 rabbit monoclonal antibody (cat. no. 14220; dilution, 1:1,000; Cell Signaling Technology, Inc.). Signals for each protein expression were detected with electrochemiluminescence reagents (GE Healthcare Life Sciences, Little Chalfont, UK) according to the manufacturer's protocol.
Target genes of miR-144
TargetScanHuman 7.0 (http://www.targetscan.org/) and miRBase (http://www.mirbase.org/) microRNA databases were used to identify the target genes of miR-144.
Construction of reporter plasmids and the luciferase reporter assay
To construct a luciferase reporter plasmid, a full-length fragment of the mTOR 3′UTR was subcloned into pmirGLO dual-luciferase miRNA target expression vector (Promega Corporation, Madison, WI, USA) located 5′ to the firefly luciferase. The nucleotide sequences of the constructed plasmids were confirmed by DNA sequencing. For luciferase reporter assays, MG-63 cells (5×104/well) were seeded into a 96-well plate and then co-transfected via Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) with the primer GLO-mTOR 3′UTR or Rictor 3′UTR (Promega Corporation) construct and miR-144-mimics or scramble control (Ambion; Thermo Fisher Scientific, Inc.). Assays were performed 48 h after transfection using the Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol. The firefly luciferase signals were normalized to the Renilla luciferase signals. Transfections were repeated three times in independent experiments.
Statistical analysis
Quantitative data are expressed as the mean ± standard error. Statistical analysis was performed by using one-way analysis of variance followed by the least significant difference test. Statistical analysis was performed with GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-144 expression is decreased in osteosarcoma cells
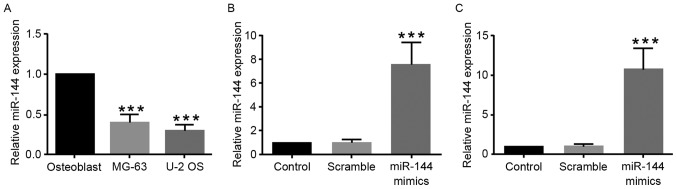
To investigate whether miR-144 regulates osteosarcoma progression, the expression level of miR-144 was examined in osteosarcoma cell lines (MG-63 and U-2 OS). Compared with the normal osteoblast cells, miR-144 expression levels were significantly reduced in osteosarcoma cell lines (Fig. 1A). To investigate the biological role of miR-144 in regulating osteosarcoma cells, miR-144 mimics or scramble control were transfected into MG-63 or U-2 OS cells. As shown in Fig. 1B and C, the expression level of miR-144 was significantly increased in miR-144-mimic-transfected cells compared with control cells or scramble-transfected cells.
Figure 1.
Expression level of miR-144 in osteosarcoma cells and osteoblasts. (A) Relative expression of miR144 in osteosarcoma cells and osteoblasts detected by reverse transcription-quantitative polymerase chain reaction. (B and C) Effects of transfection of miR-144-mimics in (B) MG63 and (C) U-2 OS cells. ***P<0.01, n=3. miR, microRNA.
