Abstract
Background
Computer vision may aid melanoma detection.
Objective
Compare melanoma diagnostic accuracy of computer algorithms to dermatologists using dermoscopic images.
Methods
Cross-sectional study using 100 randomly-selected dermoscopic images (50 melanomas,44 nevi,6 lentigines) from an international computer vision melanoma challenge dataset (n=379), along with individual algorithm results from twenty-five teams. We used five methods (non-learned and machine learning) to combine individual automated predictions into “fusion” algorithms. In a companion study, eight dermatologists classified the lesions in the 100 images as benign or malignant.
Results
Average sensitivity and specificity of dermatologists in classification was 82% and 59%. At 82% sensitivity, dermatologist specificity was similar to the top challenge algorithm (59% v. 62%,p=0.68) but lower than the best-performing fusion algorithm (59% v. 76%,p=0.02). ROC area of the top fusion algorithm was greater than the mean ROC area of dermatologists (0.86 v. 0.71, p=0.001).
Limitations
The dataset lacked the full spectrum of skin lesions encountered in clinical practice, particularly banal lesions. Readers and algorithms were not provided clinical data (e.g., age, lesion history/symptoms). Results obtained using our study design cannot be extrapolated to clinical practice.
Conclusion
Deep learning computer vision systems classified melanoma dermoscopy images with accuracy that exceeded some but not all dermatologists.
Keywords: computer vision, machine learning, melanoma, reader study, dermatologist, skin cancer, computer algorithm, International Skin Imaging Collaboration, International Symposium on Biomedical Imaging
Introduction
The early diagnosis of melanoma remains challenging.1 Estimates of the sensitivity of dermatologists for melanoma in reader studies were 70% for the Nevisense trial2 and 78% for the MelaFind trial.3 In addition, as non-physicians detect the majority of melanomas4 and population-based melanoma screening by clinicians is not recommended in the United States (US),5 there is not only interest in the development of automated image analysis algorithms to help dermatologists classify dermoscopic images, but also to aid laypersons or non-dermatology physicians in melanoma detection.6–13 To date, however, the lack of a large, public dataset of skin images has limited the ability to directly compare the diagnostic performance of competing automated image analysis approaches against clinicians.
To address this limitation, the International Skin Imaging Collaboration (ISIC) Melanoma Project created an open-access archive of dermoscopic images of skin lesions for education and research.14 Here, we describe the melanoma classification results from a challenge conducted by the ISIC Archive15 at the 2016 International Symposium on Biomedical Imaging (ISBI) involving 25 competing teams.16 We further performed a companion reader study with eight experienced dermatologists on a subset of images; these results served as a reference comparator to the automated algorithm approaches.
Materials and Methods
IRB Approval
Institutional Review Board approval was obtained at Memorial Sloan Kettering and the study was conducted in accordance with the Helsinki Declaration.
ISBI 2016 Melanoma Detection Challenge Dataset
Details of the challenge tasks, evaluation criteria, timeline, and participation have been previously described.15,17,18 In December 2015, 1,552 lesions were chosen from ~12,000 dermoscopic images in the ISIC Archive; after excluding 273 for inadequate image quality, 1,279 lesions (248 (19.3%) melanomas and 1,031 (80.7%) nevi or lentigines) were included. Images were excluded due to poor focus or if they included multiple lesions or lesions that encompassed the entire field of view. The dataset was randomly divided into training (n=900, 19.2% melanomas) and test (n=379, 19.8% melanomas) sets. All melanomas and a majority of the nevi/lentigines (n=869, 84%) had been histopathologically examined. Non-histopathologically examined nevi (n=162) originated from a longitudinal study of children; selection from this dataset was biased to include lesions with the largest diameters and all images were reviewed by two or more dermatologists to confirm their benign nature.19 Images used in this challenge were obtained with multiple camera/dermatoscope combinations and originated from >12 dermatology clinics around the world.
