Abstract
The major drawback of the Baculovirus/Sf9 system for recombinant adeno-associated viral (rAAV) manufacturing is that most of the Bac-derived rAAV vector serotypes, with few exceptions, demonstrate altered capsid compositions and lower biological potencies. Here, we describe a new insect cell-based production platform utilizing attenuated Kozak sequence and a leaky ribosome scanning to achieve a serotype-specific modulation of AAV capsid proteins stoichiometry. By way of example, rAAV5 and rAAV9 were produced and comprehensively characterized side by side with HEK293-derived vectors. A mass spectrometry analysis documented a 3-fold increase in both viral protein (VP)1 and VP2 capsid protein content compared with human cell-derived vectors. Furthermore, we conducted an extensive analysis of encapsidated single-stranded viral DNA using next-generation sequencing and show a 6-fold reduction in collaterally packaged contaminating DNA for rAAV5 produced in insect cells. Consequently, the re-designed rAAVs demonstrated significantly higher biological potencies, even in a comparison with HEK293-manufactured rAAVs mediating, in the case of rAAV5, 4-fold higher transduction of brain tissues in mice. Thus, the described system yields rAAV vectors of superior infectivity and higher genetic identity providing a scalable platform for good manufacturing practice (GMP)-grade vector production.
Keywords: AAV, gene therapy, obesity
Kondratov et al. describe a new insect cell-based production platform utilizing attenuated Kozak sequence and a leaky ribosome scanning to achieve a serotype-specific modulation of AAV capsid proteins stoichiometry. The established system yields rAAV vectors of superior infectivity and higher genetic identity, providing a scalable platform for GMP-grade vector production.
Introduction
Recombinant adeno-associated virus (rAAV) is extensively used as a vector for gene therapy/DNA vaccine delivery, but a scale-up production of highly infectious rAAVs for clinical trials remains a challenging proposition. AAV capsid consists of three capsid proteins, viral protein (VP)1, VP2, and VP3, derived via an alternatively splicing and differential codon usage of a single capsid gene in the AAV genome. The VP3 sequence is common between all three splice variants, and VP2 and VP1 have N-terminal longer sequences, with the unique region of VP1 containing a phospholipase domain A2 critical for virus infection.1 The exact amounts of VP1/VP2/VP3 in the capsid are unknown but estimated to be 1/1/10, based on densitometry analyses of the capsid proteins resolved on SDS-PAGE.2, 3, 4 Moreover, it appears that there is no defined VP1/VP2/VP3 stoichiometry, and that the assembly is stochastic such that the relative amount of VP1/VP2/VP3 that is incorporated in the capsid depends mainly on their relative expression levels.5 Therefore, the design of a capsid proteins expression unit in a given rAAV production system is essential for the assembly of biologically potent gene therapy vectors.
Toward this goal, one of the original scalable systems utilized a suspension culture of Sf9 insect cells co-infected with three recombinant baculoviruses derived from A. californica multicapsid nucleopolyhedrovirus (AcMNPV) encoding, respectively, rAAV transgene cassette, AAV rep, and cap helper genes.6 Most of the AAV serotypes produced in this system, however, were characterized by low transduction efficiencies compared with HEK293-derived vectors because of a suboptimal content of VP1 capsid protein and its phospholipase A2 activity.7, 8, 9, 10 This shortcoming resulted from the capsid gene helper vector design utilizing a non-canonical ACG initiation codon for VP1 to induce a leaky ribosome scanning.6 Even though other groups resolved the problem to some extent utilizing a different initiation codon CUG11 or artificial intron,12 the solutions appeared to lack the flexibility necessary for a serotype-specific sequence adjustment. Below, we introduce a novel system of regulation of a relative VP1/VP2/VP3 composition via adjustable leaky ribosome scanning.
In cells of mammalian origin, a P40-driven transcript in AAV undergoes splicing to produce two spliced mRNA variants encoding VP1 or VP2/PV3 capsid proteins, respectively. Because in the baculovirus/Sf9 system the polh promoter is substituted for the P40/intron sequence, the regulation by splicing is not available, necessitating the utilization of alternative regulation of the VP1 expression via leaky ribosome scanning. A consensus sequence GCCRCCAUGGC (R = A or G) is considered to be an optimal mammalian translation initiation site (TIS), also known as Kozak sequence.13 Any deviation from this sequence would increase leaky scanning of the VP1 AUG and initiation of translation from the in-frame downstream VP2 ACG or VP3 AUG codons, thus changing the VP1/VP2/VP3 stoichiometry. In the current paper, we describe an approach for rationally modulating the ratios of VP1/VP2/VP3 capsid composition in a Baculovirus/Sf9 system to derive particles with a higher VP1/VP2 content, resulting in significantly greater biological potency even compared with HEK293-derived vectors.
To characterize this advanced production platform more broadly, we conducted a next-generation sequencing (NGS) analysis of encapsidated DNA manufactured by two methods: a conventional triple-plasmid co-transfection of HEK293 and single baculovirus expression vector (BEV) infection of Sf9 cell line incorporating stably integrated rep/cap helper genes. Direct side-by-side NGS analysis of the rAAV cassettes manufactured by two platforms revealed higher precision of viral DNA packaging in insect cells encapsidating significantly less contaminating DNA.
Results
Design of AAV5 and rAAV9 Capsid Genes
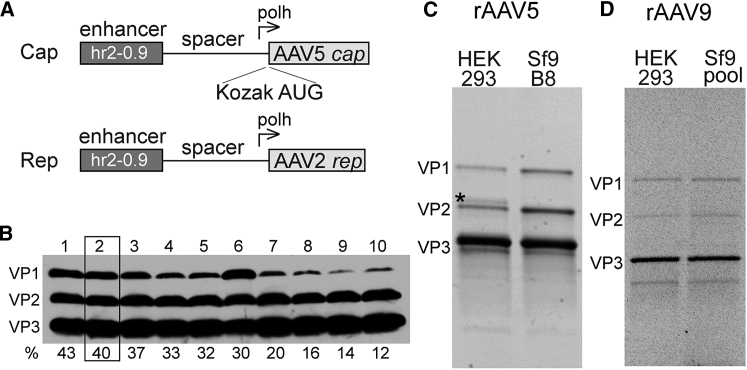
To increase a leaky ribosome scanning, we proposed to utilize a canonical AUG codon preceded by attenuated Kozak sequence. Randomly modifying nucleotides up- or downstream of the AUG would not be a realistic approach because the complexity of the possible TIS sequences spanning the relevant stretch of eight residues is 65,536 possible permutations. Moreover, the consensus Kozak sequence appears to be different for yeast,14 higher plants,15 invertebrates,16 or vertebrates.17 Therefore, one way to rationalize the screening of attenuated TISs was to utilize the empirical heatmap of all possible mammalian TIS permutations derived by Noderer et al.18 whereby all possible combinations of TISs were assigned “initiation efficiency” values relative to the consensus Kozak sequence.
We have selected an arbitrary range of 12% to 43% initiation efficiency and tested 10 mutants differing by 2%–4% increments. All 10 AAV5 VP1 tested TISs are shown in Table S1 and were used in the context of cap-expressing helper plasmids (Figure 1A) to derive BSR pooled cell lines as described previously7 and to assess their capsid compositions. As expected, and with only a few notable exceptions (e.g., Figure 1B, lane 6), the relative VP1 content gradually decreased, while closely following theoretical value declines of relative TIS efficiencies. This correlation supports the original hypothesis that leaky scanning translation initiation can be utilized to modulate VP1 content. To further identify the optimal VP1/VP2/VP3 stoichiometries, we assayed the infectious and overall titers of the respective 10 rAAV5 vectors. To produce infectious rAAV5 vector, we generated the BEV-encoding CMV-β actin (CBA)-driven GFP (pTR-Bac-UF26; Figure S1A). Side-by-side comparison revealed that the construct with 40% TIS relative efficiency (Figure 1B, lane 2) was superior compared with other constructs producing the highest yield (Figure S2A). Thus, this particular capsid gene-containing helper construct incorporating the attenuated TIS, UGUUUUAUGUC (Table S1), was chosen to derive a producer cell line. In a similar manner, 12 AAV9 VP1 plasmid constructs with attenuated Kozak sequences were screened (Table S1; Figure S2B). For AAV9, the optimal sequence among those tested was UAGUGUAUGGC, constituting 45% of relative TIS efficiency.
Figure 1.
Capsid Protein Composition of rAAV Vectors Produced in Sf9 Cells
(A) Design of rep and cap genes expression cassettes. (B) Direct correlation of rAAV5 VP1 protein expression and its relative VP1 Kozak TIS efficiency: western blotting analysis of capsid proteins isolated from 10 separate cell lines incorporating stably integrated cap expression cassettes. The relative TIS efficiencies (%) for each capsid VP1 gene construct are shown below the respective lane. (C) Capsid proteins compositions of rAAV5 purified from HEK293 and Sf9 cells: SDS-protein gel analysis of double iodixanol-purified rAAV5-GFP, directly visualized with shortwave UV photoactivation (stain-free technology; Bio-Rad). Asterisk (*) denotes a slower migrating band often observed in rAAV5 samples purified from HEK293 cells, and that was excluded from VP2 quantification analysis. (D) Capsid proteins compositions of rAAV9 purified from HEK293 and Sf9 cells. Analysis is the same as in (C). See also Table S1 and Figure S2.
