Abstract
Interleukin-1 receptor 8 (IL-1R8, also known as single immunoglobulin IL-1R-related receptor, SIGIRR, or TIR8) is a member of the IL-1 receptor (ILR) family with distinct structural and functional characteristics, acting as a negative regulator of ILR and Toll-like receptor (TLR) downstream signaling pathways and inflammation1. NK cells are innate lymphoid cells which mediate resistance against pathogens and contribute to the activation and orientation of adaptive immune responses2–4. NK cells mediate resistance against hematopoietic neoplasms but are generally considered to play a minor role in solid tumor carcinogenesis5–7. Here we report that IL-1R8 serves as a checkpoint for NK cell maturation and effector function. Its genetic blockade unleashes NK-cell mediated resistance to hepatic carcinogenesis, hematogenous liver and lung metastasis and cytomegalovirus infection.
Several lines of evidence suggest that IL-1R8 interferes with the association of TIR module-containing adaptor molecules with signaling receptor complexes of the ILR or TLR family, tuning downstream signaling, thus negatively controlling inflammatory and immune responses and T helper (TH) cell polarization and functions1,8. Moreover, IL-1R8 is the co-receptor of IL-1R5/IL-18Rα for IL-37, and is required for the anti-inflammatory activity of this human cytokine9. Deregulated activation by ILR or TLR ligands in IL-1R8-deficient mice has been associated with exacerbated inflammation and immunopathology, including selected cancers, or autoimmune diseases10.
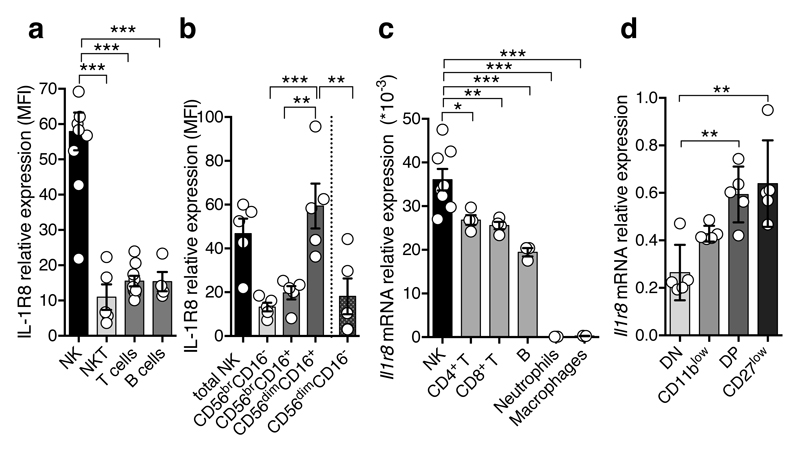
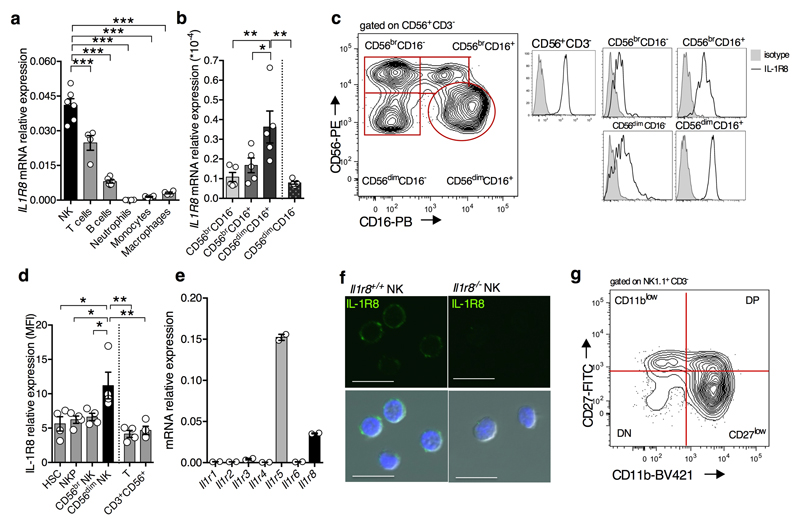
IL-1R8 is widely expressed10. However, we found strikingly high levels of IL-1R8 mRNA and protein in human NK cells, compared to other circulating leukocytes and monocyte-derived macrophages (Fig. 1a, Extended Data Fig. 1a). IL1R8 mRNA levels increased during NK cell maturation11 (Extended Data Fig. 1b) and surface protein expression mirrored transcript levels (Fig. 1b, Extended Data Fig. 1c). IL-1R8 expression was detected at low level in bone marrow pluripotent haematopoietic stem cells and NK cell precursors and was selectively upregulated in mature NK cells and not in CD3+ lymphocytes (Extended Data Fig. 1d).
Figure 1. Expression of IL-1R8 in human and murine NK cells.
(a, b) IL-1R8 protein expression in human primary NK cells and other leukocytes (a) and NK cell maturation stages (b).
(c, d) Il-1r8 mRNA expression in murine primary NK cells and other leukocytes (c) and in sorted splenic NK cell subsets (c).
*p < 0.05, **p < 0.01, ***p < 0.001 One-way ANOVA. Mean ± SEM.
Murine NK cells expressed significantly higher levels of Il1r8 mRNA, compared to other leukocytes (Fig. 1c) and relative to other ILRs (Extended Data Fig. 1e, 1f). In line with the results obtained in human NK cells, Il1r8 mRNA level increased during the 4-stage developmental transition from CD11blowCD27low to CD11bhighCD27low12 (Fig. 1d, Extended Data Fig. 1g).
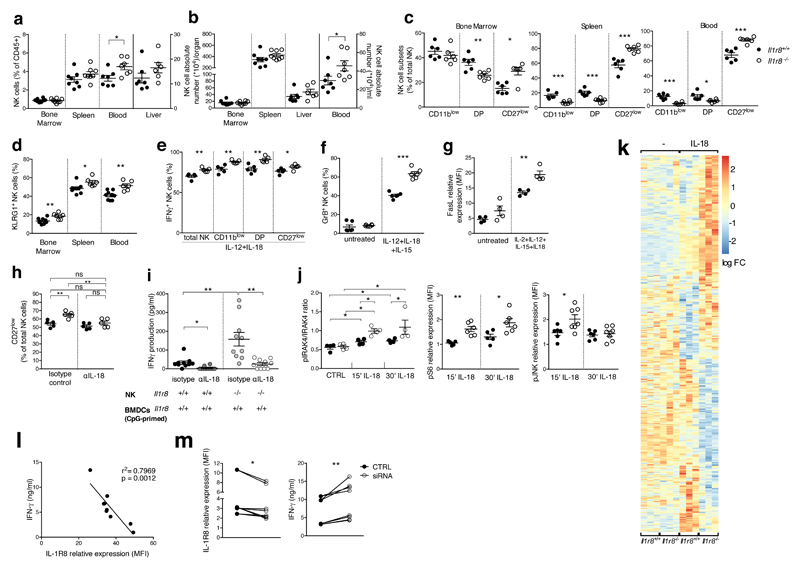
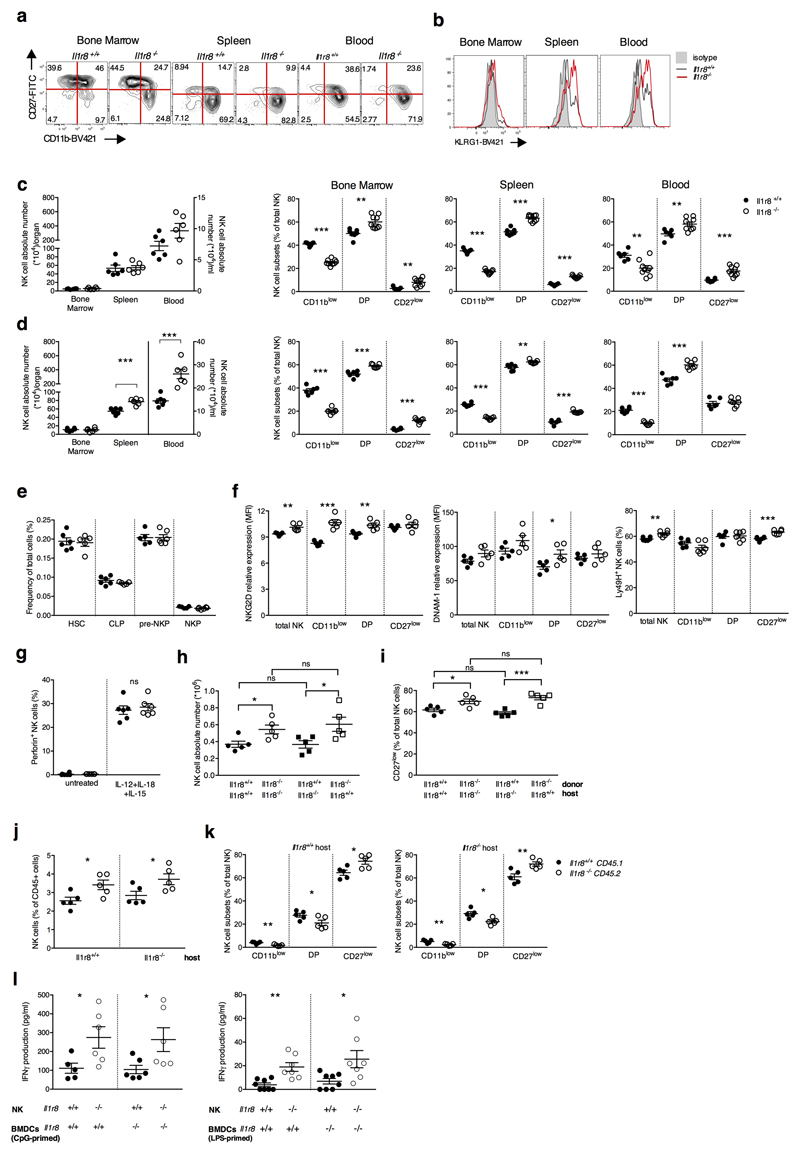
To assess the role of IL-1R8 in NK cells, we took advantage of IL-1R8-deficient mice. Among CD45+ cells, the NK cell frequency and absolute numbers were significantly higher in peripheral blood of Il1r8-/- compared to Il1r8+/+ mice and slightly increased in liver and spleen. (Fig. 2a, 2b). In addition, the frequency of the CD11b high CD27low and KLRG1+ mature subset was significantly higher in Il1r8-/- mice compared to Il1r8+/+ mice in BM, spleen and blood, indicating a more mature phenotype of NK cells13 (Fig. 2c, 2d, Extended Data Fig. 2a, 2b).