Twenty-five teams participated in the challenge, all of which used deep learning, a form of machine learning that uses multiple processing layers to automatically identify increasingly abstract concepts present in data. Computer algorithms were ranked using average precision, which corresponds to the integral under a precision-recall curve [which depicts positive predictive value (i.e., proportion of positive results that are true positives) and sensitivity (i.e., proportion of positive results that are correctly identified)], and the final results and rankings are publically available.15,18
Reader study
A reader study was performed on 50 randomly selected melanomas (31 invasive, 19 in situ) and 50 benign neoplasms (44 nevi, 6 lentigines) from the 379 test images. Non-histopathologically examined benign lesions were excluded from this image set. The median (range) Breslow depth for the invasive melanomas was 0.70 (0.10 – 2.06) mm. Eight experienced dermatologists from four countries were invited on May 13 2016 and all agreed to participate. The mean (range) number of years of (a) post-residency clinical experience and (b) use of dermoscopy among readers was 13 (3–31) and 13.5 (6–27) years, respectively, and all had a primary clinical focus on skin cancer. For each dermoscopic image, readers: (a) classified the lesion (benign v. malignant) and (b) indicated management (biopsy or observation/reassurance). Readers were blinded to diagnosis and clinical images/metadata. There were no time restrictions and participants could complete evaluations over multiple sittings.
Automated Predictions
Here we report the performance of the five top-ranked individual algorithms of the ISBI 2016 Challenge on the reader set of 100 dermoscopic images. In addition, we implemented five methods of fusing all automated predictions from the 25 participating teams in the ISBI challenge into a single prediction. These methods included two non-learned approaches (prediction score averaging and voting) and three machine learning methods: greedy ensemble fusion20, linear binary support vector machine (SVM), and non-linear binary SVM (histogram intersection kernel) (see Supplementary materials for full description of fusion approaches).21 Test set images that were not involved in the reader study (n=279) were used to train fusion methods; fusion algorithms were ranked by average precision on the reader set of 100 images.
Statistical Analysis
Main outcomes and measures were sensitivity, specificity, and area under the receiver operating characteristic (ROC) curves. Sensitivity in classification was defined as the percentage of melanomas that were correctly scored as malignant. Sensitivity in management decision was defined as the percentage of melanomas that were correctly indicated for biopsy. Specificity in classification was defined as the percentage of benign lesions that were correctly scored as benign. Specificity in management decision was defined as the percentage of benign lesions that were correctly indicated for observation/reassurance.
Computers submitted predictions between 0.0 and 1.0, with 0.5 used as a dichotomous threshold in the ISBI Challenge: values ≤0.5 were benign and values >0.5 to 1.0 were malignant. For analyses here, we considered scores closer to 0 to indicate a higher probability of a benign diagnosis and scores closer to 1 to indicate a higher probability of malignancy.
As ground truth data provided to participants in the ISBI 2016 challenge was restricted to classification (benign v. malignant) and did not include management data (biopsy v. observation/reassurance), we chose classification performance as the primary outcome. To inform clinical practice, however, we also compared management decisions of dermatologists to computer classification performance as an exploratory outcome; another rationale for reporting management decision performance was that most studies comparing human readers to computers have used management decision, and not classification, as the primary outcome. Our primary comparison between readers and computers was specificity at average dermatologist sensitivity; the secondary comparison between readers and computers was ROC area of the algorithm and mean ROC area of the dermatologists.
Descriptive statistics such as relative frequencies, means and standard deviations were used to describe the dermatologist lesion classifications and management decisions for each evaluation. Overall percent agreement, kappa and intraclass correlation were used to evaluate reader responses for lesion classification and management. Levels of inter-rater agreement were evaluated as percent agreement and multi-rater kappa. In addition, patterns of agreement for the dermatologist assessments were evaluated on the lesion level, where lesions were classified as having unanimous agreement between readers, or not.
Levels of classification and management accuracy were calculated for each individual reader and for the readers as a group. To describe the study sample, and to provide comparisons between measures of diagnostic performance between readers and algorithms, two-sample tests for proportions along with chi-square tests were used. In addition, regression models for binary outcomes using a general estimating equations approach with a log link and an exchangeable correlation structure were used. In these models, readers were considered a covariate, allowing for between reader comparisons of accuracy and stratified analyses. The exchangeable correlation structure was used to adjust the standard error estimates for the potential of clustered observations within readers. In addition, receiver operating characteristic (ROC) curves were estimated for the individual readers and for the readers as a group. Comparisons of area under the ROC curves were performed to assess differences in reader performance and to make comparisons between the reader and algorithm performance. For dichotomous predictions, area under ROC curves is equivalent to the average of sensitivity and specificity. Alpha level was set at 5% and all presented p-values are two-sided. All analyses were performed with StataSE v14.1 (Stata Corporation, College Station, TX, USA).