Characterization of the rep2/cap5 Stable Cell Line
Individual cell lines were derived using Rep2- and Cap5-expressing plasmids devoid of Rep-binding elements (RBEs).8 Cap5 helper contained the following attenuated TIS: UGUUUUAUGUC. Five individual cell lines were propagated and tested as described earlier.7 One cell line, dubbed B8, showing the highest yield of rAAV5–UF26 was chosen for further characterization. The following parameters were investigated.
VP1/VP2/VP3 Stoichiometry
The capsid composition of rAAV5-GFP purified from HEK293 cells by sequential double iodixanol gradient deviated from the theoretical value of 1/1/10 for VP1/VP2/VP3 in that it contained a lower VP1 content (Figure 1C). Moreover, this capsid incorporated an additional VP2 capsid protein with slightly higher MW. In contrast, the Sf9 B8-derived vector was composed of higher levels of VP1 and VP2 (Figure 1C). For rAAV9, the capsid composition of Sf9-derived vector was almost identical to that of the vector manufactured in HEK293 cells (Figure 1D). Unlike rAAV5, both viral samples appear to incorporate more VP1 than VP2 and also to contain the bands smaller than VP3, which could be a product of a capsid-specific proteolysis.19
To quantify the exact numerical values, we plotted the densities of the respective capsid bands for rAAV5 as areas under the curve (AUCs) (Figure S3). The following values (averages of two independent experiments, adjusted to a common denominator of VP3 = 10) were derived: HEK293 VP1/VP2/VP3 = 0.2/0.5/10; Sf9 B8 VP1/VP2/VP3 = 0.7/1.7/10. To validate the calculated stoichiometry, we analyzed rAAV5 capsid composition by mass spectrometry using MALDI-TOF.
MALDI-TOF
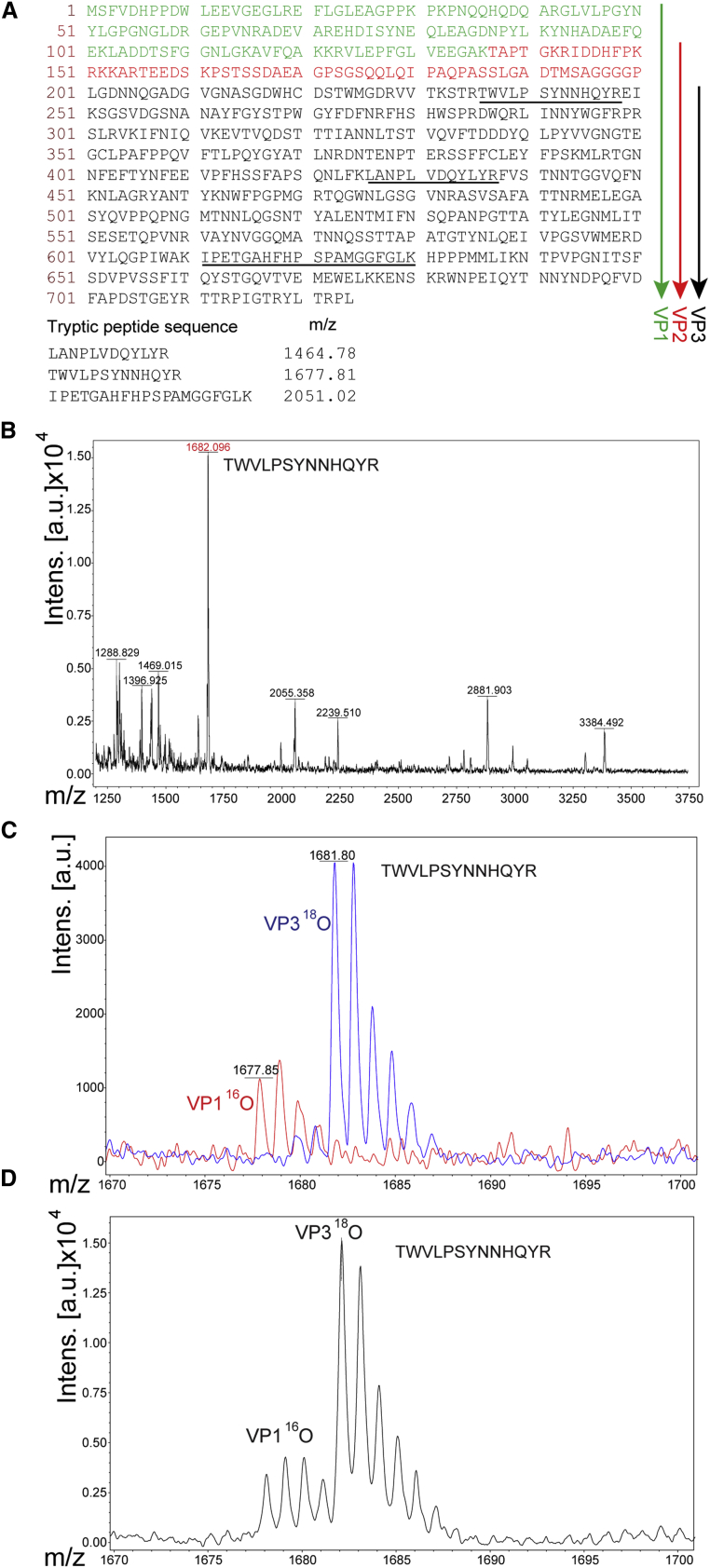
AAV VPs constituting the virion shell share their VP3 C termini (Figure 2A, shown in black). VP1 (green) and VP2 (red) unique N termini are relatively small compared with the common shared VP3 domain, creating a challenge to decipher relative stoichiometry by analyzing their unique tryptic peptides. Therefore, a differential 16O/18O labeling approach was used to discriminate identical peptides but derived from VP1, VP2, or VP3. When heavy-oxygen stable isotope water H218O is used during the trypsin digestion, two 18O atoms are exchanged at the C-terminal carboxyl group of the tryptic peptides, shifting the mass by 4 Da.20, 21 This shift between the identical peptides derived from two different proteins (e.g., VP1 or VP3) allows for the identification and quantitation of the proteins.
Figure 2.
MALDI-TOF Analysis of the AAV5 VP1, VP2, and VP3 Capsid Proteins Stoichiometry
(A) Amino acid sequence of the AAV5 capsid with VP1 unique N termini marked in green, unique VP2 in red, and common VP3 C termini in black. The downward arrows indicate the respective proteins. Tryptic peptides selected for MS analysis are underlined and shown below their respective observed masses. (B) MALDI-TOF-MS spectrum of all tryptic peptides of rAAV5 digested in H218O. The red highlighted peptide is one representative out of three analyzed. (C) Two overlaid MALDI-TOF MS spectra of the same tryptic peptide TWVLPSYNNHQYR originating from the VP1 gel band digested with trypsin prepared in 16O water (red trace) or from the VP3 gel band digested with trypsin prepared in 18O water (blue trace). 18O water incorporates two 18O atoms on the C terminus of the peptide, thus shifting the mass by 4 atomic mass units (amu). These digestion products were spotted/analyzed separately and the spectra overlaid to show the complete incorporation of two 18O into the VP3 peptide. (D) Isotopic “fingers” of the same peptide derived from VP1 or VP3 after the digestion products were mixed at 1:1 ratios to calculate the relative content. See also Tables S2 and S3 and Figure S3.
After conducting the pilot digestion, three peptides (Figure 2A) in the VP3 region were chosen to quantitate the relative abundances of each protein. A complete incorporation of two 18O atoms was confirmed by running the VP3/VP1 and VP2/VP3 samples separately. Then, the full MALDI spectrum of all the VP3 digestion tryptic products in H218O was analyzed (Figure 2B). All of the peptides showed a 4 Da mass shift from the predicted peptide molecular weights, confirming two 18O atoms are incorporated. If the mass shift observed is only 2 Da, then either the reaction is not complete or back exchange from incomplete trypsin inactivation was occurring.20 Figure 2C shows an overlay of the tryptic peptide TWVLPSYNNHQYR from either VP1 (red trace) or VP3 (blue trace) spotted separately and analyzed by MALDI-TOF. The distinct mass shift of 4 Da for the VP3 sample proved a complete 18O atom exchange with no evidence of a back exchange. Figure 2D is a representative MALDI-TOF spectra of VP3 digested with H218O and mixed 1:1 with VP1 digested in H216O. The peak areas were integrated and used to calculate the abundance of each protein.
Three unique peptides (Figure 2A) per each protein were analyzed, each in three replicate MALDI-TOF runs (Tables S2 and S3). This allowed for a high confidence quantification of their relative abundance.22 The following values (average of three independent experiments, adjusted to a common denominator of VP3 = 10) were derived: HEK293 VP1/VP2/VP3 = 0.4/0.5/10; Sf9 B8 VP1/VP2/VP3 = 1.1/1.7/10. These numbers were remarkably consistent with the AUC densities described above.
