Abstract
Desmosomes are macromolecular cell-cell junctions that provide adhesive strength in epithelial tissue. Desmosome function is inseparably linked to structure, and it is hypothesized that the arrangement, or order, of desmosomal cadherins in the intercellular space is critical for adhesive strength. However, due to desmosome size, molecular complexity, and dynamics, the role that order plays in adhesion is challenging to study. Herein, we present an excitation resolved fluorescence polarization microscopy approach to measure the spatiotemporal dynamics of order and disorder of the desmosomal cadherin desmoglein 3 (Dsg3) in living cells. Simulations were used to establish order factor as a robust metric for quantifying the spatiotemporal dynamics of order and disorder. Order factor measurements in keratinocytes showed the Dsg3 extracellular domain is ordered at the individual desmosome, single cell, and cell population levels compared to a series of disordered controls. Desmosomal adhesion is Ca2+ dependent, and reduction of extracellular Ca2+ leads to a loss of adhesion measured by dispase fragmentation assay (λ = 15.1 min). Live cell imaging revealed Dsg3 order decreased more rapidly (λ = 5.5 min), indicating that cadherin order is not required for adhesion. Our results suggest that rapid disordering of cadherins can communicate a change in extracellular Ca2+ concentration to the cell, leading to a downstream loss of adhesion. Fluorescence polarization is an effective bridge between protein structure and complex dynamics and the approach presented here is broadly applicable to studying order in macromolecular structures.
Introduction
Many critical cellular functions including nuclear transport, ATP synthesis, and cell adhesion are orchestrated by large, multiprotein assemblies (1, 2, 3). These macromolecular complexes are defined by their protein composition, architecture, and cellular localization. Importantly, how proteins are arranged within each complex confers function. This protein arrangement can be described in two ways: spatial organization, which defines the location of proteins within a complex; and orientational organization, which defines the order or particular arrangement of proteins relative to one another. Generation of order and disorder is a mechanism central to the assembly, disassembly, and function of many macromolecular complexes (4, 5, 6).
Cell junctions are macromolecular complexes that have roles in tissue integrity, communication, and barrier function. Desmosomes are epithelial cell-cell junctions that play a critical role in the skin and heart, where they are essential for maintaining tissue integrity (7). Disruption of desmosomal function results in a number of human diseases, including skin blistering diseases, cancers, and cardiomyopathies (8, 9). The dynamics of desmosome assembly, disassembly, and the transition between calcium-dependent and independent adhesion play a central role in many processes including keratinocyte migration during wound healing, embryogenesis, disease pathogenesis, and tissue remodeling (7, 10, 11, 12, 13).
Structurally characterizing desmosomes presents a challenge, due to their biochemical intractability, complex structure spanning two cells, and functional dynamics. The organization of proteins in the plaque has been determined by electron microscopy (EM) and superresolution stochastic optical reconstruction microscopy (14, 15). Through these methods, differences in protein spatial organization have been correlated with changes in adhesion. However, the dynamics of desmosome structure, including protein ordering, as it relates to function are still poorly understood.
The desmosomal adhesive junction is symmetrical across the intermembrane space, with each cell contributing half the complex. Desmosomal cadherins (desmogleins and desmocollins) are single-pass transmembrane glycoproteins with extracellular domains that interact to form the adhesive interface (16). This adhesion involves trans-binding through a strand swap, where a conserved tryptophan is inserted into a hydrophobic pocket of the opposing cadherin (17, 18). Desmosomal cadherins are characterized by their calcium-dependent structure, with calcium binding sites between cadherin repeat domains. The intracellular plaque proteins create a dense network linking the cadherin cytoplasmic tails to the intermediate filament cytoskeleton (19). Each desmosome contains many copies of each of these proteins, resulting in an overall structure ∼0.5 μm in diameter (20) with the plaque extending between 100 and 115 nm (14, 15) into the cytoplasm.
Because cadherins are the proteins responsible for spanning neighboring cells, understanding their structure and organization is central to our understanding of adhesion. The extracellular domain of the desmosomal cadherins has largely been hypothesized to be ordered, based on EM (10, 21) and in vitro characterization of individual cadherin structure (22). Evidence for order includes the dense midline seen in EM and the periodicity of electron densities by tomography (10, 21, 23, 24, 25). Taken together, this suggests that desmosomes are ordered and that the cadherins are potentially arranged in a crystalline-like manner. However, not all studies reach a similar conclusion and conflicting results describe the cadherins as a disordered “tangled knot” (26).
One drawback to the methods used to study desmosome ultrastructure is that they require the use of fixed and processed cells or tissue, and therefore cannot capture structural dynamics. To address this and characterize order of the adhesive interface in living cells, we turned to fluorescence polarization. Fluorescence polarization has been used to study single molecule dynamics (22, 27), diffusion (28), binding events (29, 30) and, when combined with microscopy, organization of biological structures (31, 32, 33). Fluorescence polarization is a powerful tool to study the order and disorder of proteins in macromolecular complexes including yeast septins (34), nuclear pore complex (4, 35), MHC complex (36, 37), and the cytoskeleton (5, 38, 39).
Herein, we present an excitation resolved fluorescence polarization microscopy approach to measure order of the desmosomal cadherin desmoglein 3 (Dsg3) in living cells. First, we introduce the approach we developed to address the question of order versus disorder in desmosomes and establish the upper and lower bounds on detecting order with simulations. We then show that the extracellular domain of Dsg3 is ordered at the single desmosome, single cell, and population level, in contrast to a series of disordered controls. Finally, we reveal the dynamics of Dsg3 order loss relative to loss of adhesion, demonstrating a direct relationship between order and function. Our results confirm the relevance of cadherin ordering to desmosome structure and adhesive strength.
Materials and Methods
Cell culture
The human immortalized keratinocyte cell line HaCaT (Addex Bio, San Diego, CA) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM L-glutamine, 10% FBS, 100 U/mL Pen/Strep, and 2.5 μg/mL Amphotericin B and maintained at 37°C and 5% CO2. For imaging, cells were seeded into eight-well No. 1.5 coverslip bottom dishes (Ibidi, Madison, WI), transfected, and imaged in supplemented FluoroBrite DMEM (Thermo Fisher Scientific, Waltham, MA). For fragmentation assays, cells were grown to confluence in 12-well tissue culture plates.
Cloning and constructs
Dsg3-ΔEA-GFP was cloned using PCR and mutagenesis to insert the EGFP sequence between residues K499 and L615 of mouse Dsg3 (UniProt: O35902). Dsg3-link-GFP was cloned with the same methods, with the flexible linker ID(GGGGS)×5TG ligated between the C-terminus of mouse Dsg3 and N-terminus of EGFP. Desmoplakin-GFP was a gift from Kathleen Green (Addgene plasmid No. 32227). The GFP was replaced with mCherry using PCR, to create DP-mCherry. Mem-GFP is EGFP with the N-terminal palmitoylation motif, LCC. All cloning was performed by the Emory Cloning Core (Emory University) and verified by sequencing.
