Abstract
Summary:
Hybridization-based target enrichment in combination with genome skimming (Hyb-Seq) is becoming a standard method of phylogenomics. We developed HybPhyloMaker, a bioinformatics pipeline that performs target enrichment data analysis from raw reads to supermatrix-, supertree-, and multispecies coalescent-based species tree reconstruction. HybPhyloMaker is written in BASH and integrates common bioinformatics tools. It can be launched both locally and on a high-performance computer cluster. Compared with existing target enrichment data analysis pipelines, HybPhyloMaker offers the following main advantages: implementation of all steps of data analysis from raw reads to species tree reconstruction, calculation and summary of alignment and gene tree properties that assist the user in the selection of “quality-filtered” genes, implementation of several species tree reconstruction methods, and analysis of the coding regions of organellar genomes.
Availability:
The HybPhyloMaker scripts, manual as well as a test data set, are available in https://github.com/tomas-fer/HybPhyloMaker/. HybPhyloMaker is licensed under open-source license GPL v.3 allowing further modifications.
Keywords: Target enrichment, phylogenomics, genome skimming, species tree, locus selection
Introduction
Hybridization-based target enrichment in combination with genome skimming (Hyb-Seq) is becoming a standard method of phylogenomics (in plants see, for example, the works by Mandel et al., Weitemier et al., and Nicholls et al.1–3; see also the works by Lemmon and Lemmon, Heyduk et al.4,5 for a general overview of genome subsampling methods). Up to now, two well-documented data analysis pipelines have been published: PHYLUCE6 and HybPiper7. PHYLUCE has been developed and optimized for working with ultraconserved elements (UCEs)8,9, but it performs poorly in case of targeted sequences in the form of multiple exons per gene, which are common targets in plant phylogenetics due to the paucity of UCEs10; for locus selection in plants, see, for example, the works by Weitemier et al. and Nicholls et al.2,11. PHYLUCE applies a very stringent filter on potentially paralogous loci. This might result in a severe loss of loci in case one is working with multiple targeted exons per gene. The often multiple contigs per gene after de novo read assembly are interpreted by PHYLUCE as an indication of paralogy, and the respective loci are rejected from phylogenetic reconstruction. This can result in a dramatic decrease in potentially orthologous and phylogenetically informative data2. The alternative pipeline, HybPiper, is able to handle not only the exonic probe sequences but also the intronic flanking regions, and it identifies and separates putative paralogs. However, apart from the identification of putative paralogs, there are no further criteria for locus selection, and gene and species tree reconstruction as well as the reconstruction of organellar phylogenies are not part of HybPiper.
Therefore, phylogeneticists using exonic probe sequences lack a straightforward and well-documented bioinformatics pipeline that performs target enrichment data analysis from raw reads to species trees, including quality filtering of raw reads, read assembly, alignment of loci, evaluation of missing data and phylogenetic utility of loci, phylogenetic reconstruction in form of gene/species trees and concatenation as well as phylogenetic reconstruction from organellar data. Especially plastid reads are often obtained in sufficient quantity as part of the off-target reads (eg, 2%12; 5%13). Incongruence between the nuclear and plastid trees often gives evidence of hybridization events, and both these data sets are usually used in phylogenetics. Here, we present our pipeline HybPhyloMaker, which carries out all of these tasks.
