Abstract
Growing evidence supports the importance of protein S-glutathionylation as a regulatory post-translational modification with functional consequences for proteins. Discoveries of redox-state-dependent protein kinase S-glutathionylation have fueled discussion of redox-sensitive signaling. Following previously published experimental evidence for S-glutathionylation induced deactivation of the Arabidopsis thaliana kinase BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR-LIKE KINASE 1 (BAK1), we investigated the consequences of S-glutathionylation on the equilibrium conformational ensemble of BAK1 using all-atom molecular dynamics simulations. We found that glutathionylation of C408 allosterically destabilizes the active-like state of BAK1 and stabilizes an inactive conformation known to recur in protein kinases. Glutathionylation of C408 also has structural consequences throughout the BAK1 kinase domain, whereas glutathionylation of C353 in the N-lobe and C374 near the ATP-binding site have few notable effects on BAK1 compared with the unmodified protein. Our results suggest an allosteric mechanism for inhibition of BAK1 by C408 S-glutathionylation, and more generally, support the notion of protein kinase S-glutathionylation as a means of redox signaling in plant cells.
Introduction
Reversible post-translational modifications are an essential mechanism for control of protein function in biochemical networks, allowing a cell to modulate protein function rapidly in response to changes in its metabolic state or in the local environment (1, 2). The phosphorylation-dephosphorylation cycle of serine, threonine, and tyrosine residues catalyzed by protein kinases and phosphatases, respectively, is a well-studied example of this mode of control. Protein phosphorylation allows for feedback, oscillation, and multistability at the network scale as well as integration of multiple signals in a single protein through the possibility of a multisite phosphorylation code (1, 2, 3).
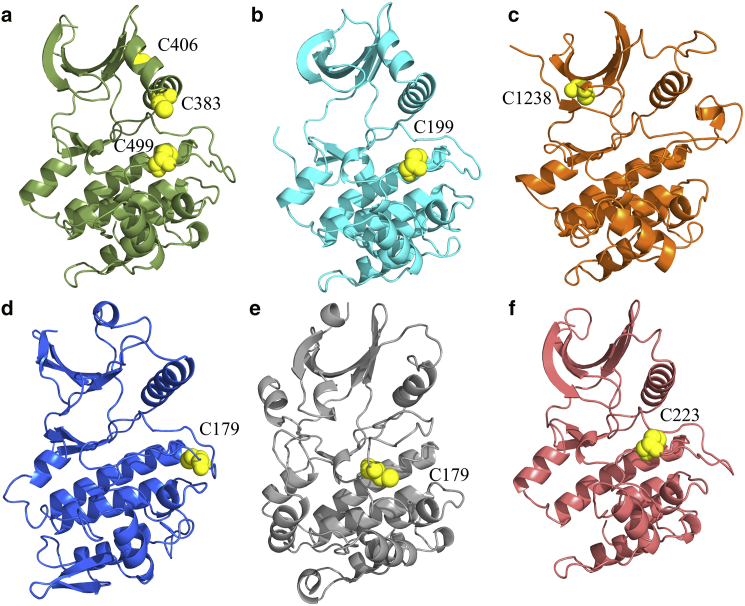
Growing evidence supports the role of S-glutathionylation in control of protein function in response to oxidative stress (4, 5). Glutathione (GSH) is a tripeptide consisting of a glutamate bound through a γ-peptide linkage to a conventional cysteine-glycine dipeptide (Fig. 1 b). GSH is present in nearly all known life forms, and the enzymes responsible for its synthesis are known to be essential for normal function in several animals and plants (6, 7). GSH is able to modulate protein function by forming a disulfide bond with cysteine residues (S-glutathionylation) under oxidative conditions (Fig. 1 b), a process believed to be catalyzed in both directions by glutaredoxin enzymes (4). S-glutathionylation plays a role in control of numerous enzymes (8), including protein kinases (9, 10, 11, 12, 13, 14, 15, 16). Glutathionylation of human AMP-activated protein kinase on C299 and C304 within the catalytic α-subunit C-terminal tail increases enzymatic activity (14, 15). On the other hand, glutathionylation of rat protein kinase C-α (9) (Fig. 2 a), mouse cyclic AMP-dependent protein kinase on C199 (10) (Fig. 2 b), rat mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 on C1238 (11) (Fig. 2 c), mouse IκB kinase subunit-β on C179 (12) (Fig. 2 d), rat Ca2+/calmodulin-dependent kinase I on C179 (13) (Fig. 2 e), and mouse p90-ribosomal S6 kinase 1 (p90-RSK1) on C223 (16) (Fig. 2 f) all reduce enzymatic activity.
Figure 1.
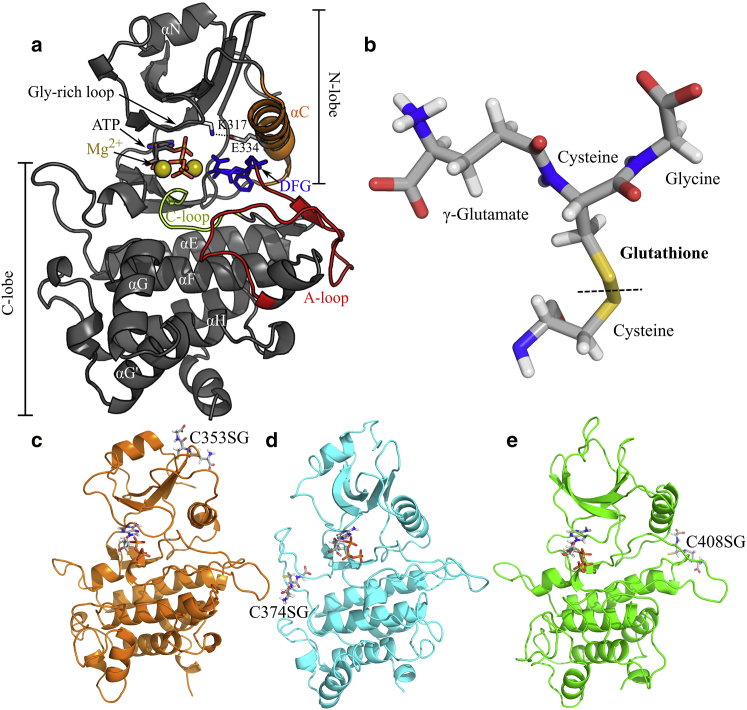
BAK1 S-glutathionylation. (a) Structural and functional features of a protein kinase. (b) Structure of glutathione in a mixed disulfide with a cysteine residue. Glutathione consists of a glutamate connected through a γ-peptide bond to a cysteine-glycine dipeptide. BAK1 structures taken from simulation with (c) C353, (d) C374, and (e) C408 glutathionylated. ATP is shown in the ATP-binding pocket for all three systems. To see this figure in color, go online.
