Abstract
Small activating RNAs (saRNAs) are short double-stranded oligonucleotides that selectively increase gene transcription. Here, we describe the development of an saRNA that upregulates the transcription factor CCATT/enhancer binding protein alpha (CEBPA), investigate its mode of action, and describe its development into a clinical candidate. A bioinformatically directed nucleotide walk around the CEBPA gene identified an saRNA sequence that upregulates CEBPA mRNA 2.5-fold in human hepatocellular carcinoma cells. A nuclear run-on assay confirmed that this upregulation is a transcriptionally driven process. Mechanistic experiments demonstrate that Argonaute-2 (Ago2) is required for saRNA activity, with the guide strand of the saRNA shown to be associated with Ago2 and localized at the CEBPA genomic locus using RNA chromatin immunoprecipitation (ChIP) assays. The data support a sequence-specific on-target saRNA activity that leads to enhanced CEBPA mRNA transcription. Chemical modifications were introduced in the saRNA duplex to prevent activation of the innate immunity. This modified saRNA retains activation of CEBPA mRNA and downstream targets and inhibits growth of liver cancer cell lines in vitro. This novel drug has been encapsulated in a liposomal formulation for liver delivery, is currently in a phase I clinical trial for patients with liver cancer, and represents the first human study of an saRNA therapeutic.
Keywords: saRNA, RNA activation, small RNA, liver cancer, hepatocellular carcinoma, HCC, CEBPA, C/EBP
Voutila et al. report the development and mechanism of action of the first small activating RNA drug to reach human clinical trials. This novel therapeutic, MTL-CEBPA, upregulates the expression of C/EBPα for the treatment of cirrhotic liver cancer.
Introduction
RNA activation (RNAa) was first described in 2006, where it was reported that short double-stranded RNAs targeted to the promoter region of a gene can activate its transcription.1 These small activating RNAs (saRNAs) have since been shown to activate a wide variety of genes in several mammalian species.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 Although similar to RNA interference (RNAi) in that it is mediated by short RNAs and requires Argonaute-2 (Ago2), RNAa is distinct in its kinetics and ability to selectively induce transcriptional elongation of a target gene in the nucleus.12 The further molecular mechanisms that distinguish RNAa from RNAi continue to be investigated, such as the identification of CTR9 and RHA as necessary cofactors for saRNA activity and the role of RNA polymerase II.13 This technology provides a new research tool for selective gene activation, but also a novel therapeutic approach for diseases in which endogenous gene expression has been downregulated through mutation or transcriptional/translational repression.
Hepatocellular carcinoma (HCC) is most commonly caused by chronic liver damage due to cirrhosis from hepatitis virus infection, alcohol abuse, or non-alcoholic fatty liver disease.14, 15 Although surgical resection is the preferred treatment for HCC, only 10%–25% of tumors are resectable, with a 5-year recurrence rate of up to 80%.16 The standard of care treatment for advanced HCC is the multikinase inhibitor sorafenib, which has a median survival increase of just 2.8 months and a less than 5% response rate.17 There thus remains a critical unmet need for treatment of patients with HCC who are ineligible for tumor resection.
The CCAAT/enhancer-binding protein alpha (CEBPA) gene encodes C/EBP-α, a basic-leucine zipper class transcription factor that is critical for the differentiation and function of liver and adipose tissue as well as the myeloid lineage.18 Deletion of the CEBPA gene in the liver results in dysregulation of liver-specific transcription factors and impaired hepatocyte maturation.19 A rat model of HCC as well as a retrospective analysis of human HCC samples shows that C/EBP-α is downregulated in HCC and associated with poor survival.20, 21 This suggests that chronic liver disease leading to HCC may cause a dysregulation of the liver-specific transcriptional network, contributing to tumorigenesis or exacerbating the poor liver function seen in HCC.22 C/EBP-α has also been described as a tumor suppressor, leading to mitotic arrest through activation of p21 and repression of E2Fs and cyclin-dependent kinases (CDKs).23 Indeed, CEBPA knock-in mice show partial protection from HCC,24 showing that upregulation of C/EBP-α activity has a potential to not only improve liver function, but also limit HCC growth. Because impaired liver function is a common complication of HCC limiting the use of surgical resection,25 activation of the C/EBP-α pathway is an attractive therapeutic target for saRNA, with the potential to improve normal liver function while inhibiting tumor growth.