Implementation
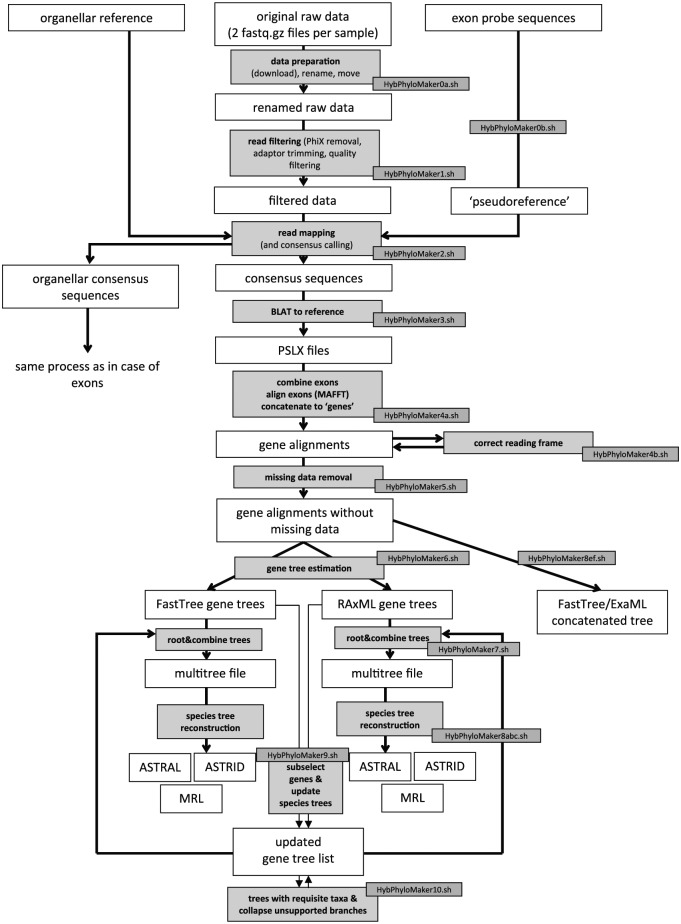
HybPhyloMaker consists of 11 major BASH scripts (HybPhyloMaker0-10) that integrate common bioinformatics tools of high-throughput sequencing and phylogenomics. These scripts perform the various steps of data analysis as separate modules within a particular directory structure that is created by them. HybPhyloMaker has a command line interface and can be run both locally and on a high-performance computer cluster. The modular BASH scripts of HybPhyloMaker enable flexible use. All steps are described in detail in Figure 1.
Figure 1.
HybPhyloMaker processing steps. Input data and intermediate results are displayed in white boxes, modification steps are shown in gray boxes. Each modification step is performed by a particular HybPhyloMaker script (small gray boxes).
Data preparation for phylogenetic tree reconstruction
HybPhyloMaker requires two types of input files: (1) paired-end Illumina reads in form of two gzipped FASTQ files per sample and (2) sequences of the probes that were used for target enrichment (FASTA file). The script HybPhyloMaker0 prepares the raw reads for HybPhyloMaker use. It gives the paired-end raw reads a unique label, sorts them according to the HybPhyloMaker-specific directory structure, and creates the reference sequence (“pseudoreference”) for the subsequent reference-guided assembly of the enriched nuclear loci: the probe sequences are concatenated and separated by a string of several hundreds of Ns each (400 Ns are recommended for 2 × 150 bp [base pairs] reads).
PhiX read removal, adapter trimming, quality filtering, and duplicate read removal are done with HybPhyloMaker1, using Bowtie 214, SAMtools15, bam2fastq16, Trimmomatic17, and FastUniq18. In a subsequent step, reads are mapped to a “pseudoreference” that was created from the probe sequences with HybPhyloMaker0. Read mapping is performed with HybPhyloMaker2 using Bowtie 2 or BWA19, and the consensus sequence is called either with OCOCO20 or Kindel21, which is also implemented in HybPhyloMaker2. The consensus sequence is called according to adjustable majority. It results in the reconstruction of the most abundant sequence, which is considered to be the ortholog, as paralogs are usually not enriched in similar quantities compared to orthologs due to a higher sequence dissimilarity to the probe sequences (see Supplement Figure 1). A similar approach was used in recent publications3,13.
