Abstract
Nevus spilus is a melanocytic neoplasm characterized by a tan macular background punctuated by multiple hyperpigmented macules or papules that represent various types of nevi. These include junctional and compound nevi, Spitz nevi, and rarely blue nevi. We report a unique case of widespread, multiple nevi spili giving rise to agminated Spitz nevi and congenital-pattern compound nevi that underwent genetic analysis to further characterize the mutational profile of this rare entity.
Keywords: nevus spilus, genomics, HRAS, Spitz nevus, congenital nevus
Introduction
Nevus spilus (speckled lentiginous nevus) is a benign melanocytic neoplasm composed of a tan macule containing darker macules or papules within its border. While they are considered congenital lesions, they may not appear until early childhood. Lesions range from small and isolated to giant or segmental, and may be associated with heritable syndromes. There are two clinically distinct variants. Nevus spilus maculosus is flat with evenly distributed macular hyperpigmentation likened to a polkadot pattern, and may be associated with phacomatosis pigmentovascularis. Nevus spilus papulosus is larger, with irregularly distributed larger papules in a “star map” pattern, and may be associated with phakomatosis pigmentokeratotica1. Recent genetic studies have revealed two phenotypically and genetically distinct types of nevus spilus papulosus.2 Small, single lesions as well as large lesions associated with phakomatosis pigmentokeratotica have been shown to contain the activating HRAS G13R mutation.3 Agminated Spitz nevi arising in nevus spilus have recently been shown to have the same activating HRAS mutations as the nevus spilus, as well as additional amplification of HRAS, leading to hypothesize that they arise from the same postzygotic clone as the nevus spilus, but with the additional development of a second “hit” mutation3. Larger, often segmental lesions containing large congenital-pattern nevi are referred to as nevus-spilus-type congenital melanocytic nevi (CMN). These have been shown to