Abstract
Controversy has long surrounded the question of whether spontaneous lateral demixing of membranes into coexisting liquid phases can organize proteins and lipids on micron scales within unperturbed, living cells. A clear answer hinges on observation of hallmarks of a reversible phase transition. Here, by directly imaging micron-scale membrane domains of yeast vacuoles both in vivo and cell free, we demonstrate that the domains arise through a phase separation mechanism. The domains are large, have smooth boundaries, and can merge quickly, consistent with fluid phases. Moreover, the domains disappear above a distinct miscibility transition temperature (Tmix) and reappear below Tmix, over multiple heating and cooling cycles. Hence, large-scale membrane organization in living cells under physiologically relevant conditions can be controlled by tuning a single thermodynamic parameter.
Introduction
Scientists have invested decades of effort into probing the lipid and protein composition of cell membranes for evidence of heterogeneity, which has the potential to control protein sorting, signal transduction, and other processes (1). Aside from several important exceptions, especially in yeast (e.g., (2, 3, 4, 5, 6, 7, 8, 9, 10)), that extensive body of work has implied that the length scale of compositional heterogeneity in the membranes of unstimulated cells is limited to nanoscales, especially in plasma membranes of mammalian cells (reviewed in (11, 12, 13, 14)). Submicron domains in membranes gained notoriety as “rafts,” and, more recently, as dynamic, short-lived “platforms” (12, 15). These concepts are controversial because both terms are loosely or inconsistently defined, and because nanoscale domains are, at best, challenging to observe directly. In contrast to cell membranes, model lipid membranes spontaneously demix on large (μm) length scales into two well-defined liquid phases. This demixing follows thermodynamic principles (16, 17). The concept of phase separation is subject to established, quantitative rules that enable rigorous verification of predictions. These rules apply equally well to simple bilayer membranes composed of only three types of lipids, the complex bilayer membranes of giant plasma membrane vesicles blebbed from cells, and phase-separated cytoplasmic droplets recently implicated across a variety of cell biological activities (18, 19, 20).
Tantalizing hints have been reported since the 1960s that living membranes are capable of separating into coexisting liquid phases, just as model membranes are. Pioneering experiments investigated the vacuole, the lysosomal organelle of budding yeast. Using freeze fracture electron microscopy (EM), Moor and Mühlethaler (2) found that vacuole membranes of unfixed yeast contained domains depleted of large proteins. Domains in vacuole membranes are physiologically regulated: large proteins are randomly distributed across vacuolar membranes in the logarithmic phase of yeast growth, whereas protein-depleted domains appear when yeast are in the stationary phase (as nutrients are exhausted and the rate of cell division slows) (3, 4, 6, 7, 9). In many cases, the domains in vacuole membranes are ∼200 nm or larger, and are therefore resolvable using conventional optical microscopy (6).
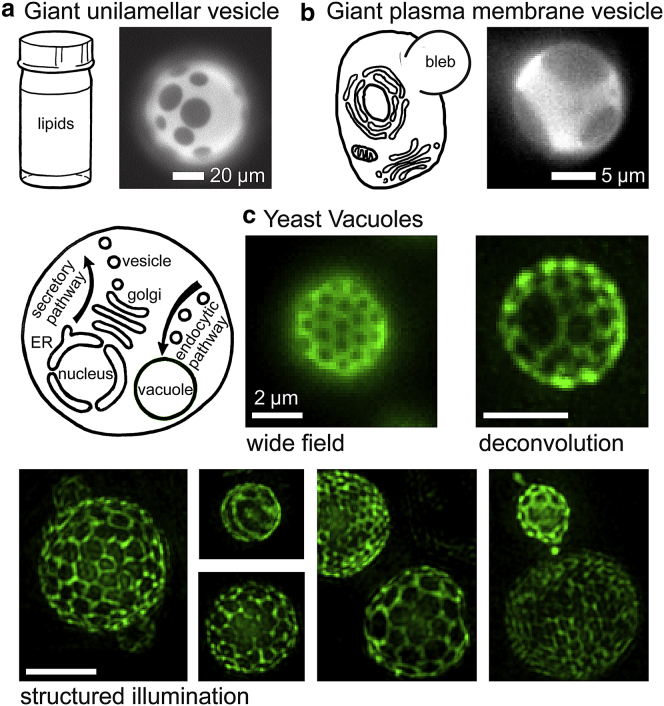
The visual similarity of domains that form in both synthetic and cell-derived model membranes (Fig. 1, a and b, respectively) to domains that form in yeast vacuole membranes in vivo (Figs. 1 c and S2) is striking. In Fig. 1 c, contrast between the two domain types is provided by green fluorescent protein (GFP)-tagged Vph1 (Vph1-GFP), an integral membrane subunit of the vacuolar proton ATPase. Elegant experiments revealed that Vph1 is one of many proteins and lipids that segregate to one of the two domain types in vacuole membranes of living cells (6). In recent work, the live cell results with Vph1 were unambiguously linked to the early freeze fracture EM observations by immunogold labeling of freeze fracture replicas (9). In other words, freeze fracture EM and optical microscopy of intact vacuoles in living cells observe exactly the same membrane structures.
Figure 1.
