Abstract
Background:
Psoriasis is a complex autoimmune disease caused by the interaction of genetic and environmental factors. PTPN22 gene polymorphism has been reported to affect psoriasis susceptibility; however, no data are available for Middle Eastern populations.
Objective:
The aim of this study was to investigate the association of PTPN22 (1858C/T) R620W polymorphism with psoriasis in a Saudi cohort.
Methods:
Saudi subjects (n = 306) including patients with psoriasis (n = 106) and matched controls (n = 200) were studied for PTPN22 variants using tetra-primer amplification refractory mutation system-polymerase chain reaction method. The frequencies of alleles and genotypes of PTPN22 (1858C/T) polymorphism were compared between patients and controls.
Results:
The frequency of CT genotype of PTPN22 (1858C/T) polymorphism was significantly higher, whereas that of CC genotype was lower in patients with psoriasis than in controls (P < .001, relative risk [RR] = 7.151). The homozygous genotype TT was absent in both the patients and healthy controls. The frequency of allele T encoding tryptophan (W) was significantly increased (P < .001, RR = 5.76), whereas that of allele C encoding arginine (R) decreased in psoriasis cases as compared with controls (P < .001, RR = 0.173) indicating that individuals carrying allele T are more susceptible to psoriasis than noncarriers.
Conclusions:
PTPN22 (1858C/T) polymorphism is positively associated with susceptibility of psoriasis in Saudis and can be developed as biomarker for evaluating psoriasis risk. However, further studies on PTPN22 polymorphism in larger samples from different geographical areas and ethnicity are warranted.
Keywords: psoriasis, PTPN22, polymorphism, genetic susceptibility, Saudi
Introduction
Psoriasis is a chronic, complex autoimmune disease with characteristic reddish patches covered by silvery-white scales. It is quite common and affects approximately 120 to 180 million people worldwide.1 The prevalence of psoriasis varies significantly depending mainly on race, geographic location, genetics, environmental factors, and ethnicity.2–7 In Saudi Arabia, its prevalence is 1% to 5% of all skin disorders.8 Psoriasis occurs in both sexes and most of the patients (75%) develop disease before the age of 40 years. Its cause is multifactorial, involving both genetic and environmental factors. Psoriasis depicts a number of characteristics which are also found in other autoimmune diseases suggesting a common causal pathway or mechanism.9,10
The molecular studies on psoriasis have helped to understand the pathogenesis of psoriasis and comorbidities,11 and a number of gene loci have been associated with susceptibility/severity of psoriasis.2,12 These association studies suggested that the pathogenesis of psoriasis involves skin barrier function, innate and adaptive immunity, gene-gene, and gene-environment interactions. As psoriasis is highly heritable and gene loci identified till date do not fully account for it, many genetic as well as environmental factors and their interaction might be involved in the etiopathogenesis.13
It has been confirmed that psoriasis is at least partially mediated by activated T cells.14,15
Typically, the inflammatory process is followed by hyperproliferation of the epidermis, leading to terminal differentiation of keratinocytes. Protein tyrosine phosphatase nonreceptor 22 (PTPN22) gene is located on chromosomes 1p13.3 to p13.1 and encodes a lymphoid-specific phosphatase (Lyp). Lyp is an intracellular protein tyrosine phosphatase and is physically bound through proline-rich motif to the SH3 domain of the C-terminal Src kinase (Csk), which suppresses kinases mediating T-cell activation.16 Although Lyp functions as a negative regulator of T cells,17 however, it also plays a role in B-cell signaling.18 It targets various signaling intermediates involved in T-cell receptor signaling and may work at various levels in signaling cascade.
A single-nucleotide polymorphism (SNP) in exon 14 of the PTPN22 gene 1858C/T (rs2476601) has been associated with a number of autoimmune diseases and considered as a risk factor due to significant production of autoantibodies.19–28 However, a few inconsistent reports on the association of PTPN22 gene polymorphism and psoriasis are available, and further testing in different ethnic cohorts is required to make sure whether these findings represent genuine associations. In this study, we investigated the association of variants of PTPN22 (1858C/T) (R620W) with the susceptibility to psoriasis in Saudi patients.