Figure 2. NK cell differentiation and function in IL-1R8-deficient mice.
(a, b) NK cell frequency and absolute number among leukocytes in Il1r8+/+and Il1r8-/- mice.
(c, d) NK cell subsets (c) and KLRG1+ NK cells (d).
(e-g) IFNγ (e), Granzyme B (f) and FasL (g) expression in stimulated NK cells.
(h) Splenic CD27low NK cell frequency upon IL-18 in vivo depletion.
(i) IFNγ production by Il1r8+/+ and Il1r8-/- NK cells upon co-culture with CpG-primed Il1r8+/+ DCs and IL-18 blockade.
(j) IRAK4, S6 and JNK phosphorylation in NK cells upon stimulation with IL-18.
(k) RNA-seq analysis of resting and IL-18-activated NK cells. Differentially expressed (p<0.05) genes are shown. FC: fold change.
(l) Correlation between IL-1R8 expression and IFNγ production in human peripheral blood NK cells.
(m) IL-1R8 expression and IFNγ production in human NK cells 7 days after transfection with control siRNA or IL-1R8-specific siRNA in duplicate.
(a-l) *p < 0.05, **p < 0.01, ***p < 0.001 between selected relevant comparisons, two-tailed unpaired Student’s t test or Mann-Whitney test; (k) r: Pearson correlation coefficient; Mean ± SEM.
The enhanced NK cell maturation in Il1r8-/- mice occurred already at 2 and 3 weeks of age, whereas the frequency of NK precursors was similar in Il1r8-/- and Il1r8+/+ BM, indicating that IL-1R8 regulated early events in NK cell differentiation, but did not affect the development of NK cell precursors (Extended Data Fig. 2c-e)12.
We next investigated whether IL-1R8 impacted on NK cell function. The expression of the activating receptors NKG2D, DNAM-1 and Ly49H was significantly upregulated in peripheral blood Il1r8-/- NK cells (Extended Data Fig. 2f). IFNγ and Granzyme B production and FasL expression were more sustained in IL-1R8-deficient NK cells upon ex-vivo stimulation in the presence of IL-18 (Fig. 2e-g, Extended Data Fig. 2g). The frequency of IFNγ+ NK cells was higher in Il1r8-/- total NK cells and in all NK cell subsets. Thus, IFNγ production was enhanced independently of the NK cell maturation state. Analysis of competitive bone marrow chimeras revealed that IL-1R8 regulates NK cell differentiation in a cell-autonomous way (Extended Data Fig. 2h-k). Along the same line, co-culture experiments of NK cells with LPS- or CpG-primed dendritic cells (DCs) showed that Il1r8-/- NK cells produced higher IFNγ levels irrespectively of the DC genotype (Extended Data Fig. 2l).
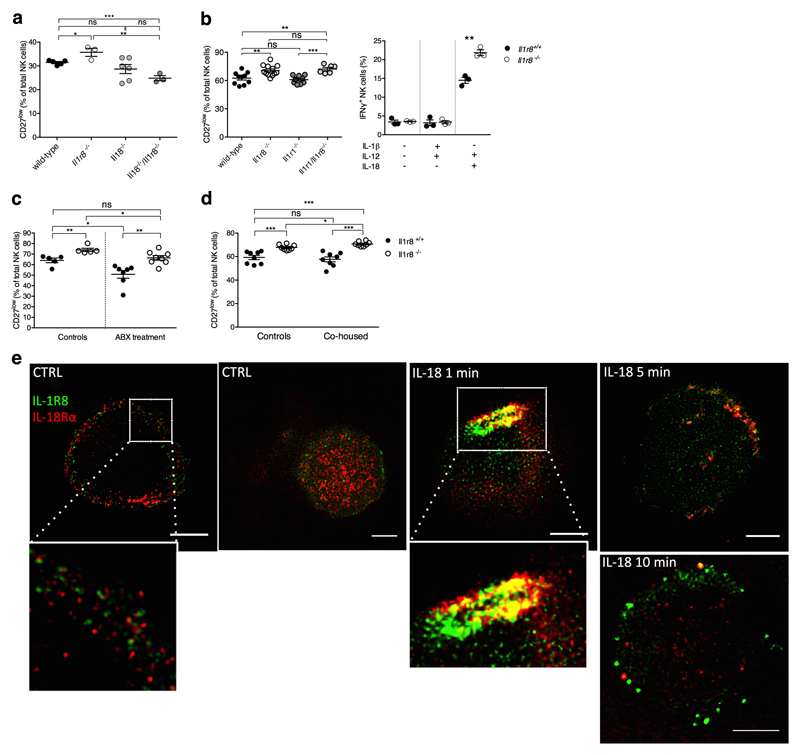
IL-18 is a member of the IL-1 family, which plays an important role in NK cell differentiation and function1,14. Enhanced NK cell maturation and effector function in Il1r8-/- mice was abolished by IL-18 blockade or genetic deficiency but unaffected by IL-1R1-deficiency (Fig. 2h, 2i, Extended Data Fig. 3a, 3b). Cohousing and antibiotic treatment had no impact, thus excluding a role of microbiota15 in the phenotype of Il1r8-/- mice (Extended Data Fig. 3c, 3d).
The results reported above suggested that IL-1R8 regulated the IL-18 signaling pathway in NK cells and indeed, increased phospho-IRAK4/IRAK4 ratio was induced by IL-18 in Il1r8-/- NK cells compared to wild type NK cells, indicating unleashed early signaling downstream of MyD88 and myddosome formation (Fig. 2j), consistently with the proposed molecular mode of action of IL-1R81,9,16. Indeed, by stimulated emission depletion (STED) microscopy, we observed clustering of IL-1R8 and IL-18Rα (Extended Data Fig. 3e), in line with previous studies9. IL-1R8-deficiency also led to enhanced IL-18-dependent phosphorylation of S6 and JNK in NK cells, suggesting that IL-1R8 inhibited IL-18-dependent activation of the mTOR and JNK pathways (Fig. 2j), which control NK cell metabolism, differentiation and activation17,18.
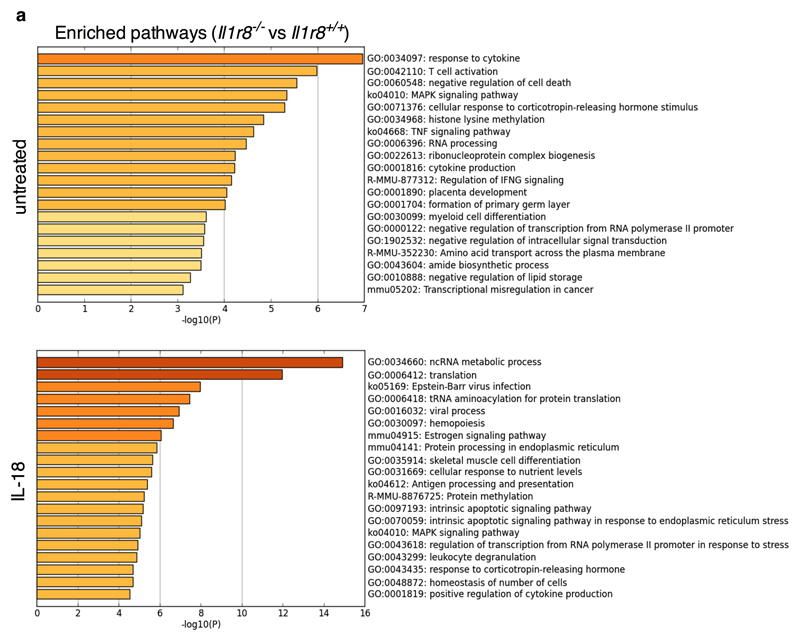
To obtain a deeper insight into the impact of IL-1R8 deficiency on NK cell function and on the response to IL-18, RNA-seq analysis was conducted. IL-1R8 deficiency had a profound impact on the resting transcriptional profile of NK cells and on top on responsiveness to IL-18 (Fig. 2k, Extended Data Fig. 4a and Supplementary Table 1). The profile of IL-1R8 deficient cells includes activation pathways (e.g. MAPK), adhesion molecules involved in cell-to-cell interactions and cytotoxicity (ICAM-1), and increased production of selected chemokines (CCL4). The latter may represent an NK cell based amplification loop of leukocyte recruitment, including NK cells themselves.
