Abstract
mRNA vaccines are rapidly emerging as a powerful platform for infectious diseases because they are well tolerated, immunogenic, and scalable and are built on precise but adaptable antigen design. We show that two immunizations of modified non-replicating mRNA encoding influenza H10 hemagglutinin (HA) and encapsulated in lipid nanoparticles (LNP) induce protective HA inhibition titers and H10-specific CD4+ T cell responses after intramuscular or intradermal delivery in rhesus macaques. Administration of LNP/mRNA induced rapid and local infiltration of neutrophils, monocytes, and dendritic cells (DCs) to the site of administration and the draining lymph nodes (LNs). While these cells efficiently internalized LNP, mainly monocytes and DCs translated the mRNA and upregulated key co-stimulatory receptors (CD80 and CD86). This coincided with upregulation of type I IFN-inducible genes, including MX1 and CXCL10. The innate immune activation was transient and resulted in priming of H10-specific CD4+ T cells exclusively in the vaccine-draining LNs. Collectively, this demonstrates that mRNA-based vaccines induce type-I IFN-polarized innate immunity and, when combined with antigen production by antigen-presenting cells, lead to generation of potent vaccine-specific responses.
Keywords: mRNA vaccines, nonhuman primates, antigen-presenting cells
mRNA vaccines have been proven to be suitable and efficient against pandemic pathogens. However, the immune processes after mRNA vaccination that lead to robust responses remain elusive. Now in Molecular Therapy, Liang et al. (2017) define the target cells and immune responses at the vaccination sites and in the lymph nodes that result in vaccine immunity.
Introduction
Nucleic acid vaccines based on plasmid DNA, viral vectors, or mRNA are being evaluated for several clinical applications including cancer and allergy.1, 2 Recently, the mRNA platforms have also shown strong potential as vaccines against infectious diseases.3, 4, 5, 6, 7, 8 Modified mRNA vaccines are attractive because they offer precision in antigen design, good tolerability, and broad immune responses with a highly scalable manufacturing platform. mRNA vaccines circumvent the challenges posed by pre-existing or post-vaccination immunity against viral vector platforms and overcome the need for multiple high doses and delivery devices needed for DNA vaccines. Furthermore, mRNA vaccine against Zika virus infection conferred sterilizing immunity in mice7, 8 and rhesus macaques,7 which demonstrates the potential to offer protection against emerging pandemic infections. The potential of rapid mRNA vaccine development and production was demonstrated by the ability to produce a self-amplifying mRNA (SAM) vaccine platform for pre-pandemic influenza just eight days after release of the viral gene sequence.6
mRNA vaccines can be refined by modifications to the mRNA chemistry that modulate mRNA stability, innate immune activation and the level of translation.9, 10 Such modified mRNA constructs have shown promise for regenerative11, 12 and cancer therapies.2, 13 In addition to mRNA modification, the choice of the delivery formulation enables the use of low doses of mRNA without compromising the vaccine responses and avoiding the need for adjuvants. Formulation of mRNA with liposomes2 or protamine14 has been shown to stimulate RNA sensors like TLR7 in mouse dendritic cells (DCs). More recently, lipid nanoparticles (LNPs) have been tested in clinical therapy trials as delivery systems for RNA.15 We recently showed that mRNA encoding hemagglutinin (HA) of H10N8 (A/Jiangxi-Donghu/346/2013) influenza A virus encapsulated in LNP induces potent and sustained immune responses in mice, ferrets, and cynomolgus macaques.16
The innate immune mechanisms by which mRNA vaccines generate potent adaptive immunity are largely unknown. Characterization of the specific target cells responsible for translation of the mRNA vaccine at the site of administration or in the draining lymph nodes (LNs) is lacking as well as the type of cellular activation of target cells. In this study, we therefore confirmed the immunogenicity of the LNP/H10 mRNA formulation in rhesus macaques that received a homologous prime and boost immunization by either the intradermal (i.d.) or the intramuscular (i.m.) route. We thereafter applied flow cytometric, confocal microscopy and gene expression analyses to decipher key innate immune responses induced locally at the injected sites (skin and muscle) and the draining LNs. For the studies, we administered fluorescently labeled LNP with mRNA encoding the fluorescent protein, mCitrine, to identify the distinct antigen-presenting cell (APC) subsets translating vaccine mRNA after LNP uptake. Lastly, we demonstrate priming of vaccine-specific T cell responses in vaccine-draining LNs as a consequence of activated APCs producing the mRNA-encoded vaccine antigen. Collectively, the data describe the local immune events after administration of LNP/mRNA in vivo from the injection site to the draining LN to increase our understanding of how mRNA vaccines target key APCs to generate vaccine-specific T cell and B cell responses.
