Abstract
Circular non-coding RNAs are found to play important roles in biology but are still relatively unexplored as a structural motif for chemically regulating gene function. Here, we investigated whether small interfering RNA (siRNA) with a circular structure can circumvent off-target gene silencing, a problem often observed with standard linear duplex siRNA. In the present work, we, for the first time, synthesized a series of circular siRNAs by cyclizing two ends of a single-stranded RNA (sense or antisense strand) to construct circular siRNAs that were more resistant to enzymatic degradation. Gene silencing of GFP and luciferase was successfully achieved using these circular siRNAs with circular sense strand RNAs and their complementary linear antisense strand RNAs. The off-target effect of sense strand RNAs was evaluated and no cross off-target effects were observed. In addition, we successfully achieved longer gene-silencing efficiency in mice with circular siRNAs than with linear siRNAs. These results indicate the promise of circular siRNAs for overcoming off-target effects of siRNAs and enhancing the possible long-term effect of siRNA gene silencing in basic research and drug development.
Keywords: oligonucleotides, circular siRNA, off-target effect, gene regulation, RNA digestion, circular oligonucleotides, non-coding RNA, long-term effect
Graphical Abstract
Introduction
Circular RNAs have been found in many biological systems.1, 2, 3 They are mostly reported to function as molecular “sponges” for microRNAs (miRNAs) and have many unknown biological activities. Small interfering RNA (siRNA), as one of many non-coding RNAs, has been an important tool in gene expression regulation and as a therapeutic agent in drug development.4, 5 Although the cleavage of target mRNA mediated by siRNA is highly sequence specific, the unintended “off-target” silencing of endogenous genes is also observed in cells, which limits further applications of siRNAs.6, 7 For example, the antisense strand of an intercellular cell adhesion molecule-1 (ICAM-1) siRNA could normally regulate gene expression of target intercellular adhesion molecule-1, whereas the sense strand of the same ICAM-1 siRNA could knock down the expression of tumor necrosis factor receptor 1.6 The off-target effects of siRNAs have also been shown to induce apoptosis in many cell lines.7 Thus, RNAi-induced gene-silencing activity must be carefully evaluated due to the potential for off-target effects with siRNA.
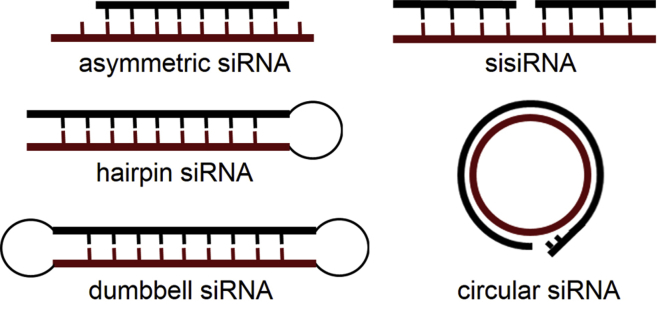
Many chemical modifications of siRNA have been investigated in an attempt to reduce off-target effects. siRNAs modified with 2′OMe RNA, locked nucleic acid, unlocked nucleoside analogs, 5-nitroindole-modified nucleotide, terminal methylation, and backbone phosphorothioate were achieved.8, 9, 10, 11, 12, 13, 14 These modifications may prevent the loading and processing of sense strand RNA to lower the off-target effect of siRNAs. Different compositions were also applied for optimization of RNAi gene silencing (Figure 1). A small segmented interfering siRNA (sisiRNA) was designed, which contains an intact antisense strand and two shorter complementary 10–12 nt sense strand RNAs (Figure 1). Alternatively, a 16-nt siRNA or 15-nt asymmetric interfering RNA (aiRNA) was developed (Figure 1).15, 16, 17, 18, 19 These shorter sense strand RNAs were less efficient for use as guide RNAs, and the off-target effects caused by these sense strand RNAs were reduced. However, the efficiency and/or stability of these siRNAs was somewhat sacrificed, probably due to the easier degradation of RNAs. siRNA or small hairpin RNA (shRNA) with dumbbell structures were also developed by cyclizing two strands of siRNA duplexes for siRNA gene silencing (Figure 1). Xi et al. reported that circular dumbbelled shRNAs were more potent in siRNA gene silencing than their open-ended counterpart, likely due to their enhanced stability to nuclease degradation. However, reduction of the off-target effect was not achieved.20, 21, 22, 23 In this paper, we present results on the silencing activity of circular siRNAs in comparison to classical linear siRNAs by the use of fluorescent cellular and animal (mice) models. The studies aim to develop siRNA silencers, overcoming off-target effects and enhancing the long-term effect of gene silencing in basic research and drug development.
Figure 1.
Overview of Different Strategies for Modified siRNAs to Reduce the Off-Target Effect Caused by Sense Strand
Results and Discussion
Synthesis and Characterization of Circular siRNA
Building on previous achievements with caged circular antisense oligonucleotides by our laboratory and others,24, 25, 26, 27, 28, 29, 30 we sought to develop a circular siRNA strategy for reducing off-target effects and short effect time. Cyclization of the 5ʹ and 3ʹ ends of the sense or antisense strand of siRNA was achieved to form a 21-mer RNA ring. Then, a linear complementary RNA strand was hybridized with the circular RNA to form a circular siRNA duplex (Figure 1). Two ends of single-stranded RNA with the 5′ terminal phosphate group were joined through the formation of a phosphodiester bond using T4 RNA ligase (Figure S1).31 The resulting circular RNAs were further purified by native PAGE based on the fact that linear and circular RNA move at different rates in the gel (Figure S2). The single-stranded circular RNAs were then collected and desalted, followed by characterization using electrospray ionization-mass spectrometry (ESI-MS) under the negative ion mode (Table S2).