Dsg3-ΔEA-GFP ribbon diagram
Diagraming of Dsg3-ΔEA-GFP was performed in the software “UCSF Chimera” using Dsg3 (EC1–4) (PDB: 5EQX) and GFP (PDB: 1EMB). The 18-amino-acid linker (VPDFNENCPSVVLEKMVS) between the end of PDB: 5EQX (EC4 of Dsg3) and the start of GFP is composed of the EC4-EC5 Dsg3 linker region and the start of PDB: 1EMB. The linker secondary structure was predicted using PredictProtein, the PROFphd method (https://www.predictprotein.org/) (40). The C-terminal of PDB: 1EMB is linked to the amino acids TGMDELYKAAIGLILLGLLMLLLAPLLLL from Dsg3, where TGMDELYK is part of the EC5 structure predicted to be α-helical by the same method used above. AAIGLILLGLLMLLLAPLLLL is the constitutive transmembrane domain of Dsg3, which PredictProtein correctly identified with the PHDhtm method.
The orientation of the chimeric protein was determined by embedding the transmembrane domain in a lipid bilayer and orienting EC1 such that it could engage in trans-binding. GFP was constrained by Dsg3, the plasma membrane, and the linker locations.
Transfection
HaCaT cells were transfected at 50% confluent using Lipofectamine 3000 (Thermo Fisher Scientific) following manufacturer’s instructions. DNA concentration was optimized to 70 ng per 1 cm2. Cells were imaged 40–48 h post-transfection.
Calcium switch
For imaging under low Ca2+ conditions, cells were switched from supplemented FluoroBrite DMEM (Thermo Fisher Scientific) with ∼3 mM Ca2+ to ∼0.03 mM Ca2+ as described by Wilson (41) while on the microscope stage.
Fluorescence polarization microscopy
Fluorescence polarization microscopy was conducted using a Ti Eclipse microscope (Nikon, Tokyo, Japan), equipped with a motorized stage, stage-top incubator to maintain 37°C and 5% CO2 (INUBG2SF-TIZB; Tokai Hit USA, Bala Cynwyd, PA) and a 60 × 1.49 NA objective. The 488-nm laser (Coherent, Santa Clara, CA) excitation passed through a cleanup polarizer, half-wave plate, and lens to focus on the back focal plane (ThorLabs, Newton, NJ). Rotation of the half-wave plate with a motorized mount (PRM1Z8; ThorLabs) controlled the orientation of the excitation polarization. Images were captured with an ORCA-Flash 4.0 v2 CMOS camera (Hamamatsu, Hamamatsu City, Japan). The system was controlled with the software NIS Elements (Nikon). Images were acquired at 0°, 45°, 90°, and 135° excitation polarization.
Cell fixation and antibody staining
Fixation and labeling protocols are described in Stahley et al. (15) with a 1-h incubation in primary antibody and 30-min incubation in secondary antibody. Primary antibody was anti-desmoplakin (DPI/II clone 2.15; Abcam, Cambridge, UK).
Fragmentation assay
Confluent cell sheets were dissociated by incubating with 2 U/mL Dispase II (Roche, Indianapolis, IN) in DMEM for 35–45 min. The intact monolayers were then transferred to 1.7 mL Eppendorf tubes. Media was removed from the cell monolayer and replaced with 500 μL of either normal (∼3 mM Ca2+) or low (∼0.03 mM Ca2+) media and incubated for 2–30 min before agitation with an orbital shaker at 350 RPM for 2 min. The fragments were immediately fixed by adding 500 μL 16% paraformaldehyde and manually counted using a dissection microscope.
Order factor derivation
The probability of excitation of a fluorophore can be expressed as a dot product between the transition dipole moment, described in a spherical coordinate system by the azimuthal angle α and polar angle β, and the polarization of excitation light in the imaging plane (ω). The square of this product is proportional to the intensity of the fluorophore:
| (1) |
To determine the order factor, we first normalized the data pixel-by-pixel where Iω is the intensity from an individual pixel Iω = (I0, I45, I90, I135), as follows:
| (2) |
Using the normalized intensities, we then calculated the amplitude of intensity modulation, which we define as the order factor:
| (3) |
Data analysis
A custom-order factor image analysis program (Polarized Order Detection Software) was written in the software MATLAB (The MathWorks, Natick, MA). Raw images were first corrected for uneven laser illumination and polarization-dependent intensity changes with the mean of three correction images, obtained by imaging a fluorescent slide at each polarization (Chroma Technology, Bellows Falls, VT). Processed images were then corrected for photobleaching using fluorescence decay constants determined from control experiments with constant excitation polarization (GFP, 0.003; mCherry, 0.06) (34). Next, a binary mask was generated to identify desmosomes. Corrected images were masked automatically using a morphological algorithm that first detects punctate objects with the MATLAB function “imopen” and a median filter. Next, the binary mask is created using the threshold from MATLAB’s “graythresh” followed by “bwareaopen”. In cases of high background variance, the threshold was set manually. Pixel-by-pixel order factor was calculated within the binary mask for corrected images using the theoretical relationships described above in Eqs. 1, 2, and 3.
Computational modeling
Using the relationships described above, mathematical modeling was performed in MATLAB using Monte Carlo method simulations of Poisson-distributed noise with 100 repeats. To determine the relevant signal-to-background ratio (S/B) range for simulations, intensity from pixels outside of the desmosome binary mask were averaged to calculate background for each image. The pixel signals within the desmosomal region were averaged to obtain a signal estimate for each image. The range of modeled signal levels were taken from the range of experimental values. The three background levels represent an average background over all data and 1 SD above and below the average.
To determine a threshold above which the order factor can be interpreted to mean true biological order, we modeled disorder at signal levels taken as the lowest signal pixel of each image, which represents the most noise-dominated signal values. The maximum calculated order factor from each simulation represents the worst-case false positive for order. The threshold for disorder was set at 2 SDs above the mean of the distribution of maximum order factors resulting from a disordered system.
We calculated a projected order factor for the time-lapse experiments to verify that the change in measured order factor in low calcium time course experiments was not due to protein loss and decreased S/B. First, simulations were conducted at the average S/B levels of every time point. Next, the order factor pixel values from t0 were matched to a theoretical orientation. In subsequent simulations, order factor values from these orientations and experimental signal and background levels were averaged to determine the theoretical order factor.
Statistics
All statistical analysis was done using GraphPad Prism (GraphPad Software, La Jolla, CA). One-way ANOVA was used to test statistical significance across multiple experimental conditions (Fig. 3). Two-way ANOVAs were done on all data comparing two or more experimental conditions across time points (Fig. 5). Two-way ANOVA with Sidak’s multiple comparisons was used to determine the significance between experimental data and projected order at individual time points (Fig. 5 c). Curve fits were generated using GraphPad Prism’s nonlinear curve fitting functions for the exponential curves as indicated (Figs. 4 and 5).
Figure 3.
