8.1 Introduction
The Herpesviridae is a broad and complex family of large DNA viruses that establish lifelong latent infections in their respective hosts (Knipe and Howley 2013). Individual members of this family display distinct patterns of tissue tropism, with a subset routinely invading the nervous system following peripheral infection (Smith 2012). The neuroinvasive herpesviruses belong to at least two genera of the Alphaherpesvirinae subfamily that include the human pathogens herpes simplex virus types 1 and 2 (HSV-1 and HSV-2; simplex virus genus) and varicella zoster virus (VZV; Varicellovirus genus). An additional member of the varicelloviruses is a pathogen of veterinary significance, pseudorabies virus (PRV), which serves as a model for severe neuroinvasive disease (Pomeranz et al. 2005; Mettenleiter 2008). These viruses share the remarkable ability to routinely enter the nervous system in an immunocompetent host in the absence of overt physical trauma (i.e., animal bites or syringe punctures). How this is achieved is largely unknown, but it is an astonishing example of efficacious neural gene delivery. The most common outcome of herpesvirus neuroinvasion is the establishment of a lifelong latent infection in the ganglia of the peripheral nervous system (PNS). Periodic reactivation from the latent state results in production of new infectious particles that exit the nervous system to replicate at exposed body surfaces and transmit to new hosts. This review discusses the assembly and egress of the alphaherpesvirus infectious particle, working off the framework that the virion is a clockwork consisting of a collection of poised mechanisms that act sequentially to move viral genomes between the nuclei of the epithelial and neuronal cells.
8.2 The Herpesvirus Virion
Viruses belonging to the Alphaherpesvirinae subfamily encode 80 or more proteins, approximately half of which serve as structural components of the virion (Knipe and Howley 2013). The virion core consists of a 125 nm diameter icosahedral capsid encasing a linear dsDNA genome. The capsid is initially assembled around a protein scaffold, which is lost when the genome is translocated into the capsid through a unique portal vertex, a process that is more typical of some bacteriophage than other mammalian viruses (McGeoch et al. 2006; Brown and Newcomb 2011). The capsid is surrounded by a protein matrix referred to as the tegument, and this massive structure is enveloped in a lipid bilayer that is decorated by an assortment of glycoproteins (Fig. 8.1).
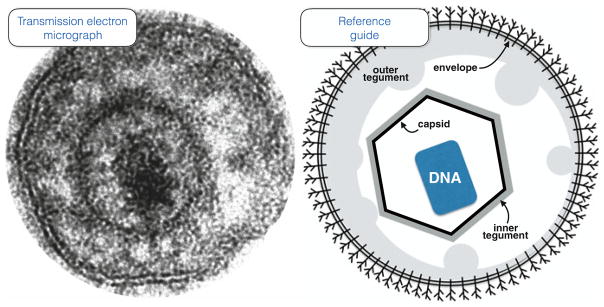
Fig. 8.1.
A direct view of an extracellular PRV particle. Conventional transmission electron microscopy is the most common method of imaging herpesvirus ultrastructure and affords a direct view of a single viral particle (left). An illustration of the particle’s component architecture is provided as reference (right). The particle is dehydrated due to processing of the sample in preparation for imaging, with the DNA genome appearing condensed within the capsid while the tegument is partially separated into two layers commonly referred to as inner and outer tegument. The two leaflets of the envelope membrane are clearly visible and includes a complex assortment of transmembrane proteins (only a single type is illustrated for simplicity). While this view hints at an asymmetric distribution of the outer tegument, asymmetry is more effectively visualized in unfixed particles such as those captured in vitreous ice for cryoEM/ET imaging and native particles imaged by ensemble mapping. The capsid is 125 nm across
Observing the virion structure in detail is challenging because it is too large and irregular for x-ray crystallography. Average composite pictures of the capsid have been produced by imaging multitudes of cryopreserved viral particles by electron microscopy (cryoEM) and offer detailed views of the topology of the capsid shell but cannot resolve many structural details of the tegument and envelope because these component architectures generally lack the symmetry needed for composite averaging (Schrag et al. 1989; Zhou et al. 2000; Homa et al. 2013). The one exception is the small proportion of tegument mass immediately adjacent to the capsid surface, which through direct interactions with the capsid is confined to icosahedral symmetry where it is detected exclusively at capsid vertices (Fig. 8.2) (Zhou et al. 1999; Huet et al. 2016). The tegument-capsid link is ascribed to the large tegument protein (viral protein 1/2, VP1/2), which is anchored to the capsid by the pUL25 minor capsid protein such that a small portion of its carboxyl terminus (<100 aa) is thought to contribute to pUL25 linear surface densities that radiate from the capsid vertices and also consist of the pUL17 minor capsid protein (Huet et al. 2016; Uetz et al. 2006; Coller et al. 2007; Trus et al. 2007; Pasdeloup et al. 2009; Dai et al. 2014; Fan et al. 2014). However, the extent of the contribution of VP1/2 to the observed densities is debated, with a major globular density that sits atop the pentons being ascribed to either an additional aspect of VP1/2 (Huet et al. 2016; Cardone et al. 2012) or to pUL25 (Dai et al. 2014). In either case, the vertices clearly serve as platforms for VP1/2 attachment and tegument assembly in general, but it is also important to consider that the mass at the vertices is visible in cryoEM reconstructions because it is symmetric. The beautiful capsid depictions produced by cryoEM are artificial composites that offer high-resolution views of symmetric capsid components but hide asymmetric features such as the portal vertex and possibly additional sites of tegument interaction. In this regard, tegument mass at sites of twofold symmetry is hinted at the Kaposi’s sarcoma-associated herpesvirus (KSHV), a gammaherpesvirus that possesses a capsid-tegument arrangement similar to the Alphaherpesvirinae, but this mass likely occupies only a small subset of these sites across the capsid surface (Dai et al. 2014). Either the alphaherpesviruses lack tegument attachments at capsid twofold symmetry sites, the tegument is anchored to only a small subset of these sites that have escaped detection by composite averaging, or tegument attachment at these sites is unstable within intact virions. Another hint at asymmetric capsid-tegument interactions is VP1/2, which has a virion copy number that is greater than should be expected by occupation at vertices alone (i.e., 5 copies per vertex predict 60 copies per virion) (Table 8.1). Finally, although the VP1/2-pUL25 link is the only defined capsid-tegument interaction so far identified, other tegument proteins are suggested to directly interact with the capsid including a protein of interest for this review: pUL16 (Oshima et al. 1998; Meckes et al. 2010). As will be discussed, tegument links to the capsid apart from the vertices may contribute to dynamic alterations in virion architecture during infection.
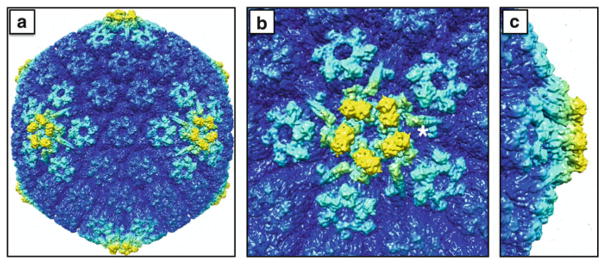
Fig. 8.2.