Figure 2.
Overview of kinase S-glutathionylation. (a–f) Locations of S-glutathionylation sites likely to be on kinase catalytic domains with experimentally demonstrated functional consequences. All homology models were made using the Swiss Model webserver (63). (a) Homology model of mouse protein kinase C-α with C383, C406, and C499 shown. As the site of S-glutathionylation was not determined for protein kinase C-α, the cysteine residues with the lowest pKa values (C383: 10.25, C406: 9.97, C499: 9.74) predicted using PropKa 3.1 (45, 46) were chosen for display. (b) Crystal structure of mouse cyclic AMP-dependent protein kinase (PDB: 1J3H (64)) with experimentally determined S-glutathionylation site C199 shown. (c) Homology model of rat mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 with experimentally determined S-glutathionylation site C1238 shown. (d) Homology model of mouse IκB kinase subunit-β with experimentally determined S-glutathionylation site C179 shown. (e) Crystal structure of rat Ca2+/calmodulin-dependent kinase I (PDB: 1A06 (65)) with missing residues modeled using Modeler (66) and experimentally determined S-glutathionylation site C179 shown. (f) Homology model of rat p90-ribosomal S6 kinase 1 with experimentally determined S-glutathionylation site C223 shown. To see this figure in color, go online.
Recently, in vitro experiments demonstrated that the Arabidopsis thaliana leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR-LIKE KINASE 1 (BAK1) interacts with the glutaredoxin GRXC2, catalyzing glutathionylation of BAK1 cysteine residues and reducing BAK1 phosphotransferase activity (17). Three potential glutathionylation sites were identified, namely C353, C374, and C408 (Fig. 1, c–e), although single and double mutations of these residues to serine suggested that C353 and C408 are the dominant sites. However, the mechanistic details of how glutathionylation can reduce BAK1 activity far from the active site remain unclear.
The structural characteristics of a catalytically competent kinase have been studied at length, and include formation of a salt bridge between a conserved glutamate-lysine pair (K-E salt bridge; E334 and K317 in BAK1), a folded αC helix, an unfolded activation loop, an inward-flipped phenylalanine in the DFG motif, and an ATP molecule bound to the ATP-binding site (18, 19, 20) (Fig. 1 a). By monitoring these structural features, we can roughly map the protein kinase conformation we observe to a qualitative estimate of phosphotransferase activity. For example, if the αC helix is completely unfolded and the K-E salt bridge is broken, the conformation is likely inactive. By comparison, if another conformation exhibits a folded αC helix and a formed K-E salt bridge in conjunction with a properly positioned DFG motif and activation loop, it is likely catalytically competent. We will refer to these structural features of kinases as activation-relevant collective variables. In our previous work on the BAK1 core kinase domain, we found that the αC helix displays considerable disorder (21). This disorder could represent a regulatory mechanism where association of BAK1 with another leucine-rich repeat receptor-like kinase promotes folding of the αC helix in a similar manner to the metazoan epidermal growth factor receptor (21, 22, 23).
To determine the molecular mechanisms of BAK1 inhibition by glutathionylation and to gain insight into the site-specific function of BAK1 glutathionylation, we performed extensive molecular dynamics (MD) simulations on the BAK1 core kinase domain, singly glutathionylated on C353, C374, and C408 (BAK1-C353SG, BAK1-C374SG, and BAK1-C408SG, respectively). Additionally, we used simulations of the nonglutathionylated BAK1 core kinase domain (BAK1-SH) from our previous work for comparison (21). Using the Kullback-Leibler (KL) divergence, a measure of similarity between probability distributions borrowed from information theory, we quantified the global effects of glutathionylation on the BAK1 conformational ensemble and found little change upon C353 or C374 glutathionylation, in contrast to the large shifts in BAK1-C408SG behavior from the nonglutathionylated kinase. We found little change in the free-energy landscapes of BAK1-C353SG or BAK1-C374SG and observed that S-glutathionylation of C408 eliminates the active-like state of BAK1 and stabilizes an outward-swung αC helix. As two of the three discussed BAK1 cysteine residues appear to be conserved, to a certain extent, in the A. thaliana kinome, glutathionylation may function similarly in regulation of other protein kinases during shifts in cellular redox state.
Methods
MD simulations
All simulations were performed using the AMBER 14 MD package (24) and the CHARMM 36 force field (25, 26, 27) on the Blue Waters petascale computing facility. All simulation systems were set up using the VMD (28) plugin Psfgen 1.6 and converted to Amber format using the CHAMBER tool (29).
All simulations of BAK1-SH were taken from our previous work (21), which we describe in the following two paragraphs. Starting coordinates for the BAK1-SH kinase domain were taken from available crystal structures (PDB: 3TL8 (chains A, D, G, and H) (30), 3UIM (31), and 3ULZ (32)). Starting structures were solvated in water boxes with dimensions of ∼90 Å × 70 Å × 63 Å with TIP3P model molecules (33). Sodium and chloride ions were added to neutralize the charge of all systems and bring salt concentration to ∼150 mM. An ATP molecule with two magnesium ions bound, taken from previous simulations, was inserted into the binding pocket for all structures in place of the modified adenosine-phosphate molecules used in crystallization, aligned with the adenosine ring of the corresponding ATP analog. All systems were subjected to 10,000 steps of energy minimization and were equilibrated for 8–10 ns in an NPT ensemble at 300 K and 1 atm, maintained using Langevin dynamics and a Berendsen barostat. Simulations were performed using a 2-fs time step, periodic boundary conditions, particle mesh Ewald electrostatics (34), and constraints of hydrogen-containing bonds using the SHAKE algorithm (35, 36). Equilibrated structures were then equilibrated for another 10 ns to obtain average dihedral angle potential energies for calculation of accelerated MD (AMD) (37) parameters (Table S1) according to (38).