We previously designed a CEBPA saRNA that showed increased expression of hepatocyte-specific factors, such as albumin, hepatocyte nuclear factor (HNF)4α, and HNF1α, and inhibited tumor growth in a rat model of HCC.11 Here, we describe the development of this saRNA into a clinical candidate, and demonstrate that its activity is an on-target mechanism consistent with RNAa. The final saRNA, CEBPA-51, has been formulated in a NOV340 SMARTICLE (MTL-CEBPA) and is currently in phase I clinical trials for the improvement of liver function in patients with HCC.26
Results
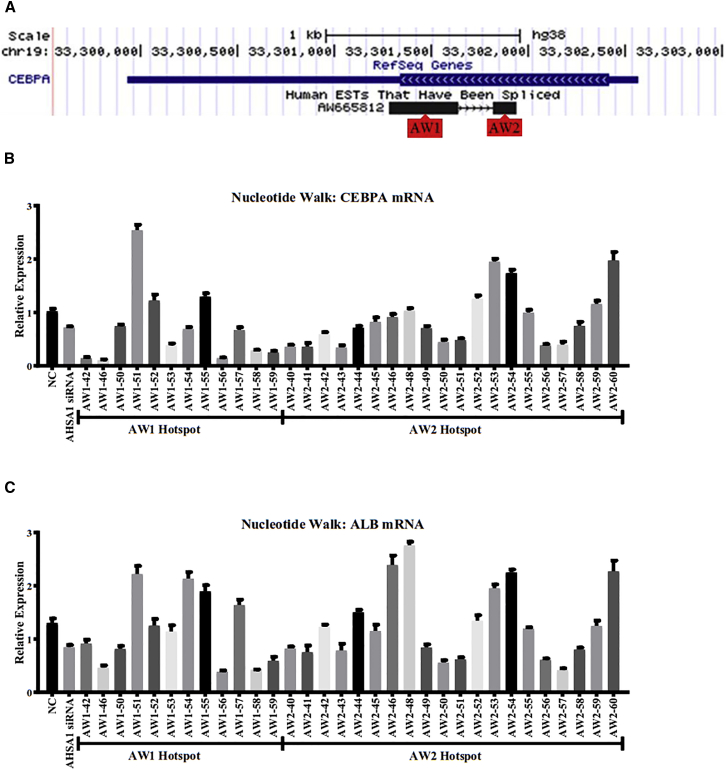
Our saRNA bioinformatics algorithm, described previously,7 identified several hotspots of putative saRNA activity at the CEBPA gene locus. Two of these hotspots were within the coding region of CEBPA where a noncoding RNA (GenBank: AW665812) overlaps the gene in the antisense orientation relative to CEBPA mRNA (Figure 1A). We synthesized a series of saRNA oligonucleotides to perform a nucleotide walk across these two hotspots, called AW1 and AW2 (Table S1). These candidate CEBPA saRNAs were tested by transfection into the human HCC cell line HepG2, and their ability to upregulate CEBPA and C/EBP-α target gene albumin18 mRNA was measured (Figures 1B and 1C). This screen identified 4 sequences that upregulated both CEBPA and albumin mRNA >1.5-fold. We chose the sequence AW1-51 for further development as a clinical candidate because it had the highest CEBPA mRNA upregulation (2.5-fold) and was in the same hotspot as our previously published CEBPA saRNA. Transfection of increasing concentrations of AW1-51 showed a clear dose response of CEBPA mRNA upregulation, with an EC50 of 5.36 nM under the conditions tested in this assay (Figure S1A). Under the same conditions, a CEBPA small interfering RNA (siRNA) had an IC50 of 0.05 nM (Figure S1B).
Figure 1.
Nucleotide Walk on CEBPA saRNA Hotspots in HepG2 Cells
(A) Schematic showing location and orientation of CEBPA mRNA and antisense transcript GenBank: AW665812, with approximate locations of AW1 and AW2 hotspots. (B) Expression of CEBPA mRNA for each sequence transfected at 50 nM relative to mock transfected cells. (C) Expression of albumin mRNA for each sequence transfected at 50 nM relative to mock transfected cells. Error bars represent SEM.
To test whether upregulation of CEBPA mRNA by AW1-51 led to an increase in functional C/EBP protein, we used a C/EBP luciferase reporter assay. Transfection of AW1-51 in HepG2 cells caused a significant increase in luciferase activity (Figure 2A), indicating an increase in functional C/EBP activity.
Figure 2.
Mechanism of Action of AW1-51 saRNA in HepG2 Cells
(A) Relative luciferase activity representing C/EBP protein activity after transfection with 10 nM AW1-51 saRNA. (B) qPCR for CEBPA mRNA on nascent transcripts isolated from nuclear run-on after 10 nM AW1-51 saRNA transfection. (C) qPCR for CEBPA mRNA after transfection with 10 nM AW1-51 duplexes with a 5′ inverted abasic modification on the indicated strand. (D) qPCR for CEBPA mRNA after transfection with 10 nM AW1-51 saRNA with mutations in the seed region. (E) qPCR for CEBPA mRNA after transfection with 10 nM AW1-51 saRNA with mutations in the center of the duplex. (F) qPCR for GenBank: AW665812 RNA after transfection with 10 nM AW1-51. Statistical significance shown for the indicated condition compared to NC transfection: *p < 0.05; **p < 0.01; ***p < 0.001. Error bars represent SEM.
An increase in steady-state mRNA could be due to enhancing mRNA stability, whereas true saRNA activity requires activation of transcription of target gene mRNA. To determine if AW1-51 activates CEBPA transcription, we measured nascent CEBPA mRNA transcription in a nuclear run-on experiment. After transfection of AW1-51 in HepG2 cells, nascent CEBPA mRNA rose 3-fold compared to control transfected cells (Figure 2B), indicating activation of transcription.
To determine which strand of the AW1-51 duplex was responsible for saRNA activity, we used a 5′ inverted abasic modification on each strand individually to block strand loading into Ago2.27 As expected, a 5′ inverted abasic modification on both strands completely negated saRNA activity, but when only the sense strand (SS) was modified, CEBPA mRNA was upregulated 2.5-fold (Figure 2C). This indicates that the antisense strand (AS) is the guide strand loaded into Ago2. We then sought to test if this is a true on-target mechanism by introducing mutations to the seed region of the saRNA. Introducing a single mutation into position 3 or 4 of the seed region reduced saRNA activity to below statistical significance, whereas additional mutations caused a complete loss of activity (Figure 2D). A duplex composed of the scrambled AW1-51 sequence was also not active (Figure 2D).