We first evaluated the enzymatic stability of the circular siRNA duplex with a circular RNA and its linear complementary RNA. Using the same antisense (GAS, GUUCACCUUGAUGCC GUUCUU) and sense (GS, GAACGGCAUCAAGGUGAACUU) sequences targeting GFP, circular siRNA (c-GAS/GS, G for GFP) and linear siRNA (GAS/GS), after annealing with two complementary strands, were treated with RNase A. When incubated with RNase A at 37°C, the positive control siRNA (GS/GAS) started to degrade in the first 2 hr (Figure S3). However, we observed only trace degradation for the circular siRNA duplex (c-GAS/GS) in comparison to the linear siRNA (GAS/GS), which exhibited a more stable nature of circular RNA. These in vitro studies predict greater longevity of circular siRNA in many biological environments.
Evaluation of GFP Gene Expression Regulation with Circular siRNAs with Circular Sense or Antisense RNAs
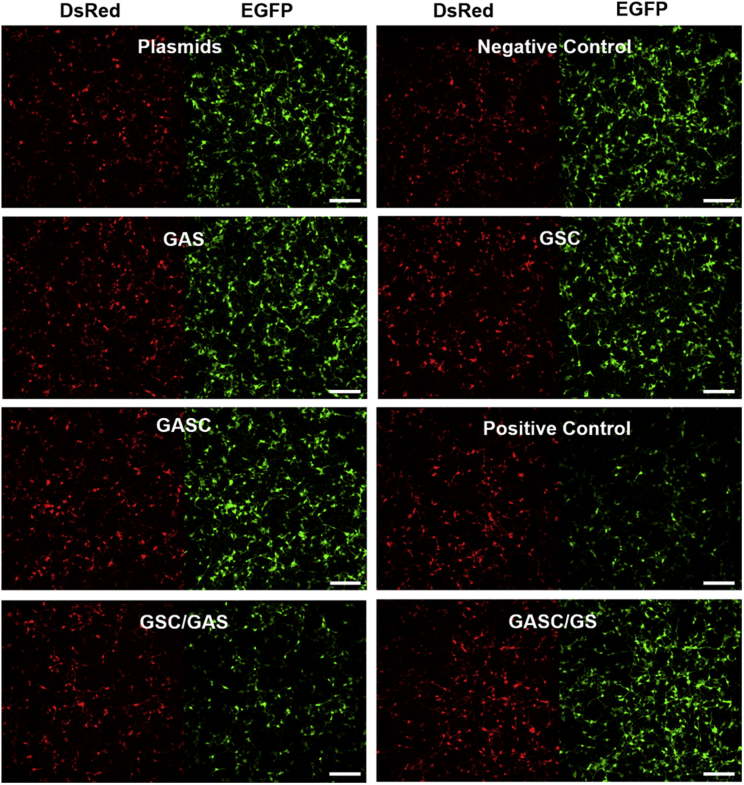
To test the effect of circular siRNAs on cellular gene expression, we first designed circular siRNAs targeting the GFP gene. The sequence of siRNA (GS/GAS) was chosen to target GFP gene expression while using RFP expression as an internal control. HEK293A cells were cotransfected with pEGFP-N1 vector and pDsRed2-N2 vector, together with circular siRNAs (c-GS/GAS and c-GAS/GS), linear siRNAs (GS/GAS and NC with random sequence) (Table S1), single-stranded RNAs (GS and GAS), and single-stranded circular RNAs (c-GS and c-GAS), respectively. After 4 hr transfection, the media was replaced with fresh media, and the cells were incubated for another 20 hr. Then, the cells were imaged (Figure 2). Single-strand RNAs (GAS or c-GS and c-GAS) had no effect on GFP gene expression as well as c-GAS/GS with circular antisense RNA and linear sense RNA (image for linear GS was not shown). Only c-GS/GAS with circular sense RNA and linear antisense RNA downregulated GFP gene expression, which was similar to the linear positive control siRNA (GS/GAS). The mean values of GFP fluorescence were quantified by flow cytometry (Figure S4). The average GFP fluorescence intensity of the transfected cells with RFP expression as control was used to represent the level of gene silencing. All the data were then normalized to the cells that were transfected only with both GFP and RFP plasmids. As shown in Figure S4, the average value of GFP fluorescence intensity was reduced to 40% for cells transfected with linear control siRNA (GS/GAS) under our assay conditions. No gene silencing was observed for cells transfected with the single-stranded RNA (linear or circular) or negative control siRNA (NC). Similarly, circular siRNA (c-GAS/GS) with a linear sense strand and circular antisense strand also functioned similarly to the negative control siRNA, with little downregulation of GFP gene expression. However, circular siRNA (c-GS/GAS) with a linear antisense strand and circular sense strand could effectively silence GFP expression as well as the positive linear control siRNA (GS/GAS).
Figure 2.
Knockdown of GFP Gene in HEK293A Cells with or without a Different Combination of Circular RNAs or siRNAs with RFP as Internal Control
GFP was excited at 488 nm and RFP was excited at 561 nm. Scale bar, 200 μm.