To generate a diverse set of starting structures for unbiased MD simulation, we performed AMD sampling for BAK1-SH, initiating 25 independent simulations from the result of equilibrating each crystal structure for an aggregate 8.6 μs (Table S2). The final round of AMD sampling was clustered using the k-means algorithm in the space of the distance between the most distal side-chain nitrogen in the lysine and carbon in glutamate within the conserved K-E salt bridge and αC helix root-mean-square deviation from the crystal structures into 100 states, and the nearest neighbors of the cluster centroids were chosen as starting structures for unbiased simulation. These structures were used to initiate 100 independent unbiased simulations for an aggregate ∼30 μs (Table S2).
The BAK1-SH structures produced by AMD were similarly used as starting structures for simulations of BAK1-C353SG and BAK1-C374SG. To prepare the initial BAK1-C353SG and BAK1-C374SG structures from BAK1-SH AMD conformations, we took the same 100 cluster centroid nearest neighbors and added a randomly rotated GSH to the appropriate cysteine, and then performed energy minimization and equilibration steps before beginning production runs in the same manner as for BAK1-SH. Total BAK1-353SG and BAK1-374SG unbiased simulation times were 30 and 26.8 μs, respectively (Table S2).
For BAK1-C408SG, separate AMD sampling was done due to the possibility of high-energy structures resulting from adding GSH to C408 in BAK1-SH AMD conformations. Eight rotational conformations of GSH attached to C408 of a single BAK1 crystal structure (PDB: 3TL8, chain A) (30) were initially set up, minimized, and equilibrated in the same manner as described for BAK1-SH. The same procedure used for generating BAK1-SH structures to initiate unbiased sampling was used for three rounds of AMD sampling (1.9 μs starting from 8 structures, 5.9 μs starting from 100 structures, and finally, 6.0 μs starting from 100 structures) (Table S2). After each round, the trajectories were clustered using the same metrics used for BAK1-SH, and starting structures for the subsequent sampling round were generated using cluster centroid nearest neighbors. Finally, 150 cluster centroid nearest neighbors were chosen from the final round of AMD sampling and were used to run 150 independent, unbiased MD simulations for an aggregate 45 μs (Table S2).
Markov state model construction
All trajectory analysis was done using MDTraj 1.7 (39) except where otherwise noted, whereas clustering and Markov state model (MSM) construction was done using MSMBuilder 3.6 (40). We aligned each frame of every trajectory to a BAK1 crystal structure (PDB: 3TL8, chain A) and calculated the root-mean-square deviation of the N-terminal lobe and C-terminal lobe separately, with respect to the crystal structure. We chose a lag time for each model by plotting implied timescales of MSMs built with increasing lag times, and choosing the lag time at which the timescales began to plateau, to improve the validity of the Markov assumption for our models (Figs. S1–S4). We then used the Osprey variational cross-validation package to choose the number of clusters, ranging from 100 to 500, that maximized the mean cross-validation score of the MSM generalized matrix Raleigh quotient, calculated from five equal partitions of the data into training and test sets (41). The chosen lag time and number of clusters for each MSM are shown in Table S3.
Free-energy landscape construction
The free-energy landscapes for each of the four systems were constructed by building normalized two-dimensional histograms from the conformations within each state of the corresponding MSM in the space of the inverse distance between the glutamate δ-carbon atom and the lysine side-chain nitrogen in the K-E salt bridge, and the α helical content of the αC helix. The α-helical content was calculated according to the NAMD 2.11 collective variable “alpha” (42), as detailed in (21). State histograms ( for state i) were weighed by the MSM equilibrium probability () of the corresponding state to estimate MSM-weighted probabilities, which were then used to estimate free energy as follows:
| (1) |
where R is the gas constant and T is the temperature. For each system, the lowest free energy was used as a reference, and set to zero.
Local KL divergence
We used the local KL divergence measure introduced in (43), defined as follows for residue i:
| (2) |
This is equivalent to the sum of relative entropies between the reference () and test () probability distributions discretized into N bins (where ) over the ϕ, ψ, and dihedral angles of residue i ( for glycine and alanine residues, and for all others). Following (43), we calculated a bootstrap distribution of KL divergences for each system to test the null hypothesis that the KL divergence between designated test and reference ensembles is no greater than expected from variability within the reference ensemble. This is accomplished using the bootstrap KL divergence
| (3) |
and the mean bootstrap KL divergence
| (4) |
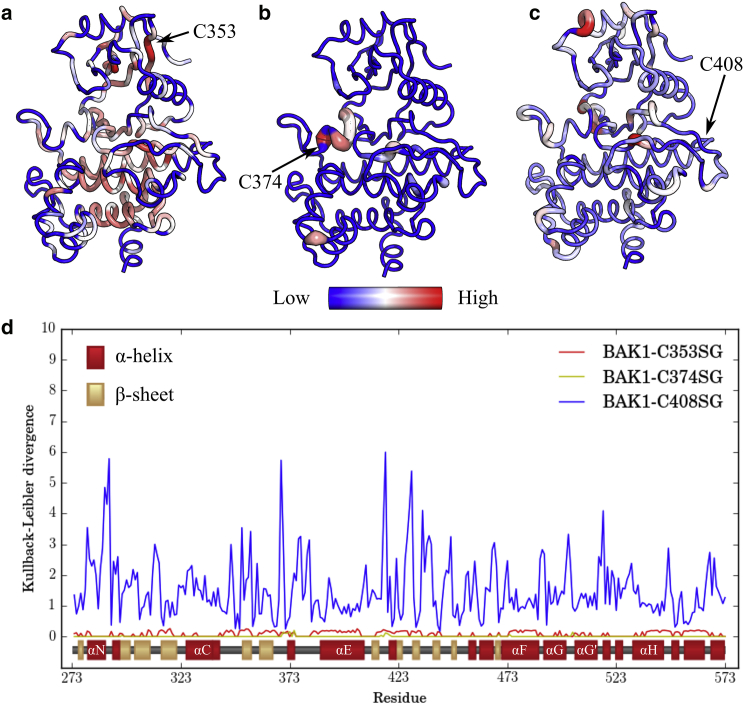
where the set of reference trajectories have been split up into blocks (), and is obtained by taking the average local KL divergence for residue i measured between a set of half the blocks (with an index b) of the reference trajectories and the set of remaining blocks (, the complement of the set of blocks b) over all combinations of blocks. We would like to note that our definition is a slight departure from the original as we have made a true KL divergence between the sets of reference ensemble blocks (compare Eqs. 3 and 4 with Eq. 26 in (43)). We can then obtain the probability, p, of obtaining a value greater than , calculated between ensembles, from the values calculated within the reference ensemble, and reject the null hypothesis if for some choice of (in this study, we used ). If we are unable to reject the null hypothesis, we set , where is the corrected KL divergence for residue i. Otherwise, we reject the null hypothesis and set . Additionally, we weighed each frame by the equilibrium MSM population of the corresponding state in creating the one-dimensional histograms, to reduce bias introduced by initiating trajectories from non-Boltzmann distributions. We have implemented the local KL divergence in Python and validated the code on a toy model (code freely available at https://github.com/ShuklaGroup/kl_divergence). To visualize population shifts in the dihedral angles of BAK1 with glutathionylation, we used the B-factor putty function in Open Source PyMOL 1.83 (44) to show the local KL divergence for each residue (Fig. 3, a–c).