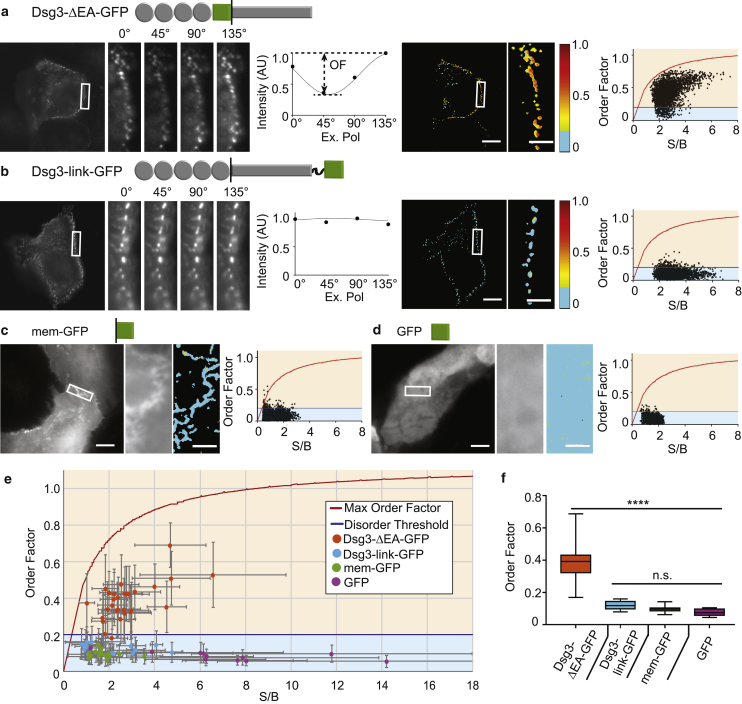
The extracellular domain of Dsg3 is ordered. Shown here are HaCaT cells expressing (a) Dsg3-ΔEA-GFP and (b) Dsg3-link-GFP. The average intensity image and individual ROIs (white rectangle) at each excitation polarization are shown. The intensity from a single pixel is plotted as a function of excitation polarization and fit to the sinusoid (Eq. 1) (solid line). Desmosome order factor is shown as a masked heatmap. Pixel-by-pixel order factor is plotted as a function of S/B. (c and d) Given here is an average intensity image of the HaCaT cell expressing (c) mem-GFP and (d) GFP. Average intensity and order factor heatmap are shown for the ROI. The pixel-by-pixel order factor is plotted over S/B. (a–d) Scale bars, 10 μm; ROI scale bars, 2 μm. (e) Shown here is the mean order factor as a function of mean S/B for individual cells expressing each construct (error bars: SD) compared to the theoretical maximum order factor (red line) and disorder threshold (blue line). (f) Box plots of cell population order factor; full range (whiskers) are given with the median (line) and 25–75 percentile range (box). (∗∗∗∗p < 0.0001; ns = no significance, by one-way ANOVA.) To see this figure in color, go online.
Figure 5.
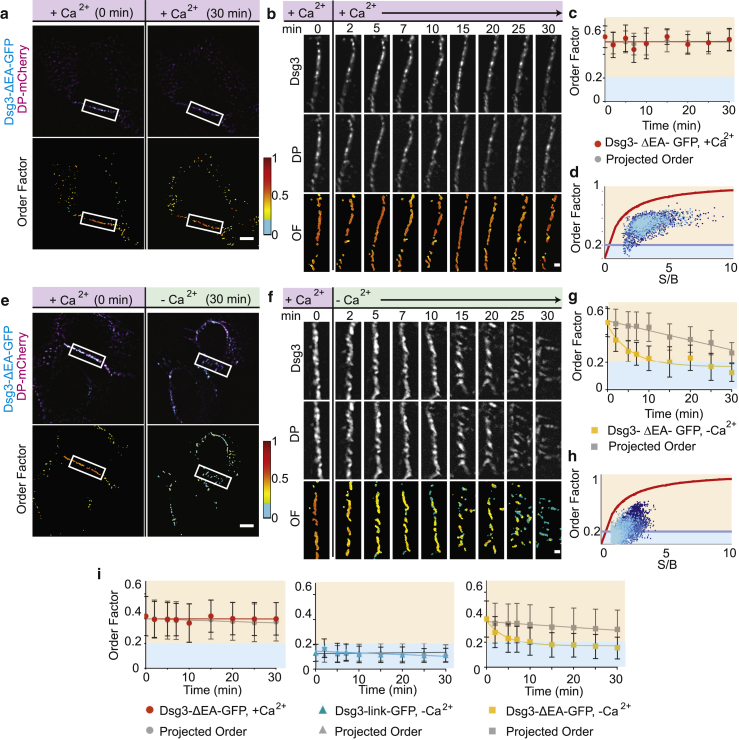
Reduction of Ca2+ results in loss of order concurrent with loss of adhesion. (a) HaCaT cells were transfected with Dsg3-ΔEA-GFP (cyan) and DP-mCherry (magenta). Cells were imaged before (0 min) and after an exchange of normal Ca2+ media. Shown are intensity and order factor images for time = 0 and 30 min. Scale bars, 5 μm. (b) Shown here is ROI time lapse of Dsg3-ΔEA-GFP, DP-mCherry, and order factor. Scale bars, 1 μm. (c) Given here is a plot of Dsg3-ΔEA-GFP order factor (red; mean ± SD) and the projected order factor (gray; mean ± SD) as a function of time. (d) Shown here is a pixel-by-pixel order factor plotted as a function of S/B for the ROI over the time course (time progresses from dark to light blue). (e) HaCaT cells were transfected with Dsg3-ΔEA-GFP (cyan) and DP-mCherry (magenta). Intensity and order factor images are shown for time = 0 and 30 min after switch from normal (∼3 mM) to low (∼0.03 mM) Ca2+ media. Scale bars, 5 μm. (f) Shown here is the time lapse of cell border ROI of Dsg3-ΔEA-GFP, DP-mCherry, and order factor showing dynamics after calcium switch. Scale bars, 1 μm. (g) Plot of Dsg3-ΔEA-GFP order factor (yellow; mean ± SD) and the projected order factor (gray; mean ± SD) from the ROI are shown as a function of time. Order factor was fit to an exponential decay (solid line) with the equation (R2 = 0.94). (h) Pixel-by-pixel order factor was plotted as a function of S/B for the ROI over the time course (time progresses from dark to light blue). (i) Population average and projected order factor were plotted as a function of time. The average Dsg3-ΔEA-GFP switch to normal calcium (red; n = 6 cells) and average Dsg3-link-GFP order factor after switch to low calcium (blue; n = 4 cells) were fit by linear regression (solid lines). The average Dsg3-ΔEA-GFP order factor switch to low Ca2+ (n = 8 cells) was fit with an exponential decay (R2 = 0.97). To see this figure in color, go online.
Figure 4.
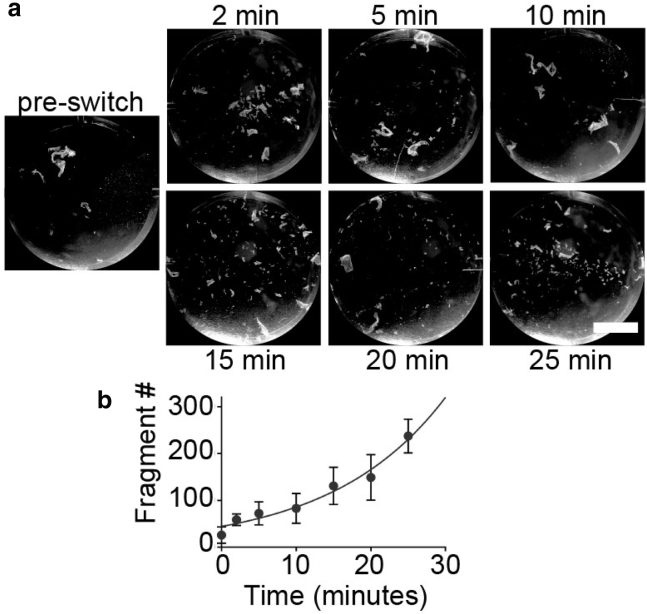
Dynamics of loss of cell adhesion. (a) Given are representative images of HaCaT cell sheets after fragmentation assay at indicated times after calcium switch. Scale bars, 10 mm. (b) Given here is a plot of the fragment count as a function of time after calcium switch (mean ± SD; n = 8). The trendline shows nonlinear (exponential) fit (R2 = 0.97).