These results indicated that gene-silencing activity of c-GS/GAS siRNA with circular sense strand RNA was nearly identical to linear positive control siRNA. In the case of c-GAS/GS siRNA with circular antisense strand RNA, RNAi-induced gene silencing was almost completely absent under the same assay conditions. For circular siRNA (c-GS/GAS) with a linear antisense guide strand, the linear antisense strand RNA was still accessible for binding to cellular mRNA within the RNA-induced silencing complex (RISC) complex. Further RNAi machinery was then triggered, leading to target mRNA cleavage.32 As expected, this did not happen for c-GAS/GS with circular antisense RNA. To test the hypothesis that the linear GAS RNA is released from its circular (c-GS) partner, the relative binding stability of circular and linear duplexes was investigated through gel shift assay. As shown in Figure S5, 5ʹ fluorescein isothiocyanate (FITC)-labeled 21-mer DNA (DNA-FITC: GTT CAC CTT GAT GCC GTT CTT-5ʹ FITC) at the fixed concentration was annealed with a set of increasing concentrations of single-stranded complementary circular (or linear) RNA, with the ratio of RNA to DNA-FITC varying from 0.25 to 8. The amount of DNA-FITC in the free and bound RNA bands was then quantified. The binding data were fit to determine the equilibrium dissociation constant Kd of target DNA-FITC with circular and linear RNAs using a hyperbola function by the non-linear curve fitting method of GraphPad PRISM.33 The affinity of circular RNA for probe DNA-FITC (Kd = 1.2 × 10−5 mol/L) was ∼17-fold weaker than for the linear RNA-DNA-FITC duplex (Kd = 7.1 × 10−7 mol/L), as evidenced by the easy displacement of the circular RNA by a competing linear complementary strand. These results were consistent with the higher thermostability of the linear duplex than the corresponding circular duplex, which made it more favorable for the complementary linear antisense strand to leave its circular partner and bind to the linear mRNA target. The binding switch suggested that circular siRNA with linear antisense strand RNA was still capable of forming the RISC complex with mRNA to knock down the target gene. The gel shift assay also showed that the circular RNA and oligonucleotide probe (c-GS/DNA-FITC) form two bands on the native PAGE gel (Figure S5). This suggested that the circular siRNA duplex adopts two conformers (e.g., kinked and fork conformations), which were similar to recently proposed structures of circular double-stranded DNA (dsDNA).34
Evaluation of Firefly Luciferase Gene Expression Regulation with Circular siRNAs Containing a Different Length of Complementary Linear RNAs
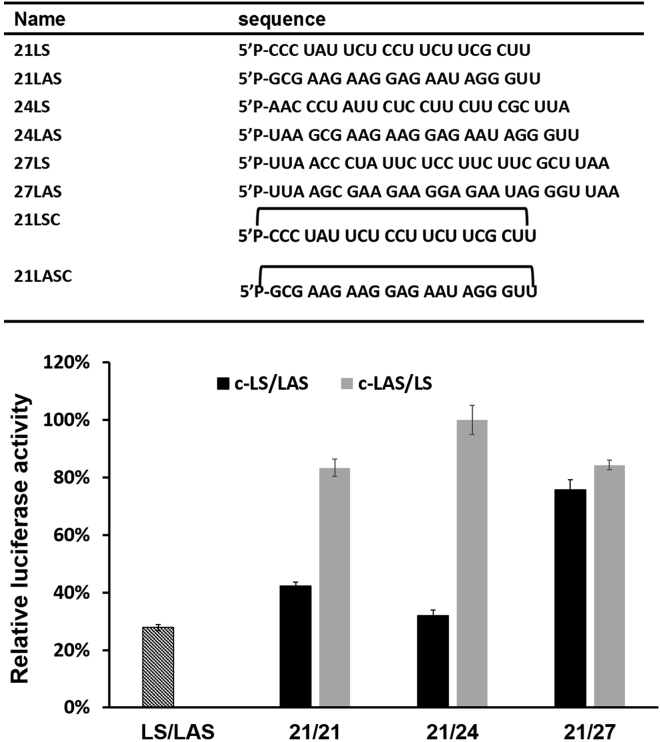
Based on the observations above, only the sense strand circular siRNA (c-S/AS) with a linear antisense RNA can be used to efficiently knock down gene expression and reduce off-target effects caused by sense strand RNA. To test the generality of gene knockdown with our circular siRNAs, we designed two sets of circular siRNAs for targeting the firefly luciferase gene with the Renilla luciferase gene as an internal control,35 and their gene silencing potencies were evaluated (Figure 3). One set was prepared with the circular sense strand RNA (c-21LS, L for firefly luciferase), with different lengths of complementary linear antisense RNA (21LAS, 24LAS, and 27LAS). The other set was prepared with the circular antisense strand RNA (c-21LAS), with a different length of linear sense strand RNAs (21LS, 24LS, and 27LS). All circular siRNA duplexes were formed with two conformers, corresponding to two bands observed in native PAGE analysis (Figure S6). These siRNA duplexes were then transfected into HEK293A cells together with a firefly luciferase reporter vector (FireflyXs) and Renilla luciferase vector (Renilla). 24 hr after transfection, the cells were lysed to quantify the levels of both luciferases and then the firefly luciferase level was normalized to Renilla luciferase for each well.
Figure 3.
RNA Sequences for Firefly Luciferase and Knock Down of Luciferase Expression in HEK293A Cells with Different Compositions of Circular siRNAs (c-21LS/21LAS, c-21LS/24LAS, c-21LS/27LAS, c-21LAS/21LS, c-21LAS/24LS, and c-21LAS/27LS) Using the Vector Renilla as Control
Mean values and SD values were performed in triplicate.