Full/Empty Particles Ratios
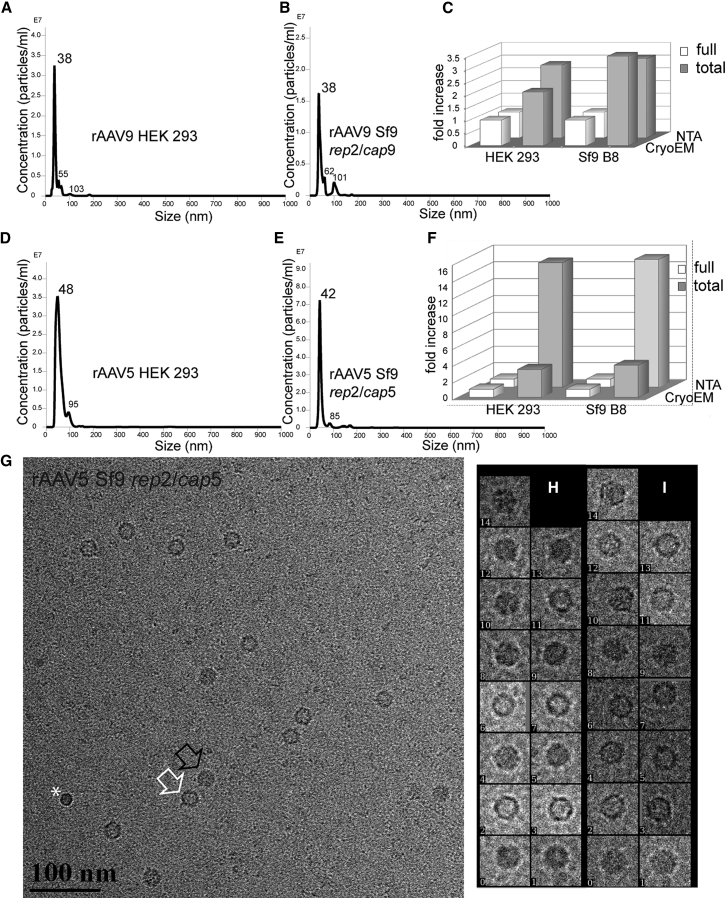
This parameter was investigated to determine whether either of the two manufacturing platforms produces higher ratios of empty particles, a potential source of untoward immune response and a technical challenge during vector purification. rAAV5 and rAAV9 were purified by one-step chromatography over monospecific antibody affinity resins, AVB for rAAV5 and POROS CaptureSelect for rAAV9. After purification, the vector genome particle titers were assayed using quantitative competitive (QC)-PCR. The total particle titers of rAAV5 and rAAV9 vectors were assayed using nanoparticle tracking analysis (NTA) of rAAV capsids decorated with gold nanoparticles. This approach utilizes the electrostatic attraction between a highly scattering material such as gold nanoparticles and the viral capsid. The resulting gold-labeled virus particles scatter enough light to be visualized and tracked by the optical system (Figure 3; Movie S1), enabling the use of NTA to measure the size and concentration of AAV. Interestingly, even though the average sizes of rAAV9 particles for both platforms were identical (major peak of 38 nm; Figures 3A and 3B), the average sizes of rAAV5 from HEK293 and from Sf9 B8 were different (48 nm versus 42 nm; Figures 3D and 3E). Of note, these sizes are the approximations of the virus/nano-gold particle complexes, which, in turn, are functions of viral particle charge.
Figure 3.
Quantification of Full-to-Total AAV Particles
(A–F) Nanoparticle tracking analysis (NTA) of rAAV9 (A–C) or rAAV5 (D–F) manufactured in HEK293 cells (A and D) or Sf9 cells (B and E). (A–D) A graphic representation of finite track length adjustment (FTLA) algorithm, an average of three independent video captures of rAAV/nano-gold particle complexes, each recorded for 30 s for each sample (see also Movie S1). (C and F) The calculated ratios of full to total particles for each preparation analyzed by NTA (back row) or CryoEM (front row) are shown. The numbers next to the peaks show the calculated rAAV/nano-gold particle complexes sizes. The smaller peaks of larger diameters represent aggregated dimers and trimers of rAAV particles. Calculated ratios of DNA-containing versus total number of AAV particles in the respective viral stocks are displayed. (G–I) CryoEM analysis of rAAV9 and rAAV5. (G) A representative CryoEM image of Sf9-derived rAAV5 is shown. An empty particle is indicated by a white outlined arrow, a full particle by a black arrow, and white asterisk indicates an air bubble. (H and I) Representative zoomed-in images quantified using EMAN2 software are shown.
After calculating the ratio of total to full particles, these values for HEK293-derived rAAV9 were 2.8 and for Sf9-derived rAAV9 were 3.1 (Figure 3C). Similarly, for rAAV5, the ratios were not significantly different: 14.8 versus 15.2 (Figure 3F).
The NTA was independently validated using cryo-electron microscopy (CryoEM), as shown in Figures 3G–3I. The quantification (Figures 3C and 3F) confirmed that ratios of total to full particles are similar for rAAV5 and rAAV9 manufactured in HEK293 and Sf9 B8.
Transduction Efficiency In Vitro
An rAAV transgene cassette incorporating luciferase and mApple reporter genes (pTR-UF50-BC;23 Figure S1B) was used to generate the respective BEV to infect the Sf9 B8 cell line to test transduction efficiency. The same plasmid was also used to generate rAAV by a triple-plasmid co-transfection of HEK293 cells. Both rAAV5-Luc-mApple vectors, purified and titered side by side, were used to infect a HeLa-derived C12 cell line.24 Fluorescence-activated cell sorting (FACS) analysis (Figure S4A) revealed significantly higher (∼5-fold) transduction efficiency for the Sf9-produced rAAV5 vector. For rAAV9, there was no significant difference between two samples of rAAV9 (Figure S4B). The data from both experiments are consistent with the protein content.
Transduction Efficiency In Vivo
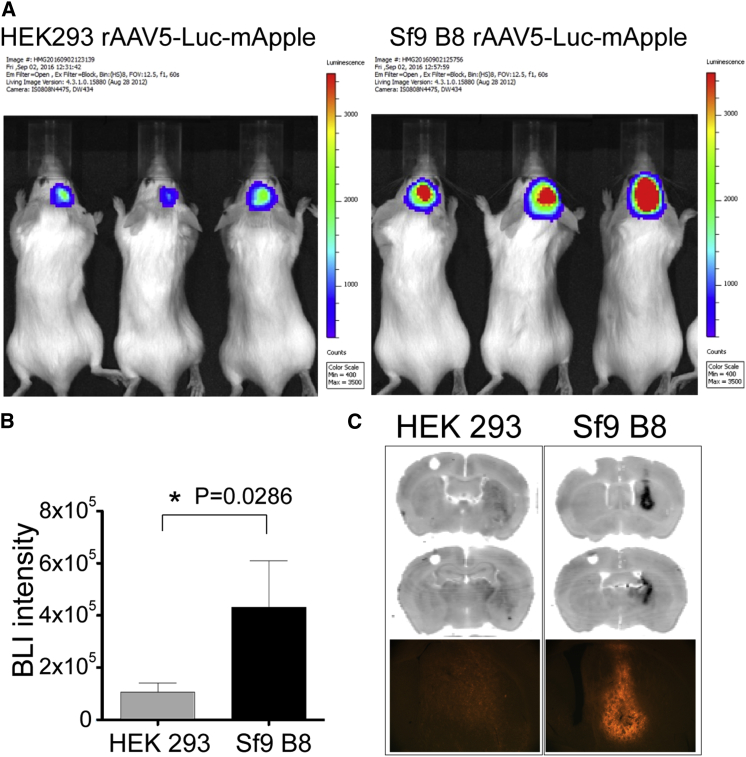
rAAV5-Luc-mApple produced in HEK293 and Sf9 B8 cells were assayed in vivo in the brain (striatum) of mice. Three weeks post-injection, transduction efficiency was determined by bioluminescence (BLI) (Figure 4A). Animals injected with Sf9 B8-derived AAV5 showed 4-fold higher BLI signal intensity than HEK293-derived AAV5 (4.3 × 105 ± 1.8 × 105 and 1.1 × 105 ± 3.5 × 104, respectively; Figure 4B). Similarly, fluorescent analysis of brain sections revealed a stronger mApple signal in Sf9 B8-derived samples compared with HEK293 (Figure 4C).
Figure 4.
Luciferase and mApple Expression after Striatal Injection of HEK293- or Sf9 B8-Derived rAAV5
(A) Heatmap images show bioluminescence (BLI) signal detected 3 weeks after AAV injections. The pseudo-color scale represents the intensity of light emitted in number of counts. Max and Min are the maximum and the minimum number of counts, respectively. (B) The graph shows the BLI intensity expressed as the total number of counts as mean ± SEM (n = 4 per group). Mann-Whitney test analysis show a significant difference between groups (*p = 0.0286, one-tailed test). (C) mApple fluorescence in the coronal brain sections as detected by a variable mode laser scanner (black-and-white [B/W] images) or fluorescence microscope (color image). See also Figures S1 and S4.
NGS Analysis of rAAV5 Vectors Produced in HEK293 and Sf9 Cells
Having established an improved OneBac system for the production of higher-potency rAAV5-based vectors, we conducted a comparative NGS analysis of the encapsidated single-stranded DNA (ssDNA; pTR-Bac-UF26; Figure S1A) manufactured by a standard triple co-transfection protocol in HEK293 cells, or by the single BEV-UF26 infection of a stable rep2cap5 Sf9 cell line B8. The purpose of the NGS analysis was to characterize rAAV cassette-specific as well as collaterally packaged contaminated DNA species, thus establishing a preferred platform meeting the demands of good manufacturing practice (GMP)-grade rAAV vector production. All NGS libraries were prepared, sequenced, and analyzed in duplicates. The general workflow is depicted in a flowchart (Figure S5). The following parameters have been analyzed.