The outer capsid surface within an extracellular viral particle. The cryoEM density maps are colored to highlight structures that are elevated from the capsid surface: (a) full capsid; (b) top down view of a vertex; (c) side view of a vertex. The pentonal vertices and surrounding peripentonal hexons of the capsid (light cyan) are composed of the major capsid protein (VP5) and, in the case of the hexons, the hexon tip protein (VP26). The elongated structures radiating outward from the penton (one of which is marked with an adjacent asterisk) are composed of two minor capsid proteins, pUL17 and pUL25, and serve as binding sites for the large tegument protein, VP1/2, which also likely contributes to this density. Five copies of a globular structure (yellow) are interdigitated above the VP5 subunits in the underlying penton, and have been proposed to be either an additional portion of the VP1/2 tegument protein or the globular domain of the pUL25 minor capsid protein (see text). In addition to being the most elevated aspects of the capsid surface, the densities highlighted in yellow are absent from nascent capsids assembled in infected cell nuclei. The images were produced using the Chimera software package and EMD-6387
Table 8.1.
Estimates of protein copy numbers in C-capsids and virions
| Study | Heine 1974 | Handler 1996 | Newcomb 2006 | Trus 2007 | Newcomb 2012 | Homa 2013 | Bohannon 2013 | |
|---|---|---|---|---|---|---|---|---|
| Source material | HSV-1 virions | HSV-1 virions | HSV-1 C-capsids | HSV-1 C-capsids | HSV-1 virions | PRV C-capsids | PRV virions | |
| Approach | Predicted | Bulk | Bulk | Bulk | Bulk | Bulk | Bulk | Single particle |
| Capsid | ||||||||
| pUL6 | 12 | |||||||
| pUL17 | 60 | 36 | 99 | |||||
| pUL25 | 60 | 75 | 82 | 179 | 143 | 71 | ||
| pUL18 (VP23) | 640 | 1480 | 632 | |||||
| pUL19 (VP5) | 955 | 845 | ||||||
| pUL35 (VP26) | 900 | |||||||
| pUL38 (VP19c) | 320 | 785 | 423 | |||||
| Tegument | ||||||||
| pUL16 | 85 | |||||||
| pUL36 (VP1/2) | 60 | <150 | 152 | 56 (Ct) 107 (Nt) |
||||
| pUL37 | 188 | 104 | ||||||
| pUL46 (VP11/12) | 505 | 197 | ||||||
| pUL47 (VP13/14) | 1640 | 1266 | 455 | |||||
| pUL48 (VP16) | 686 | 342 | ||||||
| pUL49 (VP22) | 824 | 679 | 170 | |||||
| pUS3 | 30 | |||||||
| Envelope | ||||||||
| gB | 375 | 3200 | ||||||
| gC | 545 | 4900 | ||||||
| gD | 34000 | 68 | ||||||
| gH | 52000 | |||||||
| gM | 17 | |||||||
The best views of the majority of the tegument and envelope have been achieved by tomographic cryoEM imaging of single viral particles (cryoET), which reveal that the tegument is accumulated predominantly at one capsid vertex and gradually recedes around the capsid such that the opposing vertex is mostly devoid of asymmetric tegument mass (Grunewald et al. 2003; Maurer et al. 2008). While identifying the protein compositions of these densities is currently beyond the resolution limits of cryoET, a molecular framework of the tegument and envelope substructures has begun to emerge by superresolution light microscopy. By fusing virion protein constituents to fluorescent proteins or binding them to fluorescent antibodies, the source of fluorescence emissions within virions can be pinpointed using two related and complementary approaches: ensemble mapping and stochastic optical reconstruction microscopy (STORM) (Bohannon et al. 2013; Laine et al. 2015). Whereas ensemble mapping allows for examination of native virions and can effectively resolve asymmetric distributions of the bulk mass of a given protein species within individual virions to 7 nm resolution, current STORM analysis provides an approximation of the average radial distance from the capsid that a particular protein species resides in extracted virions based on indirect immunofluorescence and composite averaging. The authors of the latter approach determined that more of VP1/2 is in close proximity to the capsid surface than can easily be accounted for by its carboxyl-terminal pUL25 binding site, which coincides with prior cryoEM reconstructions that indicated a potential second VP1/2-capsid interaction that occurs with the major capsid protein, VP5, at the penton vertices (Cardone et al. 2012; Laine et al. 2015). Despite the potential contribution of a VP1/2-VP5 linkage, ensemble mapping has achieved sufficient resolution to conclude that pUL25 and VP1/2 are present at each of the 12 capsid vertices (Bohannon et al. 2013). This observation is notable given that one vertex is occupied by the portal, consisting of 12 copies of pUL6, instead of a penton comprised of VP5 as is the case at the other 11 vertices (Newcomb et al. 2001; Trus et al. 2004; Chang et al. 2007). Given this, the topology of VP1/2 at the portal vertex may be distinct from its conformation at the 11 pentonal vertices. Two additional findings are compatible with this concept. First, the VP16 tegument protein, which is a direct binding partner of VP1/2 (Vittone et al. 2005; Ko et al. 2010), is predicted to occupy only 11 vertices based on ensemble mapping (Bohannon et al. 2013). This is in contrast to a second VP1/2-binding protein, pUL37 (Uetz et al. 2006; Vittone et al. 2005; Klupp et al. 2002; Mijatov et al. 2007), which is uniformly distributed around the capsid consistent with occupation at all 12 vertices (Bohannon et al. 2013). Although speculative, the absence of VP16 from one vertex is most easily explained by an inability to bind VP1/2 anchored at the portal vertex. Second, by cryoET, the profile of the vertex facing away from the asymmetric tegument appears different than the opposing vertex that is buried under the bulk of this tegument, hinting that the unique portal vertex may occupy one of these two poles (Grunewald et al. 2003).
While it is currently unknown if VP16 is specifically absent from the portal vertex, the hypothesis is compelling. The majority of the tegument is anchored to the envelope and likely connects to the capsid surface through several linkages including VP1/2-VP16 (Fig. 8.3) (Vittone et al. 2005; Ko et al. 2010; Svobodova et al. 2011). The absence of VP16 from the portal vertex would indicate that only VP1/2 adjacent to a pentonal vertex conforms to a state that binds VP16, thereby possibly providing the source of tegument asymmetry that is observed by cryoET and ensemble imaging (Grunewald et al. 2003; Maurer et al. 2008; Bohannon et al. 2013). In this hypothetical model, the large-scale virion asymmetries in the tegument and envelope are a projection of the inherent asymmetry of the capsid shell resulting from the unique portal vertex.
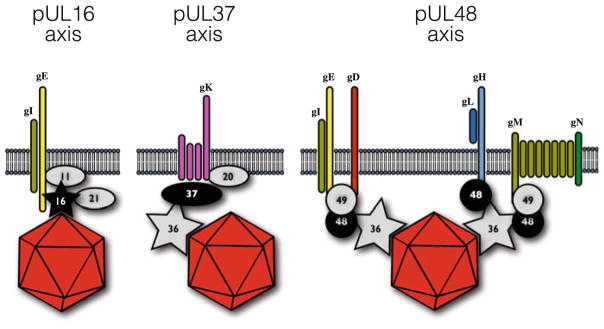
Fig. 8.3.