Figure 3.
Allosteric effects of S-glutathionylation on the BAK1 ensemble. The local KL divergence calculated for ϕ, ψ, and dihedral angles in BAK1 S-glutathionylated on (a) C353, (b) C374, and (c) C408, all with respect to the nonglutathionylated BAK1 ensemble. (a–c) KL divergence is shown in the PyMOL B-factor putty representation, where the color shows KL divergence on separate scales for each simulation set from zero (blue) to the highest KL divergence value in the set (red), to highlight differences in local KL divergence between residues within each ensemble. Because the color scales are separate, (a–c) should not be compared with one another. (d) KL divergence values by residue with secondary structure from the crystal structure shown (30, 67). To see this figure in color, go online.
pKa calculations
We used PropKa 3.1 (45, 46) to predict the pKa of cysteine sulfur atoms in structures from simulation of BAK1 with no glutathionylation. For each BAK1 MSM state, we randomly chose 10 structures and calculated the pKa for the three solvent-exposed cysteine residues (C353, C374, and C408) from each structure. The overall cysteine pKa values were calculated by taking the average pKa within each state, multiplying by the MSM equilibrium populations, and then creating kernel density estimates using the Scipy Python package (47).
Bioinformatics analysis
We created a multiple sequence alignment of the 940 known A. thaliana kinases (48) using Clustal Omega (49). Using only the section of the alignment corresponding to the BAK1 kinase domain, we calculated frequencies of cysteine residues at each alignment position to gain a qualitative insight into how general the mechanisms of allostery through S-glutathionylation, investigated in BAK1, are to the entire kinome. We expect that if the cysteine positions in BAK1 are shared with another kinase, it is more likely that the effects of S-glutathionylation on those cysteine residues will also be shared than if the cysteine positions are disparate.
Results
S-glutathionylation of C408 alters the BAK1 conformational ensemble
To assess the global effects of glutathionylation on the conformational ensemble of BAK1, we estimated discretized probability distributions for the ϕ, ψ, and dihedral angles of all residues for BAK1-SH and singly glutathionylated forms, and calculated the local KL divergence between each of the glutathionylated BAK1 ensembles and the nonglutathionylated ensemble. We chose the ϕ, ψ, and dihedral angles to allow comparison between residues with different-sized side chains and still capture some side-chain motion, although we note that glycine and alanine lack an angle. A high KL divergence for a given residue is an indication that glutathionylation has significantly altered the effective residue potential energy landscape, whereas a low or vanishing value indicates that glutathionylation has had little or no effect on the residue.
Considering relative differences in KL divergences between residues within the same ensemble, BAK1-C353SG and BAK1-C374SG are clearly perturbed most near the site of glutathionylation (Fig. 3, a and b). In this intraensemble view, distant allosteric effects are also visible for both glutathionylation sites, although the KL divergence values are generally an order of magnitude smaller than those for BAK1-C408SG (Fig. 3 d). Interestingly, the KL divergence at C408 for BAK1-C408SG is fairly small relative to most other residues in the ensemble, indicating the role of C408 as a glutathionylation site with primarily long-range effects. The residues most perturbed by S-glutathionylation of C408 are near the N-terminus of the kinase domain, at the back of the ATP-binding pocket (with the opening of the ATP-binding pocket facing the viewer, as in Fig. 3, a–c), and in the catalytic loop (Fig. 3 c). However, BAK1-C408SG KL divergences from the unglutathionylated ensemble were statistically significant for each residue, indicating a global effect of C408 glutathionylation on the structure of BAK1. These results are supported by the large change in residue fluctuations from BAK1-SH to BAK1-C408SG compared with the changes on C353 and C374 glutathionylation (Fig. S5).
S-glutathionylation of C408 promotes an inactive conformational state
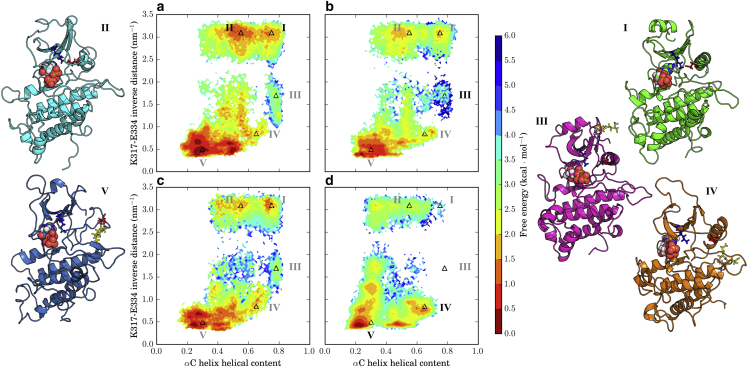
Our simulations were conducted with bound ATP and Mg2+, precluding a DFG flip, and a fully phosphorylated activation loop; consequently, the activation-relevant collective variables most likely to shift to values corresponding to an inactive kinase are the positioning and folding of the αC helix and the presence of the K-E salt bridge. To look for changes in these structural features, we estimated two-dimensional free-energy landscapes over the space of the α-helical content of the αC helix region and the inverse of the distance between the δ-carbon of E334 and the side-chain nitrogen of K317. Inverse distance was chosen to better resolve small interatomic distances. To reduce bias introduced by initiating simulations from a non-Boltzmann distribution, MSMs were constructed for each system and the free-energy landscapes were weighted by state equilibrium populations. As shown in previous work (21), BAK1-SH exhibits a large degree of structural heterogeneity in these coordinates, with a stable active-like state, (Fig. 4 a, region I), a state with a formed αC helix but a broken K-E salt bridge (Fig. 4 a, region III), and a number of metastable regions displaying high levels of αC helix disorder (Fig. 4 a, regions II, IV, and V).