Materials and Methods
Subjects
Our study subjects (n = 306) comprised 106 patients with psoriasis and 200 matched healthy controls from same ethnic population visiting Dermatology Clinic, Prince Sultan Military Medical City (PSMMC), Riyadh, Saudi Arabia. All cases and controls were biologically unrelated Saudis. They were examined and diagnosed by dermatologists as described elsewhere.2 The demographic and other information were collected using a questionnaire. The diagnosis was based on clinical findings, skin changes, and the location of the condition on the body. The Psoriasis Area and Severity Index (PASI) was determined following the work by Schmitt and Wozel.29 Patients diagnosed with arthritis prior to psoriasis were excluded from the study. All subjects gave written consent to participate in this study. Of 106 confirmed patients with psoriasis, 42 were women and 64 were men with the mean age of 37 ± 15.5 years and duration of disease ranging from 1 to 20 (mean: 9 ± 4.5) years, age of onset of diseases was 15 to 55 years with female to male ratio of 1:1.52. Among the healthy controls, 70 were women and 130 were men with the mean age of 36 ± 10 years. We calculated the power of study online: http://www.stat.ubc.ca/~rollin/stats/ssize/caco.html.
Patients or controls with any history of autoimmune disorders were excluded from the study. To minimize genetic heterogeneity, control subjects having first-degree or second-degree relative with psoriasis or any other autoimmune disorders were also excluded.
The ethical committee of PSMMC approved the protocol of the study. Inclusion criterion was that all patients must be diagnosed with plaque psoriasis for at least 1 year. Exclusion criteria for both cases and controls were coexisting inflammatory skin disease, autoimmune diseases renal/liver failure, hypothyroidism, and systemic therapy and smoking.
Peripheral blood samples collected from the patients with psoriasis and controls were brought to the laboratory in vacuum flask with ice. Genomic DNA was extracted from the blood using QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). Genomic DNA was amplified using tetra-primer amplification refractory mutation system-polymerase chain reaction (PCR) method for detection of PTPN22 (R620W) (rs2476601) polymorphism.30 Two external primers (forward outer primer: 5′-CTCACACATCAG CTTCCCAAAGTG-3′ and reverse outer primer: 5′-CAACTTTACTGATAATGTTGCTTCA ACGGA-3′) and 2 internal primers (forward inner primer: 5′-CAACCACAATAAATG ATTCAGGTGTACG-3′, reverse inner primer: 5′-ATCCCCCCTCCACTTCCTGGAT-3′) were used. Product sizes were 213 and 151 bp (base pairs) for the C and T alleles, respectively, whereas the product size of the internal control was 314 bp. The PCR was performed using 5× FIREPol Master Mix (Solis BioDyne, Tartu, Estonia) following manufacturer’s protocol. The PCR cycling conditions were as follows: 5 minutes at 95°C followed by 30 cycles of 30 seconds at 95°C, 30 seconds at 64°C, and 30 seconds at 72°C and 10 minutes at 72°C. The PCR products were electrophoresed on 2% agarose gels and photographed. We regenotyped 25% of the samples (randomly selected) to verify the initial results and ensure genotyping quality.
Statistical analysis
The genotype and allele frequencies were analyzed with the Fisher exact test using CalcFisher software. Errors due to multiple comparison tests were minimized by Bonferroni correction. Both P values are summarized in Table 1. Odds ratio interpreted as relative risk (RR), etiologic fraction (EF), and preventive fraction (PF) was calculated as described elsewhere.31,32
Table 1.
