Abstract
Although experimental evidence suggests calcium-sensing receptor (CASR) as a tumor-suppressor, the prognostic role of tumor CASR expression in colorectal carcinoma remains unclear. We hypothesized that higher tumor CASR expression might be associated with improved survival among colorectal cancer patients. We evaluated tumor expression levels of CASR by immunohistochemistry in 809 incident colorectal cancer patients within the Nurses’ Health Study and the Health Professionals Follow-up Study. We used Cox proportional hazards regression models to estimate multivariable hazard ratio (HR) for the association of tumor CASR expression with colorectal cancer-specific and all-cause mortality. We adjusted for potential confounders including tumor biomarkers such as microsatellite instability, CpG island methylator phenotype, LINE-1 methylation level, expressions of PTGS2, VDR, and CTNNB1, and mutations of KRAS, BRAF, and PIK3CA. There were 240 colorectal cancer-specific deaths and 427 all-cause deaths. The median follow-up of censored patients was 10.8 years (interquartile range: 7.2, 15.1). Compared to patients with no or weak expression of CASR, the multivariable HRs for colorectal cancer-specific mortality were 0.80 [95% confidence interval (CI): 0.55-1.16] in patients with moderate CASR expression, and 0.50 (95% CI: 0.32-0.79) in patients with intense CASR expression (p-trend = 0.003). The corresponding HRs for overall mortality were 0.85 (0.64-1.13) and 0.81 (0.58-1.12), respectively. Higher tumor CASR expression was associated with a lower risk of colorectal cancer-specific mortality. This finding needs further confirmation and if confirmed, may lead to better understanding of the role of CASR in colorectal cancer progression.
Introduction
The calcium sensing receptor (CASR) is a ubiquitously expressed G-protein coupled receptor and acts as the master regulator of the calcium homeostasis.1 CASR serves as the molecular sensor of ionized calcium (Ca2+). Binding of Ca2+ to the CASR propagates intracellular signaling cascades which are critical in both physiologic and pathologic states.2 Also, CASR has pleiotropic effects and can regulate gene expression, inflammation, cell proliferation, cell differentiation, and apoptosis.3–5 Deregulation of CASR has been implicated in different types of benign or malignant tumors of prostate, breast, parathyroid, and colon.3, 6–11
Although the underlying mechanisms are not completely understood, higher calcium intake was associated with a lower risk of colorectal cancer in most epidemiological studies.12, 13 Experimental studies reported a possible role of CASR in inhibiting proliferation of colonic epithelia.14 CASR knockout mice showed increased incidence of formation of pre-neoplastic lesion of crypt foci in the colon,15 and elevated inflammatory markers in colon epithelium, as well as more susceptibility to chemically induced colitis compared to the colons of wild type mice.15, 16 In addition, treatment with a calcimimetic sensitized colon cancer cells to the chemoprotective function of calcium and inhibited cell growth.3 Moreover, expression of CASR was decreased in advanced colorectal adenomas and undifferentiated tumors.17 Furthermore, recent evidence indicates the role of epigenetic changes such as promoter hypermethylation, histone deacetylation, and non-coding RNA involvement in the regulation of CASR expression.18, 19 Collectively, these lines of evidence indicate that CASR expression and function regulate a fine balance between proliferation, differentiation, and apoptosis.3 Despite these biological data, the association between tumor CASR expression with survival among colorectal cancer patients has not specifically been examined. We hypothesized that tumor CASR expression is associated with colorectal cancer specific survival independent of other molecular and clinical characteristics.
To test this hypothesis, we utilized two prospective cohorts of women (the Nurses’ Health Study, NHS) and men (the Health Professionals Follow-Up Study, HPFS). Because both cohorts provide detailed data on major tumor molecular features, including tumor status of microsatellite instability (MSI), CpG island methylator phenotype (CIMP), long interspersed nucleotide element-1 (LINE-1) methylation level, and KRAS, BRAF, and PIK3CA mutations, we were able to evaluate the independent association of tumor CASR expression with patient-related outcomes.
Patients and Methods
Study population
The study population consisted of the participants of two large prospective cohort studies, the NHS (N = 121,700 women observed since 1976) and HPFS (N = 51,529 men observed since 1986, HPFS). Since the baseline, participants have completed questionnaires regarding information on demographics, medical history, and lifestyle factors every two years. The study was approved by the institutional review board at Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health.
Ascertainment of cases of colorectal cancer, tumor tissue collection, and mortality measurements
Participants were asked on biennial questionnaires to report a diagnosis of colorectal cancer and other diseases. Study physicians received permission from study participants or next-of-kin (for deceased) to obtain their medical records and pathological reports on colorectal cancer. Study physicians who were unaware of tumor CASR levels and other molecular data, confirmed the incidence of colorectal cancer and recorded the information on disease stage, tumor location, and histological type of the cancer. For deceased participants with known or suspected cancer for which we have not been able to obtain medical records, we contacted the state tumor registry to confirm and classify the cancer. Colorectal cancer was defined according to the International Classification of Diseases, Ninth Revision (ICD-9).20 More than 98% of deaths were identified through the registry of National Death Index21 or by family report. Cause of death has been identified from death certificates or review of medical records. Paraffin-embedded tissue blocks have been obtained from hospitals where patients underwent respective surgeries.22 Based on the availability of tumor tissue data on CASR expression, we included a total of 809 colorectal cancer cases diagnosed up to 2008 for this study.