Because the AW1-51 AS is the guide strand loaded into Ago2, we wanted to rule out target cleavage of either non-coding RNA (ncRNA) GenBank: AW665812 or other off-target RNAs being responsible for the saRNA activity. When three mutations were introduced to the center of the AW1-51 sequence to prevent target cleavage,28 there was no significant loss of saRNA activity measured by upregulation of CEBPA mRNA (Figure 2E). Further, strand-specific reverse transcription followed by qPCR with primers flanking the saRNA target site showed that GenBank: AW665812 RNA is upregulated rather than being downregulated by AW1-51 (Figure 2F), demonstrating that cleavage is not required for CEBPA upregulation.
To further develop AW1-51 as a clinical candidate saRNA, we tested different patterns of 2′-O-methyl base modifications to prevent immune stimulation (Figure S2A). We first tested to see if these modifications affected saRNA activity. As above, a 5′ SS inverted abasic on the SS sequence does not affect saRNA activity, and two different methylation patterns were well tolerated (Figure S2B). The lack of activity of modification pattern 3 suggests that modifications to the guide strand may not be well-tolerated. These two active modified AW1-51 saRNAs were tested for TLR activation by transfection into primary human PBMCs, and one of two patterns (modification pattern 2) showed negligible tumor necrosis factor alpha (TNF-α) and interferon α (IFNα) secretion in two donors (Figure S2C). This non-immunostimulatory-modified AW1-51 saRNA was named CEBPA-51.
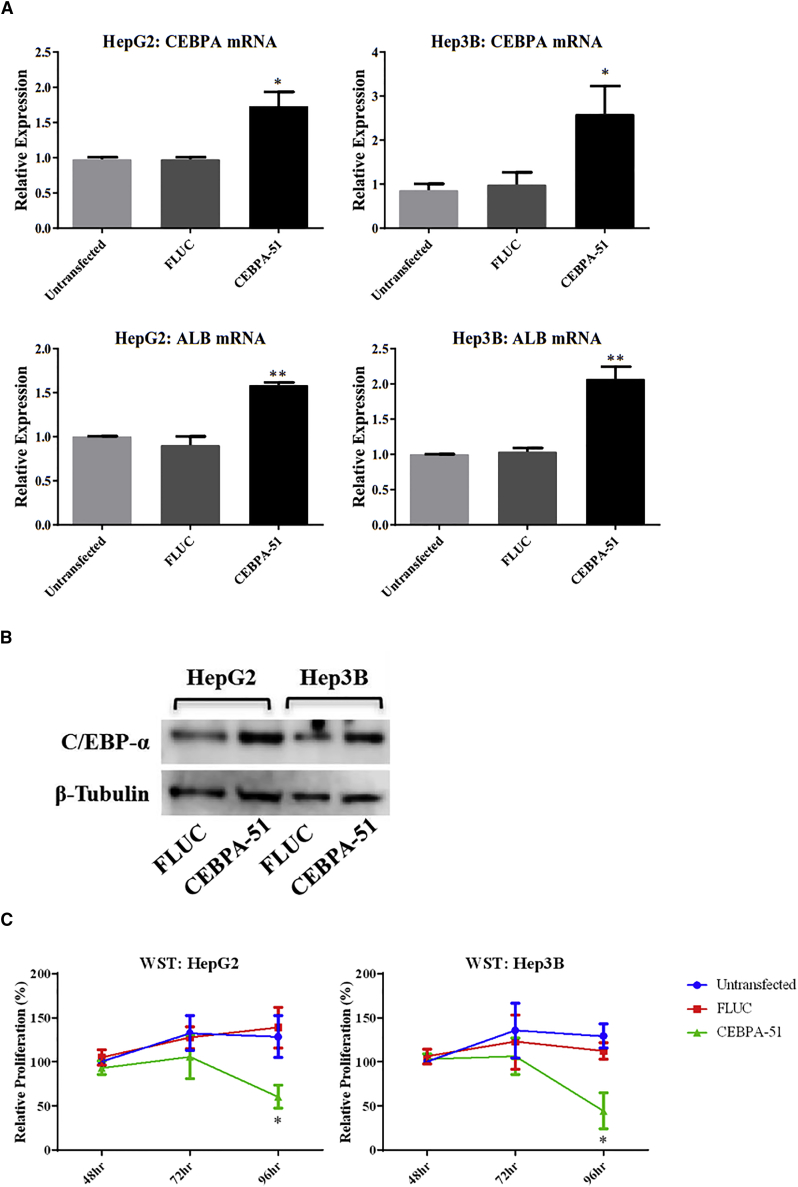
We next assessed the activity of CEBPA-51 in the HCC lines HepG2 and Hep3B. Transfection of CEBPA-51 causes 1.5- to 2.5-fold upregulation of CEBPA mRNA (Figure 3A) and a corresponding increase in C/EBP-α protein (Figure 3B) by western blot, as well as a 1.5- to 2-fold upregulation of C/EBP-α downstream target albumin mRNA in both cell lines. The effect of CEBPA-51 on HCC cell proliferation was tested by a water-soluble tetrazolium salt (WST-1) assay, showing a significant reduction in proliferation in both cell lines over a 96-hr time course (Figure 3C). Because CEBPA-51 has a canonical siRNA duplex structure, we used a bioinformatic analysis to determine if there are any predicted siRNA-like off-target effects from each strand of the CEBPA-51 duplex in the human, mouse, rat, and rhesus and cynomolgus monkey transcriptomes, as well as miRNA-like off-target effects from seed region base pairing. There were no transcripts with 0 mismatches or 1 mismatch to the AS in any of the analyzed species, and a single transcript with 1 mismatch to the SS in humans (Figure S3A). Of the 12 transcripts with 1 mismatch or 2 mismatches to either strand, we tested 6 that had known functions in liver or cancer biology for siRNA-like off-target effects. CEBPA-51 caused no significant reduction in any of these genes (Figure S3B), indicating that there is likely no off-target regulation responsible for CEBPA-51 activity consistent with the target cleavage mutation data above. The CEBPA-51 target sequence is conserved in human, non-human primates, and rodents (Figure S4A). We tested and confirmed activation of CEBPA mRNA by CEBPA-51 in CYNOM-K1 cynomolgus monkey fibroblasts and mouse embryonic fibroblasts (MEFs) (Figure S4B).