contain missense activating NRAS mutations involving Q61L, Q61H, and G13R2,4. These discoveries indicate that nevus spilus is an expression of mosaicism through post-zygotic mutations in genes involved with the Ras pathway, and thus is an example of a mosaic RASopathy5,6. We recently encountered a patient with both congenital-type and agminated Spitz nevi in a nevus spilus, and herein describe the unique histomorphologic and genetic features of this case.
Case Report
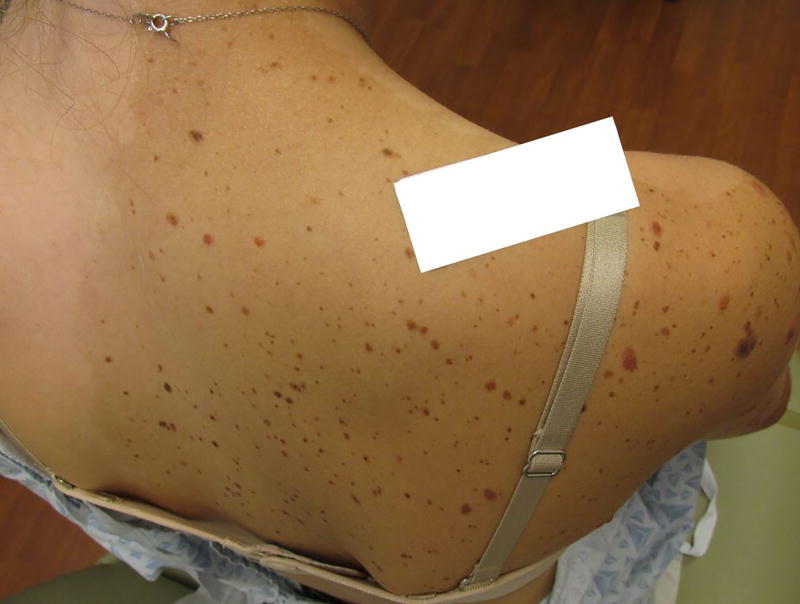
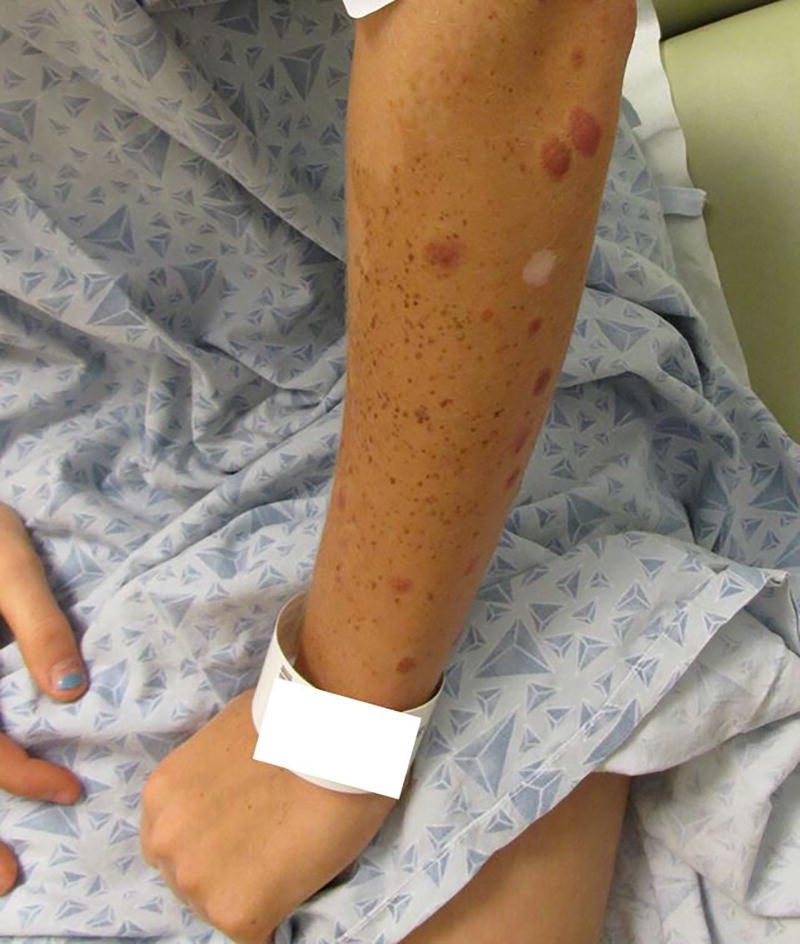
A 14 year old female presented with two large, light brown patches containing scattered dark brown macules, dark brown papules, and pink papules on both upper extremities since birth. One patch involved her right superior back, right shoulder and superior chest, and right arm and forearm (Figure 1). The second patch involved her left forearm (Figure 2). The macular brown lesions began to emerge within the tan patches around 6 months of age. By the age of 2, she had developed multiple pink papules in both patches. A few of the pink papules regressed, leaving hypopigmented residua. Prior biopsies of the pink papules were reported to show Spitz nevi. The patient had no personal history of melanoma.
Figure 1.
Extensive macular hyperpigmentation involving right shoulder and arm, studded with varying-sized and -colored papules.
Figure 2.
Macular lesion covering left forearm, containing large pink papules and small brown papules. Note hypopigmented region of prior biopsy.
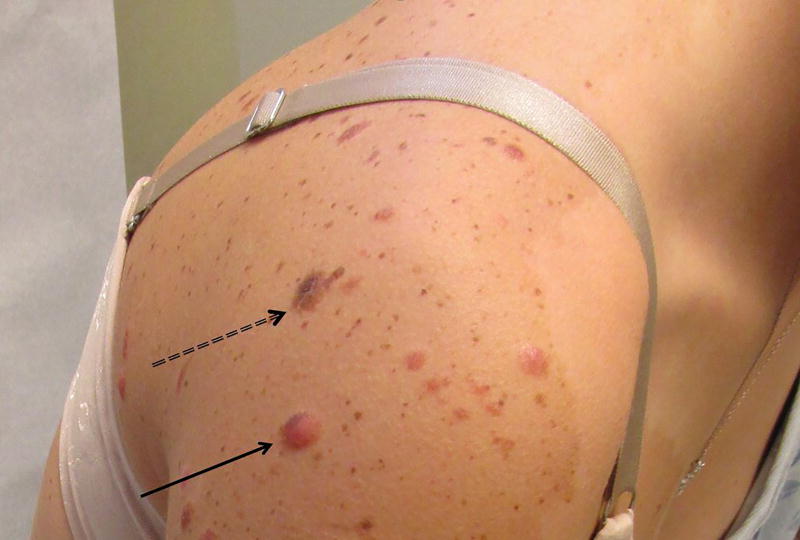
The patient underwent four biopsies of the largest, most clinically atypical lesions at our institution. The larger pink lesions (Figure 3, single arrow) were compound (2) and intradermal (1) Spitz nevi with desmoplastic features (Figure 4). The brown papule was a compound melanocytic nevus with a congenital histologic pattern (Figure 3, double dashed arrow, Figure 5). All of these nevi arose in the background of sparsely cellular lentiginous junctional melanocytic proliferation consistent with nevus spilus.