Micron-scale, coexisting liquid phases appear in membranes of synthetic and biologically derived model systems at equilibrium, and similar patterns appear on vacuole membranes of living yeast cells. (a) Giant unilamellar vesicles produced from ternary mixtures of synthetic lipids, imaged by wide-field epifluorescence microscopy. (b) Giant plasma membrane vesicles blebbed from adherent cells, imaged by standard epifluorescence. (c) Vacuoles within living yeast cells in the stationary phase of growth. Cells expressing a fluorescent vacuole membrane protein fusion (Vph1-GFP) were grown at 30°C and imaged at ambient temperature (∼22°C) using either standard wide-field epifluorescence illumination, wide-field illumination with z-sectioning followed by iterative deconvolution, or structured illumination microscopy (3D-SIM) followed by iterative deconvolution. Information on the growth and imaging procedures is in the Materials and Methods. A version of this figure without green pseudocolor appears in Fig. S1. Scale bars in (c), 2 μm. ER, endoplasmic reticulum. To see this figure in color, go online.
Two hallmarks of a membrane separating into liquid phases are 1) coalescence of domains on short timescales and 2) reversible mixing and demixing as a single thermodynamic parameter is varied. Previous attempts to observe these hallmarks in vacuole membranes have been inconclusive. Although multiple domains have been observed to merge into one larger domain, they did so on long timescales (6). Similarly, although the fractions of mixed and demixed vacuole membranes have been observed to vary with temperature changes from 30 to 40°C (by Moeller et al. in 1981), subsequent research using live yeast reported that “vacuolar structures did not visibly change when samples were visualized at a range of temperatures from 20 to 55°C” (by Toulmay and Prinz in 2013) (4, 6). Salient details of the early work are: the yeast were fixed before freeze fracture, the fraction of vacuoles exhibiting domains decreased from >75% to ∼50% when the temperature of stationary phase yeast was increased from 30 to 40°C, and that fraction recovered when temperature was returned to 30°C (4). The authors concluded that a solid (gel) phase transition had occurred (4).
Additional observations are consistent with, but do not prove, vacuole domains being due to membrane demixing into two coexisting liquid phases. 1) A variety of domain morphologies are evident in vacuole membranes, including pseudohexagonal arrays, stripes, and “half-moons” with only one large domain of each type (6, 7). 2) Under the studied growth conditions, ∼25% of vacuole membranes resembled half-moons (6). 3) On cell-free vacuoles, domains persisted after proteolytic digestion of proteins on the cytoplasmic faces of isolated vacuoles (6). 4) All labels used, which included 14 different endogenous protein markers and three lipid-sensitive probes, partitioned into one or the other of only two types of domains (6, 9). 5) Vacuoles typically contain 7–15 mol % sterol (21, 22), and the vacuole sterol content appears to increase during stationary phase (9). When ergosterol (the major sterol of yeast membranes) is depleted with drugs or genetic manipulations, domain formation is impaired (6, 7, 9). Synthetic membranes exhibit coexisting liquid phases only when one of their lipid components is a sterol such as ergosterol or cholesterol (23). 6) More broadly, several mutations that affect the lipid composition of vacuolar membranes result in the absence of membrane domains (6, 7, 9). Domain formation is linked to the availability of lipids and sterols, which are delivered to the vacuole via lipid esters stored in cytoplasmic lipid droplets, through a process called microlipophagy (7, 9, 24). Although all six of these findings are consistent with the hypothesis that vacuole domains arise through phase separation, none of them presents a direct test.
We now test key predictions of the phase-separation hypothesis. If the phases are liquids, then domains are predicted to merge and rearrange on short timescales. If a miscibility transition occurs, the membranes are predicted to reversibly demix at a constant transition temperature. We conclude that vacuole domains exhibit hallmark behaviors of phase separation.
Materials and Methods
Table S1 in the Supporting Material lists methods and conditions for each figure and movie.
Synthetic membranes
Giant unilamellar vesicles (GUVs) were electroformed (25) and imaged as previously described (18, 26). In Figs. 1, S1, S3, and S4 and Movie S8, the GUVs are composed of 35 mol % diphytanoyl-phosphatidylcholine, 35 mol % dipalmitoyl-phosphatidylcholine (Avanti Polar Lipids, Alabaster, AL), and 30 mol % cholesterol (Sigma, St. Louis, MO). The GUV in Fig. S4 is composed of 40 mol % di(13:0)-phosphatidylcholine, 20 mol % diphytanoyl-phosphatidylcholine, and 40 mol % cholesterol. All GUVs are labeled with 0.8 mol % Texas Red dipalmitoyl-phosphatidylethanolamine (Invitrogen, Carlsbad, CA).
Giant plasma membrane vesicles
The vesicle in Fig. 1 b was imaged at 10°C under control conditions in (27). Briefly, vesicles were prepared by incubating adherent RBL-2H3 cells in a buffer containing dithiothreitol and formaldehyde, and imaged on an inverted epifluorescence microscope (Olympus, Center Valley, PA) (27).
Yeast cell culture
A BY4742 derivative, MAT α his3Δ1 lys2Δ0 ura3Δ0 leu2Δ0 VPH1-GFP::HIS3MX6, was used. In general, when yeast are placed in fresh growth media, their growth follows a characteristic sequence of events. A “log phase” of rapid growth is followed by a “stationary phase” in which yeast vacuoles fuse to become as large as 5 μm in diameter (28, 29). Cultures (200 mL) were grown in synthetic complete media at 30°C in a shaking incubator for ∼20 h until the optical density of the culture was ∼1.7, using 600 nm wavelength light. The culture was then grown for an additional ∼43 h to reach the stationary phase, where the optical density using 600 nm wavelength light falls in the range of 6.8–7.8. This procedure is depicted in Fig. S5, B and C.