miR-144 inhibits proliferation of osteosarcoma cells
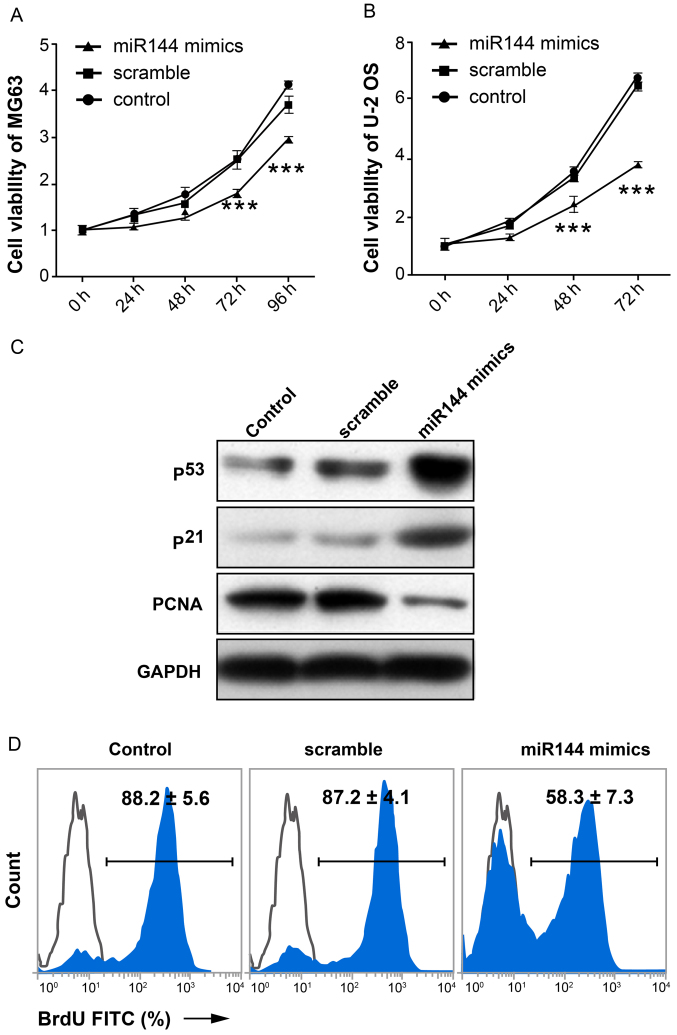
To investigate the possible effect of miR-144 on proliferation of osteosarcoma cells, MTT assay was performed. The results demonstrated that the proliferation capacity of cells in the miR-144-mimic-transfected group exhibited a significant decline compared with the scramble and control groups (P<0.01; n=3; Fig. 2A and B). The expression level of proteins associated with proliferation (p53, p21 and PCNA) in MG-63 cells is shown in Fig. 2C. p53 and p21 act as cell cycle arrest genes (17). PCNA is a biomarker of cell proliferation (18–20). The expression level of p53 and p21 in the miR-144-mimic-transfected group was markedly increased compared with the scramble and control groups. By contrast, the expression of PCNA was markedly decreased in the miR-144-mimic-transfected group compared with the other two groups. BrdU-incorporated flow cytometry showed similar data (58.3±7.3 vs. 87.2±4.1, miR-144-mimic-transfected group and scramble control, respectively; Fig. 2D). These data demonstrated that miR-144 inhibited the proliferation of osteosarcoma cells.
Figure 2.
miR-144 inhibits the proliferation of osteosarcoma cells. MTT assay demonstrates the effect of miR-144 on proliferation of (A) MG63 and (B) U-2 OS cells. (C) Western blot analysis of proteins associated with cell proliferation in MG-63 cells. p53 and p21 act as cell cycle arrest genes, while PCNA is a biomarker of cell proliferation. (D) BrdU-incorporated flow cytometry showing proliferation data. ***P<0.01, n=3. PCNA, proliferating cell nuclear antigen; miR, microRNA; BrdU, 5-bromo-2′-deoxyuridine; FITC, fluorescein isothiocyanate.
Overexpression of miR-144 promotes apoptosis of osteosarcoma cells induced by dexamethasone
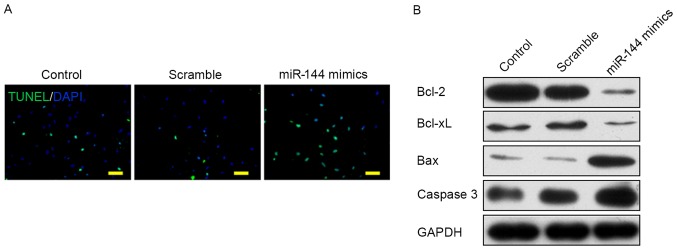
To determine the effect of miR-144 on the apoptosis of osteosarcoma cells, dexamethasone was added to the culture medium to induce apoptosis. TUNEL staining images showed a greater number of TUNEL+ cells in the miR-144-mimic-transfected group compared with the control or scramble-transfected groups (Fig. 3A). The expression level of proteins associated with apoptosis signaling pathways was examined by western blot analysis as shown in Fig. 3B. The data showed that two anti-apoptotic proteins, Bcl-2 and Bcl-xL were markedly downregulated in the miR-144-mimic group compared with the control and scramble groups. However, two pro-apoptotic proteins, Bax and caspase 3, were markedly upregulated in the miR-144-mimic-transfected group. These data showed that the overexpression of miR-144 may promote apoptosis of osteosarcoma cells induced by dexamethasone.
Figure 3.