Figure 3.
Single arrow denotes an erythematous papule that is characteristic of the patient's desmoplastic Spitz nevi. The double dashed arrow denotes the congenital-pattern compound nevus.
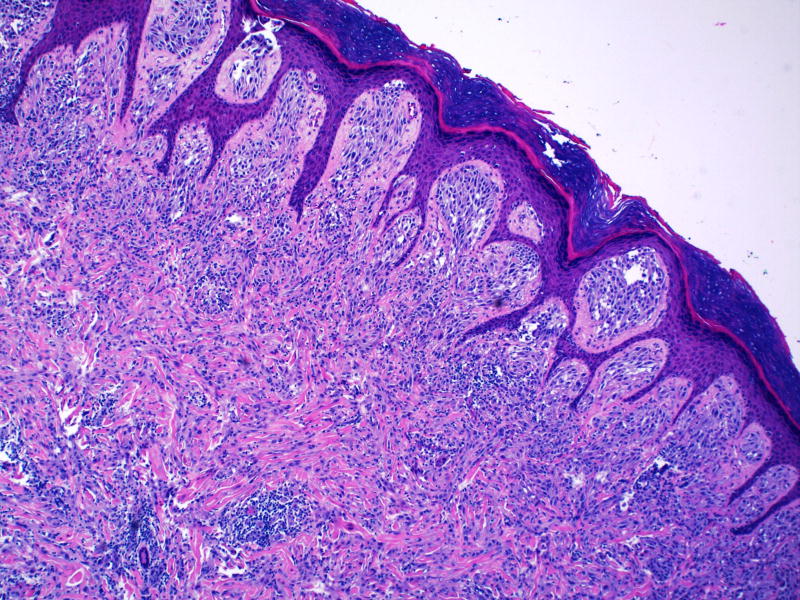
Figure 4.
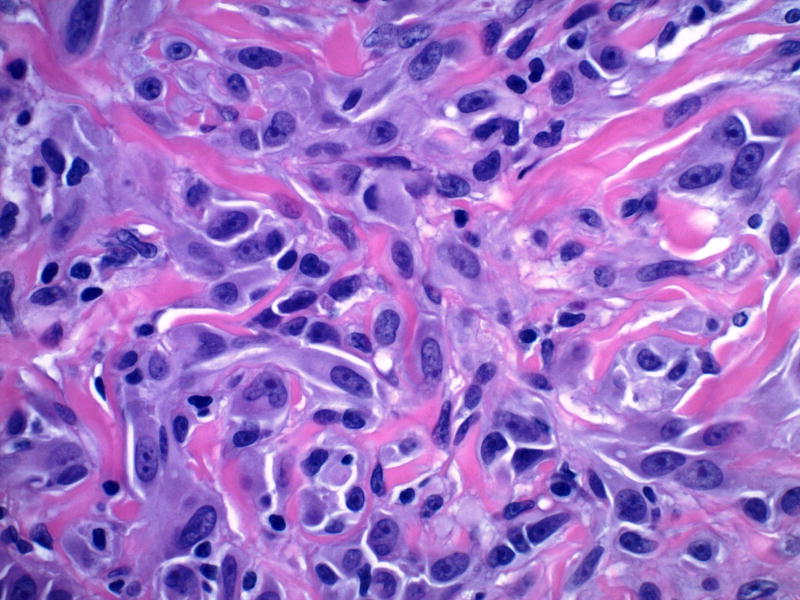
Epidermal hyperplasia overlies a compound nevus composed of oval, uniform Spitzoid melanocytes. Note cellular maturation and prominent stromal desmoplasia in deeper portions of lesion (magnification, 10×).
Figure 5.
Spitz nevus dermal component, showing large but uniform oval melanocytes without mitotic activity, surrounded by thick collagen bundles (magnification, 20×).