Figure 3.
Activity of CEBPA-51 in the HCC Lines HepG2 and Hep3B
(A) qPCR for CEBPA and ALB mRNA after transfection with 10 nM CEBPA-51. (B) Western blot for C/EBP-α after transfection with 10 nM CEBPA-51. (C) WST-1 assays over a 96-hr time course after transfection with 10 nM CEBPA-51. Statistical significance shown for CEBPA-51 compared to FLUC transfection: *p < 0.05; **p < 0.01. Error bars represent SEM.
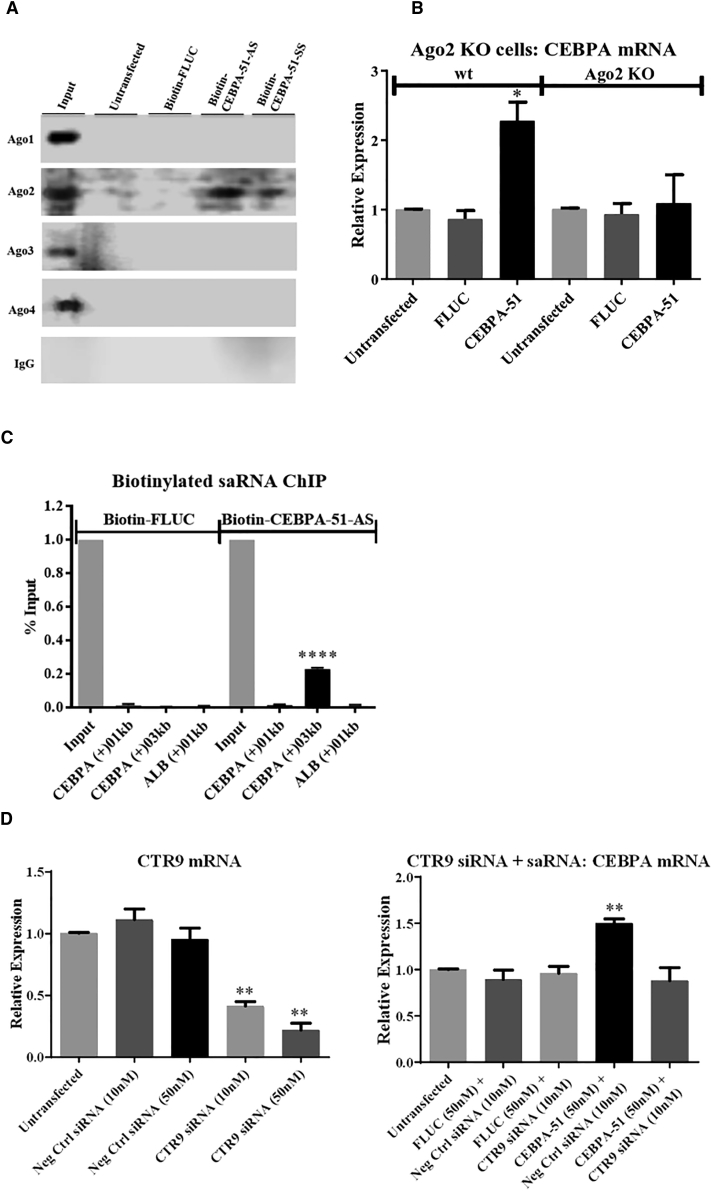
We used 3′ biotinylated SS or AS CEBPA-51 duplexes to assess the association of CEBPA-51 with Ago1–4. After transfection with 20 nM biotinylated CEBPA-51, cells were lysed and saRNA-protein complexes were purified on streptavidin beads. Subsequent western blotting showed an association of Ago2, but not Ago1, Ago3, and Ago4, to both the SS and AS strand of CEBPA-51, but not a biotinylated control oligo (Figure 4A). The importance of Ago2 for saRNA activity was confirmed by transfection of CEBPA-51 into Ago2 knockout mouse embryonic fibroblasts, where no CEBPA mRNA activation was seen. In contrast, in wild-type MEFs, a 2.3-fold activation was seen (Figure 4B). The Ago2 knockout MEFs are also negative for siRNA activity (Figure S5). We next used the biotinylated CEBPA-51 to isolate chromatin associated with CEBPA-51 after transfection. Subsequent qPCR showed a strong signal over background at the approximate genomic location of the CEBPA-51 sequence (+3 kb downstream of the transcription start site [TSS]), but not close to the TSS (Figure 4C), providing further evidence that CEBPA-51 activates CEBPA through an on-target Ago2-mediated mechanism localized to the CEBPA genomic locus. There was also no localization of CEBPA-51 at the albumin promoter (Figure 4C), and a control biotinylated oligonucleotide showed no association at the CEBPA or albumin promoters. Finally, we investigated whether activation of CEBPA mRNA by CEBPA-51 requires CTR9, a protein which has been recently shown to be part of the saRNA-induced transcriptional activation complex.13 Co-transfection of CEBPA-51 and CTR9 siRNA abolished saRNA activity, whereas co-transfection with a negative control oligo had no effect on activity (Figure 4D), providing further evidence of an saRNA mechanism.