Immunohistochemistry of CASR and other markers
We constructed tissue microarrays (TMA)22–24 from colorectal cancer blocks, and performed immunohistochemistry (IHC) assays to measure tumor CASR expression. Tissue sections were deparaffinized, rehydrated, and heated in a microwave for 15 min in Antigen Retrieval Citra Solution, pH 6 (BioGenex Laboratories, San Ramon, CA, USA). Sections were incubated with Dual Endogenous Enzyme Block (Dako, Glostrup, Denmark), followed by the treatment with Protein Block Serum-Free (Dako). Slides were then incubated for 1 hour at room temperature with a rabbit polyclonal anti-CASR antibody (ab137408; Abcam, Cambridge, MA, USA; dilution, 1:100). The primary antibody was visualized using EnVision+ System-HRP (Dako) with diaminobenzidine, and counterstained with hematoxylin. Sections processed with the replacement of primary antibody by Tris-buffered saline were used as a negative control. Immunohistochemical assessment for CASR was interpreted by a pathologist (Y. Masugi) blinded to other data. According to previously reported criteria,8 tumor CASR expression was scored as 0 (no/minimal staining), 1 (weak staining), 2 (moderately intense staining), and 3 (intense staining) based on the staining intensity in colorectal carcinoma cells (Figure 1). CASR expression was observed predominantly in the cytoplasm and membrane of colorectal carcinoma cells, but occasionally in the nucleus of tumor cells. Consistent with the previous report,8 we evaluated the intensity of cytoplasmic/membrane stain for scoring. We observed CASR expression in many types of cells, including colorectal normal epithelial cells, immune cells, endothelial cells, smooth muscle cells, and ganglion cells. A second observer (Z.R.Q), unaware of other data, examined the CASR expression in 118 tumors. The concordance between the two observers was reasonable with a weighted kappa value of 0.71 (95% CI: 0.61-0.82). As shown in our previous studies, we have already conducted IHC assays on PTGS2 (cyclooxygenase-2) using anti-PTGS2 antibody24 (clone CX229; mouse monoclonal antibody; dilution 1:300; Cayman Chemical), nuclear VDR (vitamin D receptor) using anti-VDR antibody25 (dilution 1:500; rabbit polyclonal antibody; Novus Biologicals, NBP1-19478, Littleton), and CTNNB1 (β-catenin) using anti-CTNNB1 antibody26 (clone 14; mouse monoclonal antibody; dilution 1:400; BD Transduction Laboratories).
Figure 1.
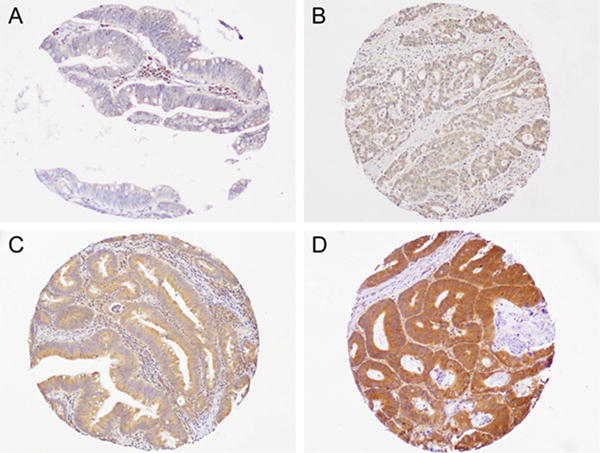
Calcium sensing receptor (CASR) expression in colorectal cancer. (A) 0 (no/minimal CASR expression), (B) 1 (weak CASR expression), (C) 2 (moderately intense CASR expression), and (D) 3 (intense CASR expression), based on the staining intensity in colorectal carcinoma cells.
Analysis of microsatellite instability, KRAS, BRAF, PIK3CA mutations, and other markers
We extracted DNA from paraffin-embedded tissue and performed PCR and pyrosequencing for BRAF (codon 600), KRAS (codons 12, 13, 61, and 146), and PIK3CA (exons 9 and 20).25, 27–30 Microsatellite instability (MSI) status was identified using a 10-marker panel (BAT25, BAT26, BAT40, D2S123, D5S346, D17S250, D18S55, D18S56, D18S67, and D18S487).28 Presence of instability in ≥ 30% of the markers was defined as MSI-high.28 We also performed analyses of long interspersed nucleotide element-1 (LINE-1) methylation31 and eight CpG island methylator phenotype (CIMP)-specific loci (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1).32 Based on the previously established criteria,33 methylation of six or more of the eight markers using the eight-marker CIMP panel was defined as CIMP-high.
Statistical analysis
Cox proportional hazard models were used to calculate the hazard ratios (HRs) and 95% confidence intervals (95% CIs) of death as a result of colorectal cancer (for colorectal cancer-specific mortality) or death as a result of any cause (for overall mortality) according to tumor CASR expression (3-tiered; no or weak, moderate, intense). We conducted the analyses in each study as well as in the combined cohorts. For the main analyses, death as a result of colorectal cancer was the primary endpoint and death due to other causes was censored. In secondary analyses, death as a result of any cause was the end point. Patients were followed up until death or 1 June 2012 for the NHS and 31 January 2012 for the HPFS, whichever occurred first. We stratified by age at diagnosis, cohort (for the pooled analysis) and disease stage, as well as further adjusted for year of diagnosis, family history of colorectal cancer, tumor differentiation, tumor location, MSI status, CIMP status, LINE-1 methylation level, PTGS2 expression, and KRAS, BRAF, and PIK3CA mutations, and nuclear expression of VDR and CTNNB1 (see Table 2 for the categorizations of these variables). There were 65 missing (7.9%) in tumor differentiation, 23 missing (2.9%) in MSI status, 65 missing (7.9%) in CIMP status, 20 missing (2.5%) in BRAF mutation, 22 (2.9%) missing in KRAS mutation, 67 missing (7.9%) in PIK3CA mutation, 21 missing (2.5%) in PTGS2 expression, 243 missing (30.1%) in nuclear VDR expression, and 34 missing (4.2%) in nuclear CTNNB1 expression. For variables with missing, we assigned a separate (“missing”) indicator variable and included those cases in the multivariate Cox models.22 We confirmed that excluding cases with a missing variable did not materially alter results (data not shown).