Vph1-GFP fusion protein
Yeast vacuoles were labeled by fusing the Vph1 protein to GFP, using homologous recombination. The fusion protein was expressed from the chromosomal VPH1 locus under the native promoter, at normal cellular copy number. The Vph1 fusions are known to retain physiological function and were previously shown to not spontaneously aggregate within the vacuole membrane, even during vacuole:vacuole docking (30). Vph1-GFP has been shown to colocalize in yeast vacuoles to the same membrane domains as the fluorescent tracer FM4-64, which partitions preferentially to the liquid-disordered phase in GUVs (6). Filipin, a dye that binds sterols, partitions preferentially to vacuole regions depleted in Vph1-GFP (6).
Yeast imaging
Yeast are typically 5–10 μm in diameter, vacuoles are 3–5 μm, and domains in vacuole membranes are often close in size to the Abbe diffraction limit (∼200 nm). Yeast were imaged by the four imaging techniques described in detail below. Image sequences collected by these techniques were then processed using a Kalman filter algorithm implemented in Image J (public domain http://rsbweb.nih.gov/ij/) to reduce detector and shot noise. For some experiments, yeast cells were adsorbed to cover slips coated with concanavalin A lectin (Elastin Products Company, Inc., Owensville, MO).
Imaging: wide-field illumination
Yeast cells were imaged with an electron-multiplying charge-coupled device camera on an Olympus IX71 fluorescence microscope as previously described, using a 60 × 1.4 NA oil immersion objective (31). To reduce noise, multiple exposures were averaged. In the wide-field micrograph in Fig. 1 c and all images of Fig. S2, eight consecutive 400 ms exposures were averaged. The micrographs in Fig. S6B were obtained by averaging four consecutive 200 ms exposures. To preserve the fidelity of images, no adjustments other than averaging and brightness levels were made (e.g., contrast was not altered).
Imaging: HILO illumination
To increase the signal-to-background ratio, yeast were imaged using highly inclined laminated optical sheet (HILO) illumination (32, 33) on a homebuilt Nikon Ti-U system with a 561-nm dipole-pumped solid-state laser (MPB Communications, Pointe-Claire, QC, Canada). A Nikon CFI Plan Apo Lambda 100 × 1.45 NA objective was used along with a dichroic quadband with 488/561/647/752 lines (Chroma) and an ET605/70m filter. Images were acquired on an electron-multiplying charge-coupled device (iXon Ultra 897, Andor) operating in frame transfer mode at 10 Hz, as described (34). To preserve the fidelity of images, no adjustments other than averaging and brightness levels were made (e.g., contrast was not altered).
Imaging: deconvolution microscopy
Iteratively deconvolved wide-field sequences of images were acquired on a DeltaVision system (GE Healthcare, Little Chalfont, UK) equipped with a CMOS camera and a 60 × 1.40 NA objective (Olympus). Cell suspensions were introduced to homemade flow chambers made from #1.5 cover slips passivated with concanavalin A. Unbound cells were washed out with depleted media taken from the supernatant of 1 mL of sample spun down at 1200 rpm for 2 min or (for hypoosmotic shock experiments) with water. Z-stacks were acquired at 200 nm spacing, usually at 0.15 s exposure per frame, and were deconvolved using SoftWorx software (GE Healthcare). From the deconvolved Z-stacks, brightest-point projections were computed. The projection time series datasets were then corrected for photobleaching using a histogram-matching algorithm implemented in ImageJ, and, finally, were Kalman filtered to reduce noise, also in ImageJ.
Imaging: structured illumination microscopy
Samples were mounted as for deconvolution and imaged using an OMX-SR instrument (GE Healthcare) equipped with a 63 × 1.42 NA objective (Olympus). The immersion oil refractive index was typically 1.516. Z-stacks were acquired at 120 nm spacing and images were deconvolved using the SoftWorx deconvolution package. Weiner spatial filter constants from 0.001 to 0.010 yielded similar reconstructions.
Imaging: standard epifluorescence
Both the synthetic GUV membranes and blebbed giant plasma membrane vesicle membrane imaged in Figs. 1, a and b, S3, and S4, and in Movie S8 were imaged as previously described (18, 35). To preserve the fidelity of images, no adjustments other than averaging and brightness levels were made (e.g., contrast was not altered).
Thermal cycling
For both in vivo and cell-free yeast vacuole samples, temperature was controlled by air from a heat gun and monitored using a calibrated thermocouple. The thermocouple tip was inserted between the cover slip and slide to make direct contact with the sample. The edges of the cover slip and slide were sealed with vacuum grease to prevent water loss due to evaporation. In experiments in which temperature data was collected, a graph is provided within the figures and, for the in vivo vacuoles, within the movies. The synthetic membrane GUV shown in Fig. S3 and Movie S8 was temperature controlled as previously described, and the temperature was cycled around its miscibility transition temperature of 46.1°C.