Figure 4.
Effects of S-glutathionylation on prevalence of the active-like state in BAK1. MSM-weighted free-energy plots projected onto the αC helix helical content and the inverse K317-E334 distance from simulations of (a) nonglutathionylated BAK1, and BAK1 S-glutathionylated on (b) C353, (c) C374, and (d) C408. Points on the plane with representative structures shown are demarcated by triangles labeled in black, and correspond to the following regions: I) BAK1-SH, active-like state; II) BAK1-SH, formed K-E salt bridge and unfolded αC helix; III) BAK1-C353SG, slightly broken K-E salt bridge and formed αC helix; IV) BAK1-C408SG, broken K-E salt bridge and partially unfolded αC helix; and V) BAK1-C408SG, broken K-E salt bridge and unfolded αC helix. To see this figure in color, go online.
The BAK1-C353SG free-energy landscape retains most features of the BAK1-SH landscape, with the notable difference that S-glutathionylation of C353 increases the free energy of the active-like state by ∼1 kcal/mol−1 (Fig. 4 b, region I), although this is unlikely to represent a significant change. The BAK1-C374SG free-energy surface is largely unchanged from the BAK1-SH surface, although it displays a reduced range of distances between K317 and E334 when the αC helix is folded (Fig. 4, b and c, region III).
In contrast, there are several clear changes between the BAK1-C408SG and BAK1-SH free-energy landscapes, most notably the near absence of both the active-like state (Fig. 4 d, region I) and the state characterized by a slightly broken K-E salt bridge (Fig. 4 d, region III) found in the BAK1-SH free-energy surface. Instead, an unfolded αC helix and a broken K-E salt bridge are favored (Fig. 4 d, region V). An additional metastable state is greatly stabilized in BAK1-C408SG (Fig. 4 d, region IV) in regions with moderate αC helix helical content (∼0.65) and moderate K317-E334 inverse distances (∼0.8 nm−1). The only viable pathway for breaking of the K-E salt bridge is through a highly unfolded αC helix (left of region II in Fig. 4 d), suggesting that states with both a moderately folded αC helix and a broken K-E salt bridge occur through unfolding and refolding of the αC helix from the active-like, state rather than a simple swinging motion.
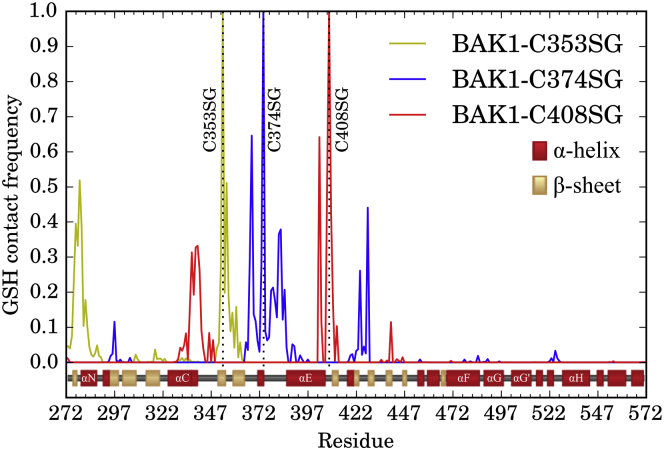
To investigate the mechanism of the structural disruption caused by C408 glutathionylation, we calculated the frequencies for each simulation set at which any heavy atom in GSH came within 4 Å of a heavy atom of each BAK1 residue (Fig. 5). Although spatial proximity between two residues is not necessarily caused by strong, direct physical interaction between them, we use it here as a simple proxy for interaction. C353SG interacts exclusively with the αN helix and β-sheets of the N-lobe, primarily at residues 273–290 and 350–365. C374SG interacts with residues around the ATP-binding pocket at residues 366–386 and 423–429, as well as R297 in the glycine-rich loop to a lesser extent. C408SG interacts most strongly with its flanking residues near the end of the αE helix around residues 405–410, the αC helix at residues 328–347, and with L440 in the activation loop to a lesser extent. It is worth noting that GSH at all three sites largely do not directly interact with both the catalytic loop (residues 416–422), containing the residues directly involved in catalysis, and the activation loop (residues 433–459), the regulatory sequence in kinases that is usually responsible for activation by phosphorylation (20).
Figure 5.
Glutathione interaction partners. Normalized contact frequencies between GSH heavy atoms and heavy atoms of all BAK1 residues with a cutoff distance of 4 Å. More explicitly, , where f denotes the normalized frequency over all trajectory frames, d() is the Euclidean distance between the two atoms with positions and , G is the set of all heavy glutathione atoms, and R is the set of all heavy atoms in a given BAK1 residue. In each simulation set, the cysteine forming a disulfide bond with GSH has a contact frequency of 1.0 and is marked with a dotted vertical line. Secondary structure from the crystal structure is shown just below the frequencies (30, 67). To see this figure in color, go online.
Cysteine thiol pKa predictions suggest low reactivity in isolation
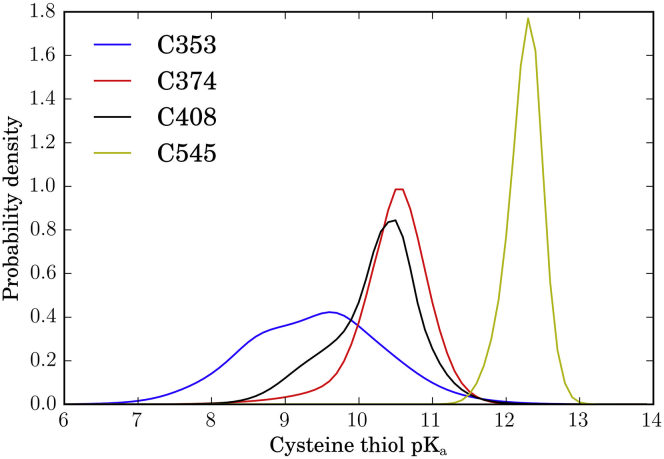
Although our simulations suggest that glutathionylation of C408 has strong allosteric effects on BAK1, it remains unclear which cysteine residues are likely to be reactive. To address this question, we estimated pKa values for randomly selected structures in each MSM state using PropKa 1.3 (45, 46), and created a pKa distribution estimate by way of a MSM population-weighted sum of state distributions (Fig. 6). We took this approach due to the fact that distinct local environments surrounding ionizable residues are known to have a strong effect on side-chain pKa values (50, 51, 52).