Table 2.
Association of tumor CASR expression with colorectal cancer specific mortality in the Nurses’ Health Study and Health Professionals Follow-Up Study
| Tumor CASR expression
|
||||
|---|---|---|---|---|
| 0-1 No/weak |
2 Moderate |
3 Intense |
p-trend | |
| Women (Nurses’ Health Study) | ||||
| N death (132) | 27 | 82 | 23 | |
| N patients (458) | 91 | 270 | 97 | |
| Model 1* | 1 (reference) | 0.69 (0.43-1.11) | 0.49 (0.27-0.90) | 0.02 |
| Model 2** | 1 (reference) | 0.79 (0.48-1.32) | 0.54 (0.29-1.03) | 0.06 |
| Men (Health Professionals Follow-Up Study) | ||||
| N death (108) | 23 | 62 | 23 | |
| N patients (351) | 65 | 191 | 95 | |
| Model 1* | 1 (reference) | 1.00 (0.59-1.69) | 0.62 (0.33-1.19) | 0.12 |
| Model 2** | 1 (reference) | 0.71 (0.39-1.28) | 0.37 (0.18-0.76) | 0.006 |
| Pooled | ||||
| N death (240) | 50 | 144 | 46 | |
| N patients (809) | 156 | 461 | 192 | |
| Model 1* | 1 (reference) | 0.82 (0.58-1.16) | 0.55 (0.35-0.85) | 0.006 |
| Model 2** | 1 (reference) | 0.80 (0.55-1.16) | 0.50 (0.32-0.79) | 0.003 |
Model 1: Cox model stratified by age groups at diagnosis (<50, 50-59, 60-69, and ≥70 years), study (for the pooled analysis), and disease stage (I, II, III, IV, and unspecified), with additional adjustment for age at diagnosis (continuous).
Model 2: We further adjusted for year of diagnosis (continuous), family history of colorectal cancer (yes, no), tumor differentiation (well differentiated, moderately differentiated, poorly differentiated, and unspecified), tumor subsite (proximal colon, distal colon, rectum and unspecified), LINE-1 (continuous), MSI, CIMP, PTGS2, and KRAS, BRAF, and PIK3CA mutations, nuclear expression of VDR and CTNNB1 (all binary).
As sensitivity analyses, we evaluated the association of tumor CASR expression with colorectal cancer-specific mortality according to disease stage (stage 1-2 versus stage 3-4), and tumor differentiation (well to moderate versus poor), as well as categorized tumor CASR expression levels into binary variables (no or weak versus moderate-to-intense expression). We assessed the interaction by including the cross-product term of two variables of interest in the Cox model and performed a Wald test.
The X2 test and analysis of variance were used to examine associations between different categorical variables and continuous variables, respectively. Two-sided hypothesis testing was used for all the comparisons. Analyses were done using SAS 9.3 (SAS institute, Cary, NC).
Results
Among 809 eligible patients with available tumor CASR data in these cohorts, 156 patients (19%) had no or weak expression, 461 (57%) had moderate expression, and 192 (24%) had intense expression (Table 1). On average colorectal cancer patients were followed 10.8 years (interquartile range: 7.2, 15.1 years), during which there were 240 colorectal cancer-specific death and 427 all-cause mortality. As shown in Table 1, moderate to intense tumor expression of CASR was associated with well to moderately differentiated tumors (p = 0.04). Additionally, no or weak CASR expression was significantly associated with CIMP-high (p = 0.02), PTGS2 negative (p < 0.001) and CTNNB1 negative (p = 0.02) tumors. The included colorectal cancer cases with tumor CASR expression data were comparable to those colorectal cancer patients without the tumor CASR data (Supplementary Table 1).
Table 1.