Cell lysis to create cell-free vacuoles
Cells were harvested in a swinging-bucket rotor (3200 × g) for 10 min at room temperature, resuspended in 0.1 M Tris (pH 9.4) and 10 mM dithiothreitol, and incubated for 10 min at 30°C. The cells were again sedimented in a swinging-bucket rotor (3200 × g) for 5 min at room temperature and then resuspended in spheroplast buffer (600 mM sorbitol, 50 mM potassium phosphate pH 7.5, and 8% v/v depleted media, saved from the first centrifugation step). Lytic enzyme (Zymolyase 20T, Seikigaku; further purified by ion exchange chromatography) was added to threefold higher concentration than in our standard vacuole prep (36) to adjust for cell wall composition in yeast grown into the stationary phase, and the cells were incubated for 1 h at 30°C. The spheroplasted cells were sedimented in a swinging-bucket rotor (3200 × g) for 5 min at 4°C. For hypoosmotic lysis, spheroplasts were resuspended in 15% ficoll buffer (10 mM Pipes-KOH pH 6.8, 200 mM sorbitol, and 15% w/v ficoll) and diethylaminoethyl-dextran was added to a final concentration of 0.005–0.01% w/v. Spheroplasts were incubated for 2 min on ice, then 3 min at 30°C. The resulting spheroplast lysates were stored on a wet ice bath for no more than 4 h before use.
Mounting cell-free vacuoles on slides
A thin agarose cushion was prepared by spin coating 55 μL of molten 0.8% (w/v) agarose in Pipes sorbitol buffer (10 mM Pipes-KOH pH 6.8 and 200 mM sorbitol) on plasma-cleaned glass cover slips. Vacuole lysates were diluted 1:10 with molten 0.8% w/v low-melt agarose in Pipes sorbitol buffer. The solution was mixed by gentle vortexing, deposited onto agarose-coated slides, and imaged by the same procedure as for living cells. Immobilization of vesicles within agarose gels does not affect the diffusion coefficient of individual lipids in vesicle membranes (37).
Osmotic gradient
A simple flow cell was constructed from two cover slips joined along their edges by spacers of double-sided tape. The bottom cover slip was coated in concanavalin A. Yeast cells in their depleted media were drawn into the flow cell by capillary action and absorbed to the concanavalin A-coated cover slip. To induce hypoosmotic swelling of the vacuole, a volume of deionized water, equal to the volume of sample loaded into the flow cell, was deposited at one end of the microfluidic chamber. Fluid was then wicked from the opposite end of the flow cell to introduce water into the flow cell.
Results and Discussion
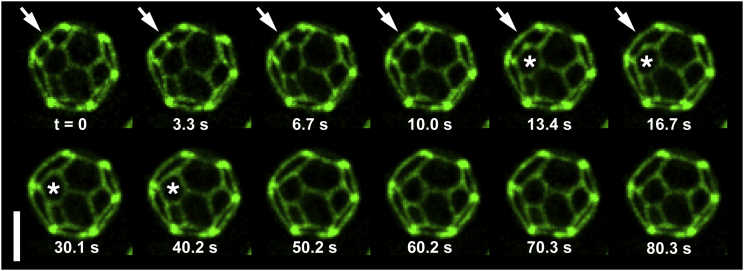
The first hallmark of liquid phases that we observe in vacuole membranes is that two domains can coalesce in timescales of seconds. In synthetic GUVs with taut membranes, liquid domains diffuse freely over the vesicle surface, collide with other domains, and coalesce as in Fig. S4 until eventually only one domain of each type remains (38). In synthetic GUVs with excess area (more area than necessary to enclose a spherical volume), domains in pseudohexagonal arrays are observed to bulge in to or out of the vesicles and are hindered from colliding (17, 39, 40, 41). Similarly, the pseudohexagonal domains of yeast vacuoles bulge inward toward the vacuole lumen (6, 9, 24) and appear to be hindered from colliding. When bulging domains are not labeled, the labeled background membrane has a faceted appearance, as in Fig. 2.
Figure 2.
Rapid coalescence of in vivo micron-scale domains in a single yeast vacuole membrane over time. Yeast cells were grown as in Fig. 1, mounted in a flow chamber, subjected to a hypoosmotic gradient, and imaged by z-sectioning and iterative deconvolution at ambient temperature (∼23°C). Maximum brightness projections of the vacuole hemisphere closest to the microscope objective are presented. Arrows denote a region where two dark domains coalesce. Stars (∗) denote a domain that changes shape from a hexagon to a pentagon, minimizing the total length of the domain interface. The scale bar represents 2 μm. Movie S1 corresponds to the above sequence, played at 10× speed. The dynamic shift in the shape of the starred domain from a hexagon to a pentagon, from seconds 13.4–40.2 above, appears in seconds 1–4 of Movie S1. To see this figure in color, go online.
We hypothesized that domain collision in vacuoles might be triggered by applying a hypoosmotic gradient to yeast cells, which causes the vacuoles inside to swell due to elevated internal turgor pressure. This was indeed the case: we capture events of domains colliding and quickly coalescing in Fig. 2 and Movie S1. We observe the reverse spontaneous process in Fig. S7 and Movie S3 (played at 30× speed) by applying a hyperosmotic gradient. During coalescence, the domain boundaries rearrange to minimize the total length of domain interfaces. This is consistent with minimization of energy arising from line tension between the two membrane phases.
Once the kinetic barrier for two domains to collide is overcome, the fastest coalescence of domains that we observe in yeast vacuoles occurs on the same timescale (seconds) as coalescence of domains that can be an order of magnitude larger in synthetic vesicles (38). These observations are consistent with the expectation that fluids in living yeast cells, namely the cytoplasm and the vacuole contents (including high concentrations of high molecular mass polyphosphate), have higher viscosities than the viscosities of fluids used in synthetic GUV preparations (42, 43, 44).