Figure 6.
Predicted distributions of BAK1 cysteine thiol pKas. We calculated the pKa of each BAK1 cysteine thiol using PropKa 3.1 (45, 46), for 10 randomly chosen structures for each MSM state from the nonglutathionylated simulations of BAK1. We created kernel density estimates (KDEs) using the Scipy package (47) for each state, with the pKa values calculated for the corresponding 10 structures. The final plots are sums of the KDEs of each state weighted by MSM equilibrium populations. Estimated means of the MSM-weighted KDEs are as follows: C353, 9.41; C374, 10.47; C408, 10.23; and C545, 12.25. To see this figure in color, go online.
All estimated mean pKa values are well above values considered reactive at physiological pH, with C353 predicted to be the most reactive (pKa 9.41), followed by C408 (pKa 10.23) and C374 (pKa 10.47) (Fig. 6). As expected, due to the fact that it is buried in the hydrophobic core of the kinase, C545 is predicted to be almost exclusively protonated (pKa 12.25). Although all other cysteine residues have sharply peaked pKa distributions, C353 has a broad, nearly bimodal distribution, suggesting a highly variable local environment (Fig. 6).
BAK1 C353 and C545 are well conserved in the A. thaliana kinome
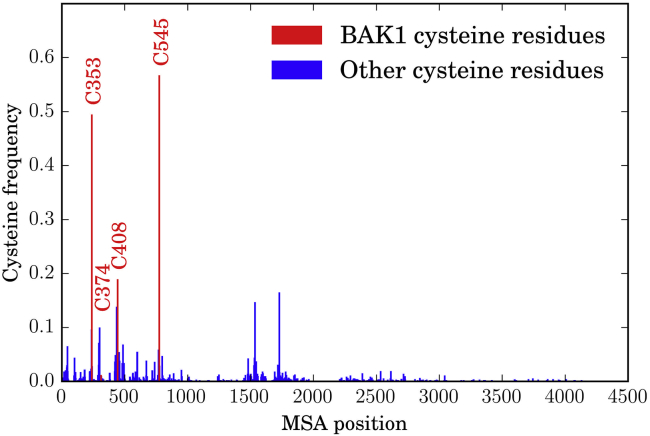
Using a multiple sequence alignment of all known A. thaliana kinase sequences, we calculated the frequency at which cysteine appears at each alignment position. Cysteine is well conserved at the alignment positions of BAK1 C353 and C545 (frequencies of 0.49 and 0.57, respectively), whereas C408 is less conserved (frequency of 0.19), and C374 is poorly conserved (frequency of 0.01); although a nearby alignment position which could be a functional equivalent in other kinases has a cysteine frequency of 0.10 (Fig. 7). Two other cysteine frequency peaks (frequencies of 0.15 and 0.16) further to the C-terminal end of the alignment are beyond the C-terminus of BAK1 in the alignment.
Figure 7.
Distribution of cysteine residues across the A. thaliana kinome. Using the 940 A. thaliana kinases identified in (48), we created a multiple sequence alignment using Clustal Omega (49). Using the region of the alignment corresponding to the BAK1 kinase domain, we calculated the frequencies of cysteine at each alignment position. All four BAK1 cysteine residues are in regions of high cysteine frequency, most notably C353 and C545. However, BAK1 C374 and C408 have nearby alignment positions with high frequency, likely corresponding to comparable positions in the contributing kinases. To see this figure in color, go online.
Discussion
We have demonstrated the dramatic effects that S-glutathionylation can have on the structure of a kinase, providing a plausible explanation for the experimentally observed effects of glutathionylation on the activity of BAK1 (17). Furthermore, although S-glutathionylation affects the activity of numerous other protein kinases (9, 10, 11, 12, 13, 14, 15, 16) (Fig. 2), to our knowledge, this is the first study addressing the molecular mechanisms of protein kinase activity modulation through glutathionylation. Our findings provide evidence that S-glutathionylation can allosterically alter the activity of protein kinases.
Our simulations suggest that glutathionylation of BAK1 C408 may be responsible for the concentration-dependent decrease in activity in the presence of GSSG and GRXC2 (17). However, we only address the effects of glutathionylation on the dynamics of the isolated kinase domain, and cannot exclude other consequences of glutathionylation relating to interactions with other proteins or other processes important for BAK1 activity not considered here. Our simulations are consistent with experiments supporting C408 and C353 as the most likely sites for GRXC2-catalyzed S-glutathionylation (17). Global changes in residue conformations as measured by the local KL divergence between each glutathionylated BAK1 simulation set, and the BAK1-SH simulations are generally an order of magnitude larger for BAK1-C408SG than BAK1-C353SG or BAK1-C374SG (Fig. 3 d). Surprisingly, most residues in BAK1-C408SG have a high KL divergence with respect to BAK1-SH, including distant residues in the C-lobe even though GSH only directly interacts with its flanking residues and the αC helix in the N-lobe.
Differences in free-energy surfaces calculated over a measure of αC helix helicity and the inverse distance between K317 and E334 for the four BAK1 constructs were consistent with calculated KL divergence values. The BAK1-C353SG and BAK1-C374SG free-energy surfaces are similar to that for BAK1-SH, although subtle differences, including an increase in the free energy of the active-like state in BAK1-C353SG, are noticeable. The level of disorder in the αC helices of BAK1-C353SG and BAK1-C374SG is largely unchanged from BAK1-SH. In contrast, the BAK1-C408SG free-energy landscape differs from BAK1-SH in several key ways. C408SG interacts directly with the αC helix, a region known to be involved in regulation of protein kinase activity (20, 53, 54, 55, 56, 57), causing the αC helix to more strongly favor both unfolded and folded but distally swung conformations. The distally-swung conformation is similar to a well-defined, metastable, inactive state found in human Src kinase, where the αC helix also swings away from the active site (53, 58, 59). In BAK1, this likely occurs due to direct interactions with GSH stabilizing the outward-swung conformation. Unlike the proposed mechanisms of formation of a Src-like inactive state in other kinases (54, 55, 56), the αC helix in BAK1-C408SG must first unfold, followed by breakage of the K-E salt bridge and refolding of the αC helix. Furthermore, the active-like state (Fig. 4, a–e, region I) is nearly eliminated in BAK1-C408SG simulations.