To investigate the role of IL-1R8 in human NK cells (Fig. 1a, 1b), we first retrospectively analyzed its expression in relation to responsiveness to a combination of IL-18 and IL-12 in normal donors. We observed an inverse correlation between IL-1R8 levels and IFNγ production by peripheral blood NK cells (r2=0.7969, p=0.0012) (Fig. 2l). In addition, IL-1R8 partial silencing in peripheral blood NK cells with small interfering RNA (siRNA) was associated with a significant increase in IFNγ production (Fig. 2m) and upregulation of CD69 expression (not shown). These results suggest that in human NK cells as in murine counterparts IL-1R8 serves as a negative regulator of activation and that its inactivation unleashes human NK cell effector function.
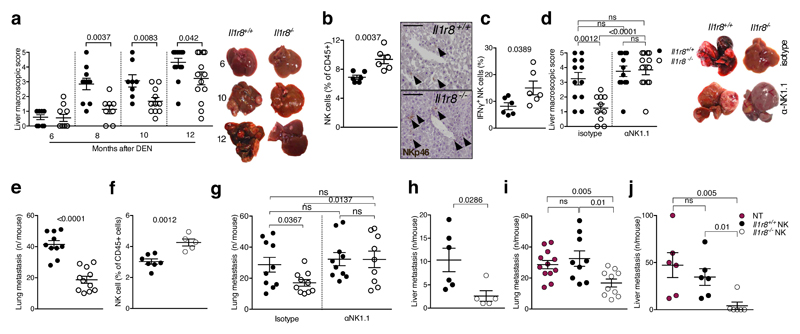
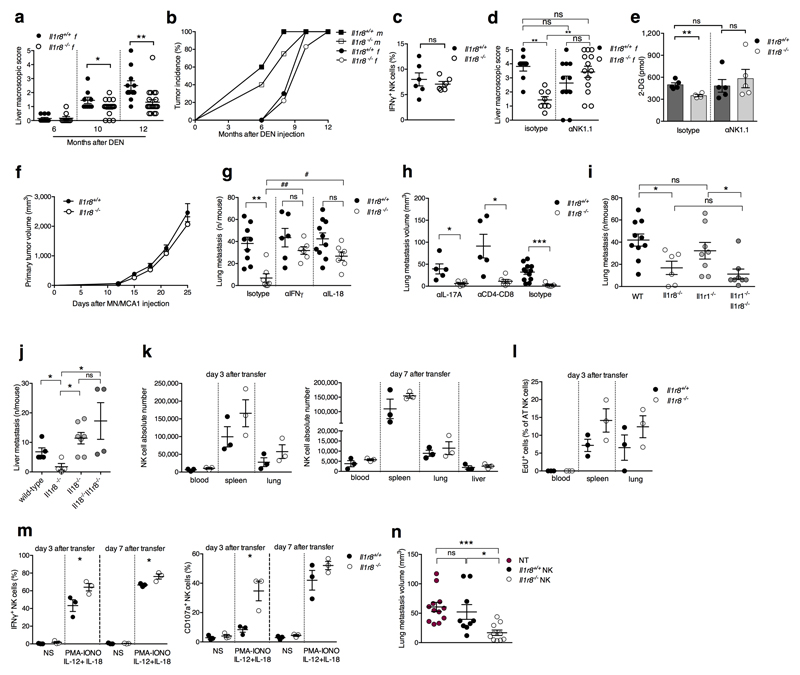
In an effort to assess the actual relevance of IL-1R8-mediated regulation of NK cells, anticancer and antiviral resistance were examined. The liver is characterized by a high frequency of NK cells19. Therefore we focused on liver carcinogenesis. In a model of diethylnitrosamine (DEN)-induced hepatocellular carcinoma (HCC), IL-1R8-deficient male and female mice20 were protected against the development of lesions, in terms of macroscopic number, size (Fig. 3a, Extended Data Fig. 5a, 5b) and histology (not shown). The percentage and absolute number of NK cells, and the percentage of IFNγ+ NK cells were higher in Il1r8-/- HCC-bearing mice (Fig. 3b, 3c, Extended Data Fig. 5c). Finally, increased levels of cytokines involved in anti-tumor immunity (e.g. IFNγ) and a reduction of pro-inflammatory cytokines associated with tumor promotion (IL-6, TNFα, IL-1β, CCL2, CXCL1) were observed (Extended Data, Table 1). Most importantly, the depletion of NK cells abolished the protection against liver carcinogenesis observed in Il1r8-/- mice (Fig. 3d and Extended Data Fig. 5d).
Figure 3. NK cell-mediated protection against liver carcinogenesis and metastasis in IL-1R8-deficient mice.
(a) Macroscopic score of liver lesions in male Il1r8+/+ and Il1r8-/- mice 6, 8, 10 and 12 months after DEN injection.
(b) Frequency and representative histological quantification of NK cell infiltrate in liver of tumor bearing mice. (20X, bar = 100µm).
(c) Frequency of IFNγ+ NK cells in liver of tumor bearing mice.
(d) Macroscopic score of liver lesions in male mice upon NK cell depletion.
(e) Number of spontaneous lung metastasis.
(f) NK cell frequency in the lungs of MN/MCA1 tumor bearing mice.
(g) Number of lung metastasis in MN/MCA1 tumor bearing mice upon NK cell depletion.
(h) Number of liver metastasis in MC38 colon carcinoma bearing mice.
(i,j) Number of lung (i) and liver (j) metastasis of Il1r8+/+ mice after adoptive transfer of Il1r8+/+ and Il1r8-/- NK cells.
(a, d) Representative images of female livers are shown.
(a-j) *p < 0.05, **p < 0.01, ***p < 0.001 between selected relevant comparisons, two-tailed unpaired Student’s t test. Mean ± SEM.
Evidence suggests that NK cells can inhibit hematogenous cancer metastasis5. In a model of sarcoma (MN/MCA1) spontaneous lung metastasis, Il1r8-/- mice showed a reduced number of hematogenous metastasis, whereas primary tumor growth was unaffected (Fig. 3e, Extended Data Fig. 5e, 5f). The frequency of total and mature CD27low NK cells was higher in Il1r8-/- lungs (Fig. 3f and not shown). Assessment of lung metastasis at sacrifice and in vivo imaging analysis (Fig. 3g, Extended Data Fig. 5e) showed that the protection was completely abolished in NK cell-depleted Il1r8-/- mice. In addition, IL-18 or IFNγ neutralization abolished or dramatically reduced the protection against metastasis observed in Il1r8-/- mice (Extended Data Fig. 5g). In contrast, depletion of CD4+/CD8+ cells or IL-17A, or deficiency of IL-1R1 (involved in TH17 development), did not affect the phenotype (Extended Data Fig. 5h, 5i).
Liver metastasis is a major problem in the progression of colorectal cancer. We therefore assessed the potential of Il1r8-/- NK cells to protect against liver metastasis using the MC38 colon carcinoma line21. As shown in Fig. 3h, Il1r8-/- mice were protected against MC38 colon carcinoma liver metastasis. In addition, IL-18 genetic deficiency abrogated the protection against liver metastasis observed in Il1r8-/- mice (Extended Data Fig. 5j), thus indicating that the IL-1R8-dependent control of MC38-derived liver metastasis occurs through the IL-18/IL-18R axis. To assess the primary role of Il1r8-/- NK cells in the cancer protection, adoptive transfer was used (Extended Data Fig. 5k-m). Adoptive transfer of Il1r8+/+ NK cells had no effect on lung and liver metastasis. In contrast, adoptive transfer of Il1r8-/- NK cells significantly and dramatically reduced the number and volume of lung and liver metastasis (Fig. 3i, 3j, Extended Data Fig. 5n). Given the natural history and clinical challenges of colorectal cancer, this observation has potential translational implications. Thus, IL-1R8 genetic inactivation unleashes NK cell mediated resistance to carcinogenesis in the liver and amplifies the anti-metastatic potential of these cells in liver and lung in a NK cell-autonomous manner.
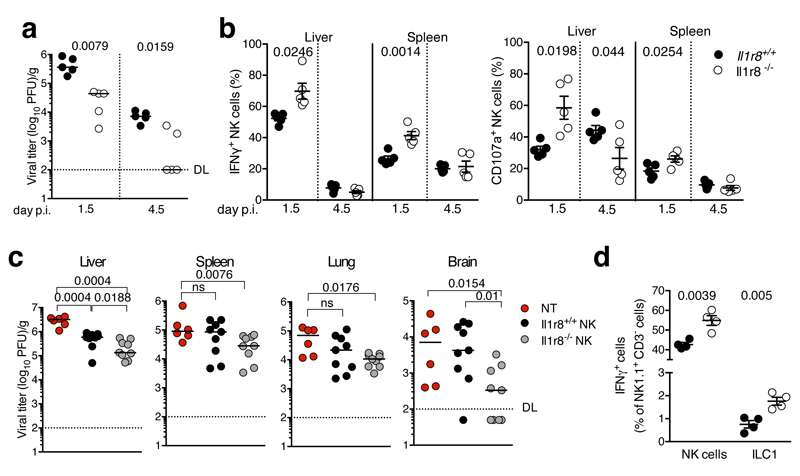
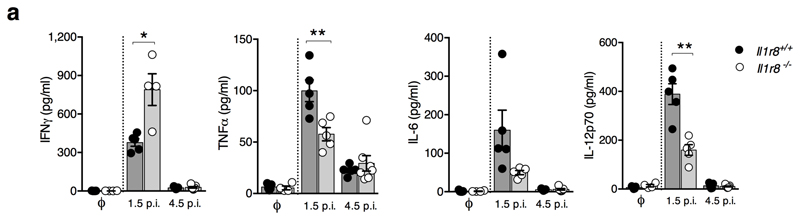
Finally we investigated whether IL-1R8 impacts on NK cell antiviral activity, focusing on murine cytomegalovirus (MCMV) infection22. As shown in Fig. 4a, liver viral titers were lower in Il1r8-/- compared to Il1r8+/+ mice, indicating that IL-1R8-deficiency was associated with a more efficient control of MCMV infection. The frequency of IFNγ+ NK cells and degranulation (i.e. frequency of CD107a+ NK cells) were significantly higher in the spleen and liver of Il1r8-/- mice on day 1.5 post infection (Fig. 4b). On day 4.5 post infection, IFNγ+ and CD107a+ NK cells were strongly reduced, in both spleen and liver, as a consequence of better control of viral spread (Fig. 4b). Consistently with a more efficient control of the infection, reduced levels of pro-inflammatory cytokines were observed in Il1r8-/- mice (Extended Data Fig. 6a). NK cell adoptive transfer experiments were performed in MCMV infected newborn mice that still do not have mature NK cells12. As shown in Fig. 4c, the adoptive transfer of Il1r8-/- NK cells conferred higher protection compared to Il1r8+/+ NK cells, with for instance 4 out of 9 mice having no detectable virus titer in the brain.