Domain coalescence is technically challenging to image, so is rarely observed. Coalescence can be identified only when domains are large enough to image without superresolution techniques, yet small enough to merge. Observation of domain coalescence in Fig. 2 required deconvolution of image stacks and reconstruction of only one hemisphere of the vacuole, the full three-dimensional image obscured the merging event. These technical challenges likely lead to overrepresentation of slow events within Fig. S6 and Movie S2 (played at 30× speed), and the previous literature (6). We speculate that slow coalescence events result because a subset of vacuole domains are associated with lipid droplets or with nuclear contact sites (7, 9, 24). Therefore, only the fastest coalescence events in vacuoles (as in Fig. 2 of the main text) are relevant for comparison with timescales in model systems.
The second hallmark of liquid phases is the existence of a miscibility transition with respect to an intrinsic thermodynamic variable such as temperature, pressure, or membrane composition, as in the schematic in Fig. S8. Cyclic changes in any one of these thermodynamic variables cause domains to reversibly appear and disappear in synthetic vesicles (16, 45). To test whether the domains in yeast vacuole membranes arise from demixing of a single liquid phase into two coexisting liquid phases, we subjected yeast to rapid temperature cycles.
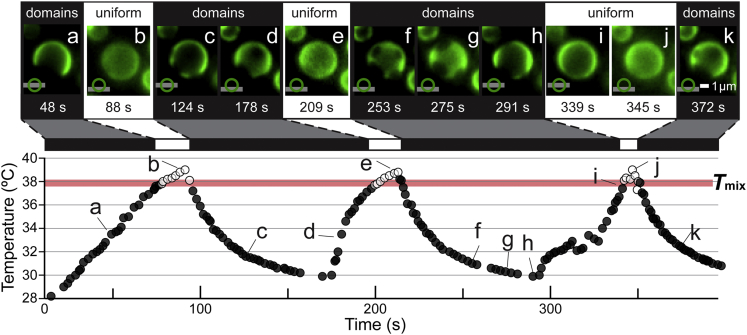
In Figs. 3 and S9 and Movies S4 and S5 (both played at 3× speed), we cycle the temperature of yeast cells in the stationary phase of cell growth. At the standard growth temperature of 30°C, yeast vacuoles exhibit dark domains on a bright background marked by GFP-Vph1 (Fig. 1 c). At temperatures above ∼37°C, the domains disappear and the membrane is uniformly labeled.
Figure 3.
Micron-scale domains in an in vivo yeast vacuole reversibly vanish and reemerge through three temperature cycles. Micrographs (a)–(k), representing 48 and 372 s, respectively, correspond to the labeled locations in the plot of temperature versus time. In the lower left-hand corner of each micrograph, a symbol illustrating the focal plane (horizontal line) at either the top, equator, or bottom of the vacuole (circle) specifies the focal plane at which the vacuole was imaged in each micrograph. The temperature at which open symbols on the graph change to filled symbols (and vice versa) is Tmix. A thick, horizontal line, labeled Tmix, is drawn to highlight the transitions; the line is not a statistical fit. Vacuoles were imaged using HILO illumination. Movie S4 corresponding to this figure appears in the Supporting Material and plays at 3× speed such that micrograph (a) appears at 16 s in Movie S4 and micrograph (k) appears at 2 min and 4 s. To see this figure in color, go online.
EM results show that large proteins within a uniformly-labeled membrane are randomly distributed across the entire membrane (9). In our experiments, domains nucleate upon cooling and are large enough to image within seconds. By comparison, transcription and protein synthesis in yeast occur on timescales of at least minutes. The ability of the vacuole membrane to abruptly and reversibly switch between two states (namely, the presence and absence of domains), at a distinct temperature, and over multiple heating and cooling cycles, is a defining feature of a phase transition.
Cell viability is not affected by the temperature cycling regime used in our experiments (Fig. S5 A; Tables S2 and S3). Moreover, individual yeast cells that exhibit domains in their vacuole membranes successfully grow and undergo mitosis when supplied with fresh nutrients (6). Within a population of yeast cells, the Tmix of the vacuole membrane varies from cell-to-cell, just as it does in plasma membrane vesicles (46), as might be expected for any biological parameter regulated by an array of biochemical and physiological variables.