Three possible links between capsid and envelope. Capsids acquire lipid envelopes by budding into intracellular membranes derived from the secretory/recycling pathways. Capsid interactions with lipid membranes are indirect and dependent upon tegument and envelope proteins. Depicted are three proposed linkages with varying degrees of support in the literature. Two tegument proteins that directly bind capsids are featured: pUL16 and pUL36 (VP1/2). Tegument proteins are indicated by their unique long gene number (i.e., UL11 is labeled as 11)
8.3 The Virion Clockwork
While the technology is nearly at hand to define the herpesvirus architecture at the molecular level, extracellular particles harvested from tissue culture media likely only represent one state of the virion. Virions form as symmetric particles (Morgan et al. 1968; Granzow et al. 1997, 2001; Chen et al. 2004) that only adopt an asymmetric appearance following release from infected cells (Newcomb and Brown 2009). The reason for this plasticity is not immediately apparent but is suggested as either a means to increase particle stability in the extracellular environment (Newcomb and Brown 2009) or as a mechanism to assist productive infection (Maurer et al. 2008). The latter suggestion is supported by the observation that virions in contact with cells frequently have an asymmetric distribution and, of particular interest, orient such that the tegument-dense pole faces away from the contacted cell (Maurer et al. 2008; Fuller et al. 1989).
Because viral particles are not metabolically active, the ability of a virion to adjust its architecture or to deliver its genetic content is inherent to the potential energy built into its architecture. The virion can be viewed as a clockwork that consists of a collection of primed mechanisms that trigger sequentially with precise timing to productively infect a cell. As with all viruses, any chance failure in this process results in a defective particle. Advances in our understanding of the molecular underpinnings of the virion architecture are beginning to hint at how triggers in the alphaherpesvirus virion may function. While it may seem unnecessary for a virus to use more than one trigger to deliver its genome into a cell, similar to a Rube Goldberg machine, research on adenovirus helps to showcase that the use of multiple independent triggering events is not unusual (Greber et al. 1993; Burckhardt et al. 2011).
Adenovirus, consisting of a non-enveloped icosahedral virion, employs a cell-surface trigger based on the removal of penton fiber from the capsid vertices, which occurs by simultaneous pulling on the penton base and fiber by their cognate cell receptors. Removing the fiber from the base primes a viral lytic protein needed for escape to the cytosol following endocytosis: akin to a pin pulled from a grenade and tossed into a clathrin-coated pit (Burckhardt et al. 2011). Following cell entry, adenovirus relies on acidification and virion proteolysis as postentry triggers to drive particle delivery to the nuclear pores and for genome injection into the nuclei (Greber et al. 1993). The sequential triggering events employed by herpesviruses are largely dissimilar from those of adenovirus and represent a different evolutionary solution to delivering the viral genome to a nucleus. First, the envelope glycoproteins participate in initial cell-surface interactions with cell-surface heparan sulfate chains linked to proteoglycans. These interactions are mediated by glycoprotein C (gC) and gB in HSV-1 and are an important step in promoting infection (Shukla and Spear 2001). These interactions also set off a cell-surface triggering event that sees the release of the pUL16 tegument protein from the capsid within the extracellular virion (Meckes and Wills 2008). The release of pUL16 is presumably just one molecular aspect of the tegument rearrangement that results in the asymmetrical ultrastructure seen in virions attached to cell surfaces, but how these events are related has not been addressed (Maurer et al. 2008; Fuller et al. 1989). Following the initial triggering event, the virus then binds its entry cellular receptor (usually by an interaction with gD, although VZV is a notable exception) and triggers membrane fusion via glycoproteins gH, gL, and ultimately gB: the fusion apparatus (Cooper and Heldwein 2015; Spear and Longnecker 2003). In the asymmetric particle, ensemble mapping indicates that one pole of the virion envelope has low occupancy of gM, a protein that inhibits membrane fusion (Klupp et al. 2000; Koyano et al. 2003; El Kasmi and Lippe 2014). Assuming that gM begins off in a symmetric radial distribution as is the case for the underlying tegument, it may help protect the virion from neutralizing antibodies (Hook et al. 2008). Upon triggering, the redistribution of the tegument and gM may make gD available for cell binding at one pole of the virion, thereby orientating the virion at the cell surface in preparation for membrane fusion and capsid delivery (Maurer et al. 2008). Oddly, in some cell types, this dual-trigger mechanism is insufficient to mediate fusion-based entry into the cytosol but instead requires acidification following endocytosis (Nicola et al. 2003, 2005). Whether acidification serves as a third trigger or replaces an aspect of gC-triggered virion asymmetry is unknown, but in either scenario, the end result of fusion is the same: the capsid and tegument are deposited into the cytosol.
Cytosolic capsids remain bound to the inner tegument proteins VP1/2, pUL37, and pUS3 but dissociate from outer tegument proteins including VP11/12, VP13/14, VP16, and VP22 (Smith 2012; Granzow et al. 2005; Luxton et al. 2005; Antinone and Smith 2010; Abaitua et al. 2012; Aggarwal et al. 2012). This partial disassembly is a hallmark of herpes virion metastability, and several proposals regarding the mechanism for triggering tegument release have been offered including phosphorylation, proteolytic cleavage, and the reducing environment of the cytosol (Newcomb and Brown 2009; Morrison et al. 1998; Delboy et al. 2008). Whether one or more of these are contributing factors, the process must be specific to the incoming particle and not impact tegument acquisition during virion assembly. Alternatively, the disassembly may occur in the virion prior to the membrane fusion event, as is the case for pUL16 (Meckes and Wills 2008). Whichever the case, the capsid-VP1/2-pUL37-pUS3 complex subsequently translocates to a nuclear pore complex where a final trigger is tripped by proteolysis of VP1/2, resulting in the viral genome exiting the capsid and being delivered into the nucleus (Table 8.2) (Jovasevic et al. 2008).
Table 8.2.
Cell-surface and post-entry alpha-herpesvirus triggers
| Trigger | Sensor | Event | |
|---|---|---|---|
| Internal transformation of virion architecture | Heparan sulfates | gC | Release of the pUL16 tegument protein from the capsid. Possibly the cause of capsid displacement with the virion to an eccentric position. |
| Fusion based entry | Nectin HVEM Modified heparin sulfates |
gD | Fusion of envelope with cell membrane deposits capsid and tegument into the cytosol. |
| Tegument release | Phosphorylation Reduction Proteases |
Likely multiple tegument proteins | Solubilizes outer tegument components and exposes the capsid and inner tegument proteins. |
| Genome injection | Nups Proteases |
Likely VP1/2 tegument protein and pUL25 capsid protein | VP1/2 is cleaved and genome is delivered through nuclear pores following release from the capsid. |
While identifying how triggering occurs is an important step in understanding how infection proceeds, it is only a small aspect of the events that follow. Our working knowledge of internal virion rearrangements, membrane fusion, tegument disassembly, and genome injection is incomplete. But to understand the viral clockwork, we first need to know how it is built. The cell biology of alphaherpesvirus assembly and egress has often been a contentious topic. Debates regarding the source of the virion envelope have raged over the years: from the inner nuclear membrane, to the trans-Golgi network, to recycling endosomes (Enquist et al. 1998; Turcotte et al. 2005; Hollinshead et al. 2012). Likewise, whether capsids exit the nucleus by way of nuclear pores has led to some notable disagreements (Leuzinger et al. 2005; Campadelli-Fiume and Roizman 2006; Mettenleiter and Minson 2006), as has the long-standing discussion of whether the envelope membrane is acquired before or after capsids are shuttled down axons following replication in neurons (Kratchmarov et al. 2012; Cunningham et al. 2013). The broader topic of alphaherpesvirus assembly and egress was comprehensively reviewed recently and provides an excellent background for the subsequent discussion of virion assembly and delivery to neurons and epithelial cells (Owen et al. 2015). Two tegument proteins that bind directly to the capsid surface will be the focus of the remainder of this review for their roles in virion assembly and potential contributions to genome delivery to new cells: VP1/2 and pUL16.