As expected, siRNAs with circular antisense strand RNA and different lengths of linear sense strand RNA did not show any knockdown of firefly luciferase, whereas siRNAs with circular sense strand RNA and different lengths of linear antisense RNAs showed gene-silencing activity of firefly luciferase in different degrees (Figure 3). By comparing circular siRNAs (c-21LS/21LAS and c-21LS/24LAS) with 21-mer and 24-mer linear antisense strand RNAs, we found that circular siRNA (c-21LS/24LAS) with the combination of 21-mer circular sense strand and 24-mer antisense strand RNAs showed better gene-silencing activity than circular siRNA (c-21LS/21LAS) with both 21-mer RNAs. This observation was probably due to the fact that the slightly longer complementary antisense strand RNA could compete to bind target mRNA more tightly and could still form an effective RISC complex with mRNA and Ago 2. 27-mer antisense RNA of the siRNA duplex (c-21LS/27LAS) should more readily compete to bind mRNA than the circular sense strand RNA partner. However, 27LAS that forms a duplex with mRNA is likely too long to stably associate with Ago 2 for further mRNA degradation36, 37 and may function more as an antisense inhibitor. These experimental data helped us to choose the optimized combination (c-21LS/24LAS and c-21LAS/24LS) for measuring on- versus off-target activity of the different siRNAs (Figure 3).
Evaluation of On- versus Off-Target Effects with Linear and Circular siRNAs
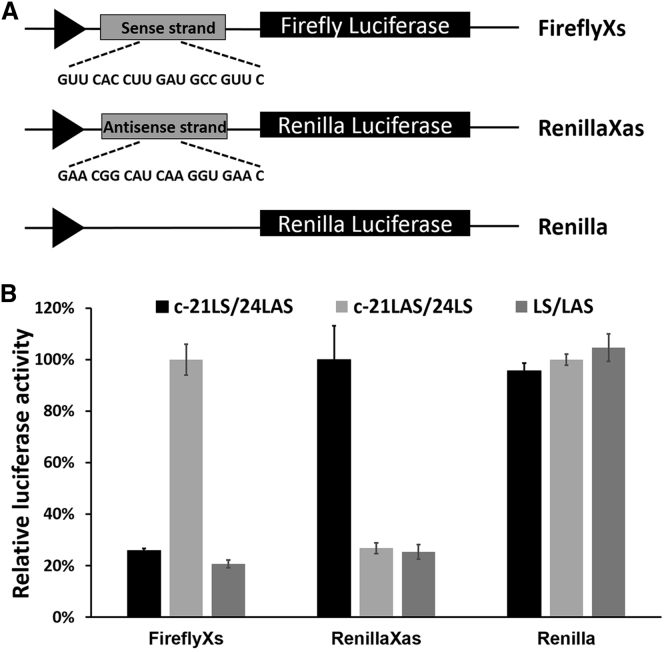
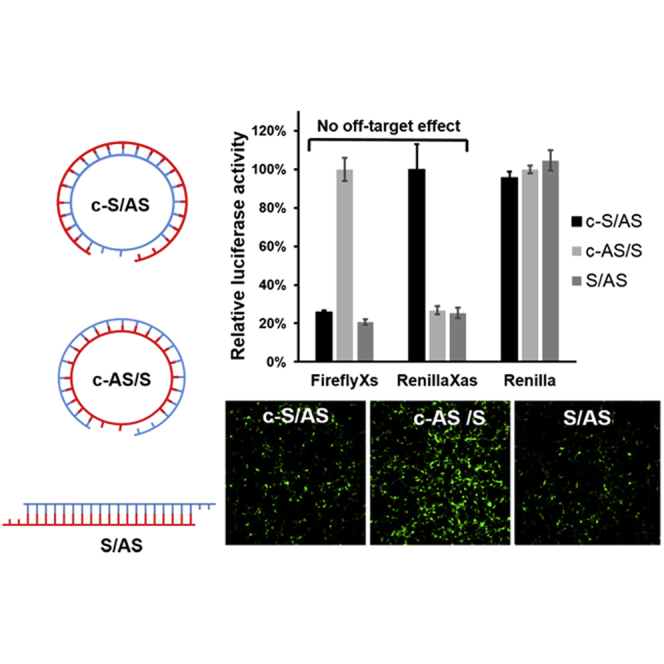
In order to investigate specific on- versus off-target effects with linear and circular siRNA, a new plasmid DNA (RenillaXas) was rationally designed and constructed (Figure 4A). This fusion reporter vector (RenillaXas) carries a target site for the sense strand of siRNA targeting FireflyXs. The circular siRNA (c-21LS/24LAS or c-21LAS/24LS) duplexes were transfected to cultured HEK293A cells together with FireflyXs luciferase vector, Renilla luciferase vector, and RenillaXas luciferase vector, respectively. As expected, circular siRNA (c-21LS/24LAS) with the linear antisense strand (24LAS) effectively knocked down FireflyXs gene expression but did not silence gene expression of Renilla and RenillaXas. Meanwhile, the circular siRNA (c-21LAS/24LS) with linear sense strand RNA (24LS) did not silence gene activity for FireflyXs and Renilla, but it was capable of knocking down expression of RenillaXas that has the linear sense strand (24LS) as the guide strand RNA (Figure 4B). Through cyclization of the sense strand in the siRNA duplex, only the linear antisense strand was available for loading into the RISC complex, thereby greatly reducing off-target effects. However, both FireflyXs and RenillaXas genes were silenced with transfection of linear siRNA (AS/LAS).
Figure 4.
Sense and Antisense Strand Activity of Linear siRNA and Circular siRNAs
(A) The reporter plasmids used to assay siRNA activity. The RenillaXas plasmid carries a target site for the sense strand of siRNA targeting FireflyXs. The control Renilla plasmid was cloned with no siRNA target. (B) Activity of linear siRNA and circular siRNAs (c-21LS/24LAS and c-21LAS/24LS) against sense (FireflyXs), antisense (RenillaXas), or control (Renilla). Mean and SD values are from experiments performed in triplicate.