With HybPhyloMaker3, the consensus sequence is fragmented into the exonic parts, which will be called contigs hereafter, and those are matched to the probe sequences using BLAT (BLAST-like alignment tool)22. Exonic multiple sequence alignments are constructed with HybPhyloMaker4a, which uses the Python script “assembled_exons_to_fastas.py”2; if an exon is missing for a particular accession, Ns are added. Also with HybPhyloMaker4a, exons are aligned using MAFFT23, and exons from the same gene are concatenated using the Perl script “catfasta2phyml.pl”24. Optionally, exon and gene alignments can be adjusted to correct the reading frame with HybPhyloMaker4b. This option later allows not only for per-exon but also for per-codon partitioning at the same time when gene trees are estimated.
With HybPhyloMaker5, the amount of missing data is calculated, and accessions as well as loci that match a user-defined threshold of missing data are retained for further analysis. In a first step, accessions that equal or exceed the maximum allowed percentage of missing data per locus are omitted from the respective loci. Then, the number of remaining accessions per locus is calculated and those loci retained that exceed the minimum allowed percentage of accessions per locus. In addition, HybPhyloMaker5 uses AMAS25, MstatX26, and trimAl27 to calculate summary statistics of properties of each locus alignment, which will in a subsequent step assist in a more stringent locus selection. Tables that summarize the amount of missing data within both the entire data set and the user-selected loci as well as histograms that show the distribution of alignment properties are provided.
Gene tree and species tree reconstruction
Gene trees are reconstructed with HybPhyloMaker6. FastTree28 and RAxML29 are the tree-building algorithms to choose from, both can be run with or without bootstrapping. FastTree is computationally less demanding than RAxML, but it tends to provide higher branch support values (based on the Shimodaira-Hasegawa test28) compared to bootstrapping in RAxML30. RAxML trees can be estimated from unpartitioned or partitioned (by exon or by codon position) data sets. In addition, HybPhyloMaker6 calculates summary statistics of properties of each gene tree, using the R script “tree_props.R” (modified from the work of Borowiec31) and the R packages ape32 and seqinr33. Alignment summary statistics, which were inferred with HybPhyloMaker5, and gene tree summary statistics are combined, and correlations among all properties are calculated and visualized with the R script “plotting_correlations.R” (modified from the work of Borowiec31). Based on those summaries and correlations, the user can optimize phylogenetic reconstruction using “quality-filtered” genes with HybPhyloMaker9. Especially saturated genes (those deviating from simple linear regression on uncorrected p-distances against inferred distances34) should be omitted from downstream analysis. However, this step is optional and users must make themselves familiar with any steps that select particular genes before applying HybPhyloMaker9. With HybPhyloMaker7, all gene trees are combined into one file and the trees optionally rooted using Newick Utilities35. Users also have the possibility to collapse unsupported branches in gene trees by specifying a minimum support value for which the branch is kept and/or subselect trees containing selected samples using HybPhyloMaker10.
Species trees are reconstructed with HybPhyloMaker8. There are several options: ASTRAL36, ASTRID37 (both coalescent summary methods), MRL38 (supertree method using matrix representation with likelihood), and maximum likelihood implemented in FastTree and ExaML39 (concatenation). In preparation of an ExaML run, the selected loci are concatenated, and gene partition information is provided by AMAS. Partition Finder 240,41 is used to find the optimal partitioning scheme.
Organellar reads
HybPhyloMaker also allows working with organellar reads that are often obtained in sufficient quantity as off-target reads (eg, 2%12; 5%13), ie, it is possible to work with organellar sequences even if one does not specifically target them. Such amount of organellar reads usually provides sufficient sequencing depth, especially for coding regions. For phylogenetic reconstruction based on organellar genomes, the user needs to provide sequences of the coding regions from the target group or from a closely related group. First, the organellar reads are extracted from the total read pool with HybPhyloMaker2 by mapping to an organellar “pseudoreference” (concatenated, coding organellar sequences that are separated by a string of several hundreds of Ns each; prepared using HybPhyloMaker0b). The resulting contigs are matched to the coding sequences with BLAT. The subsequent analysis follows the pipeline of enriched nuclear loci in most instances. Commands for processing organellar data are implemented in HybPhyloMaker2-10.