Genotype and allele frequencies of PTPN22 variants in patients with psoriasis and matched healthy controls.
| Genotype/allele | Psoriasis (n = 106) |
Control (n = 200) |
P value | RR | EFa/PF |
|---|---|---|---|---|---|
| No. (%) | No. (%) | ||||
| CC | 67 (63.21) | 185 (92.50) | <.001b,* | 0.139 | 0.622 |
| CT | 39 (36.79) | 15 (7.50) | <.001b,* | 7.151 | 0.621a |
| TT | 0 (0) | 0 (0) | — | — | — |
| C allele | 173 (81.60) | 385 (96.25) | <.001b,* | 0.173 | 0.597 |
| T allele | 39 (18.40) | 15 (3.75) | <.001b,* | 5.764 | 0.596a |
Abbreviations: EF, etiologic fraction; PF, preventive fraction; RR, relative risk.
Number of allele/genotype is represented as No. (%).
Data for EF.
Statistically significant using Fisher exact test.
P < .004—Bonferroni corrected.
Results
In this case-control study, the allele and genotype frequencies of PTPN22 (1858C/T) polymorphism in cases with psoriasis and unrelated matched controls were determined. The control/case ratio of 1.88:1 yielded a power of 90% with sample size of 106 patients with psoriasis and 200 controls assuming a prevalence of 3.2% of all dermatologic diseases. Among patients with psoriasis, the female to male ratio was 1:1.52.
The distributions of genotypes and alleles of PTPN22 (1858C/T) polymorphism in psoriasis and control groups are shown in Tables 1 and 2. The amplification of different genotypes is shown in Figure 1. In patients and controls, the genotype and allele frequency distributions of the PTPN22 (1858C/T) were in the Hardy-Weinberg equilibrium. The frequencies of alleles and genotypes of PTPN22 (1858C/T) polymorphism were significantly different in patients and control (Table 1).
Table 2.
Genotype and allele frequencies of PTPN22 variants in male and female patients with psoriasis.
| Genotype/allele | Male (n = 64) |
Female (n = 42) |
P value | RR | EFa/PF |
|---|---|---|---|---|---|
| No. (%) | No. (%) | ||||
| CC | 42 (65.62) | 25 (59.52) | .543 | 1.298 | 0.055a |
| CT | 22 (34.38) | 17 (40.48) | .543 | 0.770 | 0.069 |
| TT | 0 (0) | 0 (0) | — | — | — |
| C allele | 106 (82.81) | 67 (79.76) | .591 | 1.223 | 0.012a |
| T allele | 22 (17.19) | 17 (20.24) | .591 | 0.818 | 0.016 |
Abbreviations: EF, etiologic fraction; PF, preventive fraction; RR, relative risk.
Number of allele/genotype is represented as No. (%).
Data for EF.
Figure 1.
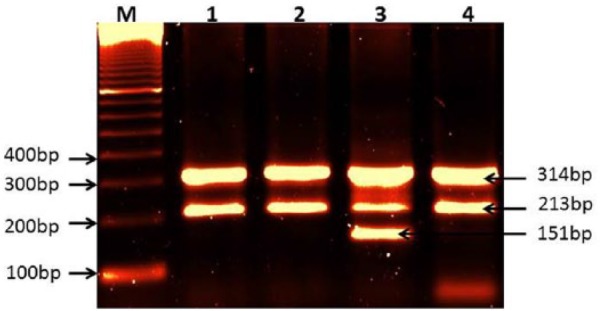
Amplification of PTPN22 (C1858T) polymorphism.
Lane M: 100-bp DNA marker; lanes 1, 2, and 4: amplification of genotype CC; lane 3: amplification of genotype CT, 213-bp band for allele C, 151-bp band for allele T, and 314-bp band for internal control.
The frequency of polymorphic allele T encoding tryptophan (W) was significantly increased, whereas that of allele C encoding arginine (R) decreased in patients as compared with controls (P < .001, P < .004—Bonferroni corrected). The frequency of heterozygous genotype (CT) was detected only in 7.5% (15/200) of healthy controls corresponding to a minor allele frequency of 0.037, whereas it was higher in patients as compared with controls (P < .001, P < .004—Bonferroni corrected). The frequency of CC genotype of PTPN22 (1858C/T) polymorphism was significantly lower (P < .001, P < .004—Bonferroni corrected) in patient group. The homozygous genotype TT was absent in both cases and controls.