Evaluation of GFP Gene Expression Regulation In Vivo with Circular siRNA
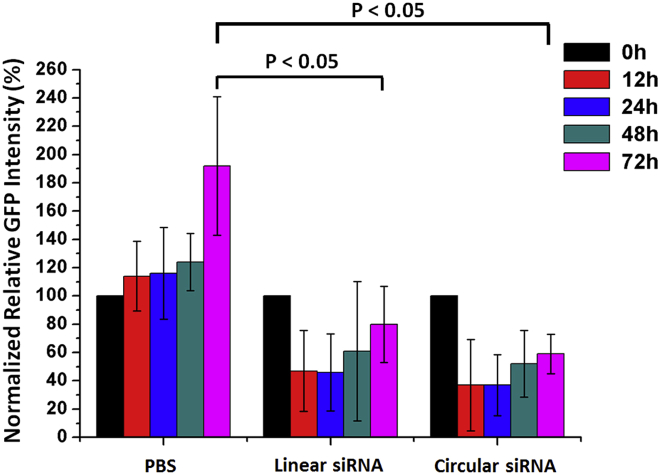
In addition to cell studies, we further applied circular siRNA for gene silencing in mice. An in vivo xenograft tumor model with U87-GFP cells was established. When the tumors with U87-GFP cells grew to ∼0.2 cm in diameter, PBS or linear/circular siRNAs were injected into the tumors, respectively. The mice were imaged, and fluorescence intensity of tumors was quantified at different time points (Figures 5, S7, and S8). As expected, GFP gene expression in tumors increased for PBS-injected mice. The mice injected with linear siRNAs observed a predicted fluorescent decrease in tumors, but the fluorescence intensity gradually recovered over 72 hr (p < 0.05). In comparison, mice tumors with circular siRNA injection downregulated GFP expression more efficiently and the value of GFP fluorescence intensity recovered more slowly (p < 0.05). Specifically, after single intratumoral injection of corresponding siRNAs, circular siRNA decreased the GFP gene expression level to a significant extent, with 59% remaining GFP fluorescence intensity in comparison to that of 80% for linear siRNA at 72 hr. If normalized to the GFP level of the PBS control group at 72 hr (Figure S9), 31% and 42% of remaining GFP level in mice tumors at 72 hr were achieved for circular siRNA and linear siRNA, respectively. Both data indicated that circular siRNA with linear antisense strand of RNA has a longer gene silencing effect for single siRNA injection. All these results indicated that the circular siRNA strategy has great potential to be developed into a new kind of siRNA drug to regulate gene expression, with a better effect in the future.
Figure 5.
Normalized GFP Fluorescence Intensity of Tumors (U87-GFP) after Single Injection of PBS, Linear siRNA, and Circular siRNA to Show the Long-Term Effect of GFP Gene Silencing with Circular siRNAs in Tumors of Mice
Mean and SD values are from experiments performed in 3–6 mice for each group.
Conclusions
We rationally designed and developed siRNA with a new circular structural motif. By transfection of circular siRNAs targeting a GFP or luciferase reporter gene into HEK293A cells, gene expression was efficiently silenced by siRNA with a circular sense strand and linear antisense strand, whereas the inverse constructs with circular antisense strand and linear sense strand did not induce RNAi gene silencing. We further confirmed that circular siRNAs could substantially reduce the off-target effect caused by the sense strand of siRNA using a reconstructed Renilla luciferase reporter gene RenillaXas. Further investigation on the duplex formation of a circular RNA with its complementary RNA strand indicated two exchanging conformations of the circular siRNA duplex. The thermodynamic stability of the circular siRNA duplex is ∼17-fold lower than the corresponding linear siRNA duplex, which should promote mRNA binding and trigger the catalytic RNAi machinery to silence gene expression. In addition, the tumor model study indicated that slow recovery of the GFP fluorescence signal using circular siRNA was observed and the gene-silencing effect in mice was achieved with 59% remaining GFP level for circular siRNAs in comparison to 80% for linear siRNAs in 72 hr for a single siRNA injection. This study highlights the transformative potential of siRNAs with a circular structure, which increase their stability against nucleases and overcome off-target effects caused by sense strand RNA while still achieving efficient RNAi gene silencing.
Materials and Methods
General Methods for Circular RNA Synthesis
A normal universal controlled pore glass (CPG) was used for RNA oligonucleotide synthesis. All the single-stranded oligonucleotides were synthesized according to the standard RNA synthesis on ABI394 with 2-tert-butyldimethylsilyl (TBDMS) RNA monomers. DNA primer oligonucleotides for construction of DNA plasmids were purchased from Sangon Biotech (Shanghai, China); unmodified RNA oligonucleotides were purchased from GenePharma (Shanghai, China). The concentrations of all single-stranded oligonucleotides were measured at 260 nm using a Thermo Scientific NanoDrop 2000 Spectrophotometer. High-performance liquid chromatography (HPLC) was performed on a Waters Alliance e2695 system equipped with a Waters Xbridge OST C18 column (2.5 μm, 10.0 × 50 mm). MS data were obtained with a Waters Xevo G2 Q-TOF spectrometer using ESI. Diethyl pyrocarbonate (DEPC)-treated water was used for all solutions and HPLC purification.