Selected clinical, pathological, and molecular characteristics among colorectal cancer patients according to tumor calcium sensing receptor (CASR) expression in the Nurses’ Health Study and Health Professionals Follow-Up Study
| Tumor CASR expression**
|
|||||
|---|---|---|---|---|---|
| Characteristics* | All cases | 0-1 No/weak |
2 Moderate |
3 Intense |
p† |
| Overall (N = 809) | 809 (100%) |
156 (19%) | 461 (57%) | 192 (24%) | 0.13 |
| Female (N = 458, NHS) | 458 (57%) | 91 (59%) | 270 (59%) | 97 (51%) | |
| Male (N = 351, HPFS) | 351 (43%) | 65 (41%) | 191 (41%) | 95 (49%) | |
| Mean age ± SD (years) | 69 ± 9 | 69 ± 9 | 69 ± 9 | 69 ± 8 | 0.41 |
| Year of diagnosis | 0.01 | ||||
| Prior to 1996 | 246 (30%) | 41 (26%) | 135 (29%) | 70 (36%) | |
| 1996-2000 | 217 (27%) | 39 (25%) | 117 (25%) | 61 (32%) | |
| 2001-2008 | 346 (43%) | 76 (49%) | 209 (45%) | 61 (32%) | |
| Family history of colorectal cancer in first-degree relative(s) | 0.16 | ||||
| No | 640 (79%) | 123 (79%) | 374 (81%) | 143 (74%) | |
| Yes | 169 (21%) | 33 (21%) | 87 (19%) | 49 (26%) | |
| Tumor location | 0.45 | ||||
| Proximal colon | 406 (50%) | 88 (56%) | 229 (50%) | 89 (46%) | |
| Distal colon | 233 (29%) | 35 (23%) | 134 (29%) | 64 (33%) | |
| Rectum | 158 (19%) | 30 (19%) | 92 (20%) | 36 (19%) | |
| Unspecified | 12 (2%) | 3 (2%) | 6 (1%) | 3 (2%) | |
| Tumor differentiation | 0.04 | ||||
| Well to moderate | 613 (82%) | 110 (77%) | 346 (82%) | 157 (88%) | |
| Poor | 132 (18%) | 33 (23%) | 77 (18%) | 22 (12%) | |
| AJCC disease stage | 0.48 | ||||
| I | 192 (24%) | 42 (27%) | 96 (21%) | 54 (28%) | |
| II | 256 (32%) | 46 (29%) | 153 (33%) | 57 (30%) | |
| III | 215 (27%) | 38 (25%) | 128 (28%) | 49 (26%) | |
| IV | 116 (14%) | 23 (15%) | 70 (15%) | 23 (12%) | |
| Unspecified | 30 (4%) | 7 (5%) | 14 (3%) | 9 (5%) | |
| MSI status | 0.07 | ||||
| MSS/MSI-low | 652 (83%) | 118 (77%) | 375 (84%) | 159 (86%) | |
| MSI-high | 134 (17%) | 36 (23%) | 73 (16%) | 25 (14%) | |
| CIMP status | 0.02 | ||||
| CIMP-low/negative | 612 (82%) | 112 (77%) | 342 (82%) | 158 (88%) | |
| CIMP-high | 132 (18%) | 34 (23%) | 77 (18%) | 21 (12%) | |
| Mean LINE-1 methylation level ± SD (%) | 62.8 ± 9.6 | 62.9 ± 10.3 | 62.6 ± 9.5 | 61.3 ± 9.1 | 0.21 |
| BRAF mutation | 0.06 | ||||
| Wild type | 671 (85%) | 123 (79%) | 384 (86%) | 164 (88%) | |
| Mutant | 118 (15%) | 32 (21%) | 64 (14%) | 22 (12%) | |
| KRAS mutation | 0.22 | ||||
| Wild type | 464 (59%) | 97 (63%) | 266 (59%) | 101 (54%) | |
| Mutant | 323 (41%) | 57 (37%) | 180 (41%) | 86 (46%) | |
| PIK3CA mutation | 0.88 | ||||
| Wild type | 627 (84%) | 129 (85%) | 357 (85%) | 141 (83%) | |
| Mutant | 115 (16%) | 22 (15%) | 65 (15%) | 28 (17%) | |
| PTGS2 (cyclooxygenase-2) expression | <0.001 | ||||
| Negative | 321 (41%) | 79 (54%) | 182 (40%) | 60 (31%) | |
| Positive | 467 (59%) | 67 (46%) | 270 (60%) | 130 (69%) | |
| Nuclear VDR expression | |||||
| Negative | 332 (58%) | 64 (63%) | 182 (57%) | 86 (57%) | 0.32 |
| Positive | 234 (42%) | 34 (37%) | 139 (43%) | 61 (43%) | |
| Nuclear CTNNB1 expression | |||||
| Negative | 418 (54%) | 90 (62%) | 243 (54%) | 85 (45%) | 0.02 |
| Positive | 357 (46%) | 58 (38%) | 199 (46%) | 100 (54%) | |
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype-specific promoters; HPFS, Health Professionals Follow-Up Study; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
There were 65 missing in tumor differentiation, 23 missing in MSI status, 65 missing in CIMP status, 20 missing in BRAF mutation, 22 missing in KRAS mutation, 67 missing in PIK3CA mutation, 21 missing in PTGS2 expression, 243 missing in nuclear VDR expression, and 34 missing in nuclear CTNNB1 expression.
0 (no/minimal staining), 1 (weak staining), 2 (moderately intense staining), and 3 (intense staining).
To compare characteristics between subgroups, we used the chi-square test for categorical variables and an analysis of variance for continuous variables.
For the colorectal-cancer specific mortality analyses, the multivariable HRs comparing patients with intense tumor CASR expression versus patients with no or weak expression were 0.54 (95% CI: 0.29-1.03, p-trend = 0.06) in the NHS, 0.37 (95% CI: 0.18-0.76, p-trend = 0.006) in the HPFS, and 0.50 (95% CI: 0.32-0.79, p-trend = 0.003) for the combined cohorts (p-interaction by gender = 0.81, Table 2). The age-adjusted results were similar to multivariable results in women but strengthened in men. Adjusting for MSI status was primarily responsible for the observed strengthened association in men because patients with intense tumor CASR expression tended to have a lower prevalence of MSI-high, which is generally associated with a better prognosis. For men and women, the observed association appeared to be similar across strata of disease stage and tumor differentiation (all p-value for interaction > 0.15, Supplementary Table 2).
For the all-cause mortality analyses, for the same comparison, the multivariate HRs (95% CIs) were 0.77 (0.48-1.25, p-trend = 0.32) for the NHS, 0.78 (0.48-1.28, p-trend = 0.29) for HPFS, and 0.81 (0.58-1.12, p-trend = 0.22) for the combined cohorts (p value for interaction by gender = 0.98, Table 3). As sensitivity analyses, we categorized tumor CASR expression into binary categories and found borderline significant association with colorectal cancer-specific mortality (HR: 0.71, 95% CI: 0.49-1.01) but not with all-cause mortality (HR: 0.84, 95% CI: 0.64-1.10, Supplementary Table 3). Additionally, we have also conducted interaction analyses between tumor CASR expression with other known prognostic markers for CRC, including MSI, CIMP, KRAS, and BRAF mutations. We did not observe any significant interactions (all p-value for interactions > 0.20).