Results
Diagnostic accuracy of dermatologists for melanoma
The average (min-max) sensitivity and specificity of the eight readers for lesion classification (i.e., benign v. malignant) was 82% (68%–98%) and 59% (34%–72%), respectively (Table 1). This corresponded to an average (min-max) ROC area of 0.71 (0.61–0.76). The average (min226 max) sensitivity for melanoma in situ and invasive melanoma was 68.4% (53%–95%) and 89.1% (75%–100%), respectively. Data describing levels of agreement among dermatologists is included in Supplementary materials.
Table 1.
Reader results
| Classification | Management | |||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | ROC Area |
Sensitivity | Specificity | ROC Area |
|
| Reader 1 | 68% | 72% | 0.70 | 72% | 68% | 0.70 |
| Reader 2 | 68% | 66% | 0.67 | 86% | 40% | 0.63 |
| Reader 3 | 98% | 54% | 0.76 | 100% | 38% | 0.69 |
| Reader 4 | 86% | 62% | 0.74 | 88% | 56% | 0.72 |
| Reader 5 | 88% | 34% | 0.61 | 90% | 30% | 0.60 |
| Reader 6 | 74% | 68% | 0.71 | 76% | 66% | 0.71 |
| Reader 7 | 82% | 58% | 0.70 | 100% | 32% | 0.66 |
| Reader 8 | 92% | 60% | 0.76 | 96% | 48% | 0.72 |
| Average | 82% | 59% | 0.71 | 89% | 47% | 0.68 |
ROC = Receiver operating characteristic
Diagnostic accuracy of computer algorithms
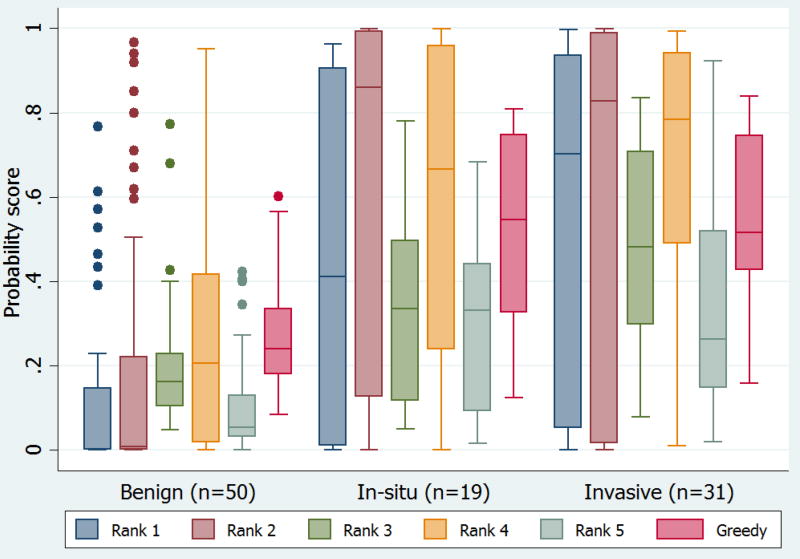
Performance of automated systems on the 100 images evaluated by the dermatologists is shown in Table 2. Ranked on average precision, greedy fusion was the top performing fusion algorithm (selected 16 algorithms for fusion from the 25 total). While non-learning methods performed similarly, more complex SVM models demonstrated a slight reduction in performance. The re-learned probabilistic SVM thresholds increased sensitivity of the corresponding systems considerably. Figure 1 illustrates the mean probability score for the top five algorithms and the best performing fusion algorithm by diagnosis.
Table 2.