Code availability
Code is available upon request.
Results
To investigate protein order in the desmosome adhesive interface, we selected a representative cadherin from the seven desmogleins and desmocollins in the family. We focused on Dsg3 because of its high expression level in the lower layers of the skin, where it is critical for keratinocyte adhesion. This function is highlighted by the skin blistering disease Pemphigus vulgaris, in which adhesion is weakened by autoantibodies targeted to the extracellular domain of Dsg3 (42, 43, 44).
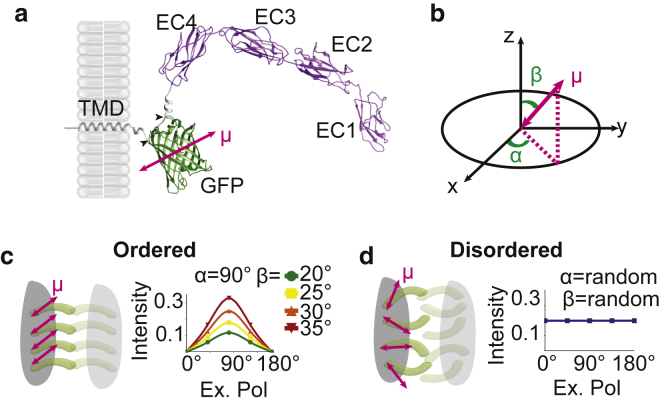
To create a polarization probe, we needed a fluorophore to report the orientation of the Dsg3 extracellular domain. We chose to replace the most membrane-proximal extracellular anchor (EA) domain of Dsg3 with GFP (Dsg3-ΔEA-GFP). We generated a ribbon diagram of the Dsg3-ΔEA-GFP ectodomain based on the crystal structures of GFP (45) and the Dsg3 extracellular domains 1–4 (EC1–4) (46), the orientation of the transition dipole moment (μ) within GFP (47), and the secondary structure predictions of the linker regions (Fig. 1 a). In Dsg3-ΔEA-GFP, GFP is tethered between the EC4 domain and the transmembrane domain. This constricted association is essential for the orientation of μ, which will be probed by polarized excitation, to reflect the protein orientation.
Figure 1.
Polarization microscopy to study desmosome protein organization. (a) Shown here is a ribbon diagram of the Dsg3-ΔEA-GFP chimeric protein. Dsg3 extracellular domains EC1-4 (purple) and GFP (green) with the EA domain deletion/GFP insertion sites are indicated by black arrowheads and the transition dipole moment (μ) by the red double-headed arrow. (b) The fluorophore transition dipole (μ; red arrow) is described by azimuthal (α) and polar (β) angles in a spherical coordinate system where x-y is the imaging plane and z is the optical axis. (c) If Dsg3 is ordered, fluorescence intensity will be modulated by the excitation polarization, resulting in a sinusoidal curve with an amplitude dependent on the polar angle as shown for β = 20°, 25°, 30°, and 35°. (d) If Dsg3 is disordered, the fluorescence intensity will be constant regardless of the excitation polarization. To see this figure in color, go online.
To determine whether proteins are ordered, we must first set up a coordinate system as a frame of reference in which to study Dsg3. The orientation of μ is defined by the azimuthal (α) and polar (tilt; β) angles in the microscope coordinate system (Fig. 1 b), where the Dsg3 extracellular domain will dictate the orientation of GFP. In a single desmosome, there will be multiple copies of Dsg3-ΔEA-GFP within a point spread function. Protein order within this volume is represented by the average of the transition dipole moments and can be measured using excitation resolved polarized fluorescence microscopy (38, 48). If Dsg3-ΔEA-GFP is ordered in a single desmosome, the fluorescence intensity will be modulated by the excitation polarization (Fig. 1 c). In contrast, if Dsg3-ΔEA-GFP is disordered, the intensity will be independent of excitation polarization (Fig. 1 d).
Theory of protein order in desmosomes
We established the theoretical capabilities of excitation resolved polarization fluorescence microscopy to distinguish between protein order and disorder in desmosomes. In our approach, images are acquired at four unique excitation polarizations (0°, 45°, 90°, and 135°) (34). To measure order, we calculate the amplitude of intensity modulation as a function of excitation polarization, which we call the order factor (Eq. 3), where is the intensity from a single excitation polarization (ω) normalized across all excitation polarizations (see Materials and Methods). Order factor represents the average dipole orientation within a pixel.
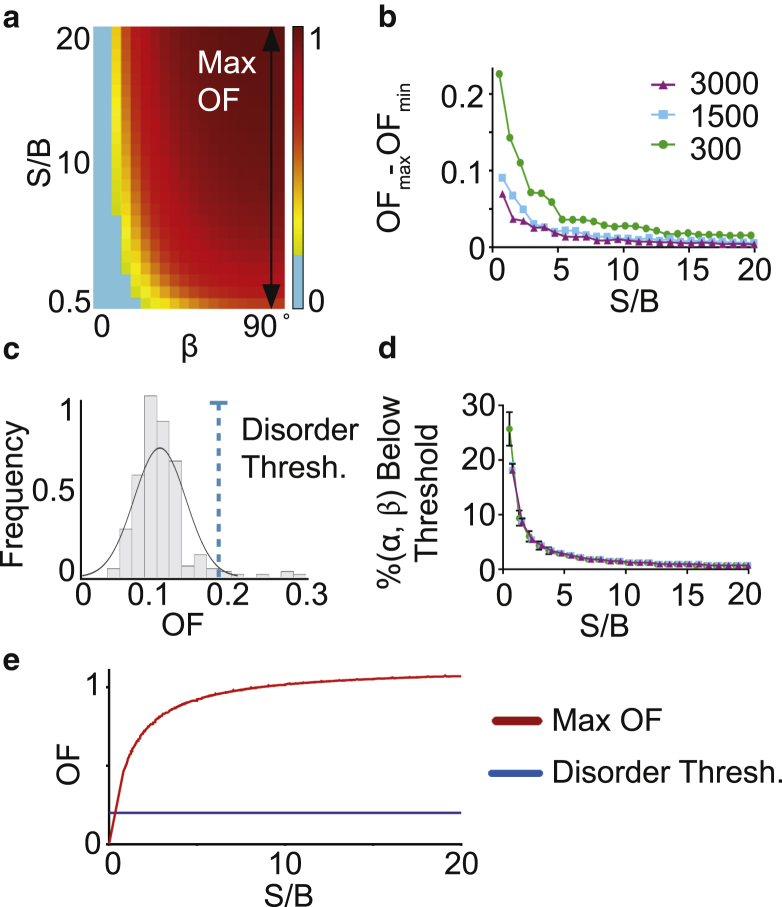
To determine the impact of experimental variables on our ability to measure order, we ran Monte Carlo simulations of raw fluorescence intensity data incorporating photon noise for all possible orientations (α, β) and a range of S/Bs (Fig. 2 a). These simulations show that order factor for an ordered system depends heavily on tilt angle (β) and S/B. Order factor at a given S/B was always maximum when the average dipole is entirely in the imaging plane (β = 90°). The dipole orientation in the imaging plane (α) has a negligible impact on order factor, and the calculations represent averages over all α (Fig. S1). To determine the impact of noise on our ability to measure order, we examined the distribution of theoretical order factors across simulations for β = 90°. At the lowest S/B, where noise is expected to have the biggest impact, the order factor variance was 0.0023 whereas the range of the distribution was 0.23 (Fig. 2 b). Both the range and the variance decreased exponentially as S/B increased. Taken together, this data demonstrates the robustness of order factor to photon noise, while illustrating the full range of order factors that can be measured from an ordered system.