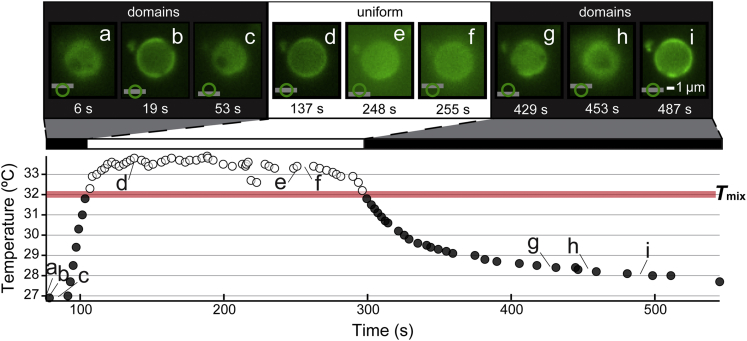
To verify that the cyclical disappearance and reappearance of vacuole domains is intrinsic to the membrane rather than originating from factors in the yeast cytoplasm, we extended the results of Fig. 3 to cell-free vacuoles. Figs. 4 and S10 and Movie S6 (played at 3× speed) show that domains in cell-free vacuoles reversibly vanish and reappear as the temperature is cycled. The location at which domains reappear depends on the speed of cycling. When temperature is quickly cycled (as occurred in Figs. 3, S10, top, and S11, rapid and Movie S7, which plays at 3× speed), domains disappear and renucleate at approximately the same positions on the vacuole. The same effect occurs when synthetic GUVs undergo rapid temperature cycling (as in Fig. S3 and Movie S8, which plays at 10× speed). This is because as the temperature increases, domains disappear through a process in which the edge blurs, as in Movie S5 (played at 3× speed); domains do not disappear by becoming continually smaller. Edge blurring is expected when labeled molecules are suddenly free to diffuse over the entire surface of the membrane. When the temperature is held above Tmix for longer periods, such that lipids and proteins have ample time to diffuse across the membrane and mix uniformly, domains nucleate at new positions, both on vacuoles (Figs. 4, S10, bottom, and S11, slow and Movie S9, which plays at 3× speed) and on synthetic vesicles (Fig. S3 and Movie S8, which plays at 10× speed). In the language of condensed matter physics, this behavior is consistent with domains arising through a mechanism of nucleation and growth rather than with fluctuations arising through a mechanism of spinodal decomposition of a membrane poised near a miscibility critical point (18, 35).
Figure 4.
Micron-scale domains in a cell-free yeast vacuole reversibly vanish and reappear through temperature cycles. This figure is one of three cycles in Fig. S10. Micrographs (a)–(i) correspond to the labeled locations in the plot of temperature versus time. To counteract effects of photobleaching of Vph1-GFP through time, brightness levels were increased in micrographs (e) and (f), and excitation intensity was increased in micrographs (g), (h), and (i). Other details are as described for Fig. 3. Movie S6 corresponding to this figure appears in the Supporting Material and plays at 3× speed; such that micrograph (a) appears at 2.1 s in Movie S6 and micrograph (i) appears at 2 min and 42.5 s. Preparation of cell-free vacuoles is described in the Materials and Methods. To see this figure in color, go online.
The location of domains in vacuole membranes does not appear to be governed by static protein scaffolds. By mass, about half of the vacuole membrane is protein, comparable to ∼70% for a plasma membrane (21). Although vacuole membranes may be associated with protein scaffolds, we observe that domains have variable sizes, move across the surface of vacuoles, smoothly and quickly coalescence into larger domains, and renucleate in new locations. In addition, domains persist after digestion of proteins on the cytoplasmic face of isolated vacuoles (6). All of these properties are inconsistent with domain edges constrained by static scaffolds.
The coalescence of small domains into larger ones followed by reorganization of domain edges within yeast vacuole membranes (as in the starred domain in Fig. 2) implies that the domains result from a miscibility phase transition rather than from the extensive cross-linking used to induce micron-scale domains in stimulated cell membranes (12), from cell polarization (47, 48, 49), from vacuolar fragmentation or deep invaginations due to hyperosmotic stress (50), or from the regulated assembly of contacts between docked yeast vacuoles (30, 51). Neither the presence nor the absence of domains is perturbed by ATP depletion (6). We can also rule out contact between lipid droplets and vacuoles as a direct driver of vacuole domain formation because only a subset of the domains in vacuole membranes correlates with the presence of docked lipid droplets (7, 9, 24). The presence of many more domains than droplets requires an additional mechanism, such as phase separation. The localized release of sterols into the vacuole membrane through microlipophagy of docked lipid droplets, via the Npc1/2 system (7, 9, 24), appears to be an indirect driver of phase separation across the entire membrane. Finally, we note that the assembly of docking domains before bilayer-bilayer fusion is consistent with, and may be driven by, membrane phase behavior (30, 51, 52).
Conclusion
Here, we show that large-scale membrane organization in yeast vacuoles is the result of demixing of the membrane into coexisting liquid phases and that this demixing is fully reversible. This mechanism operates in live cells imaged using noninvasive methods. In model vesicle membranes, demixing occurs equally upon a change in temperature or membrane composition. At constant temperature, cells may regulate membrane phase separation in response to external or internal cues. For example, the NPC1/2 cholesterol transport system controls domain formation (9). Moreover, the Slt2/Mpk1 kinase is essential for the formation of phase-separated domains in the yeast vacuole (6). Previously, we demonstrated that Slt2/Mpk1, a key node in the protein kinase C-Rho1 signaling pathway, regulates both lipid acyl chain composition and bilayer fluidity (53). The knowledge that vacuole membranes reversibly demix into coexisting liquid phases enables direct application of physical rules of membrane phase behavior established in model systems to living biological membranes. The description of domains as arising from a mechanism of phase separation rather than as less well-defined “raft-like” (9) domains provides a tractable paradigm for future investigations of the regulation and mechanisms of in vivo membrane domain partitioning.
Author Contributions
S.L.K. and A.J.M. conceived the project and supervised the experiments. S.P.R., G.E.R., A.J.M., and C.E.C. performed experiments and collected data. S.L.K., A.J.M., S.P.R., and G.E.R. designed research, wrote the article, and designed figures.
Acknowledgments
We thank Liz Manrao, Dan Nickerson, Rachael Plemel, Tyler Chozinski, Marco Howard, Lauren Gagnon, and Aaron Halpern for technical advice. Micrographs provided by Ellyn Gray and Sarah Veatch (Fig. 1 B), and Joan Bleecker (Fig. S4) are used with permission. We are grateful to Professors Linda Wordeman and Joshua Vaughan for providing microscope time.