8.4 Assembly of a Postentry Trigger
The large tegument protein, VP1/2, is required for incoming capsids to deliver their DNA content to the cell nuclei (Abaitua et al. 2012; Jovasevic et al. 2008; Roberts et al. 2009; Abaitua and O’Hare 2008). It is the only tegument protein for which a defined molecular link to the capsid has been established: the C-terminal ~60 amino acids bind to the pUL25 minor capsid protein to direct assembly on capsids (Coller et al. 2007). While additional regions of VP1/2 are proposed to bind to the capsid surface, the contributions of these regions during virion assembly have not been formally tested (Fig. 8.4) (Pasdeloup et al. 2009; Baker et al. 2006; Roberts et al. 2009). However, there are indications that truncated isoforms of VP1/2 lacking the C-terminal capsid-binding domain are incorporated into virions (Michael et al. 2006; Radtke et al. 2010) and could be partially functional (Schipke et al. 2012).
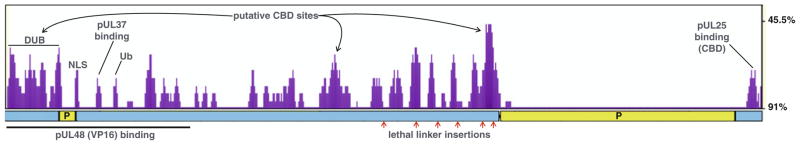
Fig. 8.4.
VP1/2 amino acid conservation plot. The plot of cumulative divergence indicates regions of the large tegument protein that are predicted functional elements under selective pressure. Evolutionary stable regions of the protein are marked by the higher peaks (purple). Overlaid on the plot are annotations of protein regions discussed in this review, including: the deubiquitinase (DUB), pUL37 binding site; ubiquitin switch site (Ub), and the C-terminal capsid binding domain (CBD). A nuclear localization signal (NLS) that is critical for infection is also indicated (Abaitua and O’Hare 2008; Abaitua et al. 2012). The locations of three additional proposed CBD’s are also indicated. Below the plot is a linear illustration of VP1/2 highlighting the two regions that are rich in proline (P) and are predicted to be disordered regions of the protein, and below this are annotations for the region of VP1/2 that interacts with VP16 by yeast-2-hybrid analysis, and six linker insertion sites that are lethal to PRV (Mohl 2010). The plot was produced with VP1/2 amino acid sequences from seven alpha-herpesviruses (BHV-1, EHV-1, EHV-4, HSV-1, HSV-2, PRV, and VZV) using a sliding window size of 25 on the eShadow web site: eshadow.dcode.org (Ovcharenko 2004)
Whether VP1/2 is loaded onto capsids in the nucleus or in the cytosol is unclear (Trus et al. 2007; Klupp et al. 2002; Roberts et al. 2009; Schipke et al. 2012; Wolfstein et al. 2006; Bucks et al. 2007; Abaitua and O’Hare 2008; Newcomb and Brown 2010). The conflicting findings may partially be due to a commonly held assumption that the bulk of DNA-filled capsids in the nucleus (aka C-capsids) represent an intermediate in virion morphogenesis. In fact, data supporting this notion is generally lacking. The alternative, that C-capsids isolated from infected cell nuclei may be defective assembly intermediates that failed to properly engage the nuclear egress machinery, would imply that studies of these capsid species may produce spurious results. Capsids destined to successfully egress from the nucleus may not be represented by those that accumulate within the nucleus. Therefore, whether egressing capsids are VP1/2 bound prior to leaving the nucleus, or only upon reaching the cytosol is not clear. VP1/2 is not required for the egress of capsids from the nucleus (Desai 2000; Fuchs et al. 2004), but it enhances the process (Luxton et al. 2006; Leelawong et al. 2012) and may contribute to a selective mechanism that favors the egress of DNA-filled capsids to the cytoplasm (Kharkwal et al. 2016).
In the cytosol, VP1/2 is essential for secondary capsid envelopment that results in the fully assembled virion, but its precise role in this process has been hard to pin down (Desai 2000; Fuchs et al. 2004; Smith and Enquist 1999). Although there is evidence that VP1/2 recruits capsids to a juxtanuclear location that contains markers consistent with the Golgi complex (Desai et al. 2008), VP1/2 is not explicitly required for capsid association with membranes (Kharkwal et al. 2015) or for recruitment of the cellular EXCRT (endosomal sorting complex required for transport)-budding machinery that is thought to contribute to secondary envelopment (Kharkwal et al. 2016). Even more quizzically, the majority of VP1/2 that is normally assembled into virions can be drastically reduced by overexpression of its C-terminal capsid-binding fragment during infection, to the extent that individual extracellular particles lack detectable amounts of full-length VP1/2 (Coller et al. 2007). Therefore, full occupancy of full-length VP1/2 on capsids is not absolutely required for virion assembly but may instead be important for the infectivity of these particles. VP1/2 interacts with at least two other tegument proteins: pUL37 and pUL48 (VP16) (Vittone et al. 2005; Ko et al. 2010; Klupp et al. 2002; Mijatov et al. 2007; Svobodova et al. 2012). Whereas interaction with pUL37 is critical for secondary envelopment (Kelly et al. 2014), the pUL48 interaction can be dispensable (Svobodova et al. 2012). Recent data suggests that pUL37 may link capsids to two envelope proteins, pUL20 and gK, that are important in secondary envelopment (Jambunathan et al. 2014; Chouljenko et al. 2016). Whereas the VP1/2-pUL37 link remains intact when virions enter cells, the VP1/2-pUL48 interaction is labile, hinting that the postentry trigger for tegument release centers around this seemingly nonessential interaction. The significance of the VP1/2-pUL48 interaction is further revealed in light particles, which are the products of cytoplasmic budding events that do not involve capsids (Szilagyi and Cunningham 1991; McLauchlan et al. 1992; McLauchlan and Rixon 1992). Importantly, the presence of VP1/2 and pUL37 in light particles is dependent upon pUL48 (Fuchs et al. 2002). The simplest interpretation of this notable finding is that there are no secondary interactions that can recruit capsids to membranes specifically by way of VP1/2 and pUL37. This brings into question the role of the pUL37/pUL20/gK interaction, which indicates that the critical function of pUL37 during secondary envelopment is something other than tethering the capsid to the membrane, and leads to the inevitable conclusion that there is an additional means by which capsids are recruited to the nascent envelope (Fig. 8.3).