Results of the International Symposium on Biomedical Imaging Challenge top five individual algorithms and fusion algorithms on the reader study dataset on 100 images evaluated by dermatologists
| Algorithm | Sensitivity | Specificity | Specificity at 82% Sensitivity |
Specificity at 89% Sensitivity |
ROC1* at 82% Sensitivity |
ROC1* at 89% Sensitivity |
ROC2** | Average Precision† |
|---|---|---|---|---|---|---|---|---|
| Rank 1 | 52% | 92% | 0.62 | 0.38 | 0.72 | 0.64 | 0.79 | 0.84 |
| Rank 2 | 60% | 80% | 0.56 | 0.40 | 0.69 | 0.65 | 0.80 | 0.83 |
| Rank 3 | 36% | 96% | 0.48 | 0.34 | 0.65 | 0.62 | 0.79 | 0.81 |
| Rank 4 | 68% | 84% | 0.50 | 0.38 | 0.66 | 0.64 | 0.80 | 0.83 |
| Rank 5 | 26% | 100% | 0.60 | 0.52 | 0.71 | 0.71 | 0.81 | 0.84 |
| Average Fusion | 46% | 92% | 0.78 | 0.66 | 0.80 | 0.78 | 0.86 | 0.86 |
| Voting Fusion | 56% | 90% | 0.82 | 0.60 | 0.82 | 0.75 | 0.86 | 0.86 |
| Greedy Fusion | 58% | 92% | 0.76 | 0.64 | 0.79 | 0.77 | 0.86 | 0.87 |
| Linear SVM Fusion | 66% | 86% | 0.68 | 0.42 | 0.75 | 0.66 | 0.82 | 0.85 |
| Non-Linear SVM Fusion | 70% | 88% | 0.68 | 0.34 | 0.75 | 0.62 | 0.82 | 0.86 |
ROC = receiver operating characteristic
ROC1 is based on dichotomizing data for response
ROC2 is based on using continuous probability generated from algorithm
Average precision is the integral under a precision-recall curve (or positive predictive value-sensitivity curve)
Figure 1. Algorithm probability scores.
Mean probability score for top five algorithms and best fusion algorithm (Greedy) by lesion diagnosis (i.e., benign nevi or lentigines, melanoma in situ, and invasive melanoma). Probability scores from computer algorithms were in the range 0 to 1, with scores closer to 0 indicating a greater probability of a benign diagnosis and scores closer to 1 indicating a greater probability of a malignant diagnosis. The upper and lower bounds of the boxed area represent the 25th and 75th percentiles, the line transecting the box is the median value, and whiskers indicate the 5% and 95% percentiles. Dots that fall outside of the whiskers indicate extreme, or outlier values.
Comparison of diagnostic accuracy of dermatologists to computers
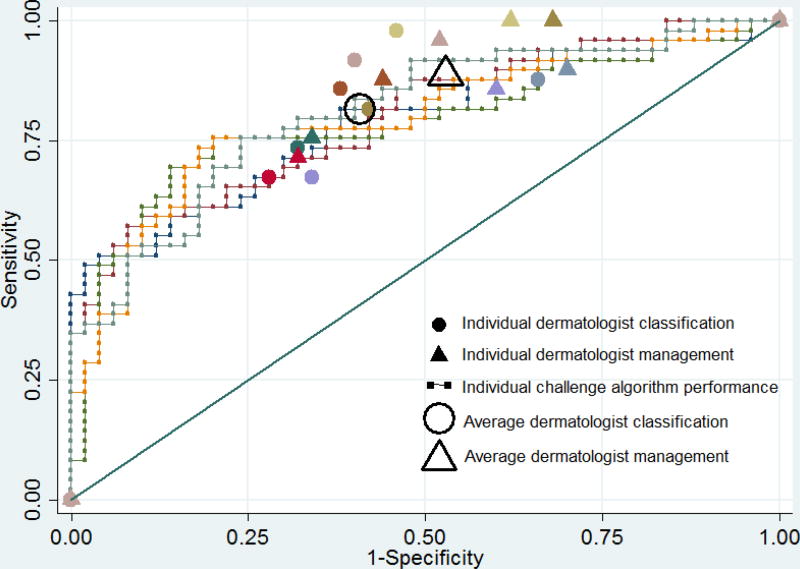
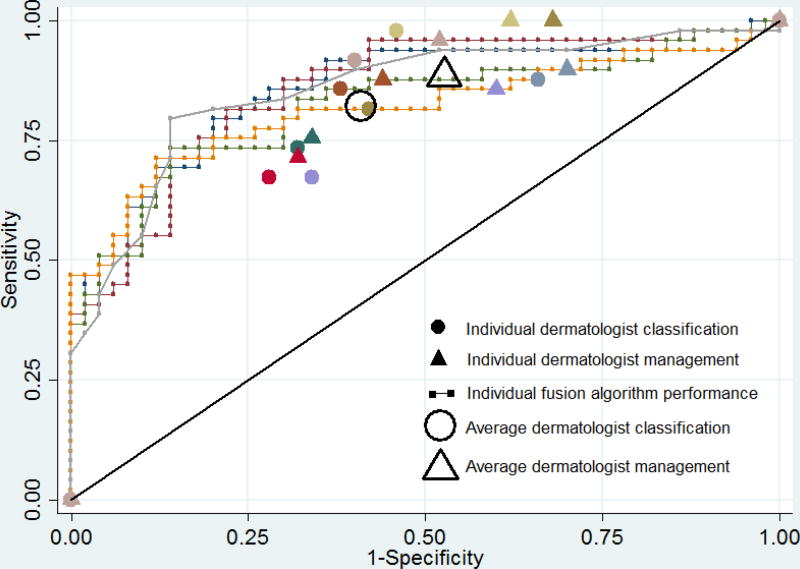