This material is based upon work supported by the National Science Foundation (NSF) Graduate Research Fellowship Program under grant DGE-1256082 to G.E.R., by NSF grant MCB-1402059 to S.L.K., by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award T32GM008268 to C.E.C., and by NIH grant GM077349 to A.J.M.
Editor: David Cafiso.
Footnotes
Scott P. Rayermann and Glennis E. Rayermann contributed equally to this work.
Eleven figures, three tables, and nine movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31072-X.
Contributor Information
Alexey J. Merz, Email: merza@uw.edu.
Sarah L. Keller, Email: slkeller@chem.washington.edu.
Supporting Material
In vivo vacuole domains merging and their subsequent rearrangement, as shown in Fig. 2. This movie plays at ∼10× speed.
In vivo vacuole domains merging and their subsequent rearrangement, as shown in Fig. S4 A. This movie plays at ∼30× speed.
In vivo vacuole domains becoming smaller, as shown in Fig. S5. The left half corresponds to the top hemisphere of the vacuole and the right half corresponds to the bottom hemisphere. This movie plays at ∼30× speed.
In vivo vacuole domains vanishing and reappearing with the temperature cycle shown in Fig. 3 as an inset in a plot of temperature through time. The movie plays at ∼3× speed.
A video of in vivo vacuole domains vanishing and reappearing with the temperature cycle shown in Fig. S7 as an inset in a plot of temperature through time. The movie plays at ∼3× speed.
A video of cell-free vacuole domains vanishing and reappearing with the temperature cycle shown in Figs. 4 and S9-bottom. The movie plays at ∼3× speed.
A video of in vivo vacuole domains vanishing and reappearing with a rapid heat cycle as shown in Fig. S10-rapid. The movie plays at ∼3× speed.
A video of GUV domains vanishing and reappearing with fast versus slow temperature ramps as shown in Fig. S11. The movie plays at ∼10× speed.
A video of in vivo vacuole domains vanishing and reappearing with a slow temperature cycle as shown in Fig. S10-slow. The movie plays at ∼3× speed.
References
- 1.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 2.Moor H., Mühlethaler K. Fine structure in frozen-etched yeast cells. J. Cell Biol. 1963;17:609–628. doi: 10.1083/jcb.17.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moeller C.H., Thomson W.W. An ultrastructural study of the yeast tomoplast during the shift from exponential to stationary phase. J. Ultrastruct. Res. 1979;68:28–37. doi: 10.1016/s0022-5320(79)90139-4. [DOI] [PubMed] [Google Scholar]
- 4.Moeller C.H., Mudd J.B., Thomson W.W. Lipid phase separations and intramembranous particle movements in the yeast tonoplast. Biochim. Biophys. Acta. 1981;643:376–386. doi: 10.1016/0005-2736(81)90082-1. [DOI] [PubMed] [Google Scholar]
- 5.Spira F., Mueller N.S., Wedlich-Söldner R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012;14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 6.Toulmay A., Prinz W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013;202:35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C.-W., Miao Y.-H., Chang Y.-S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 2014;206:357–366. doi: 10.1083/jcb.201404115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murley A., Sarsam R.D., Nunnari J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 2015;209:539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuji T., Fujimoto M., Fujimoto T. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife. 2017;6:e25960. doi: 10.7554/eLife.25960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murley A., Yamada J., Nunnari J. Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 2017;216:2679–2689. doi: 10.1083/jcb.201610032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons K., Gerl M.J. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 12.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 13.Levental I., Veatch S. The continuing mystery of lipid rafts. J. Mol. Biol. 2016;428(24 Pt A):4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft M.L. Sphingolipid organization in the plasma membrane and the mechanisms that influence it. Front. Cell Dev. Biol. 2017;4:154. doi: 10.3389/fcell.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 16.Veatch S.L., Keller S.L. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 17.Baumgart T., Hess S.T., Webb W.W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 18.Honerkamp-Smith A.R., Cicuta P., Keller S.L. Line tensions, correlation lengths, and critical exponents in lipid membranes near critical points. Biophys. J. 2008;95:236–246. doi: 10.1529/biophysj.107.128421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brangwynne C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013;203:875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 21.Schneiter R., Brügger B., Kohlwein S.D. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinser E., Sperka-Gottlieb C.D., Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beattie M.E., Veatch S.L., Keller S.L. Sterol structure determines miscibility versus melting transitions in lipid vesicles. Biophys. J. 2005;89:1760–1768. doi: 10.1529/biophysj.104.049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moeller C.H., Thomson W.W. Uptake of lipid bodies by the yeast vacuole involving areas of the tonoplast depleted of intramembranous particles. J. Ultrastruct. Res. 1979;68:38–45. doi: 10.1016/s0022-5320(79)90140-0. [DOI] [PubMed] [Google Scholar]
- 25.Angelova M.I., Soléau S., Bothorel P. Preparation of giant vesicles by external AC electric fields. Prog. Colloid Polym. Sci. 1992;89:127–131. [Google Scholar]