Synthesis and Purification of Circular RNAs
Following the procedures shown in Figure S1, Chemical Phosphorylation Reagent II (CPR II) was coupled at the 5′ terminal of RNA sequences according to the procedure of normal RNA synthesis at ABI 394 RNA synthesis. The dimethoxyltrityl (DMT) group was saved for better purification using DMT-on mode. Synthesized oligonucleotides were then cleaved from CPG and deprotected using 0.3 mL concentrated ammonium hydroxide at room temperature for 24 hr. The solution was then centrifuged at 2,500 rpm for 5 min to remove CPG. The supernatant was collected and concentrated using a Thermo Scientific Savant SPD2010 SpeedVac Concentrator. The obtained residue was redissolved in 100 μL DMSO, and then 100 μL TEA‧3HF was added to remove the TBDMS-protecting group. After the mixture was shaken at 65°C for 2 hr for quick deprotection, the solution was cooled in an ice bath and isopropoxytrimethylsilane (300 μL) was added to consume extra fluoride. Butyl alcohol (1.0 mL) and NaOAc (3 M, 50 μL) was then added to the above solution, and the mixed solution was further stored at −80°C for 1 hr. After centrifugation and removal of supernatant, the precipitated white solid was washed with ether twice and collected for further purification.
The above white solid dissolved in DEPC water was subject to HPLC purification using a reverse-phase HPLC column (C18) under the following conditions: buffer A, 0.1 M TEAB (0.05 M triethylammonium bicarbonate buffer, pH 8.5); buffer B, acetonitrile; 0%–50% buffer B in 30 min and 50%–100% buffer B in 5 min; and running temperature, 60°C. Due to the quick deprotection condition, most of the DMT group was saved, which made the required oligonucleotide quite different in elution time from the shorter side effect oligonucleotides during the HPLC separation. The main peak was collected and the solution was concentrated using a Thermo Scientific Savant SPD2010 SpeedVac Concentrator. The above separated oligonucleotides (∼10 nmol) were dissolved in 400 μL 20% acetic acid solutions at 25°C for 1 hr to remove the DMT group. The oligonucleotide was dried down and then ammonium hydroxide was added. It was left at room temperature for 15 min to achieve complete elimination of the side chain to the 5′-phosphate of RNA oligonucleotide. The solution was concentrated using a concentrator, and 5′-phosphate single-strand RNA was obtained.
The solid was dissolved in water to make the final 100 μM single-strand RNA solution. The final composition of the reaction mixture to circularize the single-strand RNA was as follows: 7 μL RNA solution, 1 μL 10 mM ATP, 1 μL 10 × reaction buffer, and 1 μL T4 RNA ligase (10 U/μL). The solution 10 μL/tube was placed in PCR at 4°C for 12 hr. After mixing these liquids together, the crude products were mixed with 6 × RNA loading buffer (0.25% bromophenol blue and 30% glycerol in DEPC-treated water). The solution (18 μL/well) was loaded into 20% native polyacrylamide PAGE (1 mm thick) gels. The gels were then electrophoresed at 220 V for 50 min using 1 × Tris-borate-EDTA (TBE) buffer (pH 8.2). For each preparative gel, the two-side sample lanes of the gel were cut and stained with 1 × SYBR Gold (Invitrogen) and then imaged. The images were printed out according to the same size of the gel, which made it possible to mark the location of the gels without SYBR Gold stain. The gel zones at the marked location were cut, crumbled into tiny particles, and immersed into a 1 × TBE buffer at 37°C overnight. After filtration out of the solid particles, the solutions of the RNA were desalted and concentrated using Millipore-Amicon Ultra-0.5 mL Centrifugal Filters (cutoff = 3,000). The collected products were frozen dried to remove water and gave the final circular single-stranded RNAs.
RNA oligonucleotides were characterized using ESI-MS, and single-stranded oligonucleotides (∼0.2 nmol) were dissolved in water/acetonitrile (50:50, 20 μL) containing 1% triethylamine to make a final concentration of 10 μM. The solutions were then analyzed with a Waters Xevo G2 Q-Tof spectrometer with ESI in the negative ion mode (Figure S10). The molecular weight of the circular RNA was 18 less than the linear one due to the condensation reaction.
The circular single-stranded RNAs were dissolved in 1 × PBS buffer to make the 6-μmol stock solution. A 10-μL stock solution was mixed with an equal amount of the complementary RNA with 5′-phosphate modification to form the circular siRNA. The siRNA was annealed by heating to 85°C for 5 min and was subsequently cooled to room temperature for at least 1 hr for further use.
Gel Shift Analysis
Gel shift analysis was designed to determine the binding ability of DNA to circular single-stranded RNA. A fixed concentration of 5′-FITC-modified 21-mer DNA (DNA-FITC: 5′ FITC-GUUCACCUUGAUGCCGUUCUU) with increased concentrations of circular single-stranded RNA was used in this experiment. A set of DNA-FITC with increased concentrations of linear single-stranded RNA was used as control. The duplex was annealed using the condition mentioned before for siRNA. The ratio of RNA/DNA-FITC was from 0.25 to 8. The samples were mixed with 6 × RNA loading buffer and loaded into 20% native polyacrylamide PAGE gels. The gels were then electrophoresed at 220 V for 50 min using TBE buffer. A GE Healthcare Life Sciences Typhoon FLA 9500 image system was used for the gel imaging at 473-nm laser excitation.
Enzymatic Stability of Circular siRNA
Circular siRNA or control linear siRNA (3 μM, 5 μL) was incubated at 37°C in an enzyme solution to make a final concentration of 1 μmol/L (15 μL). RNase A (Beyotime Institute of Biotechnology, China) was used for enzymatic stability in this study, respectively. Aliquots of 3 μL (containing 3 pmol siRNA) were aliquoted at different time points (2, 4, 6, and 8 hr) and immediately frozen in liquid nitrogen and then stored at −80°C until assayed. 1 μL 6 × RNA-loading buffer was added to the aliquots. The samples were run on 10% native polyacrylamide gels in TBE buffer according to the procedure mentioned before.