Table 3.
Association of tumor CASR expression with overall mortality in the Nurses’ Health Study and Health Professionals Follow-Up Study
| Tumor CASR expression
|
||||
|---|---|---|---|---|
| 0-1 No/weak |
2 Moderate |
3 Intense |
p-trend | |
| Women (Nurses’ Health Study) | ||||
| N death (218) | 40 | 133 | 45 | |
| N patients (458) | 91 | 270 | 97 | |
| Model 1* | 1 (reference) | 0.74 (0.51-1.09) | 0.75 (0.47-1.19) | 0.24 |
| Model 2** | 1 (reference) | 0.77 (0.52-1.14) | 0.77 (0.48-1.25) | 0.32 |
| Men (Health Professionals Follow-Up Study) | ||||
| N death (209) | 38 | 111 | 60 | |
| N patients (351) | 65 | 191 | 95 | |
| Model 1* | 1 (reference) | 1.00 (0.67-1.48) | 0.97 (0.63-1.51) | 0.89 |
| Model 2** | 1 (reference) | 0.93 (0.61-1.42) | 0.78 (0.48-1.28) | 0.29 |
| Pooled | ||||
| N death (427) | 78 | 244 | 105 | |
| N patients (809) | 156 | 461 | 192 | |
| Model 1* | 1 (reference) | 0.87 (0.66-1.13) | 0.85 (0.62-1.17) | 0.36 |
| Model 2** | 1 (reference) | 0.85 (0.64-1.13) | 0.81 (0.58-1.12) | 0.22 |
Model 1: Cox model were stratified by age groups at diagnosis, study (for the pooled analysis), and cancer stage, with additional adjustment for age at diagnosis.
Model 2: We further adjusted for year of diagnosis, family history of colorectal cancer, tumor differentiation, tumor subsite, LINE-1, MSI, CIMP, PTGS2, and KRAS, BRAF, and PIK3CA mutations, nuclear expression of VDR and CTNNB1.
Discussion
In this large cohort of patients with colorectal cancer, we found that higher tumor CASR expression was associated with a modestly lower risk of colorectal cancer-specific mortality in both men and women. This association was independent of various clinical and molecular variables such as disease stage, tumor differentiation, and tumor molecular features. In contrast, tumor CASR expression was not associated with all-cause mortality, indicating a potential role of CASR in colorectal cancer progression and pathogenesis.
In line with our findings, in vitro studies of colorectal cancer cell lines showed that higher CASR expression was positively associated with differentiation and apoptotic markers but inversely associated with proliferation markers.3, 34, 35 Specifically, CASR/parathyroid hormone double knock-out mice showed significant up-regulation of markers of proliferation (e.g. increase in MKI67 (Ki-67) labeling index) and down-regulation of markers of differentiation (e.g., decrease in CTNNB1 Ser-552 phosphorylation). However, restoration of CASR function considerably suppressed the tumorigenesis phenotype of colorectal cancer cells, suggesting CASR’s increased apoptotic and differentiation potential.3 Hence, CASR appeared to act as a tumor suppressor and a master regulator for maintaining normal intestinal cell turn over and homeostasis.15, 36
We observed loss of CASR expression is significantly prevalent in the poorly differentiated tumors. This observation suggests a potential role of CASR in colorectal cancer pathogenesis and was supported by previous experimental studies. For example, intestinal epithelial cells loss of CASR expression demonstrated aggressive and highly malignant features such as lack of differentiation and adhesion of epithelial cells and absence of localization of E-cadherin at the cell surface.37, 38 In addition, colonic epithelia isolated from intestine specific and global CASR knockout mice demonstrated increased proliferation and increased WNT/CTNNB1 (β-catenin) signaling.15, 39 Also, stimulation of the CASR increased the expression of BMP2 (bone morphogenetic protein-2),40 which induces growth suppression and enhances chemo-sensitivity of human colorectal cancer cells.41 Furthermore, CASR activation stimulated paracrine secretion of WNT5A from myofibroblasts and expression of ROR2 in epithelial cells.42 CASR-mediated Wnt5a/Ror2 interaction increased epithelial differentiation and reduced expression of the receptor for tumor necrosis factor 1.15, 39, 43 Collectively, our finding along with the experimental studies suggest that the modulators of CASR such as CASR agonists, including allosteric agonists, may be useful in therapeutic management of colorectal cancer,5, 44, 45 which requires further investigation.
We also observed higher prevalence of CIMP-high in tumors with no or weak CASR expression. In addition to genomic instability, epigenetic instability leads to irregular methylation of tumor suppressor genes. Hypermethylation of particular regions within the CpG islands encompassing the CASR gene promoter 2 was shown in 25% of neuroblastoma primary tumors and was associated with decreased CASR messenger RNA expression and several predictors of poor outcome in neuroblastomas, including MYC amplification.46 One study reported that hypermethylation of CASR was detected in 69% of colorectal cancer tissues and 90% of lymph node metastatic tissues, as well as significantly correlated with decreased CASR expression.35 These results suggest that epigenetic inactivation of CASR gene might play a role in colorectal carcinogenesis. In another study of CRC patients,47 no statistically significant survival difference was observed by genotypes of CASR (A986S; rs1801725, G>T), although there was an indication that the TT genotype may be associated with a lower risk of CRC-specific mortality. It is possible that this genotype and other genotypes may reach statistical significance in genome-wide association studies with larger sample size and in other ethnicities. An alternative explanation is that the CASR regulation in CRC is independent of this single nucleotide polymorphism (SNP) and mainly affected by epigenetic changes and post-transcriptional modifications rather than genome level changes. Nonetheless, given the limited number of studies, more studies are warranted to evaluate the effect of SNPs in CASR gene on tumor CASR expression.