Figure 2.
Computational modeling and derivation of order factor. (a) Theoretical average order factor was calculated from Monte Carlo simulations at different S/B levels and β orientations averaged over all α (background = 1500 photons). The maximum order factor at each S/B occurs when μ is entirely in the imaging place (β = 90°). (b) Range of order factor (max-min) is given as a function of S/B with varying backgrounds. (c) Given here is a histogram of data from Monte Carlo simulations of disorder showing the maximum order factor resulting from experimental signal and background levels. The threshold for distinguishing order from disorder, shown here by the blue dashed line, was set at 2 SDs above the mean of this distribution. Order factors below this threshold are considered disordered and are shown in cyan on the order factor heatmap. (d) Shown here is percent of (α, β) orientations with an order factor less than the disordered threshold as a function of S/B. (e) Given here is theoretically determined maximum order factor as a function of S/B (red) and the disorder threshold (blue). To see this figure in color, go online.
Next, we determined the range of order factors likely to be measured from a disordered system, in which intensity does not depend on excitation polarization. We ran Monte Carlo simulations where (I0 = I45 = I90 = I135) and used our lowest experimentally measured signal levels as inputs to assess the worst-case signal-to-noise ratio (Fig. 2 c). These simulations were critical to determine the threshold at which order and disorder become indistinguishable. We defined the “disorder threshold” to be 2 SDs above the mean of this distribution. Order factors below this threshold (0.2) cannot be distinguished from a disordered system.
One consideration is that a subset of ordered systems will report an order factor below the disorder threshold (cyan on the order factor heatmap; Fig. 2 a) due to the dependence of order factor on dipole orientation and S/B. The percent of (α, β) orientations that have order factors below the disorder threshold is 25% at the lowest S/B and decreases rapidly to <1% as S/B increases, regardless of absolute background level (Fig. 2 d). This means that the number of false negatives or undetectable (α, β) orientations decreases as S/B improves.
From the above simulation results, we set the theoretical upper bound on order factors likely to be measured from an ordered system (red line) and the disorder threshold (blue line) (Fig. 2 e). Experimental order factors between the upper bound and the disordered threshold are the result of an ordered system.
Dsg3 extracellular domain is ordered
After establishing order factor as a metric, we applied this approach to measure order in desmosomes. We designed two Dsg3-GFP chimeric proteins: Dsg3-ΔEA-GFP to measure Dsg3 order (Fig. 1 a), and Dsg3-link-GFP as a disordered control. In Dsg3-link-GFP, the C terminus was tagged with a flexible linker followed by GFP, which allows GFP conformational freedom relative to Dsg3, i.e., the orientation of GFP does not reflect the orientation of the Dsg3 ectodomain.
To confirm that Dsg3-ΔEA-GFP and Dsg3-link-GFP localize properly to desmosomes, we transfected the constructs into HaCaT cells, a human keratinocyte cell line. HaCaT cells are an ideal immortalized cell line for the characterization of desmosome structure and function, as they are nontumorous and are a good model of primary human keratinocytes (49). The cells were fixed and labeled with antibodies to the endogenous obligate desmosomal protein desmoplakin. To remove soluble, nondesmosomal Dsg3, the cells were pre-extracted before fixation (50). Dsg3-ΔEA-GFP and Dsg3-link-GFP both colocalized with desmoplakin in punctate structures at cell-cell borders, indicating proper localization to desmosomes (Fig. S2).
To determine if the extracellular domain of Dsg3 is ordered, we imaged HaCaT cells transfected with Dsg3-ΔEA-GFP using fluorescence polarization microscopy (Fig. 3 a). As expected for an ordered system, the fluorescence intensity at cell-cell junctions was dependent on the excitation polarization. We developed a custom MATLAB image analysis program to calculate the pixel-by-pixel order factor using Eq. 3 within a binary mask defining desmosomes (Fig. 3 a; Fig. S3). We quantified this data by plotting the order factor of every pixel in a cell border region of interest (ROI) as a function of S/B. This revealed that 93% of the pixels fall within the upper and lower bounds for an ordered system. A z-stack analysis showed the order factor was not impacted by focal plane or contributions from out of focus light (Fig. S4).
Next, we imaged HaCaT cells transfected with the control probe Dsg3-link-GFP (Fig. 3 b). Although Dsg3-link-GFP localized to desmosomes, the intensity was not dependent on the excitation polarization angle, as expected for a disordered system. The order factor was below the disorder threshold for the majority of the pixels within the cell border ROI (95%). This indicates that GFP in Dsg3-link-GFP is disordered, and that desmosomal targeting alone is not sufficient to confer order.
To confirm plasma membrane targeting does not result in order, GFP with a palmitoylation motif (mem-GFP) was transfected into HaCaT cells (Fig. 3 c). The intensity of mem-GFP was not dependent on the excitation polarization, and the majority of pixels within the ROI fell below the disordered threshold (99%). This demonstrates that plasma membrane targeting is not sufficient to confer order. Finally, for a well-characterized randomly oriented control, GFP was transfected into HaCaT cells (Fig. 3 d). There was no dependence of GFP intensity on excitation polarization, and the majority of pixels within the ROI were below the disorder threshold (98%).
We next asked if the single-cell order factors were representative of the population. The whole-cell average Dsg3-ΔEA-GFP order factors cluster within the theoretical bounds we determined for an ordered system (Fig. 3 e). This population contained a mixture of cell borders, the majority between transfected and nontransfected cells (Fig. 3 a), and some between two transfected cells (Fig. S5). The order factor of borders between two transfected cells fell within the distribution of those measured for single transfected cell borders.
The average order factors for all cells expressing Dsg3-link-GFP, mem-GFP, and GFP fell below the disorder threshold, regardless of S/B. We noted a spread in the distribution of mean order factor within the Dsg3-ΔEA-GFP cell population, as would be expected for an ordered system based on our modeling. This variability can be explained by differences in orientation (α, β) and S/B, but we cannot rule out additional contributions from biological differences between cells.
Importantly, when interrogated with the binary question is the system ordered or disordered, individual cells report the same answer as the population. The Dsg3-ΔEA-GFP order factor, 0.38 ± 0.11 (n = 29), was significantly larger than those for the controls Dsg3-link-GFP (0.12 ± 0.025; n = 17), mem-GFP (0.097 ± 0.018; n = 17), and GFP (0.073 ± 0.020; n = 11) by one-way ANOVA (Fig. 3 f). Taken together, these data reveal that the extracellular domain of Dsg3 is ordered in desmosomes.
Dsg3 order decreases before loss of cell adhesion
Next, we investigated if Dsg3 order is related to adhesive function. Desmosome adhesive strength is calcium-dependent and when calcium is depleted from calcium-dependent keratinocytes, adhesion is disrupted (51). Therefore, we used a calcium switch assay where the calcium concentration is reduced from normal (∼3 mM) to low (∼0.03 mM) to disrupt desmosome function.
To quantify the average desmosome strength for a cell population and the dynamics of loss of adhesion, we employed a dispase cell fragmentation assay. Fragmentation of a confluent cell sheet resulting from applied mechanical stress increased with time in low calcium media (n = 8) (Fig. 4 a). This fragmentation is inversely proportional to adhesive strength. We quantified the rate of loss of adhesion by fitting the fragment number to an exponential growth with a rate constant of 15.1 min (R2 = 0.97) (Fig. 4 b).