The ROC area of the best fusion computer algorithm (greedy fusion) was 0.86, which was significantly greater than the mean ROC area of 0.71 of the eight readers in classification (p=0.001). Using the dermatologist mean sensitivity value for classification (82%) as the operating point on the computer algorithm ROC curves (Figures 2A and 2B), the top fusion algorithm specificity was 76%, which was higher than the average dermatologist specificity of 59% (p=0.02) and the top-ranked individual algorithm specificity of 62% (p=0.13). Using the dermatologist mean sensitivity value for management (89%) as the operating point on the computer algorithm ROC curves, the fusion algorithm specificity was 64%, which was higher than the average dermatologist specificity of 47% (p=0.02) and the top-ranked individual algorithm specificity of 38% (p=0.009). At this cut-off threshold, there was no difference between the average dermatologist specificity and the top-ranked individual algorithm (47% v. 38%, p=0.22).
Figure 2. Diagnostic accuracy of algorithms and dermatologists for melanoma on 100 image dataset.
Receiver operating characteristic curves demonstrating sensitivity and specificity for melanoma of (A) top five ranked individual algorithms and (B) five fusion algorithms, with melanoma classification and management performance of eight dermatologists indicated by small colored solid circles and triangles, respectively. Small colored solid circles and triangles of the same color indicate the performance of an individual dermatologist. The large transparent circle and triangle with black outline indicate the average diagnostic performance of dermatologists in classification and management, respectively.
Discussion
We compared the melanoma diagnostic performance of computer algorithms from an international challenge to the average performance of eight experienced dermatologists using 100 dermoscopic images of pigmented lesions. We found that individual computer algorithms have comparable diagnostic accuracy to dermatologists; at 82% sensitivity, average reader specificity was similar to the top computer algorithm. Fusion techniques significantly improved computer performance; at 82% sensitivity, the top-ranked fusion algorithm had higher average specificity than dermatologists. In our exploratory analysis using arguably the most clinically-relevant sensitivity value, the dermatologists’ mean sensitivity in management decision (89%), dermatologists had specificity similar to the top algorithm, but lower than the top fusion algorithm approach. It is worth noting that some dermatologists had higher diagnostic performance than all individual and fusion algorithms in classification and/or management.