Figure 4. NK cell-mediated antiviral resistance in IL-1R8-deficient mice.
(a) Viral titer in livers of Il1r8+/+ and Il1r8-/- infected mice.
(b) Frequency of IFNγ+ and CD107a+ NK cells of infected mice.
(c) Viral titers in newborn wild type mice upon adoptive transfer of Il1r8+/+ and Il1r8-/- NK cells (7 days post infection).
(d) Frequency of IFNγ + cells in the liver of MCMV infected mice.
(a-d) *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Mann-Whitney test (a, c) or unpaired Student’s t test (b, d). Median (a, c). Mean ± SEM (b, d). DL: detection limit. Day p.i.: day post infection.
NK cells belong to the complex, diverse realm of innate lymphoid cells23. Human and murine non-NK ILCs express IL-1R8 mRNA and protein24 (and our unpublished data; Camilla Jandus, personal communication). Preliminary experiments were conducted in an effort to assess the role of IL-1R8 in ILC function. In the MCMV infection model, Il1r8-/- ILC1 showed increased IFNγ production, but represented a minor population compared to NK cells and 1/30 of Il1r8-/- IFNγ producing cells (Fig. 4d) and therefore are unlikely to play a significant role in the phenotype. These results provide initial evidence that IL-1R8 has a regulatory function in ILCs. Further studies are required to assess its actual significance in ILC diverse populations. Collectively, these results indicate that IL-1R8-deficient mice were protected against MCMV infection and that protection was dependent on increased NK cell activation.
IL-1R8 deficiency was associated with exacerbated inflammatory and immune reactions under a variety of conditions1,10. NK cells engage in bidirectional interactions with macrophages, dendritic cells and other lymphocytes3,4,25,26. Therefore the role of NK cells in inflammatory and autoimmune conditions associated with IL-1R8 deficiency1,10 will need to be examined. IL-1R8 deficient mice show increased susceptibility to colitis and colitis-associated azoxymethane (AOM) carcinogenesis27,28. The divergent impact on carcinogenesis of IL-1R8 deficiency in the intestine and liver is likely to reflect fundamental, tissue-dictated differences of immune mechanisms involved in carcinogenesis in these different anatomical sites. In particular, high numbers of NK cells are present in the liver19 and this physiological characteristic of this organ is likely to underlie this apparent divergence.
NK cells are generally not credited to play a major role in the control of solid tumors6. Conversely there is evidence for a role of NK cells in the control of hematogenous lung metastasis5,29. The results presented here show that unleashing NK cells by genetic inactivation of IL-1R8 resulted in inhibition of liver carcinogenesis and protection against liver and lung metastasis. IL-1R8-deficient mice show exacerbated TLR and IL-1-driven inflammation10 and inflammation promotes liver carcinogenesis30. Therefore the results presented here are likely an underestimate of the potential against liver primary and metastatic tumors of removal of the NK cell checkpoint IL-1R8. Therefore, NK cells have the potential to restrain solid cancer and metastasis, provided critical, validated checkpoints such as IL-1R8 are removed and the tissue immunological landscape is taken into account.
Methods
Animals
All female and male mice used were on a C57BL/6J genetic background and 8-12 weeks-old, unless specified. Wild-type mice were obtained from Charles River Laboratories, Calco, Italy or were littermates of Il1r8-/- mice. IL-1R8-deficient mice were generated as described31. Il1r1-/- mice were purchased from The Jackson Labs, Bar Harbor ME, USA. All colonies were housed and bred in the SPF animal facility of Humanitas Clinical and Research Center in individually ventilated cages. Il1r1-/-/Il1r8-/- mice were generated by crossing Il1r1-/- and Il1r8-/- mice. Il18-/-/Il1r8-/- were generated by crossing Il18-/- and Il1r8-/- mice. Mice were randomized based on sex, age and weight.
Procedures involving animals handling and care were conformed to protocols approved by the Humanitas Clinical and Research Center (Rozzano, Milan, Italy) in compliance with national (D.L. N.116, G.U., suppl. 40, 18-2-1992 and N. 26, G.U. March 4, 2014) and international law and policies (EEC Council Directive 2010/63/EU, OJ L 276/33, 22-09-2010; National Institutes of Health Guide for the Care and Use of Laboratory Animals, US National Research Council, 2011). The study was approved by the Italian Ministry of Health (approvals n. 43/2012-B, issued on the 08/02/2012 and n. 828/2015-PR, issued on the 07/08/2015). All efforts were made to minimize the number of animals used and their suffering. In most in vivo experiments, the investigators were unaware of the genotype of the experimental groups. Sample size was defined in order to detect differences of 20% or grater between the groups (10% significance level and 80% power).
Human primary cells
Human peripheral mononuclear cells (PBMCs) were isolated from peripheral blood of healthy donors, upon approval by Humanitas Research Hospital Ethical Committee. PBMCs were obtained through a Ficoll density gradient centrifugation (GE Healthcare Biosciences). NK cells were then purified by a negative selection, using a magnetic cell-sorting technique according to the protocols given by the manufacturer (EasySep™ Human NK Cell Enrichment Kit, Stem Cell Technology). Human monocytes were obtained from peripheral blood of healthy donors by two-step gradient centrifugation, first by Ficoll and then by Percoll (65% iso-osmotic; Pharmacia, Uppsala, Sweden). Residual T and B cells were removed from monocyte fraction by plastic adherence. Monocytes were cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS), 1% L-Glutamine, 1% Pen/Strept and 100 ng/ml M-CSF (Peprotech) for 7 days in order to generate resting macrophages. T and B cells were obtained from peripheral blood of healthy donors using RosetteSep™ Human T Cell Enrichment Cocktail and RosetteSep™ Human B Cell Enrichment Cocktail (Stem Cell Technology), following the manufacturer's instructions. Neutrophils were enriched from Ficoll–isolated granulocytes, using EasySep™ Human Neutrophil Enrichment Kit (StemCell Technologies), according to the manufacturer’s instructions.
To analyse pluripotent haematopoietic stem cells (HSC) and NK cell precursors (NKP), human Bone Marrow mononuclear cells were collected from Humanitas Biobank, upon approval by Humanitas Research Hospital Ethical Committee (Authorization 1516, issued on February 26, 2016). Frozen samples were thawed and vitality was assessed by trypan blue and Aqua LIVE/Dead-405 nm staining (Invitrogen), before flow cytometry analysis.
Informed consent was obtained from all subjects.
FACS analysis
Single-cell suspensions of BM, blood, spleen, lung and liver were obtained and stained. Representative NK cell gating strategy is reported in Supplementary Fig. 1. Foxp3/Transcription Factor Staining Buffer Set (eBioscience) was used for intracellular staining of Granzyme B and Perforin. Cytofix/Cytoperm (BD Biosciences) was used for intracellular staining of IFNγ. Liver ILC1 were identified as NK1.1+CD3-CD49a+CD49b- cells. Formalin 4% and Methanol 100% were used for intracellular staining of IRAK4, pIRAK4, pS6 and JNK. The following murine antibodies were used: CD45-BV605, -BV650 or -PerCp-Cy5.5 (Clone 30-F11); CD45.1-BV650 (Clone A20); CD45.2-APC, -BV421 (Clone 104); CD3e-PerCP-Cy5.5 or -APC (Clone 145-2C11); CD19-PerCP-Cy5.5, -eFluor450 (Clone 1D3); NK1.1-PE, -APC, -eFluor450 or –Biotin (Clone PK136); CD11b-BV421, -BV450, -BV785 (Clone M1/70); CD27-FITC or –APC-eFluor780 (Clone LG.7F9); CD4-FITC (Clone RM 4-5); CD8-PE (Clone 53-6.7); KLRG-1-BV421 (Clone 2F1); NKG2D-APC (Clone CX5); DNAM-1-APC (Clone 10E5); Ly49H-PECF594 (Clone 3D10); Granzyme B-PE (Clone NGZB); Perforin-PE (Clone eBioOMAK-D); IFNγ-Alexa700 or -APC (Clone XMG1.2); CD107a-Alexa647 (Clone 1D4B); FasL-APC (Clone MFL3); Lineage Cell Detection Cocktail-Biotin; Sca-1-FITC (Clone D7); CD117-PE or -Biotin (Clone 3C11); CD127-eFluor450 (Clone A7R34); CD135-APC or –Biotin (Clone A2F10.1); CD244-PE (Clone 2B4); CD122-PE-CF594 (Clone TM-Beta1); CD49b-PE-Cy7 or Biotin (Clone DX5), CD49a-APC (Clone Ha31/8), from BD Bioscience, eBioscience, BioLegend or Miltenyi Biotec. The following human antibodies were used: CD56-PE (Clone CMSSB); CD3-FITC (Clone UCHT1); CD16-Pacific Blue (Clone 3G8); CD34-PE-Vio770 (Clone AC136); CD117-BV605 (Clone 104D2); NKp46-BV786 (Clone 9E2/NKp46); CD45-PerCP (Clone 2D1); CD19-APC-H7 (Clone SJ25C1); CD14-APC-H7 (Clone M5E2); CD66b-APC-Vio770 (Clone REA306), from BD Bioscience, eBioscience or Miltenyi Biotec. Biotinylated anti-hSIGIRR (R&D Systems) and Streptavidin-Alexa647 (Invitrogen™) were used to stain IL-1R8 in human cells. Human NKT cells were detected using PE-CD1d tetramers loaded with αGalCer (ProImmune, Oxford, UK). Antibodies to detect protein phosphorylation were as follows: p-IRAK4 Thr345/Ser346 (Clone D6D7), IRAK4, p-S6-Alexa647 Ser235/236 (Clone D57.2.2E); p-SAPK/JNK Thr183/Tyr185 (Clone 81E11), from Cell Signaling Technology. A Goat anti-Rabbit-Alexa647 secondary antibody (Invitrogen™) was used to stain p-IRAK4, IRAK4 and p-SAPK/JNK. Results are reported as mean fluorescence intensity (MFI) normalized on isotype control or fluorescence minus one (FMO). Cell viability was determined by Aqua LIVE/Dead-405 nm staining (Invitrogen) or Fixable Viability Dye (FVD) eFluor® 780 (eBioscience), negative cells were considered viable. Cells were analyzed on LSR Fortessa or FACSVerse (BD Bioscience). Data were analyzed with FlowJo software (Treestar).
Quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen) following the manufacturer’s recommendations. RNA was further purified using miRNeasy RNA isolation kit (QIAGEN) or Direct-zol™ RNA MiniPrep Plus (Zymo Research). cDNA was synthesized by reverse transcription using High Capacity cDNA archive kit (Applied Biosystems) and quantitative real-time PCR was performed using the SybrGreen PCR Master Mix (Applied Biosystems) in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). PCR reactions were carried out with 10 ng of DNA. Data were analyzed with the Δ2CT method. Data were normalized based on GAPDH, βactin or 18Ṣexpression, as indicated, determined in the same sample. Analysis of all samples was performed in duplicate. Primers were designed according to the published sequences and listed as follows: s18/S18: forward 5'-ACT TTC GAT GGT AGT CGC CGT-3', reverse 5'- CCT TGG ATG TGG TAG CCG TTT-3'; Gapdh/GAPDH: forward 5'-GCA AAG TGG AGA TTG TTG CCA T-3', reverse 5'-CCT TGA CTG TGC CGT TGA ATT T-3'; βactin/βACTIN: forward 5'- CCC AAG GCC AAC CGC GAG AAG AT-3', reverse 5'- GTC CCG GCC AGC CAG GTC CAG -3'; il1r8: forward 5'- AGA GGT CCC AGA AGA GCC AT-3', reverse 5'- AAG CAA CTT CTC TGC CAA GG-3'; IL1R8: forward 5'- ATG TCA AGT GCC GTC TCA ACG -3', reverse 5'- GCT GCG GCT TTA GGA TGA AGT-3'; il1r1: forward 5’- TGC TGT CGC TGG AGA TTG AC -3’, reverse 5’- TGG AGT AAG AGG ACA CTT GCG AA -3’; il1r2: forward 5’- AGT GTG CCC TGA CCT GAA AGA -3’, reverse 5’- TCC AAG AGT ATG GCG CCC T -3’; il1r3: forward 5’- GGC TGG CCC GAT AAG GAT -3’, reverse 5’- GTC CCC AGT CAT CAC AGC G -3’; il1r4: forward 5’- GAA TGG GAC TTT GGG CTT TG -3’, reverse 5’- GAC CCC AGG ACG ATT TAC TGC -3’; il1r5: forward 5’- GCT CGC CCA GAG TCA CTT TT -3’, reverse 5’- GCG ACG ATC ATT TCC GAC TT -3’; il1r6: forward 5’- GCT TTT CGT GGC AGC AGA TAC -3’, reverse 5’- CAG ATT TAC TGC CCC GTT TGT T -3’; 16S: forward 5’- AGA GTT TGA TCC TGG CTC AG -3’, reverse 5’- GGC TGC TGG CAC GTA GTT AG -3’.
Purification of murine leukocytes
Splenic NK cells and bone marrow neutrophils were MACS enriched according to manufacturer’s instructions (Miltenyi Biotec). Purity of NK cells was about 90% as determined by FACS. Purity of neutrophils was ≥ 97.5%. NK cells were stained (CD45-BV650, NK1.1-PE, CD3e-APC, CD11b-BV421, CD27-FITC) and sorted on a FACSAria cell sorter (BD Bioscience) to obtain high purity NK cells and NK cell populations (CD11blowCD27low, CD11blowCD27high, CD11bhighCD27high and CD11bhighCD27low). Splenic B and T lymphocytes were stained (CD45-PerCP, CD3e-APC, CD4-FITC, CD8-PE, CD19-eFluor450) and sorted. Purity of each population was ≥ 98%. Resulting cells were processed for mRNA extraction or used for adoptive transfer or co-culture experiments. In vitro-derived macrophages were obtained from bone marrow total cells. Bone marrow cells were cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS), 1% L-Glutamine, 1% Pen/Strept and 100 ng/ml M-CSF (Peprotech) for 7 days in order to generate resting macrophages. Bone marrow cells were cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS), 1% L-Glutamine, 1% Pen/Strept and 20 ng/ml GM-CSF (Peprotech) for 7 days in order to generate DCs.
Confocal microscopy
Murine splenic NK cells were MACS enriched, let adhere on poly-D-Lysine (Sigma-Aldrich) coated coverslips, fixed with 4% PFA, permeabilized with 0.1% Triton X-100, incubated with blocking buffer (5% normal donkey serum (Sigma-Aldrich), 2% BSA, 0.05% Tween). Cells were then stained with biotin-conjugated goat polyclonal anti-SIGIRR antibody or biotin-conjugated normal goat IgG as control (both R&D Systems) (10µg/ml) followed by Alexa Fluor 488–conjugated donkey anti-goat IgG antibody (Molecular Probes) and DAPI (Invitrogen). Coverslips were mounted with the antifade medium FluorPreserve Reagent (EMD Millipore) and analyzed with an Olympus Fluoview FV1000 laser scanning confocal microscope with oil immersion lens 40× (N.A.1.3).
Stimulated emission depletion (STED) microscopy
Human NK cells were enriched and let adhere on poly-D-Lysine (Sigma-Aldrich) coated coverslips, stimulated with IL-18 (50 ng/ml; 1 min, 5 min, 10 min), fixed with 4% PFA, incubated with 5% normal donkey serum (Sigma-Aldrich), 2% BSA, 0.05% Tween in PBS2+ (pH 7.4) (blocking buffer), and then with biotin-conjugated goat polyclonal anti-human IL-1R8 antibody or biotin-conjugated normal goat IgG (all from R&D Systems) and mouse monoclonal anti-IL-18Rα (Clone 70625; R&D System) or mouse IgG1 (Invitrogen), all diluted at 5µg/ml in blocking buffer, followed by Alexa Fluor 488– conjugated donkey anti-goat IgG antibody and Alexa Fluor 555 donkey anti-mouse IgG antibody (both from Molecular Probes). Mowiol was used as mounting medium. STED xyz images were acquired in a unidirectional mode with a Leica SP8 STED3X confocal microscope system. Alexa Fluor 488 was excited with a 488nm Argon Laser and emission collected from 505 to 550 nm applying a gating between 0.4 to 7ns to avoid collection of reflection and autofluorescence. Alexa Fluor 555 was excited with a 555/547nm-tuned white light laser (WLL) and emission collected from 580 to 620 nm. Line sequential acquisition was applied to avoid fluorescence overlap. The 660nm CW-depletion laser (80% of power) was used for both excitations. Images were acquired with Leica HC PL APO 100x/1.40 oil STED White objective at 572.3mAU. CW-STED and gated CW-STED were applied to Alexa-488nm and Alexa Fluor 555, respectively. Collected images were de-convolved with Huygens Professional software.
3′-mRNA Sequencing and Analysis
Splenic NK cells (from 6 mice per genotype and pooled in pairs) were purified as described above and stimulated with IL-18 (MBL) (20 ng/ml for 4 h). RNA was prepared as described above. The QuantSeq 3′mRNA-seq Library Prep Kit for Illumina (Lexogen) was used to generate libraries, which were sequenced on the NextSeq (Illumina; 75 bp PE). The fastq sequence files were assessed using the fastqc program. The reads were first trimmed using bbduk in the bbmap suite of software32 to remove the first 12 bases and a contaminant kmer discovery length of 13 was used for contaminant removal. Regions of length 20 or above with average quality of less than 10 were trimmed from the end of the read. The reads were then trimmed to remove trailing polyG and polyA runs using cutadapt33 and the quality of the remaining reads reassessed with fastqc. The trimmed reads were aligned to the mm10 genomic reference and reads assigned to features in the mm10 annotation using the STAR program34. Differential expression analysis was performed using the generalized linear model (GLM) functions in the R/bioconductor35 edgeR package36 with TMM normalization. Gene set analysis was performed using the romer37 function in the R/bioconductor package limma38. Metascape (http://metascape.org) was used to enrich genes for GO biological processes, KEGG Pathway and Reactome Gene Sets.