We next wanted to establish the baseline dynamics of desmosome order over the 30-min time frame identified in the functional assay. To provide an independent marker for desmosome identification and tracking, desmoplakin-mCherry (DP-mCherry) was cotransfected into HaCaT cells with Dsg3-ΔEA-GFP.
Comparing images from a representative cell at 0 and 30 min revealed that Dsg3-ΔEA-GFP and DP-mCherry colocalization, cell morphology, and order factor were stable when a normal calcium level was maintained (Fig. 5 a). Desmosome morphology remained unchanged, and individual colocalized puncta with consistent order factors can be identified and tracked across multiple time points (Fig. 5 b). To control for photobleaching that results in a loss of intensity, and thus a change in S/B, we modeled a projected order factor based on the order factor at 0 min, Dsg3-ΔEA-GFP intensity, and background levels throughout the time course. The projected order factor accurately predicted the experimental order factor over the time course, and the two were not significantly different at each time point by two-way ANOVA with Sidak’s comparison (p > 0.11) (Fig. 5 c). The accuracy of this prediction shows that projected order factor is robust. Pixel-by-pixel order factor remained above the disorder threshold over the 30 min (Fig. 5 d).
To measure Dsg3 order during loss of adhesion, Dsg3-ΔEA-GFP and DP-mCherry were transfected into HaCaT cells and imaged before and after calcium switch. A representative cell shows colocalization of DP-mCherry and Dsg3-ΔEA-GFP both before (t = 0 min) and after (t = 30 min) calcium switch, indicating colocalization was maintained throughout the experiment (Fig. 5 e). Morphological changes are apparent when comparing the cell before and after calcium switch, including cell rounding and altered junctions (Fig. 5 e; top). After the calcium switch, Dsg3-ΔEA-GFP shifted from ordered (t = 0 min) to disordered (t = 30 min) (Fig. 5 e; bottom).
To more closely examine the dynamics of this process, we monitored a representative ROI containing a single cell-cell border (Fig. 5 f). For the first 10 min after calcium switch, puncta are clearly identifiable at the cell border. In contrast, by 25 min the cell border has a dramatically different morphology, although Dsg3-ΔEA-GFP colocalization with DP-mCherry remains throughout. In this individual border, order factor decreased within the first 2 min after calcium switch, when puncta localization and morphology were not obviously altered. This decrease in order factor was fit to an exponential decay with a rate of 5.5 min (R2 = 0.94) (Fig. 5 g).
Desmosomal Dsg3 level can decrease during disassembly due to diffusion of the protein in the plasma membrane or protein recycling through endocytosis, both of which would lead to a decrease in fluorescence intensity and S/B (7, 52, 53). As expected during disassembly, projected order factor decreased over the time course due to the changes in S/B, although it remained above the disorder threshold (Fig. 5 g). The experimental and projected order factors exhibited distinct behaviors, with measured order factor decreasing exponentially and more rapidly than the projected linear decrease, leading us to conclude that there was a decrease in the order of Dsg3-ΔEA-GFP as a result of the calcium switch.
To quantify the transition from ordered to disordered over time, we plotted pixel-by-pixel order factor over S/B for the ROI (time progresses from dark to light blue; Fig. 5 h). The percent of individual pixel order factors below the disorder threshold increased over the time course from 0% (0 min) to 87% disordered (30 min). We further quantified order factor on a single desmosome level. This revealed possible heterogeneity in order dynamics at early time points between the desmosomes from a single cell. Loss of order from individual desmosomes is smooth on the timescale of our experiments (Fig. S6).
To confirm that changes in cell and desmosome morphology were not dependent on the construct, we imaged HaCaT cells transfected with Dsg3-link-GFP and DP-mCherry before and after calcium switch (Fig. S7). Dsg3-link-GFP colocalized with DP-mCherry at all time points. Similar morphological changes were observed to those noted in the Dsg3-ΔEA-GFP transfected cells in the calcium switch assay. However, as expected, the average order factor was consistently below the disorder threshold at all time points.
Next, we wanted to determine if order loss followed similar kinetics in the population as in single cells. For the control conditions (Dsg3-ΔEA-GFP in normal calcium media, n = 6; and Dsg3-link-GFP in low calcium media, n = 4), the average experimental and projected order factors fit a linear regression (Fig. 5 i), with no significant difference by two-way ANOVA. These results show that the population order factor was maintained throughout the time course for each control and fit the theoretical model.
In contrast, the average experimental order factor after calcium switch for Dsg3-ΔEA-GFP for a population of cells decreased exponentially (n = 8). These dynamics were significantly different from the average projected order factor at each time point demonstrating that the decrease in order factor reflects a structural change within the desmosome, not declining S/B. Loss of order was rapid, with a significant decrease in order factor between t = 0 min and t = 2 min (p = 0.0003, n = 8; by two-way ANOVA with Sidak’s multiple comparison test). To quantify the rate of Dsg3-ΔEA-GFP loss of order, the time course for the population of cells was fit to an exponential with a decay rate of 4.5 min (R2 = 0.97). The rate of loss of order is faster than the rate of fragmentation (15.1 min), showing that loss of Dsg3 order is upstream of the loss of adhesion.
Discussion
Desmosomes play a critical role in the maintenance of tissue integrity and their dysfunction is implicated in human diseases including cardiomyopathies, skin blistering diseases, and cancers (54). Therefore, the relationship between desmosome structure and adhesive function is of much interest. There is debate in the field over how the desmosomal cadherins are organized and if the organization of these proteins is important for adhesive function. Here, we applied fluorescence polarization microscopy to study desmosome structure, and found that the extracellular domain of the cadherin Dsg3 is ordered in living cells. Furthermore, we found that order is a dynamic state: order is lost when calcium is depleted, ultimately resulting in the loss of cell adhesion.
Previous work using EM to study desmosomal cadherins in the extracellular space has conflicting results. Al-Amoudi et al. (21, 24) found an ordered structure, and hypothesized the desmosomal cadherins form a repeating array. In contrast, the less-ordered “tangled knot” described by He et al. (26) fit many cadherin orientations within a single desmosome. These differences could be due to several factors including imaging method, sample fixation, tissue type, or adhesive state. In the experiments presented here, order is measured in living cells, eliminating any potential artifacts due to fixation. Desmosome adhesive state is well understood to be either calcium-dependent or hyperadhesive (calcium-independent) (10, 55). Fluorescence polarization revealed the Dsg3 extracellular domain is ordered in calcium-dependent cells.
Order factor offers a readout for desmosome structural organization, but it does not indicate the ultrastructure conveying the order. A single desmosome is close in size to the diffraction limit and many copies of Dsg3 in a single desmosome contribute to an ensemble average dipole orientation. Complete disorganization of dipoles in the x-y plane will result in an order factor below the disordered threshold. If all dipoles possess a single orientation (α, β), order factor is impacted by β (Fig. 1 c) and to a lesser extent by α (Fig. S1). Interestingly, cadherins could adopt multiple orientations in one desmosome, but still form an ordered array. Our approach is currently unable to discriminate between a single cadherin orientation, two or more distinct repeating orientations, or a circularly symmetric order (1).