Sex-wise stratification of genotyping results showed that frequencies of allele T and CT genotype were more prevalent in the female patients than the male patients without reaching statistical significance (Table 2). This statistically insignificant difference in the frequencies of alleles and genotypes of PTPN22 (1858C/T) polymorphism in male and female patients indicated that there was no effect of sex on prevalence of PTPN22 (1858C/T) polymorphism.
Discussion
The increased frequencies of allele T and genotype CT in our cases as compared with controls showed that the allele T and genotype CT may increase psoriasis susceptibility (RR = 7.151, EF = 0.621 and RR = 5.764, EF = 596, respectively). However, the lower frequencies of allele C and genotype CC in patients than controls showed that allele C and genotype CC are less susceptible to psoriasis (RR = 0.139, PF = 0.622 and RR = 0.173 PF = 597, respectively). These results suggested that PTPN22 (1858C/T) polymorphism may be associated with psoriasis susceptibility thus highlighting the role of the immune system in the pathogenesis of psoriasis in our population.
The results of this study on patients with chronic plaque psoriasis clearly indicated an association of +1858T allele of the PTPN22 gene with susceptibility to psoriasis in Saudi population similar to several other autoimmune diseases known to be associated with allele T of PTPN22 polymorphism. However, earlier studies from different geographical regions/ethnic populations on PTPN22 (+1858C/T) polymorphism and psoriasis have reported inconsistent results.33–37
A meta-analysis suggested that PTPN22 may be among the true psoriasis susceptibility-risk genes.34 Li et al35 analyzed 15 SNPs from 7 putative psoriasis-risk genes in 1448 patients with psoriasis and 1385 control subjects and suggested that PTPN22 is one of the significant risk genes for psoriasis. Although there was no significant association of PTPN22 (+1858C/T) polymorphism with psoriasis in their analysis, however, another polymorphism in PTPN22 gene (rs3789604) was found to be significantly associated with susceptibility of psoriasis. Recently, Chen and Chang33 could not find any significant association of the PTPN22 (+1858T) allele with psoriasis but they reported a strong association of PTPN22 (+1858T) allele with psoriatic arthritis (PsA) and suggested that future studies should be based on gene-environment interaction and attention should be focused on the clinical heterogeneity of the disease together with population stratification.
Earlier, Hüffmeier et al38 also found an association of PTPN22 (+1858T allele) with psoriasis and reported sex variations in susceptibility. Excluding direct link of T allele in psoriasis susceptibility in German patients, they suggested that other susceptibility determinant(s) within noncoding regions of PTPN22 or its proximity might exist and act independently as a risk factor. The +1858T allele of the PTPN22 gene has also been strongly associated with PsA, a related autoimmune disease.23,39 Recently, a study by Budu-Aggrey et al40 has suggested that several non–HLA risk loci including PTPN22 affect PsA susceptibility. They added that there is a complete genetic overlap between psoriasis and PsA susceptibility loci and about a third of people who have psoriasis will get PsA. People with severe psoriasis could have a greater chance of getting PsA. About 40% of people who get PsA have relatives with it or with psoriasis.