Computational implementation, performance, and pipeline comparison
HybPhyloMaker runs on major Linux distributions (Debian, Ubuntu, openSUSE, Fedora, CentOS, Scientific Linux) and on MacOS X. Automated installation of the numerous software packages that are required to run HybPhyloMaker (Table 1) is provided by the script “install_software.sh”; smaller scripts and utilities (Perl, Python, Java, and R) are provided with HybPhyloMaker. The cluster version of HybPhyloMaker was optimized on the Smithsonian Institution High Performance Cluster (SI/HPC) and the Czech National Grid Organization MetaCentrum NGI (http://metacentrum.cz/) but could easily be modified for running on any other computer cluster.
Table 1.
List of software that must be installed/must be present on the local computer/cluster before running HybPhyloMaker.
All the software could be automatically installed using the script “install_software.sh”. For each software, the source and the specific command for calling the software are provided, and it is indicated in which HybPhyloMaker script the particular software is used (HybPhyloMaker0-10). Software that needs to be installed/must be present on the computer/cluster is marked with “y”; if marked with “n”, it is provided with HybPhyloMaker and does not need to be installed.
We tested the performance of HybPhyloMaker using Hyb-Seq data sets from 6 samples of the plant genus Oxalis, each containing 1.3 to 1.9 million 2 × 150 bp raw reads. These Hyb-Seq libraries were enriched for 4,926 exons from 1,164 loci11. Run time, size of produced data files, and peak of RAM usage were recorded for each HybPhyloMaker script on a computer equipped with Intel Xeon E7-4860 CPU using 4 cores at 2.27 GHz and running CentOS 7.3.1611 (Supplement Table 1). In addition, we compared the number and percentage of mapped reads using Bowtie 2, BWA, and Geneious42 (Supplement Table 2).
Finally, we processed the same samples with HybPiper and PHYLUCE (Table 2). A direct comparison of steps within each of these pipelines (Table 3), regarding, eg, contig number, is not helpful in our opinion, due to different approaches and implementation of different software with noncomparable parameter settings in steps such as assembly (reference-guided versus de novo) and identification of contigs that match to the targeted sequences (as nucleotide sequences with BLAT [HybPhyloMaker], with exonerate49 [HybPiper], and with LASTZ50 [PHYLUCE]). We provide an approximate comparison of the three pipelines by recording the number of genes that were recovered (ie, with ≥25% completeness of each gene in case of HybPhyloMaker and HybPiper) and by indicating the number and percentage of putative paralogs in case of HybPiper and PHYLUCE. Filtering against missing data was not performed in PHYLUCE, thereby providing the most conservative number and percentage of recovered genes. Duplicate read removal was performed in case of HybPhyloMaker and HybPiper. In PHYLUCE, assembly of adapter- and quality-trimmed reads was performed with Velvet51 using k-mer length k = 35. Matching of contigs to probe sequences was performed with 90% minimum sequence identity.
Table 2.
Comparison of the performance of the three pipelines PHYLUCE, HybPiper, and HybPhyloMaker when processing 6 samples from the plant genus Oxalis11.