- 26.Blosser M.C., Honerkamp-Smith A.R., Keller S.L. Transbilayer colocalization of lipid domains explained via measurement of strong coupling parameters. Biophys. J. 2015;109:2317–2327. doi: 10.1016/j.bpj.2015.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray E., Karslake J., Veatch S.L. Liquid general anesthetics lower critical temperatures in plasma membrane vesicles. Biophys. J. 2013;105:2751–2759. doi: 10.1016/j.bpj.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S.C., Kane P.M. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta. 2009;1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong J. Yeast vacuoles: more than a model lysosome. Trends Cell Biol. 2010;20:580–585. doi: 10.1016/j.tcb.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Wang L., Seeley E.S., Merz A.J. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 31.Angers C.G., Merz A.J. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol. Biol. Cell. 2009;20:4563–4574. doi: 10.1091/mbc.E09-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konopka C.A., Bednarek S.Y. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 34.Chozinski T.J., Halpern A.R., Vaughan J.C. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veatch S.L., Cicuta P., Baird B. Critical fluctuations in plasma membrane vesicles. ACS Chem. Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz M.L., Merz A.J. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J. Cell Biol. 2009;185:535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lira R.B., Steinkühler J., Riske K.A. Posing for a picture: vesicle immobilization in agarose gel. Sci. Rep. 2016;6:25254. doi: 10.1038/srep25254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanich C.A., Honerkamp-Smith A.R., Keller S.L. Coarsening dynamics of domains in lipid membranes. Biophys. J. 2013;105:444–454. doi: 10.1016/j.bpj.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacia K., Schwille P., Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. USA. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamada T., Miura Y., Takagi M. Dynamic processes in endocytic transformation of a raft-exhibiting giant liposome. J. Phys. Chem. B. 2007;111:10853–10857. doi: 10.1021/jp075412+. [DOI] [PubMed] [Google Scholar]
- 41.Ursell T.S., Klug W.S., Phillips R. Morphology and interaction between lipid domains. Proc. Natl. Acad. Sci. USA. 2009;106:13301–13306. doi: 10.1073/pnas.0903825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wintersmith J.R., Zou L., Mann E.K. Determination of interphase line tension in Langmuir films. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;75:061605. doi: 10.1103/PhysRevE.75.061605. [DOI] [PubMed] [Google Scholar]
- 43.Williams S.-P., Haggie P.M., Brindle K.M. 19F NMR measurements of the rotational mobility of proteins in vivo. Biophys. J. 1997;72:490–498. doi: 10.1016/S0006-3495(97)78690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujino T., Hirota K., Tahara T. In-cell viscosity measurement using a fluorescence up-conversion microscope. Chem. Lett. 2008;37:1240–1241. [Google Scholar]
- 45.Nicolini C., Kraineva J., Winter R. Temperature and pressure effects on structural and conformational properties of POPC/SM/cholesterol model raft mixtures--a FT-IR, SAXS, DSC, PPC and Laurdan fluorescence spectroscopy study. Biochim. Biophys. Acta. 2006;1758:248–258. doi: 10.1016/j.bbamem.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Gray E.M., Díaz-Vázquez G., Veatch S.L. Growth conditions and cell cycle phase modulate phase transition temperatures in RBL-2H3 derived plasma membrane vesicles. PLoS One. 2015;10:e0137741. doi: 10.1371/journal.pone.0137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagnat M., Simons K. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wedlich-Soldner R., Altschuler S., Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 49.Makushok T., Alves P., Brunner D. Sterol-rich membrane domains define fission yeast cell polarity. Cell. 2016;165:1182–1196. doi: 10.1016/j.cell.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 50.Takatori S., Tatematsu T., Fujimoto T. Phosphatidylinositol 3,5-bisphosphate-rich membrane domains in endosomes and lysosomes. Traffic. 2016;17:154–167. doi: 10.1111/tra.12346. [DOI] [PubMed] [Google Scholar]
- 51.Fratti R.A., Jun Y., Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNally E.K., Karim M.A., Brett C.L. Selective lysosomal transporter degradation by organelle membrane fusion. Dev. Cell. 2017;40:151–167. doi: 10.1016/j.devcel.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 53.Lockshon D., Olsen C.P., Kennedy B.K. Rho signaling participates in membrane fluidity homeostasis. PLoS One. 2012;7:e45049. doi: 10.1371/journal.pone.0045049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo vacuole domains merging and their subsequent rearrangement, as shown in Fig. 2. This movie plays at ∼10× speed.
In vivo vacuole domains merging and their subsequent rearrangement, as shown in Fig. S4 A. This movie plays at ∼30× speed.
In vivo vacuole domains becoming smaller, as shown in Fig. S5. The left half corresponds to the top hemisphere of the vacuole and the right half corresponds to the bottom hemisphere. This movie plays at ∼30× speed.
In vivo vacuole domains vanishing and reappearing with the temperature cycle shown in Fig. 3 as an inset in a plot of temperature through time. The movie plays at ∼3× speed.
A video of in vivo vacuole domains vanishing and reappearing with the temperature cycle shown in Fig. S7 as an inset in a plot of temperature through time. The movie plays at ∼3× speed.
A video of cell-free vacuole domains vanishing and reappearing with the temperature cycle shown in Figs. 4 and S9-bottom. The movie plays at ∼3× speed.
A video of in vivo vacuole domains vanishing and reappearing with a rapid heat cycle as shown in Fig. S10-rapid. The movie plays at ∼3× speed.
A video of GUV domains vanishing and reappearing with fast versus slow temperature ramps as shown in Fig. S11. The movie plays at ∼10× speed.
A video of in vivo vacuole domains vanishing and reappearing with a slow temperature cycle as shown in Fig. S10-slow. The movie plays at ∼3× speed.