Figure 4.
Mechanism of Action of CEBPA-51 in HepG2 Cells
(A) Western blot after co-immunoprecipitation of Ago1–4 and biotinylated CEBPA-51. (B) qPCR for CEBPA mRNA after transfection of 10 nM CEBPA-51 wild-type and Ago2 knockout MEFs. (C) qPCR at the CEBPA locus after chromatin immunoprecipitation with biotinylated CEBPA-51. The CEBPA-51 target site is approximately 3 kb downstream of the CEBPA TSS. (D) qPCR for CTR9 mRNA after transfection with CTR9 siRNA and qPCR for CEBPA mRNA after co-transfection of CTR9 siRNA and CEBPA-51. Statistical significance shown for the indicated condition compared to FLUC transfection: **p < 0.01. Error bars represent SEM.
Discussion
The field of oligonucleotide therapeutics has primarily centered around the approach of target knockdown and inhibition. The development of siRNA, antisense oligonucleotides, and microRNA mimics has provided valuable treatment options to downregulate the expression of genes involved in disease progression,29 but there remain few options for specific upregulation of gene expression in vivo without the delivery of long synthetic mRNAs or complicated gene expression vectors. We believe that saRNAs can provide a solution for diseases where upregulation of gene expression is therapeutically beneficial, and have described here the first saRNA therapeutic to reach the clinic, CEBPA-51.
We have shown how the CEBPA-51 sequence was determined through a nucleotide walk of bioinformatically identified hotspots at the CEBPA gene. The identified saRNA, AW1-51, shows a specific dose-dependent upregulation of CEBPA mRNA, leading to an increase in functional C/EBP protein and albumin, a downstream target. This upregulation of CEBPA mRNA is from transcription of nascent mRNA, not stabilization of existing mRNA. We have also shown that this upregulation is an on-target, Ago2-mediated mechanism. Ago2 knockout cells have no CEBPA saRNA nor siRNA activity, and biotinylated CEBPA-51 saRNAs interact with Ago2 and at the expected target site 3 kb downstream of the CEBPA TSS. Interestingly, the biotinylated FLUC negative control oligo showed no association with Ago2, despite having a canonical siRNA structure. Although designed to target firefly luciferase mRNA, this duplex has been found to have no activity in cells expressing firefly luciferase (data not shown). The lack of association with Ago proteins here may be due to inefficient Ago loading or having no mRNA target in the cells 72 hr after transfection. Biotinylated CEBPA-51 was not found localized to the albumin promoter, indicating that the upregulation of albumin seen is due to C/EBP-α, not a direct interaction of the saRNA. An investigation of possible siRNA- or miRNA-like off-target effects was negative. Mutations to the seed region of the guide strand lower or negate saRNA activity, providing more evidence of an on-target sequence-dependent Ago2-mediated mechanism. However, this mechanism does not require target cleavage because cleavage-impaired AW1-51 retains activity, and the ncRNA GenBank: AW665812, despite being perfectly complementary to the AS of the saRNA, is upregulated, not downregulated, after saRNA transfection. The upregulation of this ncRNA seen here may be a general consequence of increased transcriptional activity at the CEBPA locus or it may be a regulatory component of CEBPA expression. The role of GenBank: AW665812 in CEBPA transcription is unclear and is an area of active investigation. The passenger strand of AW1-51 is also perfectly complementary to CEBPA mRNA, meaning it is possible for this duplex to act as an siRNA. The absence of any CEBPA mRNA downregulation by AW1-51 is likely due to lower internal stability of the duplex at the 5′ guide end of duplex.30 The addition of a 5′ inverted abasic modification to the passenger strand of AW1-51 to block Ago2 loading27 increased CEBPA mRNA upregulation, suggesting that there is passenger strand loading from the unmodified AW1-51 duplex. Future saRNA screens should include passenger strand abasic modifications to prevent loading and possible off-target effects. A previous study of the molecular mechanism of RNAa identified CTR9, a component of the PAF1 complex, to be an Ago2-associated cofactor required for saRNA activity.13 The PAF1 complex is a known regulator of transcription and histone modification.31 Because CEBPA-51 activity also requires CTR9, these results are consistent with previously published saRNA reports and show that the activity of CEBPA-51 is a transcriptionally driven RNAa mechanism distinct from RNAi.
The activity of CEBPA-51 has been confirmed here in two human HCC lines, where it upregulates CEBPA mRNA and protein, upregulates the downstream C/EBP-α target albumin, and inhibits cell growth. We previously showed that CEBPA saRNA also inhibits tumor growth in a rat HCC model, upregulates a range of tumor suppressor genes, and downregulates a number of oncogenic genes, such as MYC and STAT3.11 The data reported here are consistent with this publication, and upregulation of CEBPA mRNA by CEBPA-51 is conserved in rodents and non-human primates. The modifications on CEBPA-51 prevent immune stimulation, providing evidence that the saRNA activity is not a result of innate immune reaction, and increasing the safety profile for the clinic. Similar 2′-O-methyl modification of RNA has been shown previously to suppress immune stimulation of siRNA,32 consistent with the data reported here for saRNA.