Oligonucleotides and Fusion Reporter Construction
To evaluate the gene-silencing potency of an siRNA, a previously reported siQuant luciferase reporter vector was used. The siRNA (PC) is the selected siRNA with good activity in this assay. The siQuant luciferase reporter assay contains two vectors. One is the siQuant Vector, which contained cDNA encoding the firefly luciferase. The antisense strand of siRNA (21LS/21LAS) could shut down the expression of firefly luciferase through the RNAi pathway. The other vector is the pRL-TK vector, which contained cDNA encoding the Renilla luciferase. Gene expression of this vector had not been affected by siRNA (21LS/21LAS) and was used as internal control.
To evaluate the on- or off-target effects of siRNA, a target sequence of the sense strand of siRNA (21LS/21LAS) was inserted immediately after the start codon of the Renilla luciferase gene at a Nhe I-Csp45 I cloning site. The target of the sense strand GCGAAGAAGGA GAATAGGG was inserted to the pRL-TK vector after the T7 promoter. A fusion reporter vector (RenillaXas) carrying the target site of the sense strand of siRNA (21LS/21LAS) was constructed.
Cell Culture and siRNA Transfection
HEK293A cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Gibco). The cells were seeded into 24-well plates at ∼1 × 105 cells/well 1 day before transfection. siQuant vector (100 ng/well) carrying the target site of the siRNA was transfected into HEK293 cells at ∼70% confluence, together with pRL-TK control vector (50 ng/well), with or without the control siRNAs or circular siRNAs in duplex form using Lipofectamine 2000 Transfection Reagent (Invitrogen). The activity of both luciferases was determined by a Synergy HT fluorometer (BioTek, USA) and then the firefly luciferase activity was normalized to Renilla luciferase for each well. For off-target effect detection of sense strand RNA (50 ng/well), RenillaXas was used. All experiments were performed in triplicate and repeated at least twice.
For GFP gene expression experiments, the cells were seeded into 6-well plates for the flow cytometry test and a glass bottom dish (35-mm dish with 20-mm bottom well) for confocal imaging. For each transfection, 400 ng pEGFP-N1, 400 ng pDsRed2-N1, and 12 pmol siRNA (circular or linear siRNAs) were mixed with 200 μL OptiMEM for 5 min. Then, the solution was mixed with 200 μL OptiMEM containing 4 μL Lipofectamine 2000 for 20 min. The solution was added to the cells in wells/dishes, giving a final volume of 2.4 mL. Cells were incubated for 4 hr in the transfection solution, and then the culture solution was replaced with fresh DMEM for 24-hr incubation.
Confocal Microscopy
The confocal microscopic images were taken using a Nikon confocal laser scanning microscope (A1R). All images were taken using a 10 × objective lens and charge-coupled device (CCD) camera. The Hoechst 33342 fluorescence was excited at a 405-nm wavelength, GFP fluorescence was excited at 488 nm, and RFP fluorescence was excited at 561 nm. All image analysis used the NIS-Elements viewer.
Flow Cytometry
Quantification of cells with both GFP and RFP gene expression was taken using Beckman Coulter CytoFLEX Flow cytometry. In order to define the cells were transfected by both the GFP and RFP plasmids, the mean value of GFP fluorescence intensity in the upper-right quadrant, in which the cells were well transfected with the GFP and RFP plasmids, was used to find the difference between the circular siRNA and normal siRNA. CytExpert was used for all sample analyses (Figure S4).
Xenograft Tumor Fluorescent Assays
6-week-old BALB/c nude mice (Department of Laboratory Animal Science of the Peking University Health Science, Beijing, China) were subcutaneously injected in the inner thighs with U87-GFP cells (6 × 105 cells per site in a volume of 60 μL). When the tumors grew to around 0.2 cm in diameter (about 4 days), the mice were randomly divided into two groups for intratumoral injection. One group was injected with PBS on the right and linear siRNAs on the left, respectively. The other group was injected with linear siRNAs on the right and circular siRNAs on the left, respectively. For each injection, 3 nmol linear siRNA or circular siRNA (60 μL) was mixed with 20 μL transfection reagent (Entranster-in vivo; Engreen, Beijing, China) and incubated for 15 min at room temperature. All injections were in accordance with the manufacturer’s instructions. After single injection, the mice were imaged and tumor fluorescence was quantified at different time points (0/12/24/48/72 hr) using the Maestro Automated In-Vivo Imaging system.
Author Contributions
The manuscript was written through contributions of all authors. L.Z., D.L., C.C., Y.W., G.A., J.Y., and L.Y. conducted the experiments and data analysis. I.J.D. and X.T. designed the experiments, analyzed data, and wrote the paper. All authors have given approval to the final version of the manuscript.
Conflicts of Interest
No conflicts of interest need to be disclosed.
Acknowledgments
We thank Dr. Bo Xu and Ms. Yufang Sun (State Key Laboratory of Natural and Biomimetic Drugs) for help with cell imaging and flow cytometry. The work was supported by the National Natural Science Foundation of China (21422201, 21372018, and 21672015) and National Major Scientific and Technological Special Project for “Significant New Drugs Development” (Grant No. 2017ZX09303013).
Footnotes
Supplemental Information includes eleven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.007.