Overexpression of miR-144 promotes the apoptosis of osteosarcoma cells induced by dexamethasone. (A) TUNEL staining images of TUNEL+ cells (green) in three groups following treatment with dexamethasone (scale bar, 50 µm). (B) Western blot analysis of proteins associated with apoptosis signaling pathways: Bcl-2, Bcl-xL, Bax, caspase 3. Bcl-2, B-cell lymphoma-2; Bcl-xL, B-cell lymphoma-extra-large; Bax, Bcl-2-asssocaited X protein; miR, microRNA; TUNEL, terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling.
miR-144 directly targets mTOR and inhibits its expression
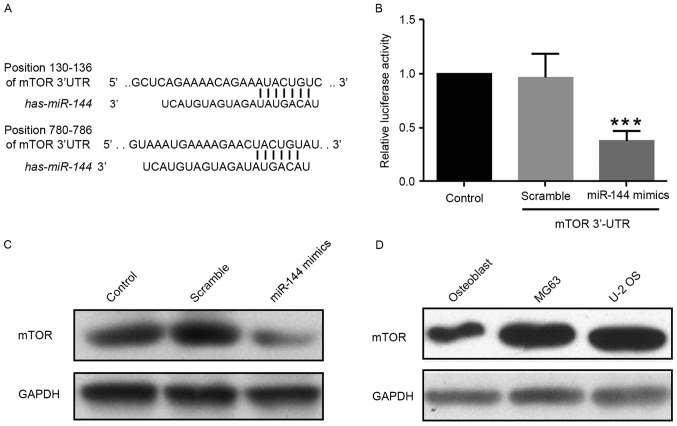
In order to identify the target genes of miR-144, candidate genes were identified using TargetScanHuman 7.0 (http://www.targetscan.org/) and miRBase (http://www.mirbase.org/) micoRNA databases. miR-144 targets the mTOR gene, an important molecule involved in tumorigenesis and cancer progression (21,22) (Fig. 4A). To confirm this prediction, a luciferase reporter vector containing the 3′UTR of mTOR [pGL3-mTOR-3′UTR-WT (wild-type)] was constructed. The reporter assay showed that miR-144 was able to significantly suppress luciferase expression (Fig. 4B). Additionally, miR-144 reduced the levels of mTOR protein in MG-63 cells (Fig. 4C). As expected, mTOR levels were markedly upregulated in MG-63 or U-2 OS cells compared with normal osteoblast cells (Fig. 4D). These data demonstrated that miR-144 inhibits endogenous mTOR expression in osteosarcoma cells.
Figure 4.
miR-144 directly targets mTOR and inhibits its expression. (A) Prediction of miR-144 targets by TargetScanHuman 7.0 (http://www.targetscan.org/) and miRBase (http://www.mirbase.org/). (B) Luciferase reporter assay to confirm the prediction. (C) Expression of mTOR protein level in MG-63 cells transfected with miR-144 mimic, control and scramble miRNA. (D) Levels of mTOR protein expression in normal osteoblast cells and osteosarcoma U-2 OS cells. ***P<0.01, n=3. mTOR, mammalian target of rapamycin; miR, microRNA; 3′UTR, 3′ untranslated region.
Discussion
MiRNAs primarily function at the post-transcription level, and they may induce downregulation of gene expression through sequence complementarity between the miRNAs and their targeted mRNA (7). miRNA sequences have properties of high conservation, tissue specificity and time sequence in different species (23,24). As described in the literature, miRNA performs an important role in the occurrence, development and metastasis of tumors (25,26). Furthermore, the abnormal expression of miRNA is associated with the occurrence of certain tumors such as gastric and breast cancer (27,28). Results of the present study revealed that miR-144 can directly target mTOR and inhibit the signaling pathway of phosphoinositide 3-kinase (PI3K)/Akt, so as to inhibit the proliferation of osteosarcoma cells and promote their apoptosis.
Significant changes in miR-144 expression have been observed in a variety of diseases, including colorectal and bladder cancer (10,12). However, the majority of studies have focused on the blood system, while fewer studies have investigated tumors. Through meta-analysis, Wang et al (29) found the presence of differentially-expressed miRNAs in osteosarcoma tissues. In particular, the differential expression of miR-144 was revealed in colorectal cancer, pancreatic cancer, thyroid cancer, laryngeal cancer and malignant mesothelioma tissues. Additionally, miR-144 was demonstrated to be downregulated in laryngeal cancer (30), cervical cancer (31), follicular thyroid carcinoma (32) and malignant mesothelioma (33). Therefore, the present study hypothesized that transfecting cells with an externally constructed vector of miRNA-144 may cause cell growth retardation and death, primarily apoptosis.