It should be noted that sampling of the BAK1-C408SG system was 1.5 times longer than any of the other systems, on top of more extensive AMD used to generate initial structures for unbiased simulation (Table S2). Thus, it is possible that the differences between BAK1-C408SG and the other systems are (unintentionally) by construction, due to the differences in sampling. However, this does not appear to be the case for several reasons. First, even with the most extensive sampling, BAK1-C408SG exhibits no breaking of the K-E salt bridge coincident with a folded αC helix, a transition well sampled by all other systems (Fig. 4). Second, BAK1-C408SG samples the active-like state far less than the other systems, which seems unlikely to be due to oversampling BAK1-C408SG with respect to the other systems. Finally, there are no high free-energy barriers impeding access to new states appearing in BAK1-C408SG, and the fringes of the areas spanned by these states are visited in other systems. If the differences in the free-energy landscapes of BAK1-C408SG and the other systems were largely due to differences in sampling, we would expect BAK1-C408SG to have explored new areas not accessible to more poorly sampled systems.
Collectively, these results suggest that modification of C408 by S-glutathionylation allosterically deactivates BAK1 through interaction with the αC helix. The αC helix is in generally believed to be a focal point for protein kinase regulation, physically linking numerous regions important for kinase function (57, 58, 59). This central positioning makes the αC helix a prime target for allosteric modulators, and possibly explains the global effects of C408 glutathionylation shown in KL divergence measurements. There is evidence for the specific importance of the αC helix in regulation of BAK1 (21), which is further supported by the apparent role of the αC helix in response to S-glutathionylation. Although the predicted C408 pKa (10.23) indicates that it is overwhelmingly protonated and therefore marginally reactive, C408 is glutathionylated in vitro (17); this inconsistency is at least partially explained by the fact that PropKa 3.1 systematically overestimates cysteine pKa values (60). Interestingly, C353 is moderately conserved in the A. thaliana kinome, present in ∼50% of the 940 known sequences, whereas C408 is present in ∼20% (Fig. 7). It is possible that the conservation of C353 is due to its function as a glutathionylation site, working to regulate kinase activity in some way not captured by our simulations, although there is currently no evidence to support this claim. The comparatively low level of conservation in C408 indicates that it is not universally essential for the function of A. thaliana protein kinases. However, the functional importance of C408 for BAK1 is supported by the decrease in catalytic activity caused by mutation of C408 to tyrosine (61). Finally, this study presents, to our knowledge, one of the first investigations of plant kinases using large-scale atomistic simulations to obtain mechanistic information at the molecular level required for engineering of plant behavior (62).
Author Contributions
K.W.B., S.C.H., and D.S. conceived the project. A.S.M. and D.S. designed research. A.S.M. performed research, analysis, and code implementation. A.S.M. wrote the manuscript with input from K.W.B., S.C.H., and D.S. A.S.M., K.W.B., S.C.H., and D.S. edited the manuscript.
Acknowledgments
We thank the Blue Waters sustained-petascale computing project for providing computing time for this study.
The Blue Waters sustained-petascale computing project is supported by the National Science Foundation (awards OCI-0725070 and ACI-1238993) and the state of Illinois.
Editor: Elizabeth Komives.
Footnotes
Five figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31074-3.
Supporting Material
References
- 1.Beltrao P., Bork P., van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 2013;9:714. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussinov R., Tsai C.J., Radivojac P. Allosteric post-translational modification codes. Trends Biochem. Sci. 2012;37:447–455. doi: 10.1016/j.tibs.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Kholodenko B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalle-Donne I., Rossi R., Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Grek C.L., Zhang J., Tew K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013;288:26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouhier N., Lemaire S.D., Jacquot J.P. The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 2008;59:143–166. doi: 10.1146/annurev.arplant.59.032607.092811. [DOI] [PubMed] [Google Scholar]
- 7.Noctor G., Mhamdi A., Foyer C.H. Glutathione in plants: an integrated overview. Plant Cell Environ. 2012;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- 8.Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward N.E., Stewart J.R., O’Brian C.A. Oxidant-induced S-glutathiolation inactivates protein kinase C-α (PKC-α): a potential mechanism of PKC isozyme regulation. Biochemistry. 2000;39:10319–10329. doi: 10.1021/bi000781g. [DOI] [PubMed] [Google Scholar]
- 10.Humphries K.M., Juliano C., Taylor S.S. Regulation of cAMP-dependent protein kinase activity by glutathionylation. J. Biol. Chem. 2002;277:43505–43511. doi: 10.1074/jbc.M207088200. [DOI] [PubMed] [Google Scholar]
- 11.Cross J.V., Templeton D.J. Oxidative stress inhibits MEKK1 by site-specific glutathionylation in the ATP-binding domain. Biochem. J. 2004;381:675–683. doi: 10.1042/BJ20040591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynaert N.L., van der Vliet A., Janssen-Heininger Y.M. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase β. Proc. Natl. Acad. Sci. USA. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kambe T., Song T., Watanabe Y. Inactivation of Ca2+/calmodulin-dependent protein kinase I by S-glutathionylation of the active-site cysteine residue. FEBS Lett. 2010;584:2478–2484. doi: 10.1016/j.febslet.2010.04.059. [DOI] [PubMed] [Google Scholar]
- 14.Zmijewski J.W., Banerjee S., Abraham E. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J. Biol. Chem. 2010;285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaus A., Zorman S., Schlattner U. Glutathione S-transferases interact with AMP-activated protein kinase: evidence for S-glutathionylation and activation in vitro. PLoS One. 2013;8:e62497. doi: 10.1371/journal.pone.0062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takata T., Tsuchiya Y., Watanabe Y. 90-kDa ribosomal S6 kinase 1 is inhibited by S-glutathionylation of its active-site cysteine residue during oxidative stress. FEBS Lett. 2013;587:1681–1686. doi: 10.1016/j.febslet.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Bender K.W., Wang X., Huber S.C. Glutaredoxin AtGRXC2 catalyses inhibitory glutathionylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem. J. 2015;467:399–413. doi: 10.1042/BJ20141403. [DOI] [PubMed] [Google Scholar]
- 18.Bojar D., Martinez J., Hothorn M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014;78:31–43. doi: 10.1111/tpj.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohmann U., Lau K., Hothorn M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 2017;68:109–137. doi: 10.1146/annurev-arplant-042916-040957. [DOI] [PubMed] [Google Scholar]
- 20.Taylor S.S., Kornev A.P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffett A.S., Bender K.W., Shukla D. Molecular dynamics simulations reveal the conformational dynamics of Arabidopsis thaliana BRI1 and BAK1 receptor-like kinases. J. Biol. Chem. 2017;292:12643–12652. doi: 10.1074/jbc.M117.792762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan Y., Eastwood M.P., Shaw D.E. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 23.Belkhadir Y., Jaillais Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015;206:522–540. doi: 10.1111/nph.13269. [DOI] [PubMed] [Google Scholar]
- 24.Case, D. A., J. T. Berryman, …, P. A. Kollman. 2015. AMBER 2015. http://ambermd.org.