There has been considerable interest in developing computer vision systems for melanoma diagnosis, but few groups have directly compared computer algorithms to human performance. In 2017, Esteva A et al trained a deep learning convolution neural network (CNN) on 129,450 images of 2,032 different diseases and reported dermatologist-level classification of skin cancer.22 In the corresponding reader studies using clinical images (33 melanomas, 97 nevi) and dermoscopy images (71 melanomas, 40 benign), the CNN had a ROC area of 0.94 and 0.91, respectively, which was superior to dermatologists.22 In 2015, Ferris et al compared the diagnostic accuracy of a computer classifier to 30 dermatology healthcare providers on a dataset of 65 lesions (25 melanomas, 32 nevi, 4 lentigines, 4 seborrheic keratoses); the computer algorithm had a sensitivity of 96% and specificity of 42.5% and the human readers, which included dermatologists, dermatology residents, and physician assistants, had a mean sensitivity of 70.8% and specificity of 58.7%.8 In 2005 Menzies et al reported on the performance of SolarScan® and included a reader study of 78 lesions (13 melanomas, 63 nevi, and 2 lentigines).11 The computer classifier had a sensitivity of 85% and specificity of 65%; this compared to a mean sensitivity and specificity of 79.5% and 50.8% for the 13 human readers. Differences in study design make comparisons of our computer algorithm results to these data challenging, highlighting the importance of creating open datasets like ours.
Compared to previous investigations, there are novel aspects to our study: (a) we compared multiple computer classifiers from around the world and an aggregated model of their performance to dermatologists, increasing the likelihood that the computer-vision results reflect the current state-of-the-art; (b) our dataset originated from more than 12 dermatology clinics, possibly increasing the generalizability of our findings; (c) the readers originated from four countries, which may have improved the generalizability of our dermatologists’ results; and (d) our dataset is public, permitting external and independent analysis and use as a future reference dataset by developers of diagnostic tools.
Our results should be interpreted with caution. A significant limitation is that our dataset did not sufficiently include: (i) the complete spectrum of skin lesions encountered in clinical practice that can mimic melanoma, including pigmented seborrheic keratoses, (ii) less common presentations of melanoma such as amelanotic, nodular, or desmoplastic types that are challenging to identify, (iii) lesions from all anatomic sites, skin types, genetic backgrounds, and age ranges, and (iv) a sufficiently representative group of benign lesions that would not typically undergo biopsy and histopathological examination. As it can reasonably be inferred that >99.9% of all benign skin lesions are routinely correctly classified by dermatologists (e.g., nevi, angiomas, seborrheic keratoses, lentigines), results obtained using our study design cannot be extrapolated to clinical practice. Our study setting was artificial as computer algorithms and readers did not have access to clinical data that might have improved diagnostic performance (e.g., age, lesion history/symptoms, etc.).23 It has also been shown that the real-world performance of a computer-based system for melanoma in the hands of non-experts is lower than that expected from experimental data; diagnostic accuracy depends on the ability of users to identify appropriate lesions for analysis.24 Finally, participants of the ISBI 2016 melanoma challenge were instructed that computer algorithms would be ranked using average precision, a metric that does not target a clinically-relevant sensitivity or specificity threshold; thus, the algorithms were not optimized for comparison to dermatologist diagnostic performance.
Our results underscore the value of public challenges conducted in open-access image resources like the ISIC Archive. This platform permits comparison of the performance of individual algorithms, as well as the type of fusion experiments presented here. Larger and more diverse collections of public, clinically-validated images are needed to advance the field of computerized lesion classification, education, and clinical decision support. A bigger 2017 ISIC Challenge represents a further step in this direction.25 In addition to providing a larger and more diversified set of images, including seborrheic keratoses, the current challenge tailors the performance metrics for comparison to the current state of clinical diagnosis.
Conclusions
In this artificial study setting without integration of clinical history, state-of-the-art computer vision systems are comparable to dermatologist diagnostic accuracy for melanoma dermoscopy images and, when using fusion algorithms, can exceed dermatologist performance in classification of some but not all dermatologists. Although these results are preliminary and should be viewed with caution, development and comparison of deep learning methods on larger, more varied datasets is likely to accelerate the potential use and adoption of computer vision for melanoma detection. Strategies for including common skin lesions that are not routinely biopsied in these datasets are critical for optimizing the generalizability of computer vision algorithms.
Supplementary Material
Capsule Summary.