The PTPN22 (1858C/T) variants are considered important non-HLA common susceptibility determinants for T-cell–mediated autoimmune diseases.41,42 The PTPN22 (1858C/T) polymorphism results into a change of arginine (R) to a tryptophan (W) at position 620 and interrupts the interaction of Csk with Lyp and stops the formation of complex ultimately suppressing the activation of T cell. It has been shown by in vitro experiments that C allele of PTPN22 binds more efficiently to Csk than T allele suggesting thereby that T cells expressing the T allele may be hyperresponsive and T-allele carriers may be prone to autoimmune disorders.20,22 Moreover, the decreased thresholds for T-cell receptor signaling in animal knockout for murine homologue of PTPN22 confirm a specific role of PTPN22 in T-cell regulation.43
In contrast, other studies reported no association of this PTPN22 (1858C/T) polymorphism with psoriasis.36,37,44–46 Although Smith et al36 found no evidence of association of PTPN22 (R620W) with psoriasis susceptibility, however, they reported an association of other 2 SNPs, namely, rs1217414 and rs3789604, in the PTPN22 gene with psoriasis. It has been suggested that genetics of psoriasis is complex and there are several susceptibility genes implicated which vary among different ethnic groups.13,47,48 In addition, the gene-environment interaction may play important role in the etiopathogenesis of the disease. Saudi being a close society with high degree of consanguinity and unique environmental conditions together with high-protein diet might have resulted into these variations in association results. Moreover, in genetic association studies where a particular factor or variant has a small effect on the risk, the sample size is instrumental. It is possible that studies reporting no association of PTPN22 (1858T) allele with some autoimmune diseases could be due to lack of power to detect a true association in these studies. Moreover, most of these studies have been performed on the white subjects, and to the best of our knowledge, no work has been done on Middle Eastern populations. This is the first report showing an association of PTPN22 (1858C/T) polymorphism with psoriasis susceptibility in Saudi patients.
In addition, these differences in the results of PTPN22 (1858C/T) polymorphism in psoriasis indicate that the causes of psoriasis may involve different pathogenic mechanisms possibly due to ethnic variations. Ethnic variations in prevalence of PTPN22 (1858C/T) polymorphism have been reported in healthy populations also. It is quite low in Asian and African populations and is virtually absent in Han Chinese.49–52 In Europe, a North-South gradient ranging between 15.5% and 2.1% has been reported in the polymorphic allele frequencies of PTPN22 (1858C/T) polymorphism.19,51
In populations where PTPN22 (1858C/T) (rs2476601) is not associated with autoimmune diseases, other functional variants of PTPN22 gene and/or SNPs that are in linkage disequilibrium with PTPN22 (1858C/T) might be involved.53,54 The polymorphisms rs2488457, rs3789604, and rs1310182 in PTPN22 gene have also been associated with several autoimmune diseases in Asian populations.12
The other possible pathway of the cause of psoriasis has been explained on the basis of the fact that there is no citrullinated keratin K1 found in skin samples from patients with psoriasis.55–58 The lack of citrullinated proteins causes excessive cornification and an inflammatory response. Psoriasis is the only disease linked to peptidylarginine deiminase (PAD) dysregulation.56–59 The PTPN22 can prevent protein citrullination by blocking PAD type 4 (PAD-4) enzymes. A molecular link between PTPN22 and PAD-4 has been reported recently.60 PTPN22 (1858C/T) polymorphism which converts R to W promotes the activation signals in lymphocytes and increases autoreactive B cells, decreases production of type I interferon by myeloid cells, and ultimately increases the risk of autoimmune diseases including psoriasis.60
Our results identify PTPN22 (1858C/T) R620W polymorphism as a susceptibility marker for psoriasis in Saudi population and suggest that polymorphism in PTPN22 may contribute to psoriasis and that can be developed as a biomarker for evaluating psoriasis risk. However, further studies on PTPN22 polymorphisms in larger samples from different geographic locations and ethnicity are warranted.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Conceived and designed the experiment: MA. Performed clinical examinations, collected demographic data: GBH, FAH. Extracted DNA, performed genotyping: SR. Analyzed the data, interpreted the results and drafted the manuscript: MA. Agree with manuscript results and conclusions: GBH, FAH, AA. Jointly developed the structure and arguments for the paper: MA, SR. Made critical revisions, supervised and approved final version: AA, MA. All authors reviewed and approved of the final manuscript.