Collateral Packaging of Contaminating DNA Sequences
The total number of reads for each sequence is listed in Tables S4 and S5. After filtering, the total number of reads assigned to an index was 757,433,116. After the alignment of reads to the referenced sequences, the coverage for rAAV cassette reached as high as 2,260,074 reads/nucleotide (nucleotide position 2,000). For collaterally packaged sequences, the coverage was significantly lower: 10,781 reads/nucleotide (nucleotide position 1,299, vector backbone) or 6,424 reads/nucleotide (nucleotide position 200, AcMNPV genome).
For both production protocols, the majority of reads were the rAAV-Bac-UF26 cassette, which accounted for 96.5% (HEK293) and 99.4% (Sf9) of all encapsidated DNA sequences (Table 1; Figures 5A and 5B). The majority of contaminated DNA in the HEK293 system was bacterial plasmid backbone (2.5%), with lower levels of the rep2/cap5 helper sequences (0.7%) and human genome DNA (0.17%), while the contaminants in the Sf9 preparation were the shuttle plasmid backbone (0.3%) and AcMNPV genome (0.2%).
Table 1.
Analysis of Specific and Collateral Packaging of rAAV5-UF26 in HEK293 and Sf9 Cells
| Reference Genome | NGS Library |
|
|---|---|---|
| PCR-free | PCR | |
| HEK293 | ||
| UF26 cassette | 96.50 | 95.24 |
| 96.59 | 95.38 | |
| Vector backbone | 2.58 | 3.49 |
| 2.52 | 3.39 | |
| pHelper | 0.04 | 0.04 |
| 0.04 | 0.04 | |
| rep2cap5 | 0.71 | 1.00 |
| 0.68 | 0.97 | |
| H. sapiens | 0.17 | 0.22 |
| 0.17 | 0.22 | |
| Sf9 | ||
| UF26 cassette | 99.38 | 99.15 |
| 99.39 | 99.20 | |
| Vector backbonea | 0.30 | 0.41 |
| 0.30 | 0.41 | |
| Baculovirus/Bacmidb | 0.29 | 0.39 |
| 0.27 | 0.34 | |
| rep2/cap5c | 0.014 | 0.003 |
| 0.015 | 0.003 | |
| S. frugiperda | 0.025 | 0.040 |
| 0.025 | 0.040 | |
Relative content of DNA in purified virions is shown as percentage of total sequences identified by NGS analysis. All NGS analyses were conducted in duplicates shown for each reference genome in two parallel rows. See also Figures S5–S7 and Tables S4 and S5.
Including GenR.
AcMNPV(strain_E2), miniF, LacZ, and KanR.
pIR-VPm11-hR2-900, pIR-rep78polHind-hr2.
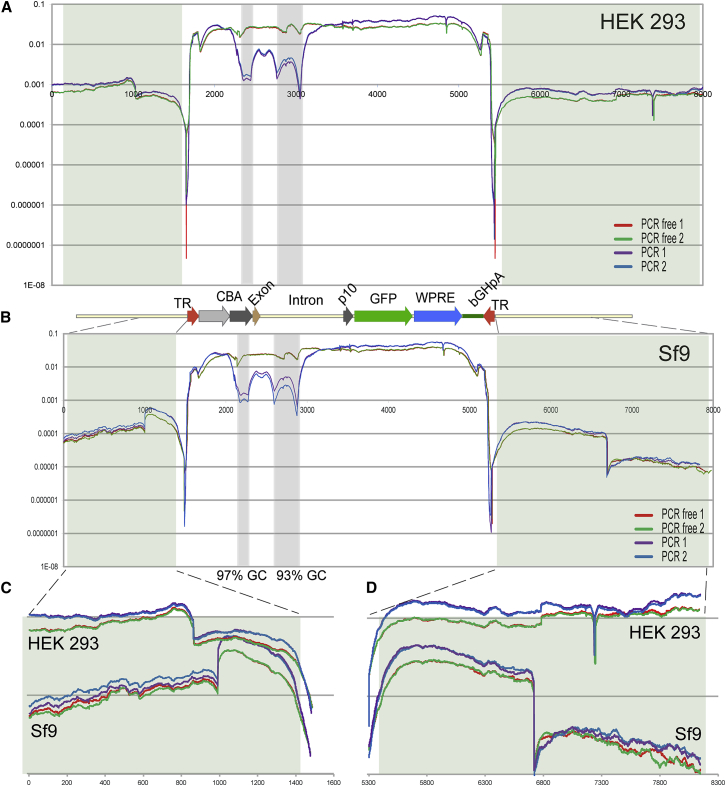
Figure 5.
Distribution of the NGS Reads of the rAAV-Bac-UF26 Cassette and Its Immediate Junctions
(A) Illumina reads coverage of rAAV5-Bac-UF26 cassette packaged in HEK293 cells numerically normalized by dividing the number of reads at each nucleotide position by the total number of reads of the referenced sequence. Sequence coverage graphs for duplicate samples are drawn in different colors: PCR-free, red and green; PCR-based, purple and blue. The drop of coverage between rAAV cassette and the adjacent sequences reflects a graphic representation of the bioinformatics analysis limitations rather than actual reduction in sequence coverage. Shown below the graph is an annotated map of referenced rAAV-Bac-UF26 cassette drawn to the scale of the sequences in (A) and (B). Sequences immediately adjacent to the rAAV cassette in the bacterial plasmid DNA are shaded (green tint). (B) Illumina reads coverage of rAAV5-Bac-UF26 cassette packaged in Sf9 B8 cells. GC-enriched sequences within rAAV cassette are shaded (blue tint). (C) Zoom from (A) and (B) of the sequences immediately adjacent to the left rAAV ITR in both HEK293 and Sf9 B8 cells. (D) Zoom from (A) and (B) of the sequences immediately adjacent to the right rAAV ITR in both HEK293 and Sf9 B8 cells.
The collaterally packaged sequences were more abundantly represented by the immediate junctions of the rAAV cassette and its respective backbones: bacterial plasmid for HEK293 cells and baculovirus genome for Sf9 cells. Notably, there was a significant difference, at least 10-fold, in the junction reads coverage between two systems whereby HEK293 cells appear to encapsidate, at much higher frequencies, bacterial plasmid backbone sequences that are more distant from both the left and right AAV terminal repeats (Figures 5C and 5D). Thus, analysis of contaminating DNA sequences suggests that the OneBac system delivers better precision and provides higher-quality rAAV vectors whereupon only 0.6% of the encapsidated vg incorporates foreign DNA as opposed to 3.5% for HEK293 cells.
rAAV Genome Coverage
Using a standard protocol, which included only eight cycles of PCR amplification step to generate NGS libraries, we identified several sequences within the CBA promoter and the downstream intron displaying at least 10-fold lower sequencing coverage compared with the rest of the rAAV cassette. Close examination revealed that these sequences are extremely guanine cytosine (GC) rich (Figures 5A and 5B). This relatively low coverage might have reflected the lower representation of these sequences in the packaged virions because of truncation during DNA replication. Alternatively, this drop represented an artifact introduced by PCR-related NGS library preparation. To exclude the second possibility, we prepared NGS libraries directly from the purified encapsidated rAAV DNA without PCR amplification. This way, the coverage of the sequences in question was restored to levels comparable with the rest of the cassette. Thus, both HEK293 and Sf9 cells support packaging of full-length rAAV cassettes with little, if any, evidence of truncation.
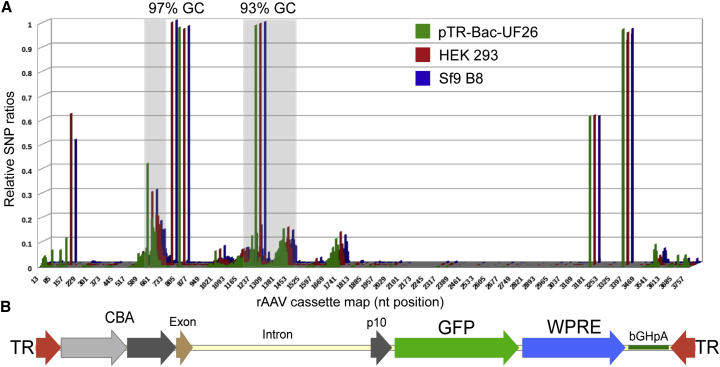
Genomic Identity of rAAV-Bac-UF26 Cassette
The high sequencing depth of AAV cassette allowed for the detailed analysis of the packaged DNA sequence identity and correlation to its respective parent bacterial plasmid. To reduce the probability of false calls and to increase the confidence of SNP analysis, we utilized only PCR-free sample NGS data. SNP variants for DNAs from both viral samples, as well as a positive control plasmid sample, displayed very similar profiles of substitutions (correlation coefficient of 0.75–0.77) (Figure 6A). Interestingly, the majority of SNPs were co-localized with non-coding sequences identified above as regions enriched in GC content (Figure 6B): chicken β-actin promoter and the intron sequence. In addition, and consistent with Lecomte et al.,25 AAV terminal repeats (TRs) displayed relatively high SNP variability.
Figure 6.