Our study has several strengths, including a large number of colorectal cancer cases from two prospective cohorts as well as extensive information on patient characteristics and various molecular features. Consequently, we were able to demonstrate an association of tumor CASR expression level with colorectal cancer-specific mortality, independent of clinical and other tumor characteristics.
In these cohorts, data on cancer treatment was limited. However, lack of such information was less likely to influence our results because such tumor CASR data were not available to the treating clinicians. We also expect minimal difference in access to the treatment in this cohort since minimal variation in socioeconomic status has been reported in both cohorts and both patient populations consisted of health care professionals.48, 49 Our result should be validated in a cohort of mixed gender that is independent of the NHS and HPFS cohorts. In addition, data on cancer recurrence and metastasis were not available in either cohort. Because 5-year survival for metastatic colorectal cancer is 5-8%, the vast majority of patients with recurrence will die from metastatic disease. Based on the comprehensive mortality data, our study has captured the vast majority of recurrences because they will die from recurrent disease at some point and we have long follow-up on these patients.
In conclusion, this large prospective study of patients with colorectal cancer suggests that increased tumor CASR expression is an independent predictor of colorectal cancer-specific mortality. These findings need further confirmation and if confirmed, may lead to better understanding of colorectal cancer pathogenesis as well as exploring potential mechanisms of preventing or modulating colorectal cancer progression by targeting CASR.
Supplementary Material
Acknowledgments
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors assume full responsibility for analyses and interpretation of these data.
Role of the Sponsors: The funders had no roles in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants [P01 CA87969, UM1 CA186107 to M.J. Stampfer; P01 CA55075, UM1 CA167552 to W.C. Willett; P50 CA127003 to C.S.F.; R01 CA137178, K24 DK098311 to A.T.C.; R01 CA205406 to K.N.; R01 CA151993, R35 CA197735 to S.O.; K07 CA190673 to R.N.; and R03 CA176717, K07 CA188126 to X.Z.]; Nodal Award (to S.O.) from the Dana-Farber Harvard Cancer Center; and by grants from The Paula and Russell Agrusa Fund for Colorectal Cancer, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. L.L. is supported by a scholarship grant from Chinese Scholarship Council and a fellowship grant from Huazhong University of Science and Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- BMI
body mass index
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- HPFS
Health Professionals Follow-Up Study
- HR
hazard ratio
- MSI
microsatellite instability
- MSS
microsatellite stable
- NHS
Nurses’ Health Study
- SNP
single nucleotide polymorphism
Footnotes
Use of standardized official symbols: We use HUGO (Human Genome Organization)-approved official symbols (or root symbols) for genes and gene products, including BMP2, BRAF, CASR, CTNNB1, KRAS, MKI67, MYC, PIK3CA, PTGS2, ROR2, VDR, WNT, and WNT5A; all of which are described at www.genenames.org. Gene names are italicized, and gene product names are non-italicized. Colloquial names are used in parenthesis following official symbols for unequivocal research communication.
Conflict of interest: The authors declare that they have no conflicts of interest.
References
- 1.Ward DT, Riccardi D. New concepts in calcium-sensing receptor pharmacology and signalling. Br J Pharmacol. 2012;165:35–48. doi: 10.1111/j.1476-5381.2011.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown EM, Pollak M, Chou YH, Seidman CE, Seidman JG, Hebert SC. Cloning and functional characterization of extracellular Ca(2+)-sensing receptors from parathyroid and kidney. Bone. 1995;17:7S–11S. doi: 10.1016/8756-3282(95)00199-n. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal A, Prinz-Wohlgenannt M, Tennakoon S, Hobaus J, Boudot C, Mentaverri R, Brown EM, Baumgartner-Parzer S, Kallay E. The calcium-sensing receptor: A promising target for prevention of colorectal cancer. Biochim Biophys Acta. 2015;1853:2158–67. doi: 10.1016/j.bbamcr.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yano S, Sugimoto T, Tsukamoto T, Chihara K, Kobayashi A, Kitazawa S, Maeda S, Kitazawa R. Association of decreased calcium-sensing receptor expression with proliferation of parathyroid cells in secondary hyperparathyroidism. Kidney Int. 2000;58:1980–6. doi: 10.1111/j.1523-1755.2000.00370.x. [DOI] [PubMed] [Google Scholar]
- 5.Tennakoon S, Aggarwal A, Kallay E. The calcium-sensing receptor and the hallmarks of cancer. Biochim Biophys Acta. 2016;1863:1398–407. doi: 10.1016/j.bbamcr.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Mamillapalli R, VanHouten J, Zawalich W, Wysolmerski J. Switching of G-protein usage by the calcium-sensing receptor reverses its effect on parathyroid hormone-related protein secretion in normal versus malignant breast cells. J Biol Chem. 2008;283:24435–47. doi: 10.1074/jbc.M801738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao J, Schneider A, Datta NS, McCauley LK. Extracellular calcium as a candidate mediator of prostate cancer skeletal metastasis. Cancer Res. 2006;66:9065–73. doi: 10.1158/0008-5472.CAN-06-0317. [DOI] [PubMed] [Google Scholar]