8.5 Assembly of a Cell-Surface Trigger
While tegument proteins other than VP1/2 are suggested to bind capsid proteins, in most cases it is not clear if these interactions are biologically relevant (Lee et al. 2008; Scholtes et al. 2010). Of particular interest, however, is the pUL16 tegument protein, which binds capsids directly and independently of VP1/2 and pUL37 (Oshima et al. 1998; Meckes et al. 2010; Nalwanga et al. 1996; Fulmer et al. 2007). Despite compelling evidence that pUL16 is a capsid-bound tegument protein during virion morphogenesis, the component of the capsid to which it binds is unknown (Meckes et al. 2010). Although yeast 2-hybrid findings suggest pUL16 may bind major capsid protein, VP5, or the hexon tip protein, VP26 (Lee et al. 2008), interactions with capsid proteins in this experimental context can be false positives (Ko et al. 2010). Furthermore, there is not much unaccounted cryoEM capsid surface density that hints at the presence of pUL16 (Fig. 8.2). There are two possible explanations for this. First, capsid-bound pUL16 may be undetectable by cryoEM due to low occupancy or because of an asymmetric distribution. Perhaps its presence was hinted at non-pentonal sites in KSHV (Dai et al. 2014). Second, pUL16 may not remain bound to capsids within virions under the conditions used for cryoEM studies (Meckes and Wills 2008).
The interaction of pUL16 with nascent capsids is complex. The protein is bound to nuclear C-capsids of HSV-2 (Oshima et al. 1998) and is present in capsid intranuclear clusters during late stages of HSV-1 infection (Nalwanga et al. 1996) but could not be detected on nuclear A-, B-, or C-capsids isolated from HSV-1-infected cells (Meckes and Wills 2007). This led the authors of the latter study to propose that pUL16 may bind to procapsids, as pUL16 was present on capsids following their egress to the cytosol. Notably, these circumstances are similar to the debate regarding the presence of VP1/2 on nuclear capsids (see above). In addition to binding capsids, pUL16 interacts with membranes through a complex interaction with pUL11 and gE, which is responsible for 70–80% of pUL16 incorporation (Meckes et al. 2010; Han et al. 2011, 2012). This complex also includes pUL21 (Klupp et al. 2005; Harper et al. 2010), which in turn recruits pUL46 (VP11/12), pUL49 (VP22), and pUS3 to the virion (Michael et al. 2007; Starkey et al. 2014). Furthermore, by the time extracellular viral particles are typically harvested from the media of infected cells, the capsid-bound population of pUL16 is no longer evident (Meckes and Wills 2007).
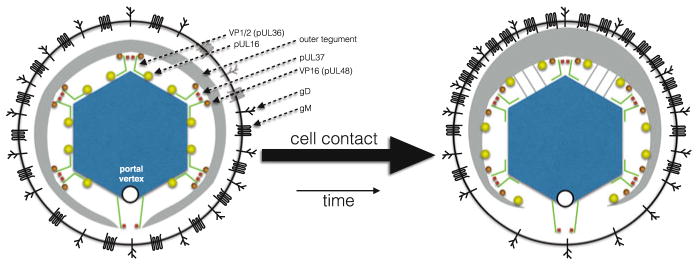
The implication of the interaction of pUL16 with the envelope is that the latter may remove and sequester pUL16 from the capsid surface. Further details of the relationship of pUL16 with capsid and envelope were obtained by treating virions with N-ethylmaleimide (NEM) (Meckes and Wills 2008). NEM reacts with cysteine residues, which interferes with the pUL16-pUL11 envelope interaction and stabilizes the pUL16-capsid interaction, indicating that the two interactions may be mutually exclusive within the virion (Meckes and Wills 2008; Yeh et al. 2008; Chadha et al. 2012). Nevertheless, the stabilized pUL16-capsid interaction is still disrupted upon contacting an uninfected cell. It is this interaction that serves as the cell-surface trigger, which corresponds to the redistribution of outer tegument into an asymmetric architecture that is visible by cryoET and ensemble mapping (Fig. 8.5) (Grunewald et al. 2003; Bohannon et al. 2013).
Fig. 8.5.
A hypothetical model of the alpha-herpesvirus dynamic architecture. Nascent virions exocytosed from cells have symmetric architectures (left), but typically are asymmetric when harvested from tissue culture media (right). By cryoET, this asymmetry manifests as a thickening of the tegument on one side of the capsid, resulting in the capsid occupying an eccentric position within the confines of the surrounding envelope. Ensemble mapping supports that the pUL16 and VP16 tegument proteins may be absent from a single vertex, which is considered to be the portal vertex in this speculative model. In contrast, the VP1/2 and pUL37 tegument proteins are projected to occupy all 12 vertices. In this model, VP1/2 is depicted to adopt different conformations at the unique portal vertex and pentonal vertices to account for the absence of VP16 at one vertex. Ensemble mapping also indicates that gD is radially distributed in the envelope, while gM is polar. How the gM asymmetry relates to underlying changes in tegument distribution is unknown: in this model, these components have been oriented together. Traditional transmission electron microscopy and cryoET reveal that virion asymmetry is prevalent upon binding to cells, and in separate studies, binding to cells was found to release pUL16 from capsids within virions. Whether the relationship between pUL16 release and tegument asymmetry is causal or coincidental is not known. In addition to cell contact, virions become asymmetric as they age and are noted to acquire additional tegument contacts across the faces of the icosahedral capsid (gray lines). It is unknown if cell-contact-dependent and age-dependent architectural changes in the virion are related, but these events demonstrate the plasticity and dynamic nature of the virion architecture. The illustration is an effort to integrate these independent observations into a simplified model based on a single underlying triggering event that could promote infection, and is intended to promote discussion
8.6 Delivery
The dissemination of a neuroinvasive herpesvirus is made more complex by the requirement for virions to achieve their triggering events in two different polarized cell types: epithelia cells and neurons. While many viruses replicate in polarized epithelia, a hallmark property of the alphaherpesviruses is the routine transmission of infection from epithelia to innervating neurons of the peripheral nervous system in the absence of tissue damage (i.e., animal bites or syringe punctures) or a compromised immune system. Implicit in this infectious cycle are three stages of virion transmission: epithelium to neuron, neuron to epithelium, and epithelium to outside world (and ultimately the epithelium of another host). Whether there are differences in the virions produced in each of these scenarios have not been thoroughly investigated, but it is the case for varicella zoster virus that only the latter results in intact cell-free virions (Chen et al. 2004).
The first hint of how epithelium-to-neuron transmission is achieved came from an unexpected finding involving the VP1/2 tegument protein. The amino terminus of VP1/2 encodes a cysteine protease that cleaves ubiquitin from proteins (aka deubiquitinase; DUB) (Kattenhorn et al. 2005; Schlieker et al. 2005, 2007). While inactivation of the DUB by genetic mutation does not substantially impair virus propagation in culture, invasion of the nervous system in vivo is lost (Böttcher et al. 2008; Lee et al. 2009). Unlike traditional neurotropic factors, the DUB is dispensable for replication and spread in the mammalian nervous system; instead the defect associated with DUB inactivation is specifically in the transmission of infection from peripheral innervated tissues to neurons. Thus, the VP1/2 DUB is a neuroinvasive determinant and yet is not a neurotropic factor.