Distribution of SNPs Identified in rAAV5-Bac-UF26 Cassette Encapsidated by HEK293 versus Sf9 B8 Cells
(A) Substitutions of four residues, A, G, C, and T, are plotted cumulatively if read in a given position relative to the reference pTR-Bac-UF26 database sequence. Relative SNPs ratios (y axes) were defined as a proportion of the total number of reads in a given position with a substitution to the total depth of sequencing in the same position. Depth of sequencing is a sum of sequence reads of a reference and alternative nucleotide in a given position excluding insertion/deletions (indels). Analysis of SNP variants included positive control, plasmid pTR-Bac-UF26 sample (green). Consequently, zero rate of SNP represents a residue identical with the database reference; positive value signifies a newly acquired SNP relatively to the parent plasmid database sequence. Regions of high GC content are shaded. (B) Annotated map of referenced rAAV-Bac-UF26 cassette drawn to the scale of the sequence in (A).
Discussion
The relatively inferior potency of insect-cell-manufactured rAAV vectors of AAV5 and AAV8 serotypes was previously documented by Urabe et al.10 and Kohlbrenner et al.9 Subsequently, we have shown that many other OneBac-derived AAV serotypes were characterized by lower infectivity compared with 293-derived AAVs.26 The unifying cause for all affected serotypes was the modified sequence of a capsid helper gene that resulted in lower content of VP1 capsid protein incorporating phospholipase A2 activity. As expected, the recommended solutions were aimed at alleviating this problem by using a different initiation codon CUG11 or artificial intron.8, 12 Even though new designs helped to increase the infectivity for some vectors, the solutions appeared to lack the flexibility necessary for a serotype-specific sequence adjustment.
Hereby, we describe a new approach to increase relative VP1/VP2 content of a capsid. This is accomplished by modifying a canonical Kozak sequence preceding the VP1 AUG start codon. As a proof of principle, we tested a range of Kozak sequences for AAV5 and AAV9 serotypes showing that the most favorable TISs were serotype specific, producing rAAV vectors that exceeded transduction efficiencies of HEK293-derived counterparts. The described approach, however, requires fine adjustment of the VP1 Kozak sequence in the narrow window of relative TIS efficiencies, which also varies for different serotypes. One of the reasons for this variation is that the VP1:VP2:VP3 ratios depend not only on VP1 TIS relative efficiency, but also on the ones of VP2 and VP3 TISs, which are also different for all serotypes (Table 2). Moreover, intentionally increasing VP1 content above a certain threshold (e.g., Figure 1B, lanes 1 and 6) appears to be counterproductive because the yield of the virus drops precipitously once the VP1:VP2:VP3 ratio shifts too far away from the theoretical value of 1:1:10. One can derive a “consensus” VP1 TIS, U(C)A(C/G)U(G)UG(U)UAUGG, with the understanding that the optimal TIS has to be identified empirically.
Table 2.
Translation Initiation Sequences Surrounding VP1, VP2, and VP3 Start Codons in AAV Serotypes
| AAV Serotype | VP1 ORF |
VP2 ORF |
VP3 ORF |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Kozak TIS, % | Down | Up | Kozak TIS, % | Down | Up | Kozak TIS, % | Down | |
| 1 | CCAGGT | ATG (130) | GC | GCTAAG | ACG | GC | ACTACA | ATG (115) | GC |
| 2 | TCAGGT | ATG (133) | GC | GTTAAG | ACG | GC | AATACG | ATG (114) | GC |
| 3 | CCAGGT | ATG (130) | GC | GCTAAA | ACG | GC | AATACA | ATG (108) | GC |
| 4 | CCAGAT | ATG (99) | AC | GGTGAG | ACG | GC | AGTGAG | ATG (80) | CG |
| 5 | GTAGTC | ATG (91) | TC | GCTAAG | ACG | GC | GATACA | ATG (114) | TC |
| 6 | TTTAAA | ATG (126) | GC | GCTAAG | ACG | GC | ACTACA | ATG (115) | GC |
| 7 | CCAGGT | ATG (130) | GC | GCTAAG | ACG | GC | GGTACA | GTGa | GC |
| 8 | CCAGGT | ATG (130) | GC | GCTAAG | ACG | GC | AATACA | ATG (108) | GC |
| 9 | TCAGGT | ATG (133) | GC | GCTAAG | ACG | GC | CTTACA | ATG (129) | GC |
| 10 | CCAGGT | ATG (130) | GC | GCTAAG | ACG | GC | GGTACA | ATG (113) | GC |
| 11 | CCAGGT | ATG (130) | GC | GCTAAA | ACG | GC | ATTGAA | ATG (81) | CG |
ORF, opened reading frame.
Designates a putative alternative initiation codon VP3 in AAV7 deduced by sequences alignment of AAV serotypes capsid genes. The relative strength of Kozak TIS is expressed as % next to the initiation codon.18
In the rAAV5 construct selected for the analysis, the numerical value of the relative VP1 content was increased from 0.2–0.4 in the HEK293-derived vector (by two independent assays) to 0.7–1.1 in Sf9 cells, i.e., increased by 3-fold on average. For VP2, these values increased from 0.5 to 1.7, a similar 3-fold increase. A concurrent increase of VP2 was one of the unpredicted effects of the relative increase of rAAV5 VP1. The N termini of both VP1 and VP2 incorporate the so-called basic region 3 (BR3; PKRKKART) representing conservative nuclear localization sequence (NLS) motif, which is necessary for AAV to deliver its genome within the nucleus and subsequently transduce the cells.27, 28, 29 It is thus not surprising that a 3-fold increase of each VP1 and VP2 increases the yield of a more infectious virus.
The data provided above do not include direct comparison of rAAV5 and rAAV9 manufactured in recently described OneBac2.08 and the current system. However, one can relate the respective capsid ratios and conclude that for these serotypes, the newly designed cap helper genes significantly improved both AAV5 and AAV9 capsid stoichiometry, which translate into higher-potency viral vectors.
Another unexpected finding was the similarity of packaging efficiencies displayed by HEK293 and Sf9 cells, which we assessed by a surrogate parameter of full-to-empty particles ratios. Using the previously identified ratio of rep/cap expression cassettes of 1:2.5 to construct a stable producer cell line,7 we have achieved similar to HEK293 packaging efficiency.
With many clinical trials under way, assessing the genetic identity of rAAV stocks manufactured by different protocols becomes a pressing regulatory issue. Many groups have reported a collateral encapsidation of sequences derived from packaging host cells,25, 30, 31 bacterial helper plasmid backbones,25, 32 helper viruses,8, 31 and wild-type (WT) AAV rep/cap sequences.25, 33 To evaluate the genetic identity of the packaged rAAV cassette, we set to conduct NGS analysis of encapsidated single-stranded viral DNA. The pilot analysis showed uneven sequence coverage of the cassettes in the GC-enriched sequences, which were almost identical for both platforms. Utilizing PCR-free protocol, we showed that the drop of coverage apparently resulted from the PCR-induced artifacts during libraries preparation and not from packaging of the truncated rAAV genomes.34, 35 The accuracy of PCR-based methods for NGS libraries preparations was questioned by several groups as not appropriate for AAV-related analysis,36 especially in GC-rich palindromes such as inverted terminal repeats (ITRs), or applied for any GC-enriched sequence.37 Consequently, NGS analysis of rAAV vector preparations should be carried out using adequate protocols.
Analysis of the genetic identity of the viral DNA derived from both platforms showed no significant differences between the encapsidated rAAV DNA in insect versus human cells. Of note, however, is a documented deviation of the sequence of the pTR-Bac-UF26 plasmid DNA pool used to transfect HEK293 cells from the sequence of this plasmid in the database. The fact of such genetic drift of a plasmid DNA, although surprising, can be explained by the following mechanism. By necessity, rAAV cassette-containing plasmids are propagated in recombination-deficient strains of E. coli to maintain the integrity of AAV ITRs. These strains lack components of pathways (endA, recB, recJ) that catalyze rearrangement and deletion of nonstandard secondary and tertiary structures. As a result, plasmid DNA accumulates mismatched bases and point mutations that otherwise would be restored by the mismatch repair system. Mismatches in DNA, if not repaired, result in a high spontaneous mutation frequency. Therefore, during transfection, mutated plasmid DNA carries its heteroduplex “imprinted” structure over into host HEK293 cell nuclei, where it is either repaired38 or replicated copying the mutation. Thus, the plasmid DNA pool in a co-transfection protocol carries over some of the mutagenesis burden of E. coli into encapsidated AAV cassettes, which could be substantial for some GC-rich sequences. Interestingly, the GFP reporter cDNA appears to have a minimal number of mutations (Figure 5A), which could reflect its synthetic “humanized” origin.39 In this regard, depleting CpG motifs from rAAV cassettes, in addition to its immunogenicity-related effects,40 might also help to increase genetic stability of a parent plasmid DNA.
Direct side-by-side NGS analysis of identical rAAV cassettes manufactured by the two platforms revealed, unexpectedly, higher precision of viral DNA packaging in insect cells, encapsidating considerably less contaminating DNA (0.6% versus 3.5%). If the same trend persists for other rAAV cassettes, a 6-fold reduction would be significant if one takes into account that US Food and Drug Administration (FDA) guidelines state, “that the level of residual cell-substrate DNA should be ≤10 ng per dose” (https://ntrl.ntis.gov/NTRL/dashboard/searchResults/titleDetail/PB2013100474.xhtml).