Results of the present study are consistent with the results of previous studies on liver cancer and non-small cell lung cancer (34,35). It is generally considered that miR-144 is able to function as a suppressor gene of tumor proliferation and affect the prognosis of patients with osteosarcoma (36,37). Studies have reported that subsequent to transfecting liver cancer HepG2 cells, the plasmids of overexpressed miR-144 are able to promote apoptosis and inhibit the occurrence of liver cancer metastasis by affecting the expression of E2F3 (34,38). Results of the present study demonstrated that overexpression of miR-144 in osteosarcoma cells is able to markedly inhibit proliferation and strengthen the apoptosis-promoting effect of dexamethasone on osteosarcoma cells. Therefore, this indicates that miR-144 may also serve as a tumor suppressor gene in osteosarcoma cells.
mTOR is an evolutionarily conserved protein kinase (39). As aberrant signaling of the mTOR pathway is associated with various malignancies, mTOR protein is considered to be a key regulatory factor for cell growth, and has gradually become a novel target for tumor therapy (40). mTOR may exert its function through participation in the PI3K/Akt/mTOR pathway, and the regulatory effect of the deregulation of the mTOR signaling pathway on tumors is primarily via inhibiting apoptosis, promoting cell survival, accelerating cell cycle transmission, enhancing angiogenesis and promoting the invasion and metastasis of tumor cells (41). An in vitro study on non-small lung cancer cell lines indicated that human papilloma virus 16 may activate the PI3K/Akt/mTOR signaling pathways to induce the expression of angiogenic cytokines, including hypoxia-inducible factor-1α, vascular endothelial growth factor and interleukin-8 (42). Zhang et al (43) used mTOR-targeted inhibition combined with antiandrogen for the treatment of prostate cancer and demonstrated that the antitumor effect was greater than the single drug, which indicated that inhibiting mTOR was able to increase the effectiveness of anticancer drugs. Iwaya et al (10) reported that downregulating miR-144 in colon cancer cells may activate mTOR signaling pathways and further, promote the proliferation of colon cancer cells and increase the sensitivity of colon cancer cells to rapamycin. Results of the present study identified that upregulating the miR-144 in osteosarcoma cells was able to downregulate the activity of mTOR signaling pathways and inhibit the proliferation of osteosarcoma cells, thereby promoting their sensitivity to dexamethasone-induced apoptosis. These results are consistent with the findings of Iwaya et al (10).
In conclusion, miR-144 may effectively inhibit the proliferation and apoptosis of osteosarcoma cells by directly targeting the mRNA 3′UTR region of mTOR genes and inhibiting the synthesis of mTOR proteins, thereby inhibiting PI3K/Akt signaling pathways. As studies on miR-144 continue, miR-144 may become a novel target for the treatment of osteosarcoma.
Acknowledgements
The authors would like to acknowledge Technical Support from the First Affiliated Hospital, China Medical University (Shenyang, China).
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Ruggieri P, Mavrogenis AF, Mercuri M. Quality of life following limb-salvage surgery for bone sarcomas. Expert Rev Pharmacoecon Outcomes Res. 2011;11:59–73. doi: 10.1586/erp.10.91. [DOI] [PubMed] [Google Scholar]
- 3.Latronico MV, Catalucci D, Condorelli G. MicroRNA and cardiac pathologies. Physiol Genomics. 2008;34:239–242. doi: 10.1152/physiolgenomics.90254.2008. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaillé J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 7.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets; Proc Natl Acad Sci USA; 2006; pp. 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33:2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 11.Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, Sileri P, Cretella M, Pallone F, Federici G, Bernardini S. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011;46:1391–1402. doi: 10.1007/s00535-011-0456-0. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT, Jegga AG, Fan GC. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49:841–850. doi: 10.1016/j.yjmcc.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang F, Huang XY, Yan DS, Zhou X, Yang DY. MicroRNA-144 over-expression induced myocytes apoptosis. Zhonghua Xin Xue Guan Bing Za Zhi. 2011;39:353–357. doi: 10.3760/cma.j.issn.0253-3758.2011.04.015. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 15.Yu M, Liu Y, Zhang B, Shi Y, Cui L, Zhao X. Inhibiting microRNA-144 abates oxidative stress and reduces apoptosis in hearts of streptozotocin-induced diabetic mice. Cardiovasc Pathol. 2015;24:375–381. doi: 10.1016/j.carpath.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology. 2002:181–182. 1–481. doi: 10.1016/s0300-483x(02)00460-2. [DOI] [PubMed] [Google Scholar]