Influences on order factor can be grouped into two categories: experimental and structural. In this work, we modeled the experimental impacts including signal-to-noise ratio and background level and showed we can measure order factor with confidence under our experimental conditions. We have also developed methods to distinguish changes in order factor brought on by structural dynamics rather than experimental factors (i.e., decreasing S/B from photobleaching or loss of protein). Several biological factors could introduce variations in cadherin dipole orientations (α, β) in a desmosome. These include, but are not limited to, nanoscale fluctuations in membrane curvature, trans-binding dynamics, protein wobble, and changes in plaque arrangement. These factors would decrease the average dipole and be detected as a decrease in the order factor.
It has been demonstrated that the tertiary structure of Dsg3 relies on calcium binding between the extracellular domains (46) to maintain rigidity, similar to the classical cadherins. Interestingly, this rigidity may not be essential to maintain adhesion. Molecular dynamics simulations by Sotomayor and Schulten (56) of classical C-cadherin suggest that trans-binding could be maintained in the absence of calcium. Experimentally, Kim et al. (57) used Förster resonance energy transfer to distinguish two phases of N-cadherin conformational changes, including a “sudden, but partial loss” of binding, after removal of calcium. Although desmosomal cadherins are similar to classical cadherins, the role of calcium in the adhesive function of Dsg3 and the direct impact of its loss on quaternary structure of the complex has been largely unexplored. We showed that order decreases within 2 min of removing calcium, indicating that Dsg3 conformation is calcium dependent in desmosomes. Intriguingly, in this time frame Dsg3 adhesive function is not yet lost. This difference in dynamics indicates that loss of order is upstream of loss of adhesion and supports the molecular dynamics showing that trans-binding is not calcium dependent (56). Our data shows there is a loss of Dsg3 order while trans-binding is maintained, followed by a loss of adhesive function similar to the two-phase dynamics measure by Förster resonance energy transfer (57). We propose that loss of cadherin order in response to calcium depletion leads to a cascade of events that culminates in the loss of trans-binding and cell adhesion.
Fluorescence polarization microscopy allows us to quantify structural dynamics in living cells. In contrast, previous methods for studying desmosome organization were entirely dependent on static, fixed samples. Implementation of fluorescence polarization involves a straightforward and inexpensive modification to an existing fluorescence microscope. The experimental and analysis methods used here do not depend on the symmetry of the complex or its orientation within a cell and can be easily applied to other cell junctions or macromolecular complexes. We believe this approach to studying desmosome structure will continue to be invaluable for exploring cadherin order in different functional states and disruption in human disease.
Conclusion
Here we establish order factor as a fast and robust metric for studying spatiotemporal structural dynamics and adhesive function of desmosomes in live cells. The order factor can be determined on an individual desmosome, cell, or population level. We anticipate this approach will become a powerful optical tool for studying the relationship between structure and function in desmosomes and other macromolecular complexes.
Author Contributions
A.L.M. and E.I.B. designed the research. E.I.B. and T.M.U. performed research. T.M.U. and S.S.R. contributed analytic tools. E.I.B., T.M.U., and S.S.R. analyzed data. All authors wrote and edited the manuscript.
Acknowledgments
We thank Andrew Kowalczyk, Khalid Salaita, Joshua Brockman, Aaron Blanchard, and Claire Atkinson for their valuable feedback and Oskar Laur with the Emory Cloning Core for generating the constructs used in this manuscript.
This work was supported by funding to A.L.M. from the National Institutes of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (R21AR066920), the National Science Foundation (NSF) IDBR grant (1353939), and the National Science Foundation (NSF) CAREER award (1553344).
Editor: David Piston.
Footnotes
Emily I. Bartle, Tara M. Urner, and Alexa L. Mattheyses's present address is Department of Cell, Developmental, and Integrative Biology, University of Alabama at Birmingham, Birmingham, Albama
Seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31071-8.
supporting material
References
- 1.Beck M., Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 2.Xu T., Pagadala V., Mueller D.M. Understanding structure, function, and mutations in the mitochondrial ATP synthase. Microb. Cell. 2015;2:105–125. doi: 10.15698/mic2015.04.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gumbiner B.M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson C.E., Mattheyses A.L., Simon S.M. Conserved spatial organization of FG domains in the nuclear pore complex. Biophys. J. 2013;104:37–50. doi: 10.1016/j.bpj.2012.11.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McQuilken M., Mehta S.B., Gladfelter A.S. Polarized fluorescence microscopy to study cytoskeleton assembly and organization in live cells. Curr. Protoc. Cell Biol. 2015;67:4.29.1–4.29.13. doi: 10.1002/0471143030.cb0429s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mouritsen O.G., Jørgensen K. Dynamical order and disorder in lipid bilayers. Chem. Phys. Lipids. 1994;73:3–25. doi: 10.1016/0009-3084(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 7.Garrod D., Chidgey M. Desmosome structure, composition and function. Biochim. Biophys. Acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Kowalczyk A.P., Green K.J. Structure, function, and regulation of desmosomes. Prog. Mol. Biol. Transl. Sci. 2013;116:95–118. doi: 10.1016/B978-0-12-394311-8.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooke M.A., Nitoiu D., Kelsell D.P. Cell-cell connectivity: desmosomes and disease. J. Pathol. 2012;226:158–171. doi: 10.1002/path.3027. [DOI] [PubMed] [Google Scholar]
- 10.Garrod D.R., Berika M.Y., Tabernero L. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J. Cell Sci. 2005;118:5743–5754. doi: 10.1242/jcs.02700. [DOI] [PubMed] [Google Scholar]
- 11.Wallis S., Lloyd S., Garrod D. The α-isoform of protein kinase C is involved in signaling the response of desmosomes to wounding in cultured epithelial cells. Mol. Biol. Cell. 2000;11:1077–1092. doi: 10.1091/mbc.11.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green K.J., Getsios S., Godsel L.M. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol. 2010;2:a000125. doi: 10.1101/cshperspect.a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt A.J., Berika M.Y., Garrod D.R. Suprabasal desmoglein 3 expression in the epidermis of transgenic mice results in hyperproliferation and abnormal differentiation. Mol. Cell. Biol. 2002;22:5846–5858. doi: 10.1128/MCB.22.16.5846-5858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North A.J., Bardsley W.G., Garrod D.R. Molecular map of the desmosomal plaque. J. Cell Sci. 1999;112:4325–4336. doi: 10.1242/jcs.112.23.4325. [DOI] [PubMed] [Google Scholar]
- 15.Stahley S.N., Bartle E.I., Mattheyses A.L. Molecular organization of the desmosome as revealed by direct stochastic optical reconstruction microscopy. J. Cell Sci. 2016;129:2897–2904. doi: 10.1242/jcs.185785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel S.D., Chen C.P., Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr. Opin. Struct. Biol. 2003;13:690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Sivasankar S. Tuning the kinetics of cadherin adhesion. J. Invest. Dermatol. 2013;133:2318–2323. doi: 10.1038/jid.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowndes M., Rakshit S., Nelson W.J. Different roles of cadherins in the assembly and structural integrity of the desmosome complex. J. Cell Sci. 2014;127:2339–2350. doi: 10.1242/jcs.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Getsios S., Huen A.C., Green K.J. Working out the strength and flexibility of desmosomes. Nat. Rev. Mol. Cell Biol. 2004;5:271–281. doi: 10.1038/nrm1356. [DOI] [PubMed] [Google Scholar]