Computer vision has shown promise in medical diagnosis
A machine learning fusion algorithm utilizing predictions from 16 algorithms exceeded the performance of most dermatologists in the classification of 100 dermoscopic images of melanomas and nevi
These results should not be extrapolated to clinical practice until validation in prospective studies
Acknowledgments
Funding/Support: This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
Codella reports being an employee and stockholder of IBM.
Scope reports being a consultant to Emerald Inc.
This study reports the efforts of the International Skin Imaging Collaboration (ISIC), the members of which can be found at http://isdis.net/isic-project/working-group-members/. We thank the leadership of the ISIC collaboration: H. Peter Soyer, MD, Dermatology Research Centre, The University of Queensland, Brisbane, Australia (Technique Working Group Co-Leader); Clara Curiel-Lewandrowski, MD, University of Arizona Cancer Center, Tucson, AZ, USA (Technique Working Group Co-Leader); Harald Kittler, MD, Department of Dermatology, Medical University of Vienna, Austria (Terminology Working Group Leader); Liam Caffery, PhD, The University of Queensland, Brisbane, Australia (Metadata Working Group Leader); Josep Malvehy, MD; Hospital Clinic of Barcelona, Spain (Technology Working Group Leader); Rainer Hofmann Wellenhof, MD, Medical University of Graz, Austria (Archive Group Leader).
The authors sincerely thank: (a) the organizing committee of The International Symposium on Biomedical Imaging (ISBI), (b) the chairs of the 2016 ISBI Grand Challenges: Bram van Ginneken, Radboud University Medical Center, NL; Adriënne Mendrik, Utrecht University, NL; Stephen Aylward, Kitware Inc., USA, and (c) all the participants of the 2016 ISBI Challenge “Skin Lesion Analysis towards Melanoma Detection.”
Abbreviations and Acronyms List
- ROC
Receiver operating characteristic
- ISIC
International Skin Imaging Collaboration
- ISBI
International Symposium on Biomedical Imaging
- IRB
Institutional Review Board
- SVM
Support vector machine
- CNN
convolutional neural network
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
IRB Statement: This research received IRB approval at Memorial Sloan Kettering Cancer Center.
Conflicts of interest: None declared.
Statement on prior presentation: Preliminary data of this study was presented at the annual meeting of the Society for Melanoma Research Congress on Tuesday November 8, 2016.
Financial Disclosure of the Authors: Marchetti, Dusza, Halpern, Marghoob, DeFazio, Yélamos, Carrera, Jaimes, Mishra, Kalloo, Quigley, Gutman, Helba, and Celebi state no financial disclosures.
References
- 1.Marghoob AA, Scope A. The complexity of diagnosing melanoma. J Invest Dermatol. 2009;129(1):11–13. doi: 10.1038/jid.2008.388. [DOI] [PubMed] [Google Scholar]
- 2.Malvehy J, Hauschild A, Curiel-Lewandrowski C, et al. Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. Br J Dermatol. 2014;171(5):1099–1107. doi: 10.1111/bjd.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monheit G, Cognetta AB, Ferris L, et al. The performance of MelaFind: a prospective multicenter study. Arch Dermatol. 2011;147(2):188–194. doi: 10.1001/archdermatol.2010.302. [DOI] [PubMed] [Google Scholar]
- 4.Brady MS, Oliveria SA, Christos PJ, et al. Patterns of detection in patients with cutaneous melanoma. Cancer. 2000;89(2):342–347. doi: 10.1002/1097-0142(20000715)89:2<342::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Bibbins-Domingo K, Grossman DC, Curry SJ, et al. Screening for Skin Cancer: US Preventive Services Task Force Recommendation Statement. Jama. 2016;316(4):429–435. doi: 10.1001/jama.2016.8465. [DOI] [PubMed] [Google Scholar]