Although the DUBs of various herpesviruses collectively have a diversity of cellular substrates (Whitehurst et al. 2009, 2012; Gastaldello et al. 2010, 2013; Inn et al. 2011; Saito et al. 2013; Wang et al. 2013; van Gent et al. 2014; Kim et al. 2016), the alphaherpesvirus neuroinvasive property proved to function by an autocatalytic mechanism that switches the virus between two invasive states (Bolstad et al. 2011; Huffmaster et al. 2015). The first invasive state promotes transmission of infection from epithelial tissues to peripheral nerves, and the second state subsequently promotes sustained retrograde transport in axons. The “ubiquitin switch” mechanism underlying this dynamic invasive process appears to trigger a change in intracellular capsid transport along microtubules. VP1/2 lacking ubiquitin modification at a lysine near the N-terminus of the protein (Fig. 8.4) moves bidirectionally on microtubules, and the addition of ubiquitin switches transport to unidirectional transport toward the minus-ends of microtubules (Huffmaster et al. 2015). The modified state is critical upon entering axon terminals, where sustained long-distance transport is required to deliver capsids to neural soma in peripheral ganglia. Viruses lacking DUB activity are locked into the axon retrograde transport state, which is incompatible with epithelium to neuron transmission. The reason for this transmission defect is unclear, but microtubule minus-ends are associated with the apical membrane in polarized epithelial cells, and constitutive minus-end transport may be incompatible with basal viral egress in the vicinity of innervating axon terminals (Bacallao et al. 1989; Meng et al. 2008). In contrast, ubiquitination is only required in neuronal cells, and the pronounced minus-end transport that ensues is consistent with the ability of VP1/2 to recruit the dynein motor (Radtke et al. 2010; Zaichick et al. 2013). One unanswered aspect of the ubiquitin switch mechanism is why the VP1/2 DUB does not prevent ubiquitination in neurons during retrograde axonal transport. It seems likely that the DUB is inactive following entry into neurons, which could be related to the postentry trigger that sees the disassembly of much of the tegument from capsids.
Following replication in neurons, viral particles transmit anterogradely within axons to axon terminals where infection spreads back to the epithelium. This represents another kind of barrier to the virus, as newly synthesized material is generally not permitted access to the axon initial segment from the neuronal soma (Song et al. 2009; Kuijpers et al. 2016). The viral effectors required to overcome the intracellular sorting barrier are the gE/gI complex and pUS9 envelope proteins (Ch’ng and Enquist 2005a, b; LaVail et al. 2007; Lyman et al. 2007, 2008; Snyder et al. 2008; McGraw et al. 2009; Howard et al. 2013), which together engage a kinesin-3 motor, KIF1A (Kramer et al. 2012; Kratchmarov et al. 2013). Once past the sorting barrier, the pUS9-KIF1A interaction becomes dispensable (Ch’ng and Enquist 2005a; Brideau et al. 2000; Daniel et al. 2015), and it is unclear if the virus particle continues to use Kif1A for long-distance anterograde axonal transport to the distal terminal (Okada et al. 1995; Lo et al. 2011; Huang and Banker 2012; Soppina et al. 2014) or switches to another motor such as a conventional kinesin (Radtke et al. 2010; Huang and Banker 2012; Nakata and Hirokawa 2003; Jacobson et al. 2006; Lee et al. 2006; Hirokawa et al. 2010). In either case, this step may not have to be directed by the virus but instead could be a default cellular process passively used by the virus.
8.7 Summary and Outlook
The past several years have seen key advances in deciphering the neuroinvasive infectious cycle of the alphaherpesviruses. These studies have not only led to the identification of virus factors promoting the robust neuroinvasive property of these viruses but the corresponding innate host barriers that the virus overcomes. Emerging evidence that the virion is a dynamic structure will likely continue to develop as the molecular details of architectural rearrangements are worked out, and their implications for epithelial and neuronal cell infections are revealed. CryoEM and cryoET advances continue to refine the virion structure, but it may be time to ask: what have we been overlooking in the application of these advanced image reconstruction methods? Is the asymmetrical extracellular particle the only relevant structure or might imaging of stabilized nascent symmetrical virions provide additional insights into herpesvirus architecture with implications for a virion clockwork? The potential preservation of the symmetrical virion configuration by N-ethylmaleimide (NEM) may be one avenue to getting at these questions. The interplay between virion structure/function, virus triggering events, neuroinvasion, and reemergence from the peripheral nervous system will undoubtedly lead to new applications in gene therapy, oncolytic vector design, and possibly effective vaccines that may one day see the end of herpes simplex keratitis, herpes simplex encephalitis, and devastating neonatal infections.
Acknowledgments
I am in debt to Dr. Fred Homa for devoting a significant amount of time responding to my email inquiries regarding herpesvirus virion structure/function and Dr. James Conway for teaching me how to use the Chimera software package. I also thank Gina Daniel for editing the manuscript. The electron micrograph was provided by Dr. Kevin Bohannon who performed imaging at the Northwestern University Center for Advanced Microscopy, which is generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. I received support from NIH grant R01 AI056346.
References
- Abaitua F, O’Hare P. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J Virol. 2008;82:5234–5244. doi: 10.1128/JVI.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaitua F, Hollinshead M, Bolstad M, Crump CM, O’Hare P. A nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore. J Virol. 2012;86:8998–9014. doi: 10.1128/JVI.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A, Miranda-Saksena M, Boadle RA, Kelly BJ, Diefenbach RJ, Alam W, Cunningham AL. Ultrastructural visualization of individual tegument protein dissociation during entry of herpes simplex virus 1 into human and rat dorsal root ganglion neurons. J Virol. 2012;86:6123–6137. doi: 10.1128/JVI.07016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinone SE, Smith GA. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol. 2010;84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EH, Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Wedemeyer WJ, Rixon FJ, Baker D, Chiu W. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLoS Comput Biol. 2006;2:e146. doi: 10.1371/journal.pcbi.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannon KP, Jun Y, Gross SP, Smith GA. Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture. Proc Natl Acad Sci U S A. 2013;110:E1613–E1620. doi: 10.1073/pnas.1221896110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad M, Abaitua F, Crump CM, O’Hare P. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J Virol. 2011;85:8738–8751. doi: 10.1128/JVI.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher S, Maresch C, Granzow H, Klupp BG, Teifke JP, Mettenleiter TC. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J Virol. 2008;82:6009–6016. doi: 10.1128/JVI.00280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau AD, Card JP, Enquist LW. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J Virol. 2000;74:834–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JC, Newcomb WW. Herpesvirus capsid assembly: insights from structural analysis. Curr Opin Virol. 2011;1:142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucks MA, O’Regan KJ, Murphy MA, Wills JW, Courtney RJ. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology. 2007;361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe. 2011;10:105–117. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Campadelli-Fiume G, Roizman B. The egress of herpesviruses from cells: the unanswered questions. J Virol. 2006;80:6716–6717. doi: 10.1128/JVI.00386-06. author replies 6717–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol. 2012;86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Enquist LW. Neuron-to-cell spread of pseudorabies virus in a compartmented neuronal culture system. J Virol. 2005a;79:10875–10889. doi: 10.1128/JVI.79.17.10875-10889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Enquist LW. Efficient axonal localization of alphaherpesvirus structural proteins in cultured sympathetic neurons requires viral glycoprotein E. J Virol. 2005b;79:8835–8846. doi: 10.1128/JVI.79.14.8835-8846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha P, Han J, Starkey JL, Wills JW. Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J Virol. 2012;86:11886–11898. doi: 10.1128/JVI.01879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Zhu Z, Gershon AA, Gershon MD. Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell. 2004;119:915–926. doi: 10.1016/j.cell.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Chouljenko DV, Jambunathan N, Chouljenko VN, Naderi M, Brylinski M, Caskey JR, Kousoulas KG. Herpes simplex virus 1 UL37 protein tyrosine residues conserved among all alphaherpesviruses are required for interactions with glycoprotein K, cytoplasmic virion envelopment, and infectious virus production. J Virol. 2016;90:10351–10361. doi: 10.1128/JVI.01202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alpha herpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RS, Heldwein EE. Herpesvirus gB: a finely tuned fusion machine. Viruses. 2015;7:6552–6569. doi: 10.3390/v7122957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham A, Miranda-Saksena M, Diefenbach R, Johnson D. Letter in response to: Making the case: married versus separate models of alphaherpes virus anterograde transport in axons. Rev Med Virol. 2013;23:414–418. doi: 10.1002/rmv.1760. [DOI] [PubMed] [Google Scholar]