References
- 1. Icen M, Crowson CS, McEvoy MT, Dann FJ, Gabriel SE, Kremers HM. Trends in incidence of adult onset psoriasis over three decades: a population-based study. J Am Acad Dermatol. 2009;60:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Harthi F, Huraib GB, Zouman A, Arfin M, Tariq M, Al-Asmari A. Apolipoprotein E gene polymorphism and serum lipid profile in Saudi patients with psoriasis. Dis Markers. 2014;2014:239645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choon SE, Lai NM, Mohammad NA, Nanu NM, Tey KE, Chew SF. Clinical profile, morbidity and outcome of adult-onset generalized pustular psoriasis: analysis of 102 cases seen in a tertiary hospital in Johor, Malaysia. Int J Dermatol. 2014;53:676–684. [DOI] [PubMed] [Google Scholar]
- 4. Ding X, Wang T, Shen Y, et al. Prevalence of psoriasis in China: a population-based study in six cities. Eur J Dermatol. 2012;22:663–667. [DOI] [PubMed] [Google Scholar]
- 5. Dogra S, Yadav S. Psoriasis in India: prevalence and pattern. Indian J Dermatol Venereol Leprol. 2010;76:595–601. [DOI] [PubMed] [Google Scholar]
- 6. Loo CH, Chan YC, Lee KQ, Palanivelu T, Tan WC. Clinical profile, morbidity and outcome of adult patients with psoriasis at a district hospital in Northern Malaysia. Med J Malaysia. 2015;70:177–181. [PubMed] [Google Scholar]
- 7. Rachakonda TD, Schupp CW, Armstrong AW. Psoriasis prevalence among adults in the United States. J Am Acad Dermatol. 2014;70:512–516. [DOI] [PubMed] [Google Scholar]
- 8. Alshobaili HA, Shahzad M, Al-Marshood A, Khalil A, Settin A, Barrimah I. Genetic background of psoriasis. Int J Health Sci (Qassim). 2010;4:23–29. [PMC free article] [PubMed] [Google Scholar]
- 9. Elder JT. Genome-wide association scan yields new insights into the immuno-pathogenesis of psoriasis. Genes Immun. 2009;10:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elder JT, Bruce AT, Gudjonsson JE, Johnston A, Stuart PE, Tejasvi T. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol. 2010;130:1213–1226. doi: 10.1038/jid.2009.319. [DOI] [PubMed] [Google Scholar]
- 11. Scarpa R, Altomare G, Marchesoni A, et al. Psoriatic disease: concepts and implications. J Eur Acad Dermatol Venereol. 2010;24:627–630. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Z, Yuan J, Tian Z, Xu J, Lu Z. Investigation of 36 non-HLA (human leucocyte antigen) psoriasis susceptibility loci in a psoriatic arthritis cohort. Arch Dermatol Res. 2016;302:71–77. doi: 10.1007/s00403-016-1706-z. [DOI] [PubMed] [Google Scholar]
- 13. Chandran V. The genetics of psoriasis and psoriatic arthritis. Clin Rev Allergy Immunol. 2013;44:149–156. [DOI] [PubMed] [Google Scholar]
- 14. Lew W, Bowcock AM, Krueger JG. Psoriasis vulgaris: cutaneous lymphoid tissue supports T-cell activation and “Type 1” inflammatory gene expression. Trends Immunol. 2004;25:295–305. [DOI] [PubMed] [Google Scholar]
- 15. Sugiyama H, Gyulai R, Toichi E, et al. Dysfunctional blood and target tissue CD4+ CD25high regulatory T cells in psoriasis: mechanism underlying unrestrained pathogenic effector T cell proliferation. J Immunol. 2005;174:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93:2013–2024. [PubMed] [Google Scholar]
- 17. Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. [DOI] [PubMed] [Google Scholar]
- 18. Arechiga AF, Habib T, He Y, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gregersen PK, Kosoy R, Lee AT, et al. Risk for myasthenia gravis maps to a (151) Pro→Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012;72:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Gene. 2004;75:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bianco B, Verreschi IT, Oliveira KC, et al. PTPN22 polymorphism is related to autoimmune disease risk in patients with Turner syndrome. Scand J Immunol. 2010;72:256–259. [DOI] [PubMed] [Google Scholar]