After patient and parental consent and IRB approval, targeted exome sequencing was performed on four samples: the nevus spilus, two desmoplastic Spitz nevi, and the compound congenital-pattern nevus. DNA was extracted using QIAamp DNA FFPE Tissue Kit according to manufacturer’s protocol (Qiagen, Valencia, CA). All samples were RNase treated, eluted in water, evaluated with the Agilent TapeStation, and quantified by Nanodrop and Qubit. Targeted DNA sequencing was performed in order to identify somatic mutations in the coding regions of 151 cancer-associated genes. Following a quantitative-PCR-based DNA quality and quantity assessment using the Agilent NGS FFPE QC Kit, 200 ng of DNA was input into the Agilent SureSelect XT ClearSeq Comprehensive Cancer Panel kit. For each tumor DNA sample, a genomic DNA library was constructed according to the manufacturer’s protocol, and the size and quality of the library was evaluated using the Agilent BioAnalyzer. Equimolar amounts of library DNA was used for a targeted enrichment using the Agilent capture baits. After quantitative PCR library quantitation and QC analysis on the BioAnalyzer, approximately 89 million 75-base paired-end sequence reads were generated using v2 chemistry on an Illumina NextSeq 500 sequencer which yielded target coverage greater than 250×.
Sequence reads were aligned to the reference human genome with the Burrows-Wheeler Aligner (BWA)7, and duplicate identification, insertion/deletion realignment, quality score recalibration, and variant identification were performed with PICARD (http://picard.sourceforge.net/) and the Genome Analysis ToolKit (GATK)8. Sequence variants were annotated to determine genic context (i.e., non-synonymous, missense, splicing) using ANNOVAR9. Additional contextual information was incorporated, including allele frequency in other studies such as 1000 Genomes, the NHLBI Exome Sequence Project, and the Exome Aggregation Consortium10, in silico function impact predictions, and observed impacts from databases like ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/), the Collection Of Somatic Mutations In Cancer (COSMIC), and The Cancer Genome Atlas (TCGA). To confirm the presence of absence of RAS mutations, sequence read alignments were manually viewed using samtools tview to observe any evidence of mutation at low frequency.
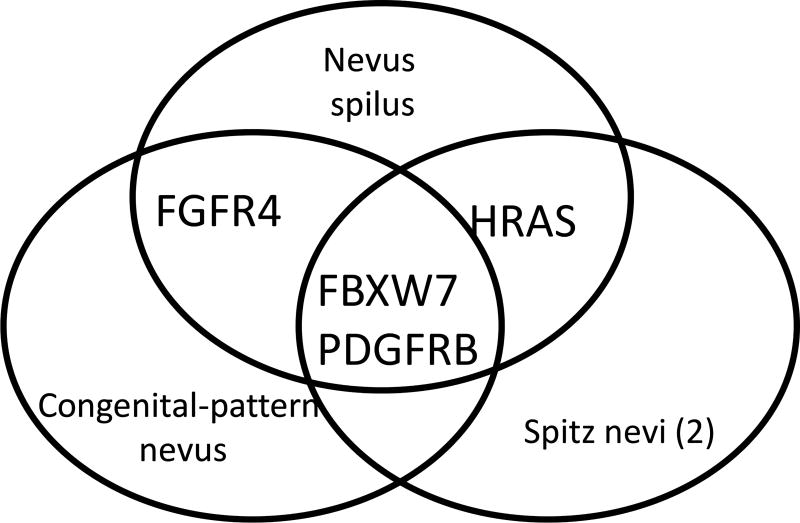
Both Spitz nevi demonstrated the same activating HRAS mutations (G13R), as well as a previously unreported variant of uncertain significance (A11S). Although a normal sample was not available for comparison, close examination of the minor allele frequency on the short arm of chromosome 11 in the region of HRAS revealed that the fraction of alternate alleles clustered around 0.33 and 0.66 in the region of the short arm, suggesting that an amplification of this portion is present. Manual examination of the nevus spilus sample revealed evidence of the G13R variant in 3.2% of reads. Neither these HRAS alterations, nor any NRAS/KRAS G12/13 or Q61, BRAF G469 or V600 were identified in the congenital-pattern nevus by either software or manual viewing. The nevus spilus and congenital-pattern nevus contained a mutation in FGFR4 (R401P) that was not found in the desmoplastic Spitz nevi. No abnormalities of NRAS were found in any of the lesions. Mutations in FBXW7 and PDGFRB were shared among all four specimens.