- Dai X, Gong D, Wu TT, Sun R, Zhou ZH. Organization of capsid-associated tegument components in Kaposi’s sarcoma-associated herpesvirus. J Virol. 2014;88:12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel GR, Sollars PJ, Pickard GE, Smith GA. Pseudorabies virus fast axonal transport occurs by a pUS9-independent mechanism. J Virol. 2015;89:8088–8091. doi: 10.1128/JVI.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delboy MG, Roller DG, Nicola AV. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J Virol. 2008;82:3381–3390. doi: 10.1128/JVI.02296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, Sexton GL, Huang E, Person S. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol. 2008;82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi I, Lippe R. HSV-1 gN partners with gM to modulate the viral fusion machinery. J Virol. 2014;89:2313–2323. doi: 10.1128/JVI.03041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW, Husak PJ, Banfield BW, Smith GA. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- Fan WH, Roberts AP, McElwee M, Bhella D, Rixon FJ, Lauder R. The large tegument protein pUL36 is essential for formation of the capsid vertex specific component at the capsid-tegument interface of HSV-1. J Virol. 2014;89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Granzow H, Klupp BG, Kopp M, Mettenleiter TC. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J Virol. 2002;76:6729–6742. doi: 10.1128/JVI.76.13.6729-6742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol. 2004;78:11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller AO, Santos RE, Spear PG. Neutralizing antibodies specific for glycoprotein H of herpes simplex virus permit viral attachment to cells but prevent penetration. J Virol. 1989;63:3435–3443. doi: 10.1128/jvi.63.8.3435-3443.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulmer PA, Melancon JM, Baines JD, Kousoulas KG. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J Virol. 2007;81:3097–3108. doi: 10.1128/JVI.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldello S, Hildebrand S, Faridani O, Callegari S, Palmkvist M, Di Guglielmo C, Masucci MG. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat Cell Biol. 2010;12:351–361. doi: 10.1038/ncb2035. [DOI] [PubMed] [Google Scholar]
- Gastaldello S, Chen X, Callegari S, Masucci MG. Caspase-1 promotes Epstein-Barr virus replication by targeting the large tegument protein deneddylase to the nucleus of productively infected cells. PLoS Pathog. 2013;9:e1003664. doi: 10.1371/journal.ppat.1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Weiland F, Jons A, Klupp BG, Karger A, Mettenleiter TC. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Klupp BG, Mettenleiter TC. Entry of pseudorabies virus: an immunogold-labeling study. J Virol. 2005;79:3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol. 2011;85:9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Chadha P, Starkey JL, Wills JW. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci U S A. 2012;109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AL, Meckes DG, Jr, Marsh JA, Ward MD, Yeh PC, Baird NL, Wilson CB, Semmes OJ, Wills JW. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol. 2010;84:2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J. 2012;31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook LM, Huang J, Jiang M, Hodinka R, Friedman HM. Blocking antibody access to neutralizing domains on glycoproteins involved in entry as a novel mechanism of immune evasion by herpes simplex virus type 1 glycoproteins C and E. J Virol. 2008;82:6935–6941. doi: 10.1128/JVI.02599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PW, Howard TL, Johnson DC. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into initial axon segments. J Virol. 2013;87:403–414. doi: 10.1128/JVI.02465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Banker G. The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic. 2012;13(4):549–564. doi: 10.1111/j.1600-0854.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet A, Makhov AM, Huffman JB, Vos M, Homa FL, Conway JF. Extensive subunit contacts underpin herpesvirus capsid stability and interior-to-exterior allostery. Nat Struct Mol Biol. 2016;23:531–539. doi: 10.1038/nsmb.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE, Smith GA. Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A. 2015;112:12818–12823. doi: 10.1073/pnas.1512559112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, Ou JH, Jung JU. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol. 2011;85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49:797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. The herpes simplex virus type-1 UL37 protein interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol. 2014;88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovasevic V, Liang L, Roizman B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol. 2008;82:3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology. 2014;454–455:67–77. doi: 10.1016/j.virol.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Kharkwal H, Shanda Furgiuele S, Smith CG, Wilson DW. HSV capsid/organelle association in the absence of the large tegument protein UL36p. J Virol. 2015;89(22):11372–11382. doi: 10.1128/JVI.01893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharkwal H, Smith CG, Wilson DW. Herpes simplex virus capsid localization to ESCRT-VPS4 complexes in the presence and absence of the large tegument protein UL36p. J Virol. 2016;90:7257–7267. doi: 10.1128/JVI.00857-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Oh SE, Kwon KM, Lee CH, Ahn JH. Involvement of the N-terminal deubiquitinating protease domain of human cytomegalovirus UL48 tegument protein in autoubiquitination, virion stability, and virus entry. J Virol. 2016;90:3229–3242. doi: 10.1128/JVI.02766-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Nixdorf R, Mettenleiter TC. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol. 2002;76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J Virol. 2005;79:1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Howley PM, editors. Fields virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. [Google Scholar]
- Ko DH, Cunningham AL, Diefenbach RJ. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2) J Virol. 2010;84:1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S, Mar EC, Stamey FR, Inoue N. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol. 2003;84:1485–1491. doi: 10.1099/vir.0.18941-0. [DOI] [PubMed] [Google Scholar]
- Kramer T, Greco TM, Taylor MP, Ambrosini AE, Cristea IM, Enquist LW. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe. 2012;12:806–814. doi: 10.1016/j.chom.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarov R, et al. Making the case: married versus separate models of alpha herpes virus anterograde transport in axons. Rev Med Virol. 2012;22(6):378–391. doi: 10.1002/rmv.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch’ng TH, Cristea IM, Enquist LW. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J Virol. 2013;87:9431–9440. doi: 10.1128/JVI.01317-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, van de Willige D, Freal A, Chazeau A, Franker MA, Hofenk J, Rodrigues RJ, Kapitein LC, Akhmanova A, Jaarsma D, Hoogenraad CC. Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron. 2016;89:461–471. doi: 10.1016/j.neuron.2016.01.022. [DOI] [PubMed] [Google Scholar]