- 22. Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. [DOI] [PubMed] [Google Scholar]
- 23. Butt C, Peddle L, Greenwood C, Hamilton S, Gladman D, Rahman P. Association of functional variants of PTPN22 and tp53 in psoriatic arthritis: a case-control study. Arthritis Res Ther. 2006;8:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinks A, Worthington J, Thomson W. The association of PTPN22 with rheumatoid arthritis and juvenile idiopathic arthritis. Rheumatology (Oxford). 2006;45:365–368. [DOI] [PubMed] [Google Scholar]
- 25. Kokkonen H, Johansson M, Innala L, Jidell E, Rantapää-Dahlqvist S. The PTPN22 1858C/T polymorphism is associated with anti-cyclic citrullinated peptide-positive early rheumatoid arthritis in northern Sweden. Arthritis Res Ther. 2007;9:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pradhan V, Borse V, Ghosh K. PTPN22 gene polymorphisms in autoimmune diseases with special reference to systemic lupus erythematosus disease susceptibility. J Postgrad Med. 2010;56:239–242. [DOI] [PubMed] [Google Scholar]
- 27. Tang L, Wang Y, Zheng S, Bao M, Zhang Q, Li J. PTPN22 polymorphisms, but not R620W, were associated with the genetic susceptibility of systemic lupus erythematosus and rheumatoid arthritis in a Chinese Han population. Hum Immunol. 2016;77:692–698. [DOI] [PubMed] [Google Scholar]
- 28. Zheng J, Ibrahim S, Petersen F, Yu X. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun. 2012;13:641–652. [DOI] [PubMed] [Google Scholar]
- 29. Schmitt J, Wozel G. The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology. 2005;210:194–199. [DOI] [PubMed] [Google Scholar]
- 30. Kouhpayeh HR, Hashemi M, Hashemi SA, et al. R620W functional polymorphism of protein tyrosine phosphatase non-receptor type 22 is not associated with pulmonary tuberculosis in Zahedan, southeast Iran. Genet Mol Res. 2012;11:1075–1081. [DOI] [PubMed] [Google Scholar]
- 31. Schallreuter KU, Levenig C, Kuhnl P, Loliger C, Hohl-Tehari M, Berger J. Histocompatability antigens in vitiligo: Hamburg study on 102 patients from Northern Germany. Dermatology. 1993;187:186–192. [DOI] [PubMed] [Google Scholar]
- 32. Svejgaard A, Platz P, Ryder LP. HLA and disease 1982—a survey. Immunol Rev. 1983;70:193–218. [DOI] [PubMed] [Google Scholar]
- 33. Chen YF, Chang JS. PTPN22 C1858T and the risk of psoriasis: a meta-analysis. Mol Biol Rep. 2012;39:7861–7870. [DOI] [PubMed] [Google Scholar]
- 34. Li Y, Chang M, Schrodi S, et al. The 5q31 variants associated with psoriasis and Crohn’s disease are distinct. Hum Mol Genet. 2008;17:2978–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, Liao W, Chang M, et al. Further genetic evidence for three psoriasis-risk genes: ADAM33, CDKAL1, and PTPN22. J Invest Dermatol. 2009;129:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith RL, Warren RB, Eyre S, et al. Polymorphisms in the PTPN22 region are associated with psoriasis of early onset. Br J Dermatol. 2008;58:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zervou MI, Castro-Giner F, Sidiropoulos P, Boumpas DT, Tosca AD, Krueger-Krasagakis S. The protein tyrosine phosphatase, non-receptor type 22 R620W polymorphism does not confer susceptibility to psoriasis in the genetic homogeneous population of Crete. Genet Test Mole Biomarkers. 2010;14:107–111. [DOI] [PubMed] [Google Scholar]