| Name and code | No. of raw reads | PHYLUCE | HybPiper | HybPhyloMaker | ||
|---|---|---|---|---|---|---|
| No. (%) of recovered loci; no filtering against missing data | No. (%) of recovered loci; no filtering against missing data | No. (%) of recovered loci; ≥25% data completeness | No. (%) of putative paralogs; ≥25% data completeness | No. (%) of recovered loci; ≥25% data completeness | ||
| Oxalis blastorrhiza J557 | 1 905 062 | 43 (3.7) | 1102 (94.7) | 1080 (92.8) | 11 (0.9) | 1160 (99.7) |
| Oxalis creaseyii J11-961 | 1 553 282 | 156 (13.4) | 1148 (98.6) | 1141 (98.0) | 14 (1.2) | 1161 (99.7) |
| Oxalis gracilis J558 | 1 306 633 | 125 (10.7) | 1147 (98.5) | 1139 (97.9) | 20 (1.7) | 1161 (99.7) |
| Oxalis helicoides J319 | 1 847 669 | 53 (4.6) | 1134 (97.4) | 1130 (97.1) | 15 (1.3) | 1161 (99.7) |
| Oxalis inconspicua J595 | 1 785 030 | 84 (7.2) | 1118 (96.0) | 1108 (95.2) | 14 (1.2) | 1163 (99.9) |
| Oxalis polyphylla J11-44 | 1 818 390 | 47 (4.0) | 994 (85.5) | 968 (83.2) | 5 (0.4) | 1161 (99.7) |
The number and percentage of genes that were recovered (ie, with ≥25% completeness of each gene in case of HybPiper and HybPhyloMaker) and the number and percentage of putative paralogs in case of HybPiper are reported. Filtering against missing data was not performed in PHYLUCE, thereby the most conservative number and percentage of recovered genes are provided. Duplicate read removal was performed in case of HybPiper and HybPhyloMaker.
Table 3.
Comparison between the major steps of PHYLUCE, HybPiper, and HybPhyloMaker.
| Step | PHYLUCE | HybPiper | HybPhyloMaker |
|---|---|---|---|
| Download from Illumina BaseSpace | No | No | Yes |
| Input | Paired-end Illumina reads | Paired-end and single-end Illumina reads | Paired-end Illumina reads |
| Adapter trimming and quality filtering of reads | Yes Illumiprocessor43; pairs with both mates surviving and orphaned reads are used |
No Adapter trimming and quality filtering of reads need to be performed before using HybPiper; pairs with both mates surviving are used as input for HybPiper |
Yes Trimmomatic; pairs with both mates surviving and orphaned reads are used |
| Duplicate read removal | No | Yes (Super deduper44) | Yes (FastUniq) |
| Assembly | De novo (Velvet; ABySS45; Trinity46) | De novo (SPAdes47) | Reference-guided (Bowtie 2/BWA; OCOCO/Kindel) |
| Identification of sequences that match to the targeted sequences | Done by matching contigs to the targeted sequences (as nucleotide sequences with LASTZ); after assembly | Before assembly: done by matching reads to the targeted sequences (as peptide sequences with BLASTX); as nucleotide sequences with BWA; After assembly: done by matching contigs to the targeted sequences with exonerate |
Done by matching contigs to the targeted sequences (as nucleotide sequences with BLAT); after assembly |
| Filtering against paralogs | Yes Paralogy is indicated if a targeted locus matches multiple contigs or if a contig matches multiple targeted loci (the respective loci are excluded) |
Yes Paralogy is indicated if a targeted locus matches multiple long-length contigs (the respective loci are flagged); separation of putative paralogs possible |
No Consensus calling after the reference-guided assembly is according to majority; this results in the reconstruction of the most abundant sequence, which is considered to be the ortholog |
| Particularly suitable for exonic probe sequences | No | Yes | Yes |
| Extraction of flanking intronic regions | No | Yes | No |
| Missing data calculation | Yes | No | Yes |
| Calculation of alignment and gene tree properties | No | No | Yes |
| Flexible handling of excluding accessions and loci | Yes | No | Yes |
| Gene tree reconstruction | No | No | Yes (RAxML, FastTree) |
| Concatenation | Yes (ExaBayesa,48; RAxML; ExaMLa) | No | Yes (FastTree, ExaMLa) |
| Species tree reconstruction | No | No | Yes (ASTRAL, ASTRID, MRL) |
| Organellar phylogeny | No | Yes (from coding sequences) |
Input file preparation.