- 8.Ahearn TU, Tchrakian N, Wilson KM, Lis R, Nuttall E, Sesso HD, Loda M, Giovannucci E, Mucci LA, Finn S, Shui IM. Calcium-Sensing Receptor Tumor Expression and Lethal Prostate Cancer Progression. J Clin Endocrinol Metab. 2016;101:2520–7. doi: 10.1210/jc.2016-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haven CJ, van Puijenbroek M, Karperien M, Fleuren GJ, Morreau H. Differential expression of the calcium sensing receptor and combined loss of chromosomes 1q and 11q in parathyroid carcinoma. J Pathol. 2004;202:86–94. doi: 10.1002/path.1489. [DOI] [PubMed] [Google Scholar]
- 10.Farnebo F, Enberg U, Grimelius L, Backdahl M, Schalling M, Larsson C, Farnebo LO. Tumor-specific decreased expression of calcium sensing receptor messenger ribonucleic acid in sporadic primary hyperparathyroidism. J Clin Endocrinol Metab. 1997;82:3481–6. doi: 10.1210/jcem.82.10.4300. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Soto G, Rocher A, Garcia-Rodriguez C, Nunez L, Villalobos C. The Calcium-Sensing Receptor in Health and Disease. Int Rev Cell Mol Biol. 2016;327:321–69. doi: 10.1016/bs.ircmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Aune D, Lau R, Chan DS, Vieira R, Greenwood DC, Kampman E, Norat T. Dairy products and colorectal cancer risk: a systematic review and meta-analysis of cohort studies. Ann Oncol. 2012;23:37–45. doi: 10.1093/annonc/mdr269. [DOI] [PubMed] [Google Scholar]
- 13.World Cancer Research Fund, American Institute for Cancer Research. Food, nutrition, physical activity, and the prevention of colorectal cancer. Washington DC: 2011. Continuous Update Project colorectal cancer report 2010 summary. [Google Scholar]
- 14.Rey O, Young SH, Jacamo R, Moyer MP, Rozengurt E. Extracellular calcium sensing receptor stimulation in human colonic epithelial cells induces intracellular calcium oscillations and proliferation inhibition. J Cell Physiol. 2010;225:73–83. doi: 10.1002/jcp.22198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLeod RJ. Extracellular calcium-sensing receptor/PTH knockout mice colons have increased Wnt/beta-catenin signaling, reduced non-canonical Wnt signaling, and increased susceptibility to azoxymethane-induced aberrant crypt foci. Lab Invest. 2013;93:520–7. doi: 10.1038/labinvest.2013.51. [DOI] [PubMed] [Google Scholar]
- 16.Cheng SX, Lightfoot YL, Yang T, Zadeh M, Tang L, Sahay B, Wang GP, Owen JL, Mohamadzadeh M. Epithelial CaSR deficiency alters intestinal integrity and promotes proinflammatory immune responses. FEBS Lett. 2014;588:4158–66. doi: 10.1016/j.febslet.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitfield JF. The calcium-sensing receptor–a driver of colon cell differentiation. Curr Pharm Biotechnol. 2009;10:311–6. doi: 10.2174/138920109787847510. [DOI] [PubMed] [Google Scholar]
- 18.Fetahu IS, Hobaus J, Aggarwal A, Hummel DM, Tennakoon S, Mesteri I, Baumgartner-Parzer S, Kallay E. Calcium-sensing receptor silencing in colorectal cancer is associated with promoter hypermethylation and loss of acetylation on histone 3. Int J Cancer. 2014;135:2014–23. doi: 10.1002/ijc.28856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fetahu IS, Tennakoon S, Lines KE, Groschel C, Aggarwal A, Mesteri I, Baumgartner-Parzer S, Mader RM, Thakker RV, Kallay E. miR-135b- and miR-146b-dependent silencing of calcium-sensing receptor expression in colorectal tumors. Int J Cancer. 2016;138:137–45. doi: 10.1002/ijc.29681. [DOI] [PubMed] [Google Scholar]
- 20.Puckett CD. The Educational Annotation of ICD-9-CM; Diseases and Procedures Tabular Lists. 1986 [Google Scholar]
- 21.Rich-Edwards JW, Corsano KA, Stampfer MJ. Test of the National Death Index and Equifax Nationwide Death Search. Am J Epidemiol. 1994;140:1016–9. doi: 10.1093/oxfordjournals.aje.a117191. [DOI] [PubMed] [Google Scholar]
- 22.Ogino S, Nosho K, Meyerhardt JA, Kirkner GJ, Chan AT, Kawasaki T, Giovannucci EL, Loda M, Fuchs CS. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 2008;26:5713–20. doi: 10.1200/JCO.2008.18.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 24.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, Qian ZR, Yamauchi M, Bertrand KA, Fitzgerald KC, Inamura K, Kim SA, Mima K, Sukawa Y, Zhang X, Wang M, Smith-Warner SA, et al. Predicted 25(OH)D score and colorectal cancer risk according to vitamin D receptor expression. Cancer Epidemiol Biomarkers Prev. 2014;23:1628–37. doi: 10.1158/1055-9965.EPI-14-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa T, Kuchiba A, Yamauchi M, Meyerhardt JA, Shima K, Nosho K, Chan AT, Giovannucci E, Fuchs CS, Ogino S. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–94. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, Mino-Kenudson M, Lauwers GY, Loda M, Fuchs CS. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun R, Nosho K, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, Wang YE, Peng S, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irahara N, Nosho K, Baba Y, Shima K, Lindeman NI, Hazra A, Schernhammer ES, Hunter DJ, Fuchs CS, Ogino S. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12:177–83. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, Giovannucci EL, Fuchs CS, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheinin Y, Kallay E, Wrba F, Kriwanek S, Peterlik M, Cross HS. Immunocytochemical localization of the extracellular calcium-sensing receptor in normal and malignant human large intestinal mucosa. J Histochem Cytochem. 2000;48:595–602. doi: 10.1177/002215540004800503. [DOI] [PubMed] [Google Scholar]