- 18.Bologna-Molina R, Mosqueda-Taylor A, Molina-Frechero N, Mori-Estevez AD, Sánchez-Acuña G. Comparison of the value of PCNA and Ki-67 as markers of cell proliferation in ameloblastic tumors. Med Oral Patol Oral Cir Bucal. 2013;18:e174–e179. doi: 10.4317/medoral.18573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Castilla C, McDonough P, Tumer G, Lambert PC, Lambert WC. Sometimes it takes darkness to see the light: Pitfalls in the interpretation of cell proliferation markers (Ki-67 and PCNA) Skinmed. 2012;10:90–92. [PubMed] [Google Scholar]
- 21.Geissler EK, Schlitt HJ, Thomas G. mTOR, cancer and transplantation. Am J Transplant. 2008;8:2212–2218. doi: 10.1111/j.1600-6143.2008.02391.x. [DOI] [PubMed] [Google Scholar]
- 22.Sabatini DM. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 23.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 25.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Tong F, Cao P, Yin Y, Xia S, Lai R, Liu S. MicroRNAs in gastric cancer: From benchtop to bedside. Dig Dis Sci. 2014;59:24–30. doi: 10.1007/s10620-013-2887-3. [DOI] [PubMed] [Google Scholar]
- 28.Serpico D, Molino L, Di Cosimo S. microRNAs in breast cancer development and treatment. Cancer Treat Rev. 2014;40:595–604. doi: 10.1016/j.ctrv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J, Yu L. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2011;129:1624–1634. doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Fu T, Wang X, Zhu W. Primary, study of miRNA expression patterns in laryngeal carcinoma by microarray. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;24:535–538. (In Chinese) [PubMed] [Google Scholar]
- 31.Tian Y, Tian Y, Luo X, Zhou T, Huang Z, Liu Y, Qiu Y, Hou B, Sun D, Deng H, et al. Identification and characterization of microRNAs related to salt stress in broccoli, using high-throughput sequencing and bioinformatics analysis. BMC Plant Biol. 2014;14:226. doi: 10.1186/s12870-014-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossing M, Borup R, Henao R, Winther O, Vikesaa J, Niazi O, Godballe C, Krogdahl A, Glud M, Hjort-Sørensen C, et al. Down-regulation of microRNAs controlling tumourigenic factors in follicular thyroid carcinoma. J Mol Endocrinol. 2012;48:11–23. doi: 10.1530/JME-11-0039. [DOI] [PubMed] [Google Scholar]
- 33.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma-A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 34.Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumour Biol. 2014;35:10759–10764. doi: 10.1007/s13277-014-2017-7. [DOI] [PubMed] [Google Scholar]
- 35.Zha W, Cao L, Shen Y, Huang M. Roles of Mir-144-ZFX pathway in growth regulation of non-small-cell lung cancer. PLoS One. 2013;8:e74175. doi: 10.1371/journal.pone.0074175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu J, Wang K, Liu D, Zhang X, Yin W. The downregulation of miR-144 is associated with the growth and invasion of osteosarcoma cells through the regulation of TAGLN expression. Int J Mol Med. 2014;34:1565–1572. doi: 10.3892/ijmm.2014.1963. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Zhou X, Wei M. MicroRNA-144 suppresses osteosarcoma growth and metastasis by targeting ROCK1 and ROCK2. Oncotarget. 2015;6:10297–10308. doi: 10.18632/oncotarget.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue J, Niu YF, Huang J, Peng G, Wang LX, Yang YH, Li YQ. miR-141 suppresses the growth and metastasis of HCC cells by targeting E2F3. Tumour Biol. 2014;35:12103–12107. doi: 10.1007/s13277-014-2513-9. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pene F, Claessens YE, Muller O, Viguié F, Mayeux P, Dreyfus F, Lacombe C, Bouscary D. Role of the phosphatidylinositol 3-kinase/Akt and mTOR/P70S6-kinase pathways in the proliferation and apoptosis in multiple myeloma. Oncogene. 2002;21:6587–6597. doi: 10.1038/sj.onc.1205923. [DOI] [PubMed] [Google Scholar]
- 41.Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): Conducting the cellular signaling symphony. J Biol Chem. 2010;285:14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang E, Feng X, Liu F, Zhang P, Liang J, Tang X. Roles of PI3K/Akt and c-Jun signaling pathways in human papillomavirus type 16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis in non-small cell lung cancer cells. PLoS One. 2014;9:e103440. doi: 10.1371/journal.pone.0103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W, Zhu J, Efferson CL, Ware C, Tammam J, Angagaw M, Laskey J, Bettano KA, Kasibhatla S, Reilly JF, et al. Inhibition of tumor growth progression by antiandrogens and mTOR inhibitor in a Pten-deficient mouse model of prostate cancer. Cancer Res. 2009;69:7466–7472. doi: 10.1158/0008-5472.CAN-08-4385. [DOI] [PubMed] [Google Scholar]