Considering current clinical trials with rAAV whereupon the doses reached as high as 6 × 1013 vg/kg,41, 42 the vector, if produced in either HEK293 cells or even in the described above system, would incorporate amounts of contaminating DNA that would exceed that recommended by FDA guidelines. However, here we show that rAAV5 vectors produced using the OneBac system are significantly more infectious and, at the same time, encapsidate less amounts of foreign DNA. Therefore, this method of production improves the potential therapeutic outcome of the vector administration by reducing its effective dose and advancing safety.
Materials and Methods
rAAV5 and rAAV9 Production in HEK293 Cells
Both rAAV5 and rAAV9 vectors were produced by a triple co-transfection procedure as described previously.43 The construction of a helper plasmid pACG2R5C encoding Rep2 and Cap5 functions was described in detail previously.44 Plasmids harboring rAAV cassettes (rAAV5-Bac-UF26 or rAAV5-UF50-BC, as well as rAAV9-Bac-UF26; Figure S1) were used in combinations with pHelper and pRep2Cap5 (or pRep2Cap9) at a 1:1:1 molar ratio. Both rAAV5 and rAAV9 were purified using sequential double-round iodixanol buoyant density centrifugations. Specifically, the first-round iodixanol fraction containing full particles was diluted with 1× PBS-MgCl2-KCl (MK) buffer 2.5-fold and used in lieu of 15% iodixanol-1 M NaCl step in a standard gradient.43
rAAV5 and rAAV9 Production in Sf9 Cells
Bac-UF26 or UF50-BC rAAV cassettes were inserted into pFastBac backbone, and the respective BEVs were derived following Bac-to-Bac system guidelines. Plaque-purified BEVs were propagated to P3, titered by a plaque assay, and used to infect Sf9-based stable cell lines harboring rep2cap5 or rep2cap9 inducible helper genes, as described previously.7 Upon harvesting, freeze-thaw lysates were treated with benzonase and clarified by a high-speed centrifugation at 20,000 × g for 30 min. Supernatants were purified as described above for 293HEK-manufactured AAV.
One-Step Purification of rAAV5 and rAAV9
rAAV5 was purified from clarified crude lysate using AVB Sepharose high-performance affinity chromatography resin (GE Healthcare), as described previously.45 rAAV9 was purified by one-step affinity chromatography using POROS CaptureSelect AAV9 resin (Thermo Fisher) from a crude lysate. Specifically, a clarified crude lysate was applied under gravity to a column containing 0.5 mL of resin; the column was then washed with 10 column volumes of 1× PBS, followed by 1 mL of elution buffer: 50 mM citrate buffer (pH 3.0)-1 M NaCl. The eluted rAAV9 was immediately neutralized by 0.1 mL 1 M Tris-HCl (pH 8.4).
CryoEM
Sample preparation for cryogenic transmission electron microscopy (cryo-TEM) was performed in the EM Core of the University of Florida’s Interdisciplinary Center for Biotechnology Research. Three-microliter aliquots of suspended virus were applied to C-flat holey carbon grids (Protochips) and vitrified using a Vitrobot Mark IV (FEI) operated at 4°C and with ∼90% humidity in the control chamber. The vitrified sample was stored under liquid nitrogen and transferred into a Gatan cryo-holder (model 626/70) for imaging. The sample was examined using a 16-megapixel charge-coupled device (CCD) camera (Gatan) on a Tecnai (FEI) G2 F20-TWIN Transmission Electron Microscope operated at a voltage of 200 kV using low-dose conditions (∼20 e/Å2). Images were recorded with a defocus of approximately 3 μm to improve contrast.
In order to determine the ratio of packaged AAV particles per sample, we manually boxed full and empty AAV particles using EMAN2 software.46 Particles with non-icosahedral shape or incorrect particle diameter were excluded from the analyses. The number of full particles for each sample was determined by manual counting and then divided by the total number of boxed particles to calculate a percentage. A total of 10 micrographs was used for each sample, and more than 100 total particles were counted for each.
rAAV Titering
Direct comparative analysis of rAAV vectors requires an accurate estimate of the respective viral titers. Four independent assays were used to titer rAAV derived from HEK293 or Sf9 cells: (1) Droplet Digital PCR (ddPCR),47 to establish a reference standard, using Bio-Rad QX200 Digital PCR System; (2) QC-PCR using iTaq Universal SYBR Green Supermix kit (1725121; Bio-Rad) and qPCR Bio-Rad CFX Connect RealTime System; (3) a PicoGreen-based protocol;48 and (4) NTA using NanoSight 300 (NS-300; Malvern Instruments, Malvern, UK), to quantify the titer and the size of rAAV particles. After establishing the conditions of each protocol, and using a reference standard derived by ddPCR, the calculated titers from protocols 2 and 3 were always within factor of 2 and were averaged to derive the working titers. Briefly, the following procedures were followed.
ddPCR
A set of dilutions, in triplicates, in the range of 1 to 103 viral genome copies per reaction was prepared using 1× lactated Ringer (LR) solution supplemented with 0.05% Pluronic F-68. ddPCR was carried out using the following primers: TM_CMV_F 5′-ATAGGGACTTTCCATTGACGTC-3′, TM_CMV_R 5′-TGATACACTTGATGTACTGCCAAG-3′, TM_CMV_Probe FAM 5′-TGGGTGGACTATTTACGGTAAACTGCC-3′ BHQ.
QC-PCR
rAAV-UF26 transgene cassette was gel-purified after SmaI digestion of the respective plasmid pTR-Bac-UF26 to derive a standard curve. The recovered DNA was diluted to the approximate concentration of 1 ng/μL, and the precise concentration was determined by QUBIT dsDNA assay. Direct QC-PCR titering of highly purified Sf9- or HEK 29-produced rAAV was performed with the same primer set and using the standard curve prepared by 10-fold serial dilutions of a reference rAAV sample (ddPCR) and the gel-purified rAAV cassette (QC-PCR).
PicoGreen-Based Assay
PicoGreen-based assay was conducted by Quanti-iT PicoGreen dsDNA Assay kit (P7589; Life Technology), using Lambda DNA standard to calibrate the standard curve as described previously.48 Optical density was measured by PerkinElmer 1420 Multilabel Counter Victor V.
NTA
Prior to the NTA analysis, the titers of the viral stocks were assessed by polyacrylamide gel (PAAG) electrophoresis. Knowing approximate titers, 50 μl of AAV stock diluted in LR buffer was added to labeling mix containing labeling buffer (20 mM citric acid [pH 3.5], 0.1% Pluronic F68, 1 mM NaCl) and gold nanoparticles (catalog no. 741949; Sigma). After 30-min incubation at room temperature (RT), gold-labeled AAV was diluted by the labeling buffer to a final concentration of 5 × 108 to 3 × 109 particles/mL. Labeling mix without AAV and AAV in the labeling buffer without gold was used as a negative control. Measurements were carried out using NS-300 instrument with the following settings: laser type, Blue488; camera level, 15; number of frames, 749; time of recording, 30 s; number of records, 3 per each data point. At least four data points generated by NS-300 were used to calculate AAV titer.
MALDI-TOF
VP1, VP2, and VP3 bands were cut out from SDS-PAAG and prepared for trypsin digestion. Capsid proteins were digested in gel slices with sequencing-grade trypsin from Promega (Madison, WI, USA) using the manufacturer-recommended protocol. Briefly, bands were trimmed as close as possible to minimize background polyacrylamide material. Gel pieces were then washed in nanopure H2O for 5 min. The wash step was repeated twice followed by two cycles of de-staining with 1:1 v/v methanol:50 mM ammonium bicarbonate for 10 min each cycle. The gel pieces were dehydrated with 1:1 v/v acetonitrile:50 mM ammonium bicarbonate. The gel slices were rehydrated and incubated with DTT solution (25 mM in 100 mM ammonium bicarbonate) for 30 min prior to the addition of 55 mM iodoacetamide in 100 mM ammonium bicarbonate solution. Iodoacetamide was incubated with the gel slices in darkness for 30 min. Gel slices were washed again with two cycles of H2O and dehydrated with 1:1 v/v acetonitrile:50 mM ammonium bicarbonate. The protease was driven into the gel pieces by rehydration, then in 12 ng/mL trypsin in 0.01% ProteaseMAX Surfactant for 5 min. The gel piece was then overlaid with 40 μL of 0.01% ProteaseMAX surfactant:50 mM ammonium bicarbonate and gently mixed on a shaker for 1 hr. The digestion was stopped by adding 0.5% trifluoroacetic acid (TFA). Mass spectrometry (MS) analysis was either immediately performed to ensure high-quality tryptic peptides with minimal non-specific cleavage or frozen at −80°C until samples could be analyzed. The 18O-labeled digest was performed the same way, except the ProteaseMAX Surfactant and trypsin were prepared in H218O. In order to prevent back exchange, we inactivated trypsin by incubation at 100°C for 15 min. VP3 was digested using H218O, whereas VP1 was digested in regular H216O; the digestion products were mixed 1:1 and analyzed by MALDI-TOF. A similar analysis was conducted for VP2/VP3 as well.