Measurement of cytokines
BD Cytometric Bead Array (CBA) mouse inflammation kit (BD) or Duoset ELISA kits (R&D System) were used to measure cytokines.
In vitro functional assays
Total murine splenocytes or enriched murine or human NK cells were cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum (FBS) 1% L-Glutamine, 1% Pen/Strept and treated with IL-2, IL-12, IL-15 (Peprotech), IL-18 (MBL), IL-1β (Peprotech) and PMA-Ionomycin (Sigma-Aldrich), as specified. FasL expression was evaluated upon treatment for 45 minutes with IL-18 (50 ng/ml), IL-15 (50 ng/ml), IL-2 (20 ng/ml) and IL-12 (10 ng/ml). IFNγ production was analysed upon 16 hours of treatment with IL-12 (20 ng/ml) and IL-18 (20 ng/ml) or IL-1β (20 ng/ml), by intracellular staining using BD Cytofix/Cytoperm™ Fixation/Permeabilization Kit, following the manufacturer's instructions, or by ELISA. Granzyme B and Perforin intracellular staining was performed upon 18 hours of stimulation with IL-12 (10 ng/ml), IL-15 (10 ng/ml) and IL-18 (50 ng/ml), using Foxp3/Transcription Factor Staining Buffer Set (eBioscience). CD107a-Alexa647 antibody was added during the 4-hour culture and analysed by flow cytometry. BD GolgiPlug™ (containing Brefeldin) and BD GolgiStop™ (containing Monensin) were added 4 hours prior to intracellular staining. PMA (50 ng/ml)- Ionomycin (1 µg/ml) were added 4 hours prior to intracellular staining, when specified.
NK-DC co-culture experiments were performed as previously described39. DCs were treated with LPS from Escherichia coli O55:B5 (Sigma-Aldrich; 1µg/ml) or CpG ODN 1826 (Invivogen; 3µg/ml) and with anti-mIL-18 neutralizing antibody (BioXCell, Clone YIGIF74-1G7; 5µg/ml) or Rat Isotype Control (BioXCell, Clone 2A3).
IFNγ and CD107a expression upon viral infection was analyzed by flow cytometry upon 4-hour treatment with BD GolgiPlug™, BD GolgiStop™ and IL-2 (500U/ml).
Phosphorylation of IRAK4, S6 and JNK was analyzed upon 15-30 minutes of stimulation with IL-18 (10 ng/ml).
Human primary NK cell transfection
Human NK cells were enriched from peripheral blood of healthy donors and transfected with Dharmacon™ Acell™ siRNA (GE Healthcare) using Accell™ delivery medium (GE Healthcare), following the manufacturer’s instructions. 1 µM SIGIRR-specific siRNA (On-Target Plus; Dharmacon, GE Healthcare) comprised 250 nM of the four following antisense sequences: I, AGU UUC GCG AGC CGA GAU CUU; II, UAC CAG AGC AGC ACG UUG AUU; III, UGA CCC AGG AGU ACU CGU GUU; IV, CUU CCC GUC GUU UAU CUC CUU (all 5’ to 3’).
Generation of bone marrow chimeras
Il1r8-/- and Il1r8+/+ mice were lethally irradiated with a total dose of 900 cGy. 2 h later, mice were injected in the retro-orbital plexus with 4x106 nucleated bone marrow cells obtained by flushing of the cavity of freshly dissected femurs from wild type or Il1r8-/- donors. Competitive bone marrow chimeric mice were generated by reconstituting recipient mice with 50% CD45.1 Il1r8+/+ and 50% CD45.2 Il1r8-/- bone marrow cells. Recipient mice received gentamycin (0.8 mg/ml in drinking water) starting 10 days before irradiation and for 2 weeks after irradiation. NK cells of chimeric mice were analyzed 8 weeks after bone marrow transplantation.
Depletion and blocking experiments
Mice were treated intraperitoneally with 200 µg of specific mAbs (Mouse anti-NK1.1, Clone PK136; Mouse Isotype Control, Clone C1.18.4; Rat anti-mIL-18, Clone YIGIF74-1G7; Rat Isotype Control, Clone 2A3; Rat anti-IFNγ, Clone XMG1.2; Rat IgG1 HRPN; Mouse anti-IL-17A, Clone 17F3; Mouse Isotype Control, Clone MOPC-21; Rat anti-CD4/CD8, Clone GK1.5/YTS; Rat Isotype Control, Clone LTF-2 (all from BioXCell)) and then with 100 µg once (anti-NK1.1) or three times (anti-IL-18, anti-IFNγ, anti-IL-17A, anti-CD4/CD8) a week for the entire duration of the experiment.
Microflora depletion
6-week-old mice were treated every day for 5 weeks by oral gavage with a cocktail of antibiotics [ampicillin (Pfizer) 10 mg/ml, vancomycin (PharmaTech Italia) 10 mg/ml, metronidazol (Società Prodotti Antibiotici) 5 mg/ml and neomycin (Sigma-Aldrich) 10 mg/ml]. Control mice were treated with drinking water. A gavage volume of 10 ml/kg body weight was delivered with a stainless steel tube without prior sedation of mice. DNA was isolated from bacterial fecal pellets with PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc.) and quantified by spectrophotometry at 260 nm. PCR was performed with 10 ng of DNA using the SybrGreen PCR Master Mix (Applied Biosystems) in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). Data were analyzed with the Δ2CT method (Applied Biosystems, Real-Time PCR Applications Guide).
Cancer models
Mice were injected intraperitoneally (i.p.) with 25 mg/kg of diethylnitrosamine (DEN, Sigma) at 15 days of age. Mice were sacrificed 6-8-10-12 months later, to analyze liver cancer. Liver cancer score was based on number and volume of lesions (0: no lesions; 1: lesion number<3, or lesion dimension <3mm; 2: lesion number<5, or lesion dimension <5mm; 3: lesion number<10, or lesion dimension <10mm; 4: lesion number<15, or lesion dimension <10mm; 5: lesion number>15, or lesion dimension >10mm). Lung metastasis experiments were performed injecting i.m. the 3-MCA derived mycoplasma-free sarcoma cell line MN/MCA1 (105 cells/mouse in 100 µl PBS)40. Primary tumor growth was monitored twice weekly, and lung metastases were assessed by in vivo imaging and by macroscopic counting at sacrifice 25 days after injection. Liver metastases were generated by injecting intrasplenically 1.5x105 mycoplasma-free colon carcinoma cells (MC38) 21. Mice were sacrificed 12 days after injection and liver metastasis were counted macroscopically. MC38 cells were received from ATCC just before use. MN/MCA1 cells were authenticated morphologically by microscopy in vitro and by histology ex vivo. Tumor size limit at which mice were sacrificed was based on major diameter (≤ 2cm).
Viral infections
Mice were injected intravenously (i.v.) with 5×105 PFU of the tissue culture (TC)-grown virus in PBS. Bacterial artificial chromosome (BAC)-derived MCMV strain MW97.01 has been previously shown to be biologically equivalent to MCMV strain Smith (VR-1399) and is hereafter referred to as wild-type (WT) MCMV41. Mice were sacrificed 1.5 and 4.5 days post infection and viral titer was assessed by plaque assay, as previously described42,43. Newborn mice were infected i.p. with 2000 PFU of the MCMV strain MW97.01 and sacrificed at day 7 post infection. Viral titer was assessed by plaque assay, as previously described42,43.
Adoptive transfer
106 Il1r8+/+ or Il1r8-/- sorted NK cells were injected i.v. in wild type adult mice 5 hours before MN/MCA or MC38 injection, or i.p. in newborn mice 48 hours after MCMV injection. Adoptively transferred NK cell engraftment, proliferative capacity and functionality (IFNγ production and degranulation after ex vivo stimulation) were assessed 3 and 7 days after injection.
In vivo proliferation was measured using Click-iT® Edu Flow Cytometry Assay Kit (Invitrogen). Edu was injected i.p. (0.5 mg/mouse), mice were sacrificed 24 hours later and cells were stained following the manufacturer’s instructions and analyzed by flow cytometry.
Immunohistochemistry
Liver frozen tissues were cut at 8 mm and then fixed with 4% PFA. Endogenous peroxidases were blocked with 0.03% of H2O2 for 5 min and unspecific binding sites were blocked with PBS + 1% FBS for 1h. Tissues were stained with polyclonal goat anti mouse NKp46/NCR1 (R&D System) and goat on mouse HRP polymer kit (GHP516, Biocare Medical) was used as secondary antibody. Reactions were developed with 3,3’-Diaminobenzidine (DAB) (Biocare Medical) and then slides were counterstained with hematoxylin. Slides were mounted with eukitt (Sigma-Aldrich). 20X images were analyzed with cell^F software (Olympus).
In vivo Imaging
After feeding with AIN-76A alfalfa-free diet (Mucedola srl, Italy) for two weeks, to reduce fluorescence background, mice were intravenously (i.v.) injected with XenoLight RediJect 2-DeoxyGlucosone (DG) (PerkinElmer) and 24 hours later 2-DG fluorescence was measured using Fluorescence Molecular Tomography system (FMT 2000, Perkin Elmer). Acquired images were subsequently analyzed with TrueQuant 3.1 analysis software (Perkin Elmer).
Data availability
The data discussed in this publication have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSEXXXXX, or from the corresponding author. Figure source data are provided.