Results
LNP/mRNA Induces Robust Vaccine Responses by Distinct Routes of Immunization
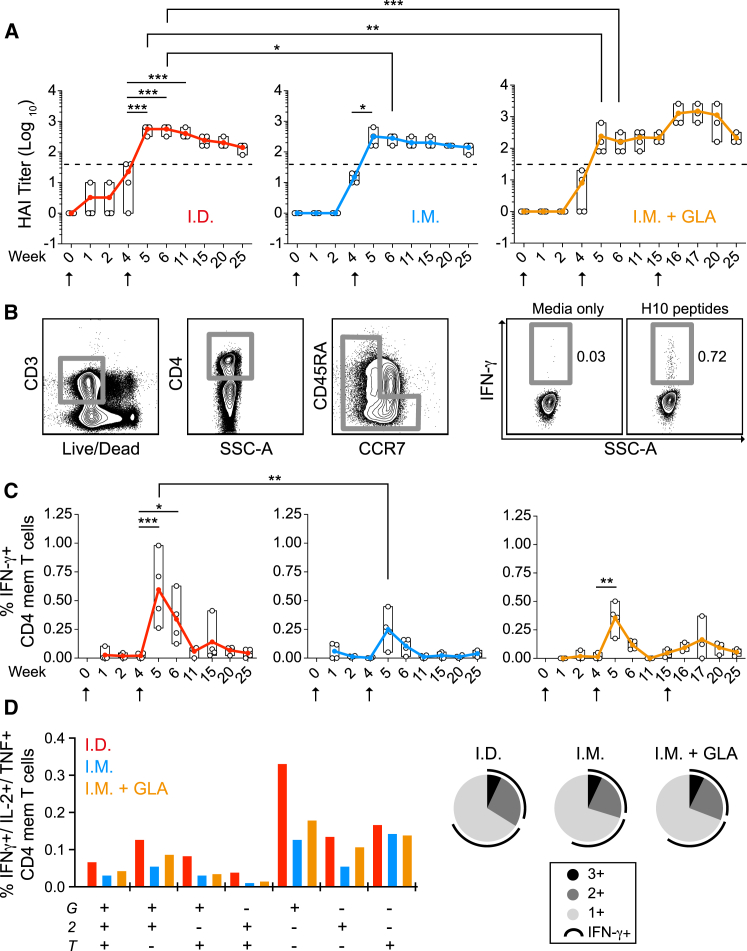
An mRNA vaccine encoding full-length HA H10 of H10N8 (A/Jiangxi-Donghu/346/2013) formulated in LNPs was first evaluated in rhesus macaques by determining the levels of neutralizing hemagglutination inhibition (HAI) titers to H10 (Figure 1A) and H10-specific CD4+ T cell responses (Figures 1B–1D). Rhesus macaques received a prime and boost immunization by either i.m. or i.d. administration. Some animals in the i.d. group had detectable HAI titers after priming, but they were below the threshold (1:40) accepted as a correlate of protection for seasonal influenza.17 The HAI titers in all groups were significantly increased following the second immunization and remained above the protective threshold for the remainder of the study (21 weeks). The titers were significantly higher in the i.d. group compared to the i.m. group for two weeks after the boost but were thereafter at similar levels. To evaluate any enhancement by the addition of an adjuvant, a third group received the vaccine i.m. together with the TLR4 agonist glucopyranosyl lipid adjuvant (GLA). However, GLA did not increase the titers indicating that the LNP/mRNA formulation itself was sufficiently immunogenic. The GLA group received a third i.m. immunization that resulted in a transient increase in HAI titers, which returned to similar levels as the other groups five weeks later (Figure 1A).