Results and Discussion
Performance and pipeline comparison
Computer performance of HybPhyloMaker is summarized in Supplement Table 1. The most time-consuming steps are read mapping and consensus calling, reconstruction of RAxML gene trees, and ExaML analysis of the concatenated and partitioned data set. The most RAM memory-demanding step is phylogenetic tree reconstruction based on the concatenated data set (both FastTree and ExaML). The largest files are FASTQ files that are generated during raw read processing and BAM files obtained in the step of read mapping. Geneious performed best among the three implemented mapping software: BWA and Bowtie 2 mapped 78% to 93% and 67% to 80% of reads that were mapped by Geneious, respectively.
HybPhyloMaker is the first data analysis pipeline for hybridization-based target enrichment data that are generated with exonic probe sequences, which performs all relevant steps from raw reads to species and organellar trees. Two alternative, well-documented data analysis pipelines are available, PHYLUCE and HybPiper, and a detailed comparison of the steps of these two pipelines with HybPhyloMaker is provided in Table 3. Major differences between them are as follows: (1) The assembly strategy (de novo in PHYLUCE and HybPiper or reference-guided in HybPhyloMaker): both assembly strategies allow for the assembly of both exonic and intronic regions. In HybPhyloMaker, this is due to the use of a reference sequence that is built from the concatenated exonic probe sequences, which are separated by a string of several hundreds of Ns each. (2) Paralog identification: both PHYLUCE and HybPiper detect putatively paralogous loci, which are either excluded from subsequent analyses (PHYLUCE) or flagged (HybPiper). In HybPhyloMaker, an adjustable majority consensus sequence is obtained. This results in the reconstruction of the most abundant sequence, which is considered to be the ortholog (Supplement Figure 1). (3) Suitability for exonic probe sequences: both HybPiper and HybPhyloMaker are tailored for exonic probe sequences, whereas PHYLUCE might exclude a large number of loci in case one works with multiple targeted exons per gene (Table 2), as in such case multiple contigs per gene are often formed, which is an indicator of paralogy in PHYLUCE. HybPiper filters putative paralogs less stringently (Table 2), as in case of multiple contigs per gene these contigs must exceed a certain minimum length threshold (>85% length of the targeted locus). (4) Extraction of flanking intronic regions: only HybPiper provides a script for that, the other pipelines obtain these intronic regions during assembly, but do not process them further. (5) Missing data calculation: PHYLUCE and HybPhyloMaker offer estimation of missing data. (6) Calculation of alignment and gene tree properties: this is only implemented in HybPhyloMaker. The alignment properties comprise number of accessions, alignment length, proportion of variable sites, proportion of parsimony informative sites, GC content, alignment entropy, and conservation distribution. Gene tree properties are as follows: average bootstrap support, average branch length, average uncorrected p-distance, clocklikeness, simple linear regression on uncorrected p-distances against inferred distances, and long-branch score. (7) Flexible handling of excluding accessions and loci: this is possible in both PHYLUCE and HybPhyloMaker. (8) Gene and species tree reconstruction: software for gene tree reconstruction is implemented in PHYLUCE and for both gene and species tree reconstruction implemented in HybPhyloMaker. HybPhyloMaker offers per-exon and per-codon partitioning. (9) Reconstruction of organellar phylogenies: only HybPhyloMaker offers their reconstruction, based on coding regions.
We consider PHYLUCE not well suitable for exclusively exonic probe sequences due to the drastic loss of potentially orthologous loci (Table 2). HybPiper has the benefits of extraction of the flanking intronic regions, which are especially needed in the reconstruction of shallow phylogenies, and identification of putative paralogs. The identification of paralogs is mainly essential (1) if putatively paralogous loci are not excluded during probe design (in such case, the identified paralogs should be excluded from phylogenetic reconstruction) and (2) if the ancestry of an allopolyploid is of interest (in such case, paralogs can be beneficial for the inference of complex reticulate relationships52,53). HybPhyloMaker treats the most abundant sequence of a locus as ortholog and does not identify putatively paralogous loci, which we consider an appropriate approach, except for the latter two cases.