Although surgical resection provides the best prognosis for long-term survival in HCC, many patients are ineligible for treatment due to poor liver function. We believe that upregulation of CEBPA can not only inhibit tumor cell growth as shown here, but also restore critical liver function in patients with advanced HCC. This is supported by our rat HCC model, showing reduction of tumor burden as well as increased serum albumin and decreased bilirubin, AST, and ALT with CEBPA saRNA delivery. The combination of this novel approach with already well-established oligonucleotide delivery vehicles like the NOV340 SMARTICLES33, 34 puts saRNA therapy in a unique position for translation to the clinic. This first saRNA therapeutic, CEBPA-51 encapsulated in the NOV340 SMARTICLES (MTL-CEBPA), is currently in clinical trials for patients with liver cancer.26
Materials and Methods
Design of saRNA Oligonucleotides
Candidate saRNA hotspots were generated using the previously described bioinformatics algorithm.7 The list of oligonucleotides used in this study can be found in Table S1. All bases are RNA, except when preceded by the following to indicate a modified base: m, 2′-O-methyl; d, DNA base; and ps, phosphorothioate. Nontargeting oligo “NC” or “MM” were used as a negative transfection control for experiments using unmodified saRNAs. An inactive siRNA targeting firefly luciferase (“FLUC”) was used as a negative transfection control for experiments using modified saRNAs. saRNAs with seed mutations relative to AW1-51 have their changed bases underlined. Biotinylated oligos were synthesized with a biotin-triethyleneglycol (TEG) spacer attached to the 3′ end of the indicated strand of the duplex.
Cell Culture and Transfection
HepG2 human hepatocellular carcinoma cells (ATCC) were grown in RPMI supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and penicillin/streptomycin in a 5% CO2 incubator. Ago2 knockout mouse embryonic fibroblasts were a kind gift from Pal Saetrom. Unless otherwise specified, for transfections, the cells were seeded at 1 × 105 cells per well in a 24-well plate and reverse transfected immediately after seeding with the indicated oligonucleotide concentration using 1 μL of Lipofectamine 2000 (Life Technologies). Cells were then forward transfected after 24 hr and collected for analysis 72 hr after seeding. The indicated Silencer Negative Control (Life Technologies) or CTR9 siRNA (Life Technologies) were used for siRNA transfections.
Luciferase Assay
HepG2 cells were transfected as indicated above with the Cignal C/EBP Luciferase Reporter kit (QIAGEN) according to the manufacturer’s protocol. Reporter plasmids were co-transfected with the indicated oligonucleotide at the reverse and forward transfection. After 72 hr, the cells were lysed with Passive Lysis Buffer and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) on a PHERAstar Plus luminescence microplate reader (BMG Labtech). C/EBP firefly luciferase activity was normalized to renilla luciferase activity.
Nuclear Run-On
CEBPA transcriptional activity was measured by nuclear run-on as previously described.35 HepG2 cells were used to determine transcriptional activity after transfection with the indicated oligonucleotide.
RNA Isolation and qPCR
RNA was isolated from cultured cells using the RNeasy Mini Kit (QIAGEN). RNA was quantitated using a Nanodrop 1000 spectrophotometer (Thermo Scientific), and 500 ng was reverse transcribed using the Quantitect Reverse Transcription Kit (QIAGEN). Relative expression levels were determined by qPCR using Quantifast SYBR Green Master Mix (QIAGEN) on an ABI 7900HT thermal cycler (Applied Biosystems). The following Quantitect Primer Assays (QIAGEN) were used: ALB_1_SG, CEBPA_1_SG, CTR9_1_SG, and GAPDH_1_SG. For relative GenBank: AW665812 transcript expression, strand-specific RT primer 5′-caagaagtcggtggacaagaa was used with the Quantitect Reverse Transcription Kit before amplification with the following primers: F, 5′-cgcagcgtgtccagttc; and R, 5′-gtggagacgcagcagaag. Relative expression was determined using the ΔΔCt method normalized to GAPDH expression.
Western Blot
Cells were harvested in a 24-well-plate format (in triplicates) for a pool of 3 wells per condition for total protein extraction. Prior to cell lysis, the wells are washed twice with cold PBS and transferred into pre-chilled tubes with the use of a cell scraper. The cells were pelleted gently at 3,500 rpm for 5 min at 4°C before addition of RIPA lysis buffer containing 50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP40, and protease inhibitor cocktail (Sigma). Cells were incubated for 10 min on ice, followed by vortexing for 2 min to allow complete cell lysis. Cell debris was then removed by centrifugation at 10,000 rpm for 10 min at 4°C. The protein supernatant was then transferred into a clean pre-chilled tube. Protein amount per sample was quantified using the RC-DC Bradford assay kit following the manufacturer’s protocol (Bio-Rad), and 50 μg of total protein was loaded for SDS-PAGE. The acrylamide gels were then transferred onto polyvinylidene fluoride (PVDF) membranes for western blotting using the following antibodies: C/EBP-α, ab40764 (Abcam); β-tubulin, ab6046 (Abcam); and anti-rabbit-HRP, 926-8011 (LI-COR).
WST-1 Growth Assays
Cells were assessed for cell metabolism using the WST-1 assay, as previously described.11 Cells were seeded at 10,000 cells per well in a 96-well plate in triplicate and transfected as described above.