- 38. Hüffmeier U, Steffens M, Burkhardt H, et al. Evidence for susceptibility determinant(s) to psoriasis vulgaris in or near PTPN22 in German patients. J Med Genet. 2006;43:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Juneblad K, Johansson M, Rantapää-Dahlqvist S, Alenius GM. Association between the PTPN22 +1858 C/T polymorphism and psoriatic arthritis. Arthritis Res Ther. 2011;13:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Budu-Aggrey A, Bowes J, Barton A. Identifying a novel locus for psoriatic arthritis. Rheumatology (Oxford). 2016;55:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregersen PK. Pathways to gene identification in rheumatoid arthritis: PTPN22 and beyond. Immunol Rev. 2005;204:74–86. [DOI] [PubMed] [Google Scholar]
- 42. Siminovitch KA. PTPN22 and autoimmune disease. Nat Genet. 2004;36:1248–1249. [DOI] [PubMed] [Google Scholar]
- 43. Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303:685–689. [DOI] [PubMed] [Google Scholar]
- 44. Criswell LA, Pfeiffer KA, Lum RF, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hinks A, Barton A, John S, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. [DOI] [PubMed] [Google Scholar]
- 46. Nistor I, Nair RP, Stuart P, et al. Protein tyrosine phosphatase gene PTPN22 polymorphism in psoriasis: lack of evidence for association. J Invest Dermatol. 2005;125:395–396. [DOI] [PubMed] [Google Scholar]
- 47. Abdelnoor AM, Al-Akl N. Factors involved in the pathogenesis of psoriasis. Adv Stud Med Sci. 2013;1:75–94. [Google Scholar]
- 48. Bowcock AM, Cookson WO. The genetics of psoriasis, psoriatic arthritis and atopic dermatitis. Hum Mol Genet. 2004;13:R43–R55. [DOI] [PubMed] [Google Scholar]
- 49. Lee HS, Korman BD, Le JM, et al. Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum. 2009;60:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee YH, Rho YH, Choi SJ, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases—a meta-analysis. Rheumatology (Oxford). 2007;46:49–56. [DOI] [PubMed] [Google Scholar]
- 51. Mori M, Yamada R, Kobayashi K, Kawaida R, Yamamoto K. Ethnic differences in allele frequency of autoimmune disease-associated SNPs. J Hum Genet. 2005;50:264–266. [DOI] [PubMed] [Google Scholar]
- 52. Zhang ZH, Chen F, Zhang XL, Jin Y, Bai J, Fu SB. PTPN22 allele polymorphisms in 15 Chinese populations. Int J Immunogenet. 2008;35:433–437. [DOI] [PubMed] [Google Scholar]
- 53. Kawasaki E, Awata T, Ikegami H, et al. Systematic search for single nucleotide polymorphisms in a lymphoid tyrosine phosphatase gene (PTPN22): association between a promoter polymorphism and type 1 diabetes in Asian populations. Am J Med Genet. 2006;140:586–593. [DOI] [PubMed] [Google Scholar]
- 54. Viken MK, Amundsen SS, Kvien TK, et al. Association analysis of the 1858CT polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun. 2005;6:271–273. [DOI] [PubMed] [Google Scholar]
- 55. György B, Tóth E, Tarcsa E, Falus A, Buzás EI. Citrullination: a posttranslational modification in health and disease. Int J Biochem Cell Biol. 2006;38:1662–1677. [DOI] [PubMed] [Google Scholar]
- 56. Ishida-Yamamoto A, Senshu T, Takahashi H, Akiyama K, Nomura K, Iizuka H. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol. 2000;114:701–705. [DOI] [PubMed] [Google Scholar]
- 57. Méchin MC, Sebbag M, Arnaud J, et al. Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. Int J Cosmet Sci. 2007;29:147–168. [DOI] [PubMed] [Google Scholar]
- 58. Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. [DOI] [PubMed] [Google Scholar]
- 59. Wang S, Wang Y. Peptidylarginine deiminases in citrullination, gene regu-lation, health and pathogenesis. Biochim Biophys Acta. 2013;1829:1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang HH, Dwivedi N, Nicholas AP, Ho IC. The W620 polymorphism in PTPN22 disrupts its interaction with peptidylarginine deiminase type 4 and enhances citrullination and NETosis. Arthritis Rheumatol. 2015;67:2323–2334. [DOI] [PubMed] [Google Scholar]