Supplemental Information
References
- 1.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 2.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 3.Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson B.L., McCray P.B., Jr. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 6.Clark P.R., Pober J.S., Kluger M.S. Knockdown of TNFR1 by the sense strand of an ICAM-1 siRNA: dissection of an off-target effect. Nucleic Acids Res. 2008;36:1081–1097. doi: 10.1093/nar/gkm630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramsen J.B., Pakula M.M., Hansen T.B., Bus C., Langkjær N., Odadzic D., Smicius R., Wengel S.L., Chattopadhyaya J., Engels J.W. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elmén J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., Wahren B., Liang Z., Ørum H., Koch T. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaish N., Chen F., Seth S., Fosnaugh K., Liu Y., Adami R., Brown T., Chen Y., Harvie P., Johns R. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler J., Stessl M., Amartey J., Noe C.R. Off-target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem. 2010;5:1344–1352. doi: 10.1002/cmdc.201000156. [DOI] [PubMed] [Google Scholar]
- 12.Chen P.Y., Weinmann L., Gaidatzis D., Pei Y., Zavolan M., Tuschl T., Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Zheng J., Lu C., Du Q., Liang Z., Xi Z. Modification of the siRNA passenger strand by 5-nitroindole dramatically reduces its off-target effects. ChemBioChem. 2012;13:1940–1945. doi: 10.1002/cbic.201200349. [DOI] [PubMed] [Google Scholar]
- 14.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 15.Hong S.W., Park J.H., Yun S., Lee C.H., Shin C., Lee D.K. Effect of the guide strand 3′-end structure on the gene-silencing potency of asymmetric siRNA. Biochem. J. 2014;461:427–434. doi: 10.1042/BJ20140407. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Z., Wu X., Liu C., Xu G., Wu Z. Asymmetric siRNA: new strategy to improve specificity and reduce off-target gene expression. Hum. Gene Ther. 2012;23:521–532. doi: 10.1089/hum.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C.I., Yoo J.W., Hong S.W., Lee S.E., Kang H.S., Sun X., Rogoff H.A., Ban C., Kim S., Li C.J. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol. Ther. 2009;17:725–732. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X., Rogoff H.A., Li C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 19.Bramsen J.B., Laursen M.B., Damgaard C.K., Lena S.W., Babu B.R., Wengel J., Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei L., Cao L., Xi Z. Highly potent and stable capped siRNAs with picomolar activity for RNA interference. Angew. Chem. Int. Ed. Engl. 2013;52:6501–6503. doi: 10.1002/anie.201301122. [DOI] [PubMed] [Google Scholar]
- 21.Abe N., Abe H., Nagai C., Harada M., Hatakeyama H., Harashima H., Ohshiro T., Nishihara M., Furukawa K., Maeda M. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjug. Chem. 2011;22:2082–2092. doi: 10.1021/bc2003154. [DOI] [PubMed] [Google Scholar]
- 22.Abe N., Abe H., Ito Y. Dumbbell-shaped nanocircular RNAs for RNA interference. J. Am. Chem. Soc. 2007;129:15108–15109. doi: 10.1021/ja0754453. [DOI] [PubMed] [Google Scholar]
- 23.Shah S., Friedman S.H. Tolerance of RNA interference toward modifications of the 5′ antisense phosphate of small interfering RNA. Oligonucleotides. 2007;17:35–43. doi: 10.1089/oli.2006.0067. [DOI] [PubMed] [Google Scholar]
- 24.Su M., Wang J., Tang X. Photocaging strategy for functionalisation of oligonucleotides and its applications for oligonucleotide labelling and cyclisation. Chemistry. 2012;18:9628–9637. doi: 10.1002/chem.201103833. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Wu L., Wang P., Lv C., Yang Z., Tang X. Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res. 2012;40:11155–11162. doi: 10.1093/nar/gks840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L., Wang Y., Wu J., Lv C., Wang J., Tang X. Caged circular antisense oligonucleotides for photomodulation of RNA digestion and gene expression in cells. Nucleic Acids Res. 2013;41:677–686. doi: 10.1093/nar/gks996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang X., Zhang J., Sun J., Wang Y., Wu J., Zhang L. Caged nucleotides/nucleosides and their photochemical biology. Org. Biomol. Chem. 2013;11:7814–7824. doi: 10.1039/c3ob41735b. [DOI] [PubMed] [Google Scholar]
- 28.Tang X. Photochemical biology of caged nucleic acids. In: Albini A., Fasani E., editors. Photochemistry. Royal Society of Chemistry; 2013. pp. 319–341. [Google Scholar]
- 29.Richards J.L., Seward G.K., Wang Y.-H., Dmochowski I.J. Turning the 10-23 DNAzyme on and off with light. ChemBioChem. 2010;11:320–324. doi: 10.1002/cbic.200900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazoe S., Shestopalov I.A., Provost E., Leach S.D., Chen J.K. Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem. Int. Ed. Engl. 2012;51:6908–6911. doi: 10.1002/anie.201201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe N., Abe H., Ohshiro T., Nakashima Y., Maeda M., Ito Y. Synthesis and characterization of small circular double-stranded RNAs. Chem. Commun. (Camb.) 2011;47:2125–2127. doi: 10.1039/c0cc04551a. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 33.Tahiri-Alaoui A., Frigotto L., Manville N., Ibrahim J., Romby P., James W. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res. 2002;30:e45. doi: 10.1093/nar/30.10.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C., Lee O.C., Kim J.Y., Sung W., Lee N.K. Dynamic release of bending stress in short dsDNA by formation of a kink and forks. Angew. Chem. Int. Ed. Engl. 2015;54:8943–8947. doi: 10.1002/anie.201502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M., Zhang L., Zhang H.Y., Xiong X., Wang B., Du Q., Lu B., Wahlestedt C., Liang Z. A universal plasmid library encoding all permutations of small interfering RNA. Proc. Natl. Acad. Sci. USA. 2005;102:2356–2361. doi: 10.1073/pnas.0401549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.