Statistical analysis
For animal studies, sample size was defined on the basis of past experience on cancer and infection models, in order to detect differences of 20% or grater between the groups (10% significance level and 80% power). Values were expressed as mean ± SEM or median of biological replicates, as specified. One-way ANOVA or Kruskal-Wallis test were used to compare multiple groups. Two-sided unpaired Student’s t test was used to compare unmatched groups with Gaussian distribution and Welch’s correction was applied in case of significantly different variance. Mann-Whitney test was used in case of non-Gaussian distribution. ROUT test was applied to exclude outliers. p≤0.05 was considered significant. Statistics were calculated with GraphPad Prism version 6, GraphPad Software.
Statistics and reproducibility
Figure 1: a, n=4 (B cells), n=5 (NKT cells), n=9 (T cells) , n=10 (NK cells) donors; b, n=5 donors; c, n=8 (NK cells) or n=4 (T cells) or n=3 (other leukocytes) mice; d, n=5 mice. b, representative experiment out of 6 performed. a, c, d, one experiment performed.
Figure 2: a, b, n=8 or n=7 (spleen, Il1r8+/+ liver) or n=6 (Il1r8-/- liver) mice; c, n= 6 mice; d, n=9 (Il1r8+/+) or n=6 (Il1r8-/-) mice; e, n=5 mice; f, n=6 mice; g, n=4 mice; h, n=5 mice; i, n=10 wells; j, n=4 (IRAK4), n=6 or n=5 (S6 Il1r8-/-) or n=7 (JNK Il1r8-/-) mice; k, n=3 mice; l, n=9 healthy donors; m, n=4 healthy donors. Representative experiments out of 3 (a, b), 5 (c), 2 (d, j), 4 (e) performed. f-m one experiment performed.
Figure 3: a, n= 8, 10, 11, 13, 14 mice; b, c, n=6 mice; d, n=10, 12, 13 mice; e, n=10, 11 mice; f, n=5, 6, 7 mice; g, n=9, 10 mice; h, n=5, 6 mice; i, n= 9, 10 or 12 mice; j, n=6 mice. Representative experiments out of 6 (e), 3 (a), 2 (d, f, g, h, i). b, c, j, one experiment performed.
Figure 4: a,b, n=5 mice; c, n=6, n=9 mice; d, n=4 mice. a, two experiments were performed; b-d, one experiment was performed.
Extended Data
Extended Data Figure 1. Expression of IL-1R8 in human and murine NK cells.
(a, b) IL-1R8 mRNA (a) expression in human primary NK cells, compared with T and B cells, neutrophils, monocytes and in vitro-derived macrophages (a) and in human primary NK cell maturation stages (CD56brCD16-, CD56brCD16+, CD56dimCD16+), and in the CD56dimCD16- subset (b).
(c) Representative FACS plot of human NK cell subsets and histograms of IL-1R8 expression in NK cell subsets.
(d) IL-1R8 protein expression in human bone marrow precursors and mature cells.
(e) IL-1 receptor family members (Il1r1, Il1r2, Il1r3, Il1r4, Il1r5, Il1r6, Il1r8) mRNA expression in murine primary NK cells isolated from the spleen.
(f) IL-1R8 protein expression in murine NK cells by confocal microscopy. Magnification bar: 10μm.
(g) Representative FACS plot of murine NK cell subsets.
(a, b, d) *p < 0.05, **p < 0.01, ***p < 0.001 One-way ANOVA. Mean ± SEM.
a, n=6 (NK and B cells) or n=4 donors; b, n=5 donors; d, n=4 donors; e, n=2 mice; f, representative images out of four collected per group.
a, b, d, e, f, one experiment performed.
Extended Data Figure 2. Phenotypic analysis of Il1r8-/- NK cells.
(a, b) Representative FACS plot of murine NK cell subsets in Il1r8+/+ and Il1r8-/- mice (a) and histograms of KLRG1 expression in NK cells.
(c, d) NK absolute number and NK cell subsets (DN, CD11blow, DP and CD27low) in bone marrow, spleen and blood of Il1r8+/+ and Il1r8-/- newborn mice at 2 (c) and 3 (d) weeks of age.
(e) Frequency of bone marrow precursors in Il1r8+/+ and Il1r8-/- mice.
(f) NKG2D, DNAM-1 and LY49H expression in peripheral NK cells and NK cell subsets of Il1r8+/+ and Il1r8-/- mice.
(g) Frequency of splenic Perforin+ NK cell subsets upon stimulation in Il1r8+/+ and Il1r8-/- mice.
(h and i) Peripheral NK cell absolute number (h) and CD27low NK cell frequency (i) in bone marrow chimeric mice upon reconstitution (9 weeks).
(j and k) Peripheral NK cell (j) and NK cell subset (k) frequency in competitive chimeric mice transplanted with 50% of Il1r8+/+ CD45.1 cells and 50% of Il1r8-/- CD45.2 cells upon reconstitution (9 weeks). Upon reconstitution a defective engraftment (12% instead of 50% engraftment) of Il1r8-/- stem cells was observed in competitive conditions.
(l) IFNγ production by Il1r8+/+ and Il1r8-/- NK cells upon co-culture with LPS- or CpG-primed Il1r8+/+ and Il1r8-/-DCs.
(c-l) *p < 0.05, **p < 0.01, ***p < 0.001 between selected relevant comparisons, two-tailed unpaired Student’s t test. Centre values and error bars represent mean ± SEM. At least 5 animals per group were used. c, d: 3 pooled experiments. e-l: one experiment was performed.
Extended Data Figure 3. Mechanism of IL-1R8-dependent regulation of NK cells.
(a) Splenic CD27low NK cell frequency in wild type, Il1r8-/-, Il18-/-, and Il18-/-/Il1r8-/- mice.
(b) Peripheral CD27low NK cell frequency in wild-type, Il1r8-/-, Il1r1-/- and Il1r8-/-Il1r1-/- mice (left) and IFNγ production by splenic NK cells after IL-12 and IL-1β or IL-18 stimulation (right).
(c, d) Splenic CD27low NK cell frequency in Il1r8+/+ and Il1r8-/- mice upon commensal flora depletion (c) and breeding in co-housing conditions (d).
(e) STED microscopy of human NK cells stimulated with IL-18. Magnification bar: 2μm.
(a-d) *p < 0.05, **p < 0.01, ***p < 0.001 between selected relevant comparisons, two-tailed unpaired Student’s t test; Centre values and error bars represent mean ± SEM. a, n= 3, 5, or 6 mice; at least 5 animals per group were used (b-d). a-d: one experiment was performed. e: representative images out of three collected from two donors.
Extended Data Figure 4. RNA-seq analysis of Il1r8+/+ and Il1r8-/- NK cells.
(a) Metascape analysis of enriched gene pathways of resting and IL-18-activated Il1r8+/+ and Il1r8-/- NK cells.
See also Supplementary Table 1 and data deposited in NCBI Gene Expression Omnibus accessible through GEO Series accession number GSEXXXXX.
Extended Data Figure 5. NK cell-mediated resistance to HCC and metastasis in IL-1R8-deficient mice.
(a) Macroscopic score of liver lesions in female Il1r8+/+ and Il1r8-/- mice 6, 10 and 12 months after DEN injection.
(b) HCC incidence in Il1r8+/+ and Il1r8-/- female and male mice.
(c) Frequency of IFNγ+ NK cells in spleen of Il1r8+/+ and Il1r8-/- tumor bearing mice.
(d) Macroscopic score of liver lesions in female Il1r8+/+ and Il1r8-/- mice upon NK cell depletion.
(e) 2-DG quantification in lungs of Il1r8+/+ and Il1r8-/- tumor bearing mice upon NK cell depletion.
(f) Primary tumor growth in Il1r8+/+ and Il1r8-/- mice (25 days after MN/MCA1 cell line injection).
(g) Number of lung metastasis in Il1r8+/+ and Il1r8-/- MN/MCA1 sarcoma bearing mice upon IFNγ or IL-18 neutralization.
(h) Volume of lung metastases in Il1r8+/+ and Il1r8-/- MN/MCA1-bearing mice upon depletion of IL-17A or CD4+/CD8+ cells.
(i) Number of lung metastases in Il1r8+/+ and Il1r8-/-, Il1r1-/-, Il1r1-/-/Il1r8-/- MN/MCA1-bearing mice.
(j) Number of liver metastasis in Il1r8+/+, Il1r8-/-, Il18-/-, Il18-/-Il1r8-/- MC38 colon carcinoma bearing mice.
(k) Il1r8+/+ and Il1r8-/- NK cell absolute number three or 7 days after adoptive transfer.
(l) In vivo Il1r8+/+ and Il1r8-/- NK cell proliferation three days after adoptive transfer.
(m) Ex vivo IFNγ production and degranulation upon 4-hour stimulation with PMA-Ionomycin, IL-12 and IL-18 in adoptively transferred Il1r8+/+ and Il1r8-/- NK cells.
(n) Volume of lung metastasis of Il1r8+/+ MN/MCA1 sarcoma bearing mice after adoptive transfer of Il1r8+/+ and Il1r8-/- NK cells.
(a, c-e, g-j, m-n) *p < 0.05, **p < 0.01, ***p < 0.001 between selected relevant comparisons, two-tailed unpaired Student’s t test or Mann-Whitney test. #p<0.05, ##p<0.01, Kruskal-Wallis and Dunn's multiple comparison test. Centre values and error bars represent mean ± SEM. a, n=9, 10, 11, 18, 21 mice; b, n=8-21 mice; c, n=6 mice; d, n= 10, 12, 13 mice; e, n=4 (Il1r8-/- isotype) or n=5; f, n=10; g, n=6, 7, 9, 10 mice; h, n=5, 6, 12 mice; i, n=6, 8, 10 mice; j, n=4, 5, 7 mice; k, l, m, n=3 mice; n, n=9, 10, 12 mice. Representative experiment out of 3 (a, b), 2 (d), 6 (f), or one (c, e, g-n) experiment performed.