Argonaute Protein Coimmunoprecipitation
HepG2 cells were seeded in 6-well plates and transfected as described above. After 72 hr, cells were cross linked with 1% formaldehyde for 15 min at 37°C in 5% incubator. Cold PBS with glycine was used to quench the formaldehyde and rinse the cells before harvest for whole cell extraction on ice using RIPA lysis buffer. Biotinylated-saRNA protein complex was immobilized using Dynabeads-Biotin Binder (Invitrogen). Following the appropriate wash cycles on a magnetic column, the eluted protein complex was then coimmunoprecipitated with anti-Ago1 (Millipore 07-599); Ago2 (Millipore 07-590); Ago3 (Abcam ab154844); or Ago4 (Abcam, ab85077). Isotype immunoglobulin G (IgG) (Santa Cruz sc0-2027) was used as a negative control. The coimmunoprecipitation complex was then immobilized using Dynabead Protein G (Thermo Fisher), and after the appropriate wash cycles on a magnetic column, samples were separated on SDS-PAGE and transferred onto a PVDF membrane for western blotting, as described above.
Chromatin Immunoprecipitation
HepG2 cells were transfected as described above. Prior to harvest, the cells were cross-linked in situ with 1% formaldehyde at 37°C for 10 min. Glycine was then added to a final concentration of 250 mM for 3 min to allow quenching of formaldehyde. Cells were washed immediately 3x with ice-cold PBS and lysed with standard RIPA lysis buffer (150 mM NaCl, 1.0% NP40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris) with 1 × 106 cells for each pull-down column. Cells were allowed to swell on ice for 10 min before chromosomal fragmentation by sonication (5x pulsed at 25% output on a Mircoson-Ultrasonic Cell disruptor XL). Cell fragments were pelleted and discarded, and the supernatant containing the fragmented chromosome was collected. Biotin immobilization was performed overnight on a rotating chamber at 4°C using a magnetic Dyna Bead Biotin Binder (Invitrogen), with the samples diluted in ChIP dilution buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl). The beads were then washed 2x with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, and 150 mM NaCl2), followed by 1x wash with high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, and 500 mM NaCl), followed by 1x wash in lithium chloride buffer (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl), and a final 2x wash in Tris-EDTA (TE) buffer (10 mM Tris-HCl and 1 mM EDTA). The biotin/saRNA complexes were then reverse cross-linked for 4 hr at 65°C with 300 mM NaCl. DNA was then purified using phenol/chloroform/isoamyl alcohol (IAA) extraction. The DNA was precipitated in 3 M sodium acetate buffer at −80°C for at least 1 hr, followed by ultracentrifugation at 4°C. The pellet was then washed with 70% ethanol, allowed to dry, and resuspended in EB buffer for amplification using RT2 SYBR Green qPCR Mastermix (QIAGEN), according to the manufacturer’s protocol, with the following EpiTect ChIP qPCR Assays (QIAGEN): CEBPA, GPH1020591(+)01A and GPH1020591(+)03A; and ALB, GPH1010055(+)01A.
Statistical Analysis
Data are displayed as the mean of triplicates ± SEM. Statistical analysis was determined using an unpaired t test, with two-tailed p values less than 0.05 considered significant.
Author Contributions
J.V., V.R., D.C.B., R.H., H.H., P.S., J.J.R., and N.A.H. conceptualized the study and designed experiments. P.S. designed the activating oligonucleotides. T.C.R. designed, performed, and analyzed the nuclear run-on experiments. J.V. and V.R. performed and analyzed the remaining experiments. P.P. and P.A. performed additional experiments for manuscript revision. J.V., V.R., D.C.B., R.H., and N.A.H. contributed to construction and writing of the manuscript. D.C.B., R.H., and N.A.H. managed the execution of the study.
Conflicts of Interest
V.R., P.S., J.J.R., and N.A.H. are shareholders of MiNA (Holdings) Limited.
Acknowledgments
Work was funded by MiNA Therapeutics Limited.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.07.018.
Contributor Information
Jon Voutila, Email: jon@minatx.com.
Nagy A. Habib, Email: nagy.habib@imperial.ac.uk.