- 35.Hizaki K, Yamamoto H, Taniguchi H, Adachi Y, Nakazawa M, Tanuma T, Kato N, Sukawa Y, Sanchez JV, Suzuki H, Sasaki S, Imai K, et al. Epigenetic inactivation of calcium-sensing receptor in colorectal carcinogenesis. Mod Pathol. 2011;24:876–84. doi: 10.1038/modpathol.2011.10. [DOI] [PubMed] [Google Scholar]
- 36.Rey O, Chang W, Bikle D, Rozengurt N, Young SH, Rozengurt E. Negative cross-talk between calcium-sensing receptor and beta-catenin signaling systems in colonic epithelium. J Biol Chem. 2012;287:1158–67. doi: 10.1074/jbc.M111.274589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhagavathula N, Hanosh AW, Nerusu KC, Appelman H, Chakrabarty S, Varani J. Regulation of E-cadherin and beta-catenin by Ca2+ in colon carcinoma is dependent on calcium-sensing receptor expression and function. Int J Cancer. 2007;121:1455–62. doi: 10.1002/ijc.22858. [DOI] [PubMed] [Google Scholar]
- 38.Singh N, Liu G, Chakrabarty S. Cellular responses to TGFbeta and TGFbeta receptor expression in human colonic epithelial cells require CaSR expression and function. Cell Calcium. 2013;53:366–71. doi: 10.1016/j.ceca.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 39.MacLeod RJ, Hayes M, Pacheco I. Wnt5a secretion stimulated by the extracellular calcium-sensing receptor inhibits defective Wnt signaling in colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2007;293:G403–11. doi: 10.1152/ajpgi.00119.2007. [DOI] [PubMed] [Google Scholar]
- 40.Peiris D, Pacheco I, Spencer C, MacLeod RJ. The extracellular calcium-sensing receptor reciprocally regulates the secretion of BMP-2 and the BMP antagonist Noggin in colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2007;292:G753–66. doi: 10.1152/ajpgi.00225.2006. [DOI] [PubMed] [Google Scholar]
- 41.Vishnubalaji R, Yue S, Alfayez M, Kassem M, Liu FF, Aldahmash A, Alajez NM. Bone morphogenetic protein 2 (BMP2) induces growth suppression and enhances chemosensitivity of human colon cancer cells. Cancer Cell Int. 2016;16:77. doi: 10.1186/s12935-016-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pacheco II, Macleod RJ. CaSR stimulates secretion of Wnt5a from colonic myofibroblasts to stimulate CDX2 and sucrase-isomaltase using Ror2 on intestinal epithelia. Am J Physiol Gastrointest Liver Physiol. 2008;295:G748–59. doi: 10.1152/ajpgi.00560.2007. [DOI] [PubMed] [Google Scholar]
- 43.Kelly JC, Lungchukiet P, Macleod RJ. Extracellular Calcium-Sensing Receptor Inhibition of Intestinal EpithelialTNF Signaling Requires CaSR-Mediated Wnt5a/Ror2 Interaction. Front Physiol. 2011;2:17. doi: 10.3389/fphys.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodland KD. The role of the calcium-sensing receptor in cancer. Cell Calcium. 2004;35:291–5. doi: 10.1016/j.ceca.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Kallay E, Bajna E, Wrba F, Kriwanek S, Peterlik M, Cross HS. Dietary calcium and growth modulation of human colon cancer cells: role of the extracellular calcium-sensing receptor. Cancer Detect Prev. 2000;24:127–36. [PubMed] [Google Scholar]
- 46.Casala C, Gil-Guinon E, Ordonez JL, Miguel-Queralt S, Rodriguez E, Galvan P, Lavarino C, Munell F, de Alava E, Mora J, de Torres C. The calcium-sensing receptor is silenced by genetic and epigenetic mechanisms in unfavorable neuroblastomas and its reactivation induces ERK1/2-dependent apoptosis. Carcinogenesis. 2013;34:268–76. doi: 10.1093/carcin/bgs338. [DOI] [PubMed] [Google Scholar]
- 47.Fedirko V, Riboli E, Tjonneland A, Ferrari P, Olsen A, Bueno-de-Mesquita HB, van Duijnhoven FJ, Norat T, Jansen EH, Dahm CC, Overvad K, Boutron-Ruault MC, et al. Prediagnostic 25-hydroxyvitamin D, VDR and CASR polymorphisms, and survival in patients with colorectal cancer in western European ppulations. Cancer Epidemiol Biomarkers Prev. 2012;21:582–93. doi: 10.1158/1055-9965.EPI-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baer HJ, Glynn RJ, Hu FB, Hankinson SE, Willett WC, Colditz GA, Stampfer M, Rosner B. Risk factors for mortality in the nurses’ health study: a competing risks analysis. Am J Epidemiol. 2011;173:319–29. doi: 10.1093/aje/kwq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jimenez M, Hu FB, Marino M, Li Y, Joshipura KJ. Prospective associations between measures of adiposity and periodontal disease. Obesity (Silver Spring) 2012;20:1718–25. doi: 10.1038/oby.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