- 25.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 26.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 29.Crowley M.F., Williamson M.J., Walker R.C. CHAMBER: comprehensive support for CHARMM force fields within the AMBER software. Int. J. Quantum Chem. 2009;109:3767–3772. [Google Scholar]
- 30.Cheng W., Munkvold K.R., Chai J. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host Microbe. 2011;10:616–626. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan L., Ma Y., Lou Z. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 2012;22:1304–1308. doi: 10.1038/cr.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou, Z. Y., L. M. Yan, Y. Y. Ma. 2012. 3ULZ: Crystal structure of apo BAK1. http://www.rcsb.org/pdb/explore.do?structureId=3ULZ.
- 33.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 34.Salomon-Ferrer R., Götz A.W., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 35.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 36.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 37.Hamelberg D., Mongan J., McCammon J.A. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 38.Wereszczynski J., McCammon J.A. Accelerated molecular dynamics in computational drug design. In: Baron R., editor. Computational Drug Discovery and Design. Springer; New York: 2012. pp. 515–524. [DOI] [PubMed] [Google Scholar]
- 39.McGibbon R.T., Beauchamp K.A., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beauchamp K.A., Bowman G.R., Pande V.S. MSMBuilder2: modeling conformational dynamics on the picosecond to millisecond scale. J. Chem. Theory Comput. 2011;7:3412–3419. doi: 10.1021/ct200463m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGibbon R.T., Pande V.S. Variational cross-validation of slow dynamical modes in molecular kinetics. J. Chem. Phys. 2015;142:124105. doi: 10.1063/1.4916292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClendon C.L., Hua L., Jacobson M.P. Comparing conformational ensembles using the Kullback-Leibler divergence expansion. J. Chem. Theory Comput. 2012;8:2115–2126. doi: 10.1021/ct300008d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.PyMOL . Schrödinger; LLC, New York: 2015. The PyMOL Molecular Graphics System. Version 1.8. [Google Scholar]
- 45.Søndergaard C.R., Olsson M.H., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 46.Olsson M.H., Søndergaard C.R., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 47.Jones E., Oliphant T., Peterson P. SciPy: open source scientific tools for Python. 2001. http://www.scipy.org/
- 48.Zulawski M., Schulze G., Schulze W.X. The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC Genomics. 2014;15:548. doi: 10.1186/1471-2164-15-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F., Wilm A., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanford C., Kirkwood J.G. Theory of protein titration curves. I. General equations for impenetrable spheres. J. Am. Chem. Soc. 1957;79:5333–5339. [Google Scholar]
- 51.Mehler E.L., Fuxreiter M., Garcia-Moreno E.B. The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins. 2002;48:283–292. doi: 10.1002/prot.10153. [DOI] [PubMed] [Google Scholar]
- 52.Jensen J.H.H., Li H., Molina P.A. Prediction and rationalization of protein pKa values using QM and QM/MM methods. J. Phys. Chem. A. 2005;109:6634–6643. doi: 10.1021/jp051922x. [DOI] [PubMed] [Google Scholar]
- 53.Jura N., Zhang X., Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H., Zhao R., Post C.B. αC helix as a switch in the conformational transition of Src/CDK-like kinase domains. J. Phys. Chem. B. 2012;116:4465–4475. doi: 10.1021/jp301628r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmieri L., Rastelli G. αC helix displacement as a general approach for allosteric modulation of protein kinases. Drug Discov. Today. 2013;18:407–414. doi: 10.1016/j.drudis.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Endres N.F., Barros T., Kuriyan J. Emerging concepts in the regulation of the EGF receptor and other receptor tyrosine kinases. Trends Biochem. Sci. 2014;39:437–446. doi: 10.1016/j.tibs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Taylor S.S., Shaw A.S., Kornev A.P. Integration of signaling in the kinome: Architecture and regulation of the αC Helix. Biochim. Biophys. Acta. 2015;1854(10 Pt B):1567–1574. doi: 10.1016/j.bbapap.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng Y., Shukla D., Roux B. Transition path theory analysis of c-Src kinase activation. Proc. Natl. Acad. Sci. USA. 2016;113:9193–9198. doi: 10.1073/pnas.1602790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shukla D., Meng Y., Pande V.S. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat. Commun. 2014;5:3397. doi: 10.1038/ncomms4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Awoonor-Williams E., Rowley C.N. Evaluation of methods for the calculation of the pKa of cysteine residues in proteins. J. Chem. Theory Comput. 2016;12:4662–4673. doi: 10.1021/acs.jctc.6b00631. [DOI] [PubMed] [Google Scholar]
- 61.Schwessinger B., Roux M., Zipfel C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall-Colon A., Long S.P., Zhu X.-G. Crops in silico: generating virtual crops using an integrative and multi-scale modeling platform. Front. Plant Sci. 2017;8:786. doi: 10.3389/fpls.2017.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Biasini M., Bienert S., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akamine P., Madhusudan, Taylor S.S. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J. Mol. Biol. 2003;327:159–171. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 65.Goldberg J., Nairn A.C., Kuriyan J. Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell. 1996;84:875–887. doi: 10.1016/s0092-8674(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 66.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 67.Berman H.M., Westbrook J., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.