Extended Data Figure 6. NK cell-mediated antiviral resistance in IL-1R8-deficient mice.
(a) Cytokine serum levels in Il1r8+/+ and Il1r8-/- infected mice (1.5 and 4.5 days post infection).
*p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student’s t test. Centre values and error bars represent mean ± SEM. n=5 mice. One experiment was performed.
Extended Data Table 1. Serum cytokine and liver enzyme levels in HCC-bearing mice.
| 6 months after DEN | 8-10 months after DEN | 12 months after DEN | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cytokine pg/mL |
Il1 r8
+/+ n=4-5* |
Il1 r8 -/-n=5 | p value |
Il1 r8
+/+ n=7-10* |
Il1 r8
-/- n=9-10* |
p value |
Il1 r8
+/+ n=3-5* |
Il1 r8
-/- n=3-5* |
p value |
| IL-23 | 173.1 ±29.12 | 247.3 ± 15.16 | 0.05 | 187.7 ± 13.47 | 343.4 ±66.29 | 0.04 | 103.7 ±26.72 | 138.6 ±37.51 | 0.47 |
| IL-12p70 | 277.6 ± 44.49 | 358.4 ± 12.44 | 0.12 | 293 ± 16.31 | 357.2 ± 34.77 | 0.13 | 152 ±20.14 | 164.9 ± 15.22 | 0.62 |
| IL-17A | 69.98 ±9.88 | 95.03 ± 6.44 | 0.07 | 56.41 ±7.46 | 102.4 ± 19.01 | 0.04 | 38.13 ± 10.39 | 45.05 ± 8.78 | 0.62 |
| IFNγ | 295 ± 72.78 | 385.4 ±48.6 | 0.32 | 357.5 ±57.63 | 593.2 ±84.33 | 0.05 | 195.4 ±65.29 | 243.3 ± 104 | 0.72 |
| IL-6 | 90.37 ± 6.45 | 67.23 ± 9.79 | 0.08 | 126.9 ± 19.52 | 69.64 ± 6.93 | 0.01 | 61.24 ± 18.05 | 42.28 ± 12.17 | 0.44 |
| IL-1β | 91.99 ±5.23 | 58.68 ±7.29 | 0.006 | 142.4 ± 28.24 | 60.35 ±4.42 | 0.01 | 47.66 ± 14.08 | 29.81 ±7.66 | 0.31 |
| TNFα | 163.5 ±7.16 | 92.06 ±21.04 | 0.01 | 194.6 ±28.03 | 100.1 ± 14.24 | 0.008 | 94.77 ± 14.24 | 57.45 ± 14.51 | 0.13 |
| CCL2 | 32.51 ± 1.54 | 24.1 ±5.64 | 0.19 | 43.97 ±7.25 | 25.42 ± 1.37 | 0.02 | 28.1 ±4.99 | 19.72 ± 1.23 | 0.14 |
| CXCL1 | 197.6 ±8.85 | 142.5 ±20.93 | 0.04 | 183.4 ± 17.75 | 123.7 ± 10.5 | 0.01 | 105.6 ±6.49 | 77.86 ± 9.64 | 0.04 |
| Liver enzymes** | |||||||||
| ALT | 142.5± 52.5 | 0.00 ± 0.00, | 0.004 | 111.7± 70.77*** | 60.0 ±35.0*** | 0.32 | 0.00 ± 0.00 | 0.00 ± 0.00 | NA |
| AST | 159.6± 39.79 | 101.0 ± 1.87 | 0.18 | 134.0± 15.28*** | 97.0 ± 8.0*** | 0.06 | 105.0± 25.45 | 89.0 ±5.1 | 0.55 |
Samples with not detectable levels were not included in the analysis.
levels are U/L.
n=5, 8 months after DEN
Supplementary Material
Acknowledgments
We thank N. Polentarutti, G. Benigni, M. Erreni, F. Colombo, V. Juranić Lisnić, D. Kvestak for technical assistance, Prof. M. Nebuloni for HCC histology, Dr. A. Doni for STED images, Dr. F. Ficara, Dr. R. Carriero and Prof. D. Mavilio for discussion.
The contribution of the European Commission (ERC project PHII-669415; FP7 project 281608 TIMER; ESA/ITN, H2020-MSCA-ITN-2015-676129), Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) (project FIRB RBAP11H2R9), Associazione Italiana Ricerca sul Cancro (AIRC IG-19014 and AIRC 5x1000 -9962), Fondazione CARIPLO (project 2015-0564), and the Italian Ministry of Health is gratefully acknowledged. MM received an EFIS short-term fellowship to perform viral infection experiments in SJo's laboratory. The authors declare no competing financial interests.
Footnotes
Author contributions
E.B. and M.M. played a key role in designing and conducting most experiments and drafted the manuscript. F.R., M.B., F.G., E.M. provided technological support in in vivo experiments. A.P., S.Ja., B.P. and G.B. contributed to the experimental design and in in vivo experiments. S.Z. contributed to RNA-seq analysis. S.Jo. and A.S. contributed to the experimental design and supervision of the study. C.G. and A.M. contributed to the experimental design and supervision of the study, and suggested the role of IL-1R8 as a novel checkpoint inhibitor of NK cells.
Author Information
Reprints and permissions information is available at www.nature.com/reprints.
Authors declare no Competing financial interests.
References
- 1.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellora F, et al. Human NK cells and NK receptors. Immunol Lett. 2014;161:168–173. doi: 10.1016/j.imlet.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 6.Stojanovic A, Cerwenka A. Natural killer cells and solid tumors. J Innate Immun. 2011;3:355–364. doi: 10.1159/000325465. [DOI] [PubMed] [Google Scholar]
- 7.Gismondi A, Stabile H, Nisti P, Santoni A. Effector Functions of Natural Killer Cell Subsets in the Control of Hematological Malignancies. Front Immunol. 2015;6:567. doi: 10.3389/fimmu.2015.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulen MF, et al. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity. 2010;32:54–66. doi: 10.1016/j.immuni.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nold-Petry CA, et al. IL-37 requires the receptors IL-18Ralpha and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal transduction. Nat Immunol. 2015;16:354–365. doi: 10.1038/ni.3103. [DOI] [PubMed] [Google Scholar]
- 10.Garlanda C, Riva F, Bonavita E, Mantovani A. Negative regulatory receptors of the IL-1 family. Seminars in immunology. 2013;25:408–415. doi: 10.1016/j.smim.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Cooper MA, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 12.Chiossone L, et al. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 14.Takeda K, et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 15.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Gong J, et al. Inhibition of Toll-like receptors TLR4 and 7 signaling pathways by SIGIRR: a computational approach. J Struct Biol. 2010;169:323–330. doi: 10.1016/j.jsb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Marcais A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat Immunol. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, et al. JNK MAP kinase activation is required for MTOC and granule polarization in NKG2D-mediated NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:3017–3022. doi: 10.1073/pnas.0712310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng H, Tian Z. Re-examining the origin and function of liver-resident NK cells. Trends Immunol. 2015;36:293–299. doi: 10.1016/j.it.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Naugler WE, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 21.Dupaul-Chicoine J, et al. The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity. Immunity. 2015;43:751–763. doi: 10.1016/j.immuni.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Lisnic B, Lisnic VJ, Jonjic S. NK cell interplay with cytomegaloviruses. Curr Opin Virol. 2015;15:9–18. doi: 10.1016/j.coviro.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih HY, et al. Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell. 2016;165:1120–1133. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellora F, et al. M-CSF induces the expression of a membrane-bound form of IL-18 in a subset of human monocytes differentiating in vitro toward macrophages. Eur J Immunol. 2012;42:1618–1626. doi: 10.1002/eji.201142173. [DOI] [PubMed] [Google Scholar]
- 26.Martin-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 27.Garlanda C, et al. Increased susceptibility to colitis-associated cancer of mice lacking TIR8, an inhibitory member of the interleukin-1 receptor family. Cancer Res. 2007;67:6017–6021. doi: 10.1158/0008-5472.CAN-07-0560. [DOI] [PubMed] [Google Scholar]
- 28.Xiao H, et al. The Toll-interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26:461–475. doi: 10.1016/j.immuni.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 30.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garlanda C, et al. Intestinal inflammation in mice deficient in Tir8, an inhibitory member of the IL-1 receptor family. Proc Natl Acad Sci U S A. 2004;101:3522–3526. doi: 10.1073/pnas.0308680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bushnell B. Bbmap: A Fast, Accurate, Splice-Aware Aligner. Ernest Orlando Lawrence Berkeley National Laboratory; Berkeley, CA (US); 2014. [Google Scholar]
- 33.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17 [Google Scholar]
- 34.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majewski IJ, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116:731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingozzi F, et al. Prolonged contact with dendritic cells turns lymph node-resident NK cells into anti-tumor effectors. EMBO Mol Med. 2016;8:1039–1051. doi: 10.15252/emmm.201506164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giavazzi R, Alessandri G, Spreafico F, Garattini S, Mantovani A. Metastasizing capacity of tumour cells from spontaneous metastases of transplanted murine tumours. Br J Cancer. 1980;42:462–472. doi: 10.1038/bjc.1980.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jonjic S, Pavic I, Lucin P, Rukavina D, Koszinowski UH. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J Virol. 1990;64:5457–5464. doi: 10.1128/jvi.64.11.5457-5464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddehase MJ, et al. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSEXXXXX, or from the corresponding author. Figure source data are provided.