- 20.Tamarin A., Sreebny L.M. An analysis of desmosome shape, size, and orientation by the use of histometric and densitometric methods with electron microscopy. J. Cell Biol. 1963;18:125–134. doi: 10.1083/jcb.18.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Amoudi A., Díez D.C., Frangakis A.S. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- 22.Boggon T.J., Murray J., Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 23.Al-Amoudi A., Castaño-Diez D., Frangakis A.S. The three-dimensional molecular structure of the desmosomal plaque. Proc. Natl. Acad. Sci. USA. 2011;108:6480–6485. doi: 10.1073/pnas.1019469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Amoudi A., Dubochet J., Norlén L. Nanostructure of the epidermal extracellular space as observed by cryo-electron microscopy of vitreous sections of human skin. J. Invest. Dermatol. 2005;124:764–777. doi: 10.1111/j.0022-202X.2005.23630.x. [DOI] [PubMed] [Google Scholar]
- 25.Rayns D.G., Simpson F.O., Ledingham J.M. Ultrastructure of desmosomes in mammalian intercalated disc; appearances after lanthanum treatment. J. Cell Biol. 1969;42:322–326. doi: 10.1083/jcb.42.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W., Cowin P., Stokes D.L. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- 27.Weber G. Rotational Brownian motion and polarization of the fluorescence of solutions. Adv. Protein Chem. 1953;8:415–459. doi: 10.1016/s0065-3233(08)60096-0. [DOI] [PubMed] [Google Scholar]
- 28.Weber G., Anderson S.R. The effects of energy transfer and rotational diffusion upon the fluorescence polarization of macromolecules. Biochemistry. 1969;8:361–371. doi: 10.1021/bi00829a050. [DOI] [PubMed] [Google Scholar]
- 29.Kierszenbaum F., Dandliker J., Dandliker W.B. Investigation of the antigen-antibody reaction by fluorescence polarization. Measurement of the effect of the fluorescent label upon the bovine serum albumin (BSA) anti-BSA equilibrium. Immunochemistry. 1969;6:125–137. doi: 10.1016/0019-2791(69)90184-0. [DOI] [PubMed] [Google Scholar]
- 30.Yin L., Stern L.J. Measurement of peptide binding to MHC class II molecules by fluorescence polarization. Curr. Protoc. Immunol. 2014;106:5.10.1–5.10.12. doi: 10.1002/0471142735.im0510s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Axelrod D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 1979;26:557–573. doi: 10.1016/S0006-3495(79)85271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampmann M., Atkinson C.E., Simon S.M. Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat. Struct. Mol. Biol. 2011;18:643–649. doi: 10.1038/nsmb.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vrabioiu A.M., Mitchison T.J. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443:466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 34.DeMay B.S., Noda N., Oldenbourg R. Rapid and quantitative imaging of excitation polarized fluorescence reveals ordered septin dynamics in live yeast. Biophys. J. 2011;101:985–994. doi: 10.1016/j.bpj.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattheyses A.L., Kampmann M., Simon S.M. Fluorescence anisotropy reveals order and disorder of protein domains in the nuclear pore complex. Biophys. J. 2010;99:1706–1717. doi: 10.1016/j.bpj.2010.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocheleau J.V., Edidin M., Piston D.W. Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophys. J. 2003;84:4078–4086. doi: 10.1016/S0006-3495(03)75133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kress A., Ferrand P., Brasselet S. Probing orientational behavior of MHC class I protein and lipid probes in cell membranes by fluorescence polarization-resolved imaging. Biophys. J. 2011;101:468–476. doi: 10.1016/j.bpj.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valades Cruz C.A., Shaban H.A., Brasselet S. Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Proc. Natl. Acad. Sci. USA. 2016;113:E820–E828. doi: 10.1073/pnas.1516811113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta S.B., McQuilken M., Tani T. Dissection of molecular assembly dynamics by tracking orientation and position of single molecules in live cells. Proc. Natl. Acad. Sci. USA. 2016;113:E6352–E6361. doi: 10.1073/pnas.1607674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yachdav G., Kloppmann E., Rost B. PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson V.G. Growth and differentiation of HaCaT keratinocytes. Methods Mol. Biol. 2014;1195:33–41. doi: 10.1007/7651_2013_42. [DOI] [PubMed] [Google Scholar]
- 42.Amagai M. Autoimmunity against desmosomal cadherins in pemphigus. J. Dermatol. Sci. 1999;20:92–102. doi: 10.1016/s0923-1811(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 43.Amagai M., Klaus-Kovtun V., Stanley J.R. Autoantibodies against a novel epithelial cadherin in Pemphigus vulgaris, a disease of cell adhesion. Cell. 1991;67:869–877. doi: 10.1016/0092-8674(91)90360-b. [DOI] [PubMed] [Google Scholar]
- 44.Saito M., Stahley S.N., Kowalczyk A.P. Signaling dependent and independent mechanisms in Pemphigus vulgaris blister formation. PLoS One. 2012;7:e50696. doi: 10.1371/journal.pone.0050696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brejc K., Sixma T.K., Remington S.J. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison O.J., Brasch J., Shapiro L. Structural basis of adhesive binding by desmocollins and desmogleins. Proc. Natl. Acad. Sci. USA. 2016;113:7160–7165. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosell F.I., Boxer S.G. Polarized absorption spectra of green fluorescent protein single crystals: transition dipole moment directions. Biochemistry. 2003;42:177–183. doi: 10.1021/bi0266535. [DOI] [PubMed] [Google Scholar]
- 48.Oldenbourg R. Polarized light microscopy: principles and practice. Cold Spring Harb. Protoc. 2013 doi: 10.1101/pdb.top078600. [DOI] [PubMed] [Google Scholar]
- 49.Sprenger A., Weber S., Dengjel J. Consistency of the proteome in primary human keratinocytes with respect to gender, age, and skin localization. Mol. Cell. Proteomics. 2013;12:2509–2521. doi: 10.1074/mcp.M112.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palka H.L., Green K.J. Roles of plakoglobin end domains in desmosome assembly. J. Cell Sci. 1997;110:2359–2371. doi: 10.1242/jcs.110.19.2359. [DOI] [PubMed] [Google Scholar]
- 51.Penn E.J., Burdett I.D., Rees D.A. Structure and assembly of desmosome junctions: biosynthesis and turnover of the major desmosome components of Madin-Darby canine kidney cells in low calcium medium. J. Cell Biol. 1987;105:2327–2334. doi: 10.1083/jcb.105.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delva E., Jennings J.M., Kowalczyk A.P. Pemphigus vulgaris IgG-induced desmoglein-3 endocytosis and desmosomal disassembly are mediated by a clathrin- and dynamin-independent mechanism. J. Biol. Chem. 2008;283:18303–18313. doi: 10.1074/jbc.M710046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calkins C.C., Setzer S.V., Kowalczyk A.P. Desmoglein endocytosis and desmosome disassembly are coordinated responses to pemphigus autoantibodies. J. Biol. Chem. 2006;281:7623–7634. doi: 10.1074/jbc.M512447200. [DOI] [PubMed] [Google Scholar]
- 54.Broussard J.A., Getsios S., Green K.J. Desmosome regulation and signaling in disease. Cell Tissue Res. 2015;360:501–512. doi: 10.1007/s00441-015-2136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Windoffer R., Borchert-Stuhlträger M., Leube R.E. Desmosomes: interconnected calcium-dependent structures of remarkable stability with significant integral membrane protein turnover. J. Cell Sci. 2002;115:1717–1732. doi: 10.1242/jcs.115.8.1717. [DOI] [PubMed] [Google Scholar]
- 56.Sotomayor M., Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys. J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim S.A., Tai C.Y., Schuman E.M. Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc. Natl. Acad. Sci. USA. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.