Compared with existing target enrichment data analysis pipelines, HybPhyloMaker offers the following main advantages:
It implements all steps of target enrichment data analysis: from raw reads to species tree reconstruction.
It provides calculation and summary of many alignment and gene tree properties that assist the user in the selection of appropriate “quality-filtered” genes for species tree reconstruction. This step is optional and users must make themselves familiar with any steps that select particular genes.
It implements several species tree reconstruction methods (ASTRAL, ASTRID, MRL) as well as concatenation (FastTree, ExaML).
It allows the analysis of the coding part of organellar genomes, ie, the analysis of a large proportion of the off-target reads, especially plastid reads.
Conclusions
HybPhyloMaker is a user-friendly pipeline that conducts the analysis of phylogenetic Hyb-Seq data sets from raw reads to species tree reconstruction. It is written in BASH and requires a priori installation of several other software packages. An install script is provided for easy installation of these software packages. HybPhyloMaker runs on major Linux distributions and MacOS X. The software is open source and available in https://github.com/tomas-fer/HybPhyloMaker/.
Supplementary Material
Acknowledgments
The authors thank Aaron Liston, Kevin Weitemier (Oregon State University), and Shannon Straub (Hobart and William Smith Colleges) for sharing ideas about Hyb-Seq data analysis.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Computational resources were provided by CESNET LM2015042 and CERIT Scientific Cloud LM2015085 under the program “Projects of Large Research, Development, and Innovations Infrastructures”. Smithsonian Institution provided funds to T.F. enabling pipeline coding. This work was supported by the Czech Science Foundation (GACR) (GA14-13541S to T.F. and 16-15134Y to R.E.S.).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: TF and RES conceived the tool and drafted the manuscript; TF coded the pipeline. Both authors read and approved the final manuscript.
References
- 1. Mandel JR, Dikow RB, Funk VA, et al. A target enrichment method for gathering phylogenetic information from hundreds of loci: an example from the Compositae. Appl Plant Sci. 2014;2:1300085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weitemier K, Straub SCK, Cronn RC, et al. Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl Plant Sci. 2014;2:1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholls JA, Pennington RT, Koenen EJ, et al. Using targeted enrichment of nuclear genes to increase phylogenetic resolution in the neotropical rain forest genus Inga (Leguminosae: Mimosoideae). Front Plant Sci. 2015;6:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lemmon EM, Lemmon AR. High-throughput genomic data in systematics and phylogenetics. Ann Rev Ecol Evol Syst. 2013;44:99–121. [Google Scholar]
- 5. Heyduk K, Stephens JD, Faircloth BC, Glenn TC. Targeted DNA region re-sequencing. In: Aransay AM, Lavín Trueba JL, eds. Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing. Berlin, Germany: Springer; 2016:43–68. [Google Scholar]
- 6. Faircloth BC. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics. 2016;32:786–788. [DOI] [PubMed] [Google Scholar]
- 7. Johnson MG, Gardner EM, Liu Y, et al. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl Plant Sci. 2016;4:1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. Ultraconserved elements anchor thousands of genetic markers for target enrichment spanning multiple evolutionary timescales. Syst Biol. 2012;61:717–726. [DOI] [PubMed] [Google Scholar]
- 9. Faircloth BC, Sorenson L, Santini F, Alfaro ME. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS ONE. 2013;8:e65923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reneker J, Lyons E, Conant GC, et al. Long identical multispecies elements in plant and animal genomes. Proc Natl Acad Sci U S A. 2012;109:E1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmickl R, Liston A, Zeisek V, et al. Phylogenetic marker development for target enrichment from transcriptome and genome skim data: the pipeline and its application in southern African Oxalis (Oxalidaceae). Mol Ecol Resour. 2016;16:1124–1135. [DOI] [PubMed] [Google Scholar]
- 12. Stephens JD, Rogers WL, Mason CM, Donovan LA, Malmberg RL. Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Amer J Bot. 2015;102:910–920. [DOI] [PubMed] [Google Scholar]
- 13. Folk RA, Mandel JR, Freudenstein JV. A protocol for targeted enrichment of intron-containing sequence markers for recent radiations: a phylogenomic example with genomic resources from Heuchera (Saxifragaceae). Appl Plant Sci. 2015;3:1500039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Handsaker B, Wysoker A, et al; 1000. Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. HudsonAlpha. bam2fastq. http://gsl.hudsonalpha.org/information/software/bam2fastq/, 2010. Accessed June 20, 2017.