Figure 1.
LNP-Encapsulated Modified mRNA Platform Elicit Strong Vaccine Immunity by Distinct Vaccination Routes
(A–D) Immunogenicity analysis of rhesus macaques after intradermal (i.d.) or intramuscular (i.m.) immunizations with LNPs containing mRNA encoding hemagglutinin of H10N8 influenza A virus (H10) alone or co-formulated with GLA adjuvant (i.m. + GLA) (n = 4/group). (A) H10-specific hemagglutination inhibition (HAI) titers. Immunizations (arrows) were given at weeks 0 and 4. The i.m. + GLA group was boosted again at week 15. Filled and open circles show mean and individual values, respectively. Dashed lines indicate the threshold for protective titers. (B) Gating strategy of cytokine+ total CD4 memory T cells. (C) Percentage of IFNγ+ CD4+ H10-specific memory T cells. (D) CD4+ T cell polyfunctionality as measured by IFNγ (G), IL-2 (2), and TNF (T) expression. Pie slivers in black, gray, and light gray indicate triple, double, or single cytokine+ cells, respectively, and arc shows IFNγ+ cells. Two-way Anova test with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001.
The i.d. group has the highest HAI titers, as well as a significantly higher number of H10-specific CD4+ T cells early after boost (Figure 1C). The majority of H10-specific CD4+ T cells produced only interferon γ (IFNγ) and polyfunctionality (i.e., production of IFNγ, interleukin-2 [IL-2], and tumor necrosis factor [TNF] or a combination thereof) was similar between the groups and was not enhanced by the addition of GLA (Figure 1D). H10-specific CD8+ T cell responses were detected in all groups after the boost but were more modest than the CD4+ T cell responses (Figure S1).
Immune Cells Are Rapidly Mobilized to the Site of LNP/mRNA Vaccine Administration
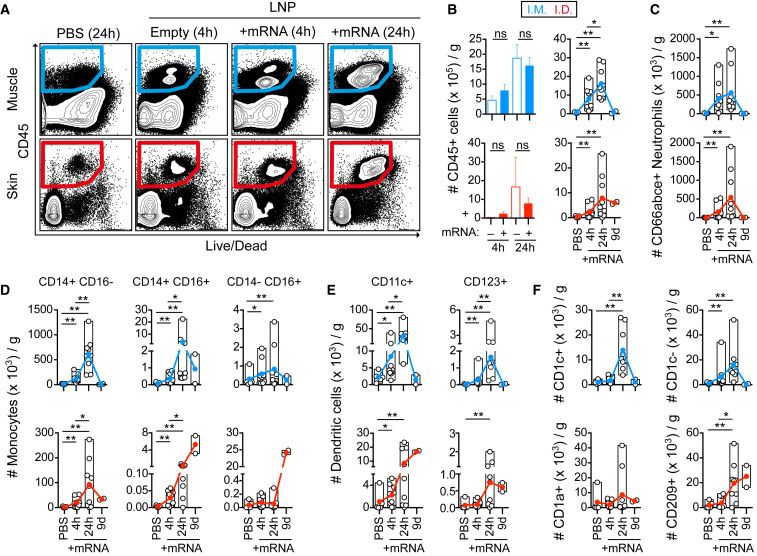
Details on the mechanisms of action by which LNP/mRNA-based vaccines generate strong vaccine responses are largely lacking. Whether distinct immunization routes lead to the targeting of different immune cells and induction of distinct innate immune responses in the context of LNP/mRNA vaccine immunity is unexplored. The sequence of immune events culminating into vaccine-induced immunity is initiated by early transient local inflammation that recruits immune cells, including APCs, to the site of delivery. To analyze the innate immune responses, such as cell infiltration, vaccine uptake, and translation of mRNA vaccine after administration, rhesus macaques received i.m. or i.d. vaccine injections of LNP with mRNA encoding the fluorescent protein mCitrine formulated in Atto655-labeled LNP. Injection of PBS or “empty” non-labeled LNP without mRNA cargo served as controls.