MALDI-TOF was performed on a Bruker Daltonics Microflex LRF mass spectrometer (Bruker Daltonics, Bremen, Germany) operated in reflectron, positive ion mode with an N2 laser. Laser power was used at the threshold level required to generate signal. The instrument was calibrated with Peptide Calibration Standard II (Bruker Daltonics), which is a mixture of angiotensin II, angiotensin I, substance P, Bombesin, ACTH clip 1–17, ACTH clip 18–39, somatostatin 28, bradykinin fragment 1–7, renin substrate tetradecapeptide porcine with a covered mass range ∼700–3,200 Da. α-Cyano-4-hydroxycinnamic acid was used as the matrix and prepared as a saturated solution in 50% acetonitrile (can)/0.1% TFA (in H2O). Allotments of 1 μL of matrix and 1 μL of sample were thoroughly mixed together; 0.5 μL of this was spotted on the target plate and allowed to dry.
In Vitro Transduction Assay
rAAV5-Bac-UF26 and rAAV9-Bac-UF26 were assayed using C12 cells24 infected with rAAVs at an MOI of 2,000 and co-infected with adenovirus type 5 (Ad5) at an MOI of 5. Forty-eight hours after infection, cells, positive for mApple fluorescence, were scored by FACS.
AAV Injections
All animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee. Four- to five-week-old female BALB/c (The Jackson Laboratory, Bar Harbor, ME, USA) mice were used for the experiments.
All surgical procedures were performed using aseptic techniques and isoflurane gas anesthesia. Brain surgeries were performed as previously described.23 Briefly, once anesthetized, mice were placed in the stereotactic frame (Kopf Instruments, Tujunga, CA, USA), and 2 μL of either HEK- or 8B-derived rAAV5-UF50-BC vectors (4.75 × 1011 vg/mL) was injected into the right striatum (coordinates: anterior-posterior −0.3 mm, lateral −2.0 mm, dorsoventral −3.0 mm), through a glass micropipette with an inner diameter of 30–40 μm at a rate of 0.5 μL/min. The needle was left in place for 5 min prior to withdrawal from the brain.
BLI Imaging
Mice were imaged as previously described.23 Twelve minutes after intraperitoneal injection of D-luciferin (15 mg/mL in PBS, 126 mg luciferin/kg body weight), BLI measurements were obtained from region-of-interest analysis using a Xenogen IVIS Lumia in vivo imaging system (PerkinElmer, Waltman, MA, USA). Three mice were imaged at the same time with a field of view of 25 cm. An imaging time of 60 s with medium binning and an F-stop of 1 were used for the camera settings. The images displayed in each dataset were normalized to the appropriate color intensity scale. The BLI data are reported as raw data, as the total number of counts reaching the charge-coupled device detector.
Brain Tissue Preparation and Fluorescence Imaging
Mice were deeply anesthetized with pentobarbital (Beuthanasia-D) and perfused through the ascending aorta with 10 mL of saline solution, followed by 10 mL of ice-cold 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (pH 7.4). Brains were removed and post-fixed overnight at 4°C in 4% PFA. Sixty-micrometer-thick coronal sections were cut on a Vibratome stage VT1000 S (Leica Microsystems, Wetzlar, Germany). mApple fluorescence was analyzed by a variable mode laser scanner (Typhoon 9200; GE Amersham, Pittsburgh, PA, USA) or using inverted microscope DMI4000 B (Leica Microsystems).
NGS Analysis
NGS was performed by UF ICBR Core using HiSeq 3000 instrument (Illumina, San Diego, CA, USA) and paired-ended sequencing. To demonstrate the reproducibility of the selected NGS protocol, we prepared all DNA samples in duplicates. Similarly, all steps of NGS library synthesis, sequencing, and bioinformatics were conducted in parallel and in duplicates. The total numbers of reads for HEK293-derived and Sf9-derived rAAV5 vectors are shown in Tables S4 and S5, respectively.
The referenced DNA for HEK293-based production included all DNA sequences that could potentially contaminate rAAV stock: H. sapiens genome sequence (HEK293, GRCh38.p9, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.35 or RefSeq assembly accession: GCF_000001405.35); adenovirus helper plasmid (pHelper, GenBank: AF369965.1); pACGr2c5 encoding AAV2 Rep and AAV5 VP proteins (rep2cap5); and the respective plasmid backbones. For Sf9-based production, the following sequences were analyzed: S. frugiperda genome (JQCY02.1.fsa_nt.gz, GenBank: JQCY00000000.2); AcMNPV genome (GenBank: KM667940.1) including synthetic construct kanamycin resistance protein (GenBank: AF012346); lacZ-alpha peptide and beta-lactamase genes (GenBank: M74750) and F-plasmid DNA complete mini-F region (GenBank: M12987.1); FastBac shuttle plasmid backbone; and sequences encoding AAV2 Rep and AAV5 VP.
Accel-NGS 2S PCR-Free DNA Library Kit (Swift Biosciences, Ann Arbor, MI, USA) containing both PCR-free and PCR-enrichment options was utilized to produce NGS libraries. Deep DNase treatment of purified rAAV particles and double-stranded (ds) DNA synthesis were performed following the protocol described by Lecomte et al.25 with minor modifications. Briefly, 4 × 1011 vg-containing particles were extensively treated by Baseline ZERO and Plasmid-Safe exonucleases, followed by Proteinase K and RNaseA treatment. DNA was purified using the Mag-Bind RxnPure Plus Kit (Omega Bio-Tek, Norcross, GA, USA) using beads:DNA ratio of 2:1. After second-strand synthesis with DNApolI, DNA was sonicated using Covaris instrument. The following settings for DNA shearing were used: target size, 400 bp; peak increment power, 175 W; duty factor, 10%; cycles per burst, 200; time, 38 s; water level, 15; water temperature, 7.6°C–7.8°C; and reaction volume, 47 μL. Profiles of sheared input dsDNA and synthesized NGS libraries are presented in Figure S6. Samples of rAAV DNA derived from Sf9 and HEK293 cells demonstrated similar profiles and quality on each stage during NGS library preparation. At all stages of library preparations, the DNA quality was monitored by Tape station (Agilent) and Qubit (Thermo Fisher Scientific). For synthesis of PCR-free NGS libraries, 220 ng of dsDNA was sheared to a target size of 200 bp option input. For PCR-enriched libraries, 10% of DNA after Ligation Step 2 was amplified for eight cycles following manufacturer’s recommendations. Finally, all libraries were purified with 1× Mag-Bind beads.
Bioinformatics Analysis
The flowchart of the bioinformatics workflow is depicted in Figure S7. Fastq files were analyzed with the dedicated open source software ContaVect v2.025 modified to be used on a supercomputing cluster in its default configuration. The ContaVect’s code was adapted to appropriately scale the execution of blastn and bwa tools for maximum efficiency and throughput in a batch environment governed by a job scheduler. One of the changes was related to the ContaVect’s limitation that would always run blastn and bwa with the number of threads equal to the total number of central processing unit (CPU) cores on a computing node. Although this assumption may be valid in some cases when used on a personal workstation in a shared computing environment, the number of threads generally has to be equal to the number of CPU cores requested for the batch job or available within a virtual machine. Otherwise, the efficiency of the program in question suffers greatly, reducing the throughput of the analysis. The new modifications still allow the use of the old model for compatibility when the thread number is set to zero in the ContaVect configuration file, but now ContaVect can run blastn and bwa with the specified number of threads. One thread is used by default. In addition, an optional configuration file has been added that can be exported into the environment via the MATPLOTLIBRC environment variable to allow ContaVect to produce graphs using matplotlib without failure when run on cluster nodes without X11 environment. With the above modifications, the analyses had been successfully completed on the UF Research Computing HiPerGator supercomputer, and the run time of ContaVect on the data was reduced from multiple days to hours. All modifications have been submitted as GitHub code pull requests to the ContaVect authors in the following repositories: https://github.com/a-slide/ContaVect and https://github.com/a-slide/pyDNA. ContaVect was run using standard settings: minimal mean quality of reads, 25; minimal quality mapping of reads, 30; and minimal size of mapping, 25. All of the described adaptations have been acknowledged and incorporated into the source code. SNP variants were retrieved from BAM files by MiniCaller software, a Java-based application utilized for the analysis of sequences with high-depth coverage (https://github.com/lindenb/jvarkit/wiki/MiniCaller). MiniCaller-generated VCF files for HEK293 or Sf9 rAAV libraries, as well as positive control libraries (each in duplicates), were aligned to the GenBank reference molecules. Parsing of VCF files was performed by Python script that generates CSV files (http://nbviewer.jupyter.org/github/a-slide/iPython-Notebook/blob/master/Notebooks/VCF_analysis.ipynb). The FastQ, VCF, and CSV files are available from the UF Research Computing Data Sharing Server: https://bio.rc.ufl.edu/secure/zolotukhin/genther.
Author Contributions
Conceptualization, O.K., S.Z.; Methodology, O.K., D.M., H.R.M.-G., O.M., J.R.A., K.B.G., M.A.-M., S.Z.; Investigation, O.K., S.M.C., H.R.M.-G., O.M., J.R.A., K.B.G., S.Z.; Writing – Original Draft, S.Z.; Writing – Review & Editing, O.K., D.M., H.R.M.-G., O.M., J.R.A., K.B.G., M.M., R.H., M.A.-M.; Supervision, S.Z.
Conflicts of Interest
S.Z. is a co-founder of Lacerta Therapeutics, a licensee of the technology described in the manuscript.