Discussion
These results augment the expanding body of knowledge concerning this rare lesion. Our patient demonstrated a nevus spilus papulosus with multiple Spitz nevi containing the same HRAS mutation that has been previously described in this lesion.
There is a growing body of literature concerning mutational profiles of Spitz nevi, congenital nevi, and nevus spilus, all of which may occur in both sporadic and syndromic settings. Spitz nevi may display a variety of mutations as well as translocations that lead to fusion proteins. These include HRAS mutation, loss of BAP1, and rearrangements of ALK, ROS1, NTRK1, RET, MET, and BRAF.11 In 2013, Sarin et al. reported the activating HRAS mutation in agminated Spitz nevi arising in a nevus spilus, which demonstrated mosaicism.12 Another case report showed the absence of HRAS and BRAF mutations in eruptive Spitz nevi.13 Common melanocytic nevi are associated with BRAF V600 E/K, while congenital nevi may demonstrate NRAS mutations, specifically at codon 61.11 Multiple congenital melanocytic nevi express NRAS Q61K and Q61R point mutations.14 Krengel et al described a congenital nevus arising in a nevus spilus that contained a novel mutation for NRAS at Q61L.2 These findings have shown that nevus spilus and nevus spilus-type congenital melanocytic nevi are two distinct variants both genotypically and phenotypically: a true nevus spilus is associated with a HRAS mutation, while nevus spilus-type CMN is associated with CMN related-mutations, such as BRAF and NRAS2.
Our patient is the first reported case of genetic analysis of both agminated Spitz nevi and a congenital-pattern compound nevus arising in a nevus spilus papulosus. Similar to prior reported cases, her Spitz nevi demonstrated an activating HRAS mutation. While this was not initially detected in the nevus spilus, low cellularity of the sample (which did not undergo microdissection) is the likely cause; low level evidence for this mutation was observed on manual examination of the nevus spilus sample. The observed allelic fractions suggested that the Spitz nevi had amplification of the HRAS gene. Although the congenital-pattern nevus was devoid of mutations typically associated with this histology, such as NRAS and BRAF, NRAS is mutations are found in 70% of congenital-pattern nevi15. Although FGFR4 overexpression has been reported in melanoma16, the specific point mutation in FGFR4 has not to our knowledge been previously reported in either melanoma, congenital-pattern nevus or nevus spilus. The significance of the common mutations in FBXW7 and PDGFRB found in all four specimens is unclear, but these most likely represent inherited mutations, as they are found with low frequency in European populations. There is currently no evidence to suggest that these mutations place the patient at risk for malignant transformation, but she continues to be monitored at 6 month intervals, and to undergo biopsy of any lesions with suspicious or changing clinical patterns.
Figure 6.
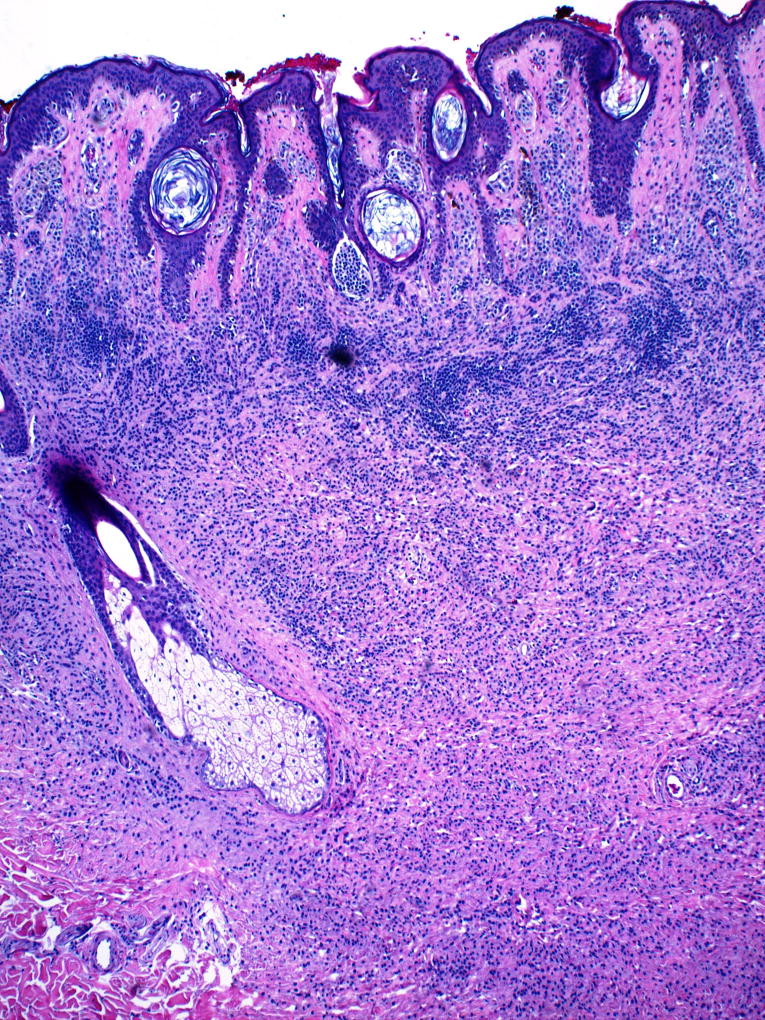
Congenital-pattern compound nevus, showing dermal component of small nested melanocytes in linear arrays, extending into deep dermis with splaying among collagen bundles (magnification, 4×).