- Laine RF, Albecka A, van de Linde S, Rees EJ, Crump CM, Kaminski CF. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat Commun. 2015;6:5980. doi: 10.1038/ncomms6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail JH, Tauscher AN, Sucher A, Harrabi O, Brandimarti R. Viral regulation of the long distance axonal transport of herpes simplex virus nucleocapsid. Neuroscience. 2007;146:974–985. doi: 10.1016/j.neuroscience.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GE, Murray JW, Wolkoff AW, Wilson DW. Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J Virol. 2006;80:4264–4275. doi: 10.1128/JVI.80.9.4264-4275.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology. 2008;378:347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Lee JI, Sollars PJ, Baver SB, Pickard GE, Leelawong M, Smith GA. A herpesvirus encoded deubiquitinase is a novel neuroinvasive determinant. PLoS Pathog. 2009;5:e1000387. doi: 10.1371/journal.ppat.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelawong M, Lee JI, Smith GA. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol. 2012;86:6303–6314. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzinger H, Ziegler U, Schraner EM, Fraefel C, Glauser DL, Heid I, Ackermann M, Mueller M, Wild P. Herpes simplex virus 1 envelopment follows two diverse pathways. J Virol. 2005;79:13047–13059. doi: 10.1128/JVI.79.20.13047-13059.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett. 2011;491:168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci U S A. 2005;102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol. 2006;80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman MG, Feierbach B, Curanovic D, Bisher M, Enquist LW. PRV Us9 directs axonal sorting of viral capsids. J Virol. 2007;81(20):11363–11371. doi: 10.1128/JVI.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman MG, Curanovic D, Enquist LW. Targeting of pseudorabies virus structural proteins to axons requires association of the viral Us9 protein with lipid rafts. PLoS Pathog. 2008;4:e1000065. doi: 10.1371/journal.ppat.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer UE, Sodeik B, Grunewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci U S A. 2008;105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J Virol. 2009;83:8315–8326. doi: 10.1128/JVI.00633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J, Rixon FJ. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol. 1992;73(Pt 2):269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- McLauchlan J, Addison C, Craigie MC, Rixon FJ. Noninfectious L-particles supply functions which can facilitate infection by HSV-1. Virology. 1992;190:682–688. doi: 10.1016/0042-6822(92)90906-6. [DOI] [PubMed] [Google Scholar]
- Meckes DG, Jr, Wills JW. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol. 2007;81:13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Wills JW. Structural rearrangement within an enveloped virus upon binding to the host cell. J Virol. 2008;82:10429–10435. doi: 10.1128/JVI.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr, Marsh JA, Wills JW. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology. 2010;398:208–213. doi: 10.1016/j.virol.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC. Encyclopedia of virology. 3. Vol. 4. Elsevier; Amsterdam: 2008. Pseudorabies virus; pp. 341–351. [Google Scholar]
- Mettenleiter TC, Minson T. Egress of alphaherpesviruses. J Virol. 2006;80:1610–1611. doi: 10.1128/JVI.80.3.1610-1612.2006. Author reply 1611–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael K, Bottcher S, Klupp BG, Karger A, Mettenleiter TC. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J Gen Virol. 2006;87:3503–3507. doi: 10.1099/vir.0.82168-0. [DOI] [PubMed] [Google Scholar]
- Michael K, Klupp BG, Karger A, Mettenleiter TC. Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21. J Virol. 2007;81:1048–1051. doi: 10.1128/JVI.01801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatov B, Cunningham AL, Diefenbach RJ. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology. 2007;368:26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Mohl BS, et al. Random transposon-mediated mutagenesis of the essential large tegument protein pUL36 of Pseudorabies virus. J Virol. 2010;84(16):8153–8162. doi: 10.1128/JVI.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Rose HM, Mednis B. Electron microscopy of herpes simplex virus. I. Entry. J Virol. 1968;2:507–516. doi: 10.1128/jvi.2.5.507-516.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Wang YF, Meredith DM. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalwanga D, Rempel S, Roizman B, Baines JD. The UL16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology. 1996;226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Time-dependent transformation of the herpesvirus tegument. J Virol. 2009;83:8082–8089. doi: 10.1128/JVI.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, McEvoy AM, Straus SE. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J Virol. 2003;77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, Hou J, Major EO, Straus SE. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol. 2005;79:7609–7616. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Oshima S, Daikoku T, Shibata S, Yamada H, Goshima F, Nishiyama Y. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch Virol. 1998;143:863–880. doi: 10.1007/s007050050338. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, et al. eShadow: a tool for comparing closely related sequences. Genome Res. 2004;14(6):1191–1198. doi: 10.1101/gr.1773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Crump CM, Graham SC. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses. 2015;7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association to the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Abaitua F, O’Hare P, McNab D, Rixon FJ, Pasdeloup D. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol. 2009;83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Murata T, Kanda T, Isomura H, Narita Y, Sugimoto A, Kawashima D, Tsurumi T. Epstein-Barr virus deubiquitinase down-regulates TRAF6-mediated NF-kappaB signaling during productive replication. J Virol. 2013;87:4060–4070. doi: 10.1128/JVI.02020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. The C-terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol. 2012;86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J Virol. 2005;79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtes LD, Yang K, Li LX, Baines JD. The capsid protein encoded by U(L)17 of herpes simplex virus 1 interacts with tegument protein VP13/14. J Virol. 2010;84(15):7642–7650. doi: 10.1128/JVI.00277-10. Epub 2010 May 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag JD, Prasad BV, Rixon FJ, Chiu W. Three-dimensional structure of the HSV1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol. 2012;66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Enquist LW. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Polcicova K, Johnson DC. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J Virol. 2008;82:10613–10624. doi: 10.1128/JVI.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, Verhey KJ. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci U S A. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol. 2014;88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova S, Bell S, Crump CM. Analysis of the interaction between the essential HSV-1 tegument proteins VP16 and VP1/2. J Virol. 2011;86:473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova S, Bell S, Crump CM. Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J Virol. 2012;86:473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilagyi JF, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell. 2007;26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte S, Letellier J, Lippe R. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol. 2005;79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- van Gent M, Braem SG, de Jong A, Delagic N, Peeters JG, Boer IG, Moynagh PN, Kremmer E, Wiertz EJ, Ovaa H, Griffin BD, Ressing ME. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathog. 2014;10:e1003960. doi: 10.1371/journal.ppat.1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol. 2005;79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang K, Li J, Zheng C. HSV-1 ubiquitin-specific protease UL36 inhibits IFN-beta production by deubiquitinating TRAF3. J Virol. 2013;87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst CB, Ning S, Bentz GL, Dufour F, Gershburg E, Shackelford J, Langelier Y, Pagano JS. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J Virol. 2009;83:4345–4353. doi: 10.1128/JVI.02195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst CB, Vaziri C, Shackelford J, Pagano JS. Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J Virol. 2012;86:8097–8106. doi: 10.1128/JVI.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- Yeh PC, Meckes DG, Jr, Wills JW. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol. 2008;82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe. 2013;13:193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]