Acknowledgments
S.Z. is supported, in part, by grant 2 R01 HL097088-05A1 from NIH/NHLBI. The authors acknowledge Dr. E. Lecomte for assisting with ContaVect v2.0 software, Dr. E. Ayuso for providing unpublished NGS data, and Dr. Moraga Amador from UF ICBR for assisting with NGS and NTA data acquisition.
Footnotes
Supplemental Information includes seven figures, five tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.08.003.
Supplemental Information
For each sample, 30 seconds video was recorded 3.
References
- 1.Girod A., Wobus C.E., Zádori Z., Ried M., Leike K., Tijssen P., Kleinschmidt J.A., Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 2.Rose J.A., Maizel J.V., Jr., Inman J.K., Shatkin A.J. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson F.B., Ozer H.L., Hoggan M.D. Structural proteins of adenovirus-associated virus type 3. J. Virol. 1971;8:860–863. doi: 10.1128/jvi.8.6.860-863.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buller R.M., Rose J.A. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J. Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snijder J., van de Waterbeemd M., Damoc E., Denisov E., Grinfeld D., Bennett A., Agbandje-McKenna M., Makarov A., Heck A.J. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J. Am. Chem. Soc. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urabe M., Ding C., Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 7.Aslanidi G., Lamb K., Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc. Natl. Acad. Sci. USA. 2009;106:5059–5064. doi: 10.1073/pnas.0810614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mietzsch M., Casteleyn V., Weger S., Zolotukhin S., Heilbronn R. OneBac 2.0: Sf9 cell lines for production of AAV5 vectors with enhanced infectivity and minimal encapsidation of foreign DNA. Hum. Gene Ther. 2015;26:688–697. doi: 10.1089/hum.2015.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohlbrenner E., Aslanidi G., Nash K., Shklyaev S., Campbell-Thompson M., Byrne B.J., Snyder R.O., Muzyczka N., Warrington K.H., Jr., Zolotukhin S. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol. Ther. 2005;12:1217–1225. doi: 10.1016/j.ymthe.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urabe M., Nakakura T., Xin K.Q., Obara Y., Mizukami H., Kume A., Kotin R.M., Ozawa K. Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J. Virol. 2006;80:1874–1885. doi: 10.1128/JVI.80.4.1874-1885.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urabe, M., Ozawa, K., Haast, S., and Hermens, W. (2015). Improved AAV vectors produced in insect cells. European Patent 2311967 A2 20110420 (EN), filed October 19, 20006, and published April 20, 2011.
- 12.Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol. Ther. 2008;16:924–930. doi: 10.1038/mt.2008.35. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 14.Cigan A.M., Donahue T.F. Sequence and structural features associated with translational initiator regions in yeast—a review. Gene. 1987;59:1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- 15.Joshi C.P., Zhou H., Huang X., Chiang V.L. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 16.Pesole G., Gissi C., Grillo G., Licciulli F., Liuni S., Saccone C. Analysis of oligonucleotide AUG start codon context in eukariotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014;10:748. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salganik M., Venkatakrishnan B., Bennett A., Lins B., Yarbrough J., Muzyczka N., Agbandje-McKenna M., McKenna R. Evidence for pH-dependent protease activity in the adeno-associated virus capsid. J. Virol. 2012;86:11877–11885. doi: 10.1128/JVI.01717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storms H.F., van der Heijden R., Tjaden U.R., van der Greef J. Considerations for proteolytic labeling-optimization of 18O incorporation and prohibition of back-exchange. Rapid Commun. Mass Spectrom. 2006;20:3491–3497. doi: 10.1002/rcm.2738. [DOI] [PubMed] [Google Scholar]
- 21.Ye X., Luke B., Andresson T., Blonder J. 18O stable isotope labeling in MS-based proteomics. Brief. Funct. Genomics Proteomics. 2009;8:136–144. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian W.J., Liu T., Petyuk V.A., Gritsenko M.A., Petritis B.O., Polpitiya A.D., Kaushal A., Xiao W., Finnerty C.C., Jeschke M.G., Inflammation and the Host Response to Injury Large Scale Collaborative Research Program Large-scale multiplexed quantitative discovery proteomics enabled by the use of an (18)O-labeled “universal” reference sample. J. Proteome Res. 2009;8:290–299. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsic D., Méndez-Gómez H.R., Zolotukhin S. High-accuracy biodistribution analysis of adeno-associated virus variants by double barcode sequencing. Mol. Ther. Methods Clin. Dev. 2015;2:15041. doi: 10.1038/mtm.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark K.R., Voulgaropoulou F., Johnson P.R. A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Ther. 1996;3:1124–1132. [PubMed] [Google Scholar]
- 25.Lecomte E., Tournaire B., Cogné B., Dupont J.B., Lindenbaum P., Martin-Fontaine M., Broucque F., Robin C., Hebben M., Merten O.W. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther. Nucleic Acids. 2015;4:e260. doi: 10.1038/mtna.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mietzsch M., Grasse S., Zurawski C., Weger S., Bennett A., Agbandje-McKenna M., Muzyczka N., Zolotukhin S., Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum. Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grieger J.C., Snowdy S., Samulski R.J. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 2006;80:5199–5210. doi: 10.1128/JVI.02723-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonntag F., Bleker S., Leuchs B., Fischer R., Kleinschmidt J.A. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 2006;80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popa-Wagner R., Sonntag F., Schmidt K., King J., Kleinschmidt J.A. Nuclear translocation of adeno-associated virus type 2 capsid proteins for virion assembly. J. Gen. Virol. 2012;93:1887–1898. doi: 10.1099/vir.0.043232-0. [DOI] [PubMed] [Google Scholar]
- 30.Allay J.A., Sleep S., Long S., Tillman D.M., Clark R., Carney G., Fagone P., McIntosh J.H., Nienhuis A.W., Davidoff A.M. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye G.J., Scotti M.M., Liu J., Wang L., Knop D.R., Veres G. Clearance and characterization of residual HSV DNA in recombinant adeno-associated virus produced by an HSV complementation system. Gene Ther. 2011;18:135–144. doi: 10.1038/gt.2010.102. [DOI] [PubMed] [Google Scholar]
- 32.Chadeuf G., Ciron C., Moullier P., Salvetti A. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol. Ther. 2005;12:744–753. doi: 10.1016/j.ymthe.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Nony P., Chadeuf G., Tessier J., Moullier P., Salvetti A. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J. Virol. 2003;77:776–781. doi: 10.1128/JVI.77.1.776-781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapranov P., Chen L., Dederich D., Dong B., He J., Steinmann K.E., Moore A.R., Thompson J.F., Milos P.M., Xiao W. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum. Gene Ther. 2012;23:46–55. doi: 10.1089/hum.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Ling C., Song L., Wang L., Aslanidi G.V., Tan M., Ling C., Srivastava A. Limitations of encapsidation of recombinant self-complementary adeno-associated viral genomes in different serotype capsids and their quantitation. Hum. Gene Ther. Methods. 2012;23:225–233. doi: 10.1089/hgtb.2012.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogné B., Snyder R., Lindenbaum P., Dupont J.B., Redon R., Moullier P., Leger A. NGS library preparation may generate artifactual integration sites of AAV vectors. Nat. Med. 2014;20:577–578. doi: 10.1038/nm.3578. [DOI] [PubMed] [Google Scholar]
- 37.Kozarewa I., Ning Z., Quail M.A., Sanders M.J., Berriman M., Turner D.J. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folger K.R., Thomas K., Capecchi M.R. Efficient correction of mismatched bases in plasmid heteroduplexes injected into cultured mammalian cell nuclei. Mol. Cell. Biol. 1985;5:70–74. doi: 10.1128/mcb.5.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zolotukhin S., Potter M., Hauswirth W.W., Guy J., Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faust S.M., Bell P., Cutler B.J., Ashley S.N., Zhu Y., Rabinowitz J.E., Wilson J.M. CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Invest. 2013;123:2994–3001. doi: 10.1172/JCI68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog R.W. A cure for hemophilia: the promise becomes a reality. Mol. Ther. 2016;24:1503–1504. doi: 10.1038/mt.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolgin E. Early clinical data raise the bar for hemophilia gene therapies. Nat. Biotechnol. 2016;34:999–1001. doi: 10.1038/nbt1016-999. [DOI] [PubMed] [Google Scholar]
- 43.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 44.Zolotukhin S., Potter M., Zolotukhin I., Sakai Y., Loiler S., Fraites T.J., Jr., Chiodo V.A., Phillipsberg T., Muzyczka N., Hauswirth W.W. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]
- 45.Smith R.H., Levy J.R., Kotin R.M. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol. Ther. 2009;17:1888–1896. doi: 10.1038/mt.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Lock M., Alvira M.R., Chen S.J., Wilson J.M. Absolute determination of single-stranded and self-complementary adeno-associated viral vector genome titers by droplet digital PCR. Hum. Gene Ther. Methods. 2014;25:115–125. doi: 10.1089/hgtb.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piedra J., Ontiveros M., Miravet S., Penalva C., Monfar M., Chillon M. Development of a rapid, robust, and universal picogreen-based method to titer adeno-associated vectors. Hum. Gene Ther. Methods. 2015;26:35–42. doi: 10.1089/hgtb.2014.120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each sample, 30 seconds video was recorded 3.