Supplemental Information
References
- 1.Li L.C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 3.Turunen M.P., Lehtola T., Heinonen S.E., Assefa G.S., Korpisalo P., Girnary R., Glass C.K., Väisänen S., Ylä-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ. Res. 2009;105:604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 4.Chu Y., Yue X., Younger S.T., Janowski B.A., Corey D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang V., Qin Y., Wang J., Wang X., Place R.F., Lin G., Lue T.F., Li L.C. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Place R.F., Huang V., Wang X., Noonan E.J., Magyar C.E., Huang J., Li L.C. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70:10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voutila J., Sætrom P., Mintz P., Sun G., Alluin J., Rossi J.J., Habib N.A., Kasahara N. Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol. Ther. Nucleic Acids. 2012;1:e35. doi: 10.1038/mtna.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Wang J., Huang V., Place R.F., Li L.C. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem. J. 2012;443:821–828. doi: 10.1042/BJ20111491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T., Li M., Yuan H., Zhan Y., Xu H., Wang S., Yang W., Liu J., Ye Z., Li L.C. saRNA guided iNOS up-regulation improves erectile function of diabetic rats. J. Urol. 2013;190:790–798. doi: 10.1016/j.juro.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Reebye V., Saetrom P., Mintz P.J., Rossi J.J., Kasahara N., Nteliopoulos G., Nicholls J., Haoudi A., Gordon M., Habib N.A. A short-activating RNA oligonucleotide targeting the islet β-cell transcriptional factor MafA in CD34(+) cells. Mol. Ther. Nucleic Acids. 2013;2:e97. doi: 10.1038/mtna.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reebye V., Sætrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portnoy V., Huang V., Place R.F., Li L.C. Small RNA and transcriptional upregulation. Wiley Interdiscip. Rev. RNA. 2011;2:748–760. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portnoy V., Lin S.H., Li K.H., Burlingame A., Hu Z.H., Li H., Li L.C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26:320–335. doi: 10.1038/cr.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perz J.F., Armstrong G.L., Farrington L.A., Hutin Y.J., Bell B.P. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 2006;45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 15.White D.L., Kanwal F., El-Serag H.B. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin. Gastroenterol. Hepatol. 2012;10:1342–1359.e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson P.J. Non-surgical treatment of hepatocellular carcinoma. HPB (Oxford) 2005;7:50–55. doi: 10.1080/13651820410024076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 18.Ramji D.P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akai Y., Oitate T., Koike T., Shiojiri N. Impaired hepatocyte maturation, abnormal expression of biliary transcription factors and liver fibrosis in C/EBPα(Cebpa)-knockout mice. Histol. Histopathol. 2014;29:107–125. doi: 10.14670/HH-29.107. [DOI] [PubMed] [Google Scholar]
- 20.Flodby P., Liao D.Z., Blanck A., Xanthopoulos K.G., Hällström I.P. Expression of the liver-enriched transcription factors C/EBP alpha, C/EBP beta, HNF-1, and HNF-4 in preneoplastic nodules and hepatocellular carcinoma in rat liver. Mol. Carcinog. 1995;12:103–109. doi: 10.1002/mc.2940120207. [DOI] [PubMed] [Google Scholar]
- 21.Tseng H.H., Hwang Y.H., Yeh K.T., Chang J.G., Chen Y.L., Yu H.S. Reduced expression of C/EBP alpha protein in hepatocellular carcinoma is associated with advanced tumor stage and shortened patient survival. J. Cancer Res. Clin. Oncol. 2009;135:241–247. doi: 10.1007/s00432-008-0448-5. [DOI] [PubMed] [Google Scholar]
- 22.Johnson P.J., Berhane S., Kagebayashi C., Satomura S., Teng M., Reeves H.L., O’Beirne J., Fox R., Skowronska A., Palmer D. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach-the ALBI grade. J. Clin. Oncol. 2015;33:550–558. doi: 10.1200/JCO.2014.57.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuster M.B., Porse B.T. C/EBPalpha: a tumour suppressor in multiple tissues? Biochim. Biophys. Acta. 2006;1766:88–103. doi: 10.1016/j.bbcan.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Tan E.H., Hooi S.C., Laban M., Wong E., Ponniah S., Wee A., Wang N.D. CCAAT/enhancer binding protein alpha knock-in mice exhibit early liver glycogen storage and reduced susceptibility to hepatocellular carcinoma. Cancer Res. 2005;65:10330–10337. doi: 10.1158/0008-5472.CAN-04-4486. [DOI] [PubMed] [Google Scholar]
- 25.Ribero D., Curley S.A., Imamura H., Madoff D.C., Nagorney D.M., Ng K.K., Donadon M., Vilgrain V., Torzilli G., Roh M. Selection for resection of hepatocellular carcinoma and surgical strategy: indications for resection, evaluation of liver function, portal vein embolization, and resection. Ann. Surg. Oncol. 2008;15:986–992. doi: 10.1245/s10434-007-9731-y. [DOI] [PubMed] [Google Scholar]
- 26.ClinicalTrials.gov. (2016). First-in-human safety and tolerability study of MTL-CEBPA in patients with advanced liver cancer. https://ClinicalTrials.gov/show/NCT02716012.
- 27.Lima W.F., Wu H., Nichols J.G., Sun H., Murray H.M., Crooke S.T. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundin K.E., Gissberg O., Smith C.I. Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 2015;26:475–485. doi: 10.1089/hum.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 31.Jaehning J.A. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broering R., Real C.I., John M.J., Jahn-Hofmann K., Ickenstein L.M., Kleinehr K., Paul A., Gibbert K., Dittmer U., Gerken G. Chemical modifications on siRNAs avoid Toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro. Int. Immunol. 2014;26:35–46. doi: 10.1093/intimm/dxt040. [DOI] [PubMed] [Google Scholar]
- 33.Reebye V., Voutila J., Huang K.-W., Muragundla A., Jayaprakash A., Vadnal P., Huber H., Habib R., Sætrom P., Rossi J. Systemic administration of a novel development candidate, MTL-CEBPA, up-regulates the liver-enriched transcription factor C/EBP-α and reverses CCl4-induced liver failure in vivo. Hepatology. 2015;62(Suppl 1):269A–270A. [Google Scholar]
- 34.Tolcher A.W., Rodrigueza W.V., Rasco D.W., Patnaik A., Papadopoulos K.P., Amaya A., Moore T.D., Gaylor S.K., Bisgaier C.L., Sooch M.P. A phase 1 study of the BCL2-targeted deoxyribonucleic acid inhibitor (DNAi) PNT2258 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014;73:363–371. doi: 10.1007/s00280-013-2361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts T.C., Hart J.R., Kaikkonen M.U., Weinberg M.S., Vogt P.K., Morris K.V. Quantification of nascent transcription by bromouridine immunocapture nuclear run-on RT-qPCR. Nat. Protoc. 2015;10:1198–1211. doi: 10.1038/nprot.2015.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.