- 17. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu H, Luo X, Qian J, et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE. 2012;7:e52249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Břinda K, Boeva V, Kucherov G. Dynamic read mapping and online consensus calling for better variant detection. arXiv.org. 2016:160509070. https://arxiv.org/abs/1605.09070. [Google Scholar]
- 21. Constantinides B. Kindel: indel-aware consensus calling. https://github.com/bede/kindel, 2017. Accessed June 20, 2017.
- 22. Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. [DOI] [PubMed] [Google Scholar]
- 24. Nylander J. catfasta2phyml. https://github.com/nylander/catfasta2phyml/, 2016. Accessed June 20, 2017.
- 25. Borowiec ML. AMAS: a fast tool for alignment manipulation and computing of summary statistics. Peer J. 2016;4:e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collet G. MstatX. https://github.com/gcollet/MstatX/, 2012. Accessed June 20, 2017.
- 27. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. [DOI] [PubMed] [Google Scholar]
- 31. Borowiec M. good_genes. https://github.com/marekborowiec/good_genes, 2016. Accessed June 20, 2017.
- 32. Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. [DOI] [PubMed] [Google Scholar]
- 33. Charif D, Lobry JR. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, eds. Structural Approaches to Sequence Evolution (Series Biological and Medical Physics, Biomedical Engineering). Berlin, Germany: Springer; 2007:207–232. [Google Scholar]
- 34. Philippe H, Forterre P. The rooting of the universal tree of life is not reliable. J Mol Evol. 1999;49:509–523. [DOI] [PubMed] [Google Scholar]
- 35. Junier T, Zdobnov EM. The Newick Utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26:1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vachaspati P, Warnow T. ASTRID: Accurate Species TRees from Internode Distances. BMC Genomics. 2015;16:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nguyen N, Mirarab S, Warnow T. MRL and SuperFine+MRL: new supertree methods. Algorithms Mol Biol. 2012;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozlov AM, Aberer AJ, Stamatakis A. ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics. 2015;31:2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. [DOI] [PubMed] [Google Scholar]
- 41. Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 2014;14:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faircloth B. Illumiprocessor: parallel adapter and quality trimming. https://illumiprocessor.readthedocs.io/en/latest/, 2013. Accessed June 20, 2017.
- 44. Petersen KR, Streett DA, Gerritsen AT, Hunter SS, Settles ML. Super deduper, fast PCR duplicate detection in fastq files. Paper presented at: Proceedings of the 6th ACM Conference On Bioinformatics, Computational Biology and Health Informatics; September 9-12, 2015; Atlanta, GA:491–492. New York, NY: ACM. [Google Scholar]
- 45. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol İ. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aberer AJ, Kobert K, Stamatakis A. ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol Biol Evol. 2014;31:2553–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris RS. Improved Pairwise Alignment of Genomic DNA [PhD thesis]. State College, PA: The Pennsylvania State University; 2007. [Google Scholar]
- 51. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Doyle JJ, Doyle JL, Rauscher JT, Brown AHD. Diploid and polyploid reticulate evolution throughout the history of the perennial soybeans (Glycine subgenus Glycine). New Phytol. 2004;161:121–132. [Google Scholar]
- 53. Kamneva OK, Syring J, Liston A, Rosenberg NA. Evaluating allopolyploid origins in strawberries (Fragaria) using haplotypes generated from target capture sequencing. BMC Evol Biol. 2017;17:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.