- 6.Celebi ME, Kingravi HA, Uddin B, et al. A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph. 2007;31(6):362–373. doi: 10.1016/j.compmedimag.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyatomi H, Oka H, Celebi ME, et al. An improved Internet-based melanoma screening system with dermatologist-like tumor area extraction algorithm. Comput Med Imaging Graph. 2008;32(7):566–579. doi: 10.1016/j.compmedimag.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Ferris LK, Harkes JA, Gilbert B, et al. Computer-aided classification of melanocytic lesions using dermoscopic images. J Am Acad Dermatol. 2015;73(5):769–776. doi: 10.1016/j.jaad.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 9.Zortea M, Schopf TR, Thon K, et al. Performance of a dermoscopy-based computer vision system for the diagnosis of pigmented skin lesions compared with visual evaluation by experienced dermatologists. Artif Intell Med. 2014;60(1):13–26. doi: 10.1016/j.artmed.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Blum A, Luedtke H, Ellwanger U, Schwabe R, Rassner G, Garbe C. Digital image analysis for diagnosis of cutaneous melanoma. Development of a highly effective computer algorithm based on analysis of 837 melanocytic lesions. Br J Dermatol. 2004;151(5):1029–1038. doi: 10.1111/j.1365-2133.2004.06210.x. [DOI] [PubMed] [Google Scholar]
- 11.Menzies SW, Bischof L, Talbot H, et al. The performance of SolarScan: an automated dermoscopy image analysis instrument for the diagnosis of primary melanoma. Arch Dermatol. 2005;141(11):1388–1396. doi: 10.1001/archderm.141.11.1388. [DOI] [PubMed] [Google Scholar]
- 12.Rajpara SM, Botello AP, Townend J, Ormerod AD. Systematic review of dermoscopy and digital dermoscopy/ artificial intelligence for the diagnosis of melanoma. Br J Dermatol. 2009;161(3):591–604. doi: 10.1111/j.1365-2133.2009.09093.x. [DOI] [PubMed] [Google Scholar]
- 13.Rubegni P, Cevenini G, Sbano P, et al. Evaluation of cutaneous melanoma thickness by digital dermoscopy analysis: a retrospective study. Melanoma Res. 2010;20(3):212–217. doi: 10.1097/CMR.0b013e328335a8ff. [DOI] [PubMed] [Google Scholar]
- 14.ISIC Archive. [Accessed September 2, 2016]; https://isic-archive.com/
- 15.ISBI 2016: Skin Lesion Analysis Towards Melanoma Detection. [Accessed September 2, 2016]; https://challenge.kitware.com/#challenge/n/ISBI_2016%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection.
- 16.ISBI 2016 Challenges: International Symposium on Biomedical Imaging: From Nano To Macro April 13–16, 2016. [Accessed September 2, 2016]; http://biomedicalimaging.org/2016/?page_id=416.
- 17.Codella N, Nguyen QB, Pankanti S, et al. Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images. IBM Journal of Research and Development. 2017;61(4/5) [Google Scholar]
- 18.Gutman D, Codella NC, Celebi E, et al. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC) 2016 arXiv preprint arXiv:1605.01397. [Google Scholar]
- 19.Scope A, Marchetti MA, Marghoob AA, et al. The study of nevi in children: Principles learned and implications for melanoma diagnosis. J Am Acad Dermatol. 2016 doi: 10.1016/j.jaad.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R, Fleury M-O, Merler M, Natsev A, Smith JR. Proceedings of the First ACM workshop on Large-scale multimedia retrieval and mining. Beijing, China: 2009. Large-scale multimedia semantic concept modeling using robust subspace bagging and MapReduce. [Google Scholar]
- 21.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2(3):1–27. [Google Scholar]
- 22.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binder M, Kittler H, Dreiseitl S, Ganster H, Wolff K, Pehamberger H. Computer-aided epiluminescence microscopy of pigmented skin lesions: the value of clinical data for the classification process. Melanoma Res. 2000;10(6):556–561. doi: 10.1097/00008390-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Dreiseitl S, Binder M, Hable K, Kittler H. Computer versus human diagnosis of melanoma: evaluation of the feasibility of an automated diagnostic system in a prospective clinical trial. Melanoma Res. 2009;19(3):180–184. doi: 10.1097/CMR.0b013e32832a1e41. [DOI] [PubMed] [Google Scholar]
- 25.ISIC 2017: Skin Lesion Analysis Towards Melanoma Detection. [Accessed December 21, 2016]; https://challenge.kitware.com/#challenge/n/ISIC_2017%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.