Figure 7.
Shared mutations found in nevus spilus, Spitz nevi and congenital nevus.
Acknowledgments
Our study received valuable assistance from the Tissue Core, Molecular Genomics and Cancer Informatics Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center, supported under NIH grant P30-CA76292.
References
- 1.Happle R. Speckled lentiginous naevus: which of the two disorders do you mean? Clinical and experimental dermatology. 2009;34(2):133–135. doi: 10.1111/j.1365-2230.2008.02966.x. [DOI] [PubMed] [Google Scholar]
- 2.Krengel S, Widmer DS, Kerl K, Levesque MP, Schiestl C, Weibel L. Naevus spilus-type congenital melanocytic naevus associated with a novel NRAS codon 61 mutation. The British journal of dermatology. 2016;174(3):642–644. doi: 10.1111/bjd.14105. [DOI] [PubMed] [Google Scholar]
- 3.Sarin KY, McNiff JM, Kwok S, Kim J, Khavari PA. Activating HRAS mutation in nevus spilus. The Journal of investigative dermatology. 2014;134(6):1766–1768. doi: 10.1038/jid.2014.6. [DOI] [PubMed] [Google Scholar]
- 4.Kinsler VA, Krengel S, Riviere JB, et al. Next-generation sequencing of nevus spilus-type congenital melanocytic nevus: exquisite genotype-phenotype correlation in mosaic RASopathies. The Journal of investigative dermatology. 2014;134(10):2658–2660. doi: 10.1038/jid.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafner C, Groesser L. Mosaic RASopathies. Cell Cycle. 2013;12(1):43–50. doi: 10.4161/cc.23108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SB, Hummel HM, Krengel S, Enk A, Haenssle HA. Mosaic RASopathies: phenotypic and genotypic differentiation of naevus spilus-type congenital melanocytic naevus and segmental naevus spilus. Journal of the European Academy of Dermatology and Venereology : JEADV. 2016;30(7):1244–1246. doi: 10.1111/jdv.13173. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiesner T, Kutzner H, Cerroni L, Mihm MC, Jr, Busam KJ, Murali R. Genomic aberrations in spitzoid melanocytic tumours and their implications for diagnosis, prognosis and therapy. Pathology. 2016;48(2):113–131. doi: 10.1016/j.pathol.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarin KY, Sun BK, Bangs CD, et al. Activating HRAS mutation in agminated Spitz nevi arising in a nevus spilus. JAMA dermatology. 2013;149(9):1077–1081. doi: 10.1001/jamadermatol.2013.4745. [DOI] [PubMed] [Google Scholar]
- 13.Gantner S, Wiesner T, Cerroni L, et al. Absence of BRAF and HRAS mutations in eruptive Spitz naevi. Br J Dermatol. 2011;164(4):873–877. doi: 10.1111/j.1365-2133.2011.10210.x. [DOI] [PubMed] [Google Scholar]
- 14.Kinsler VA, Thomas AC, Ishida M, et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol. 2013;133(9):2229–2236. doi: 10.1038/jid.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. The Journal of investigative dermatology. 2007;127(1):179–182. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 16.Streit S, Mestel DS, Schmidt M, Ullrich A, Berking C. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. British journal of cancer. 2006;94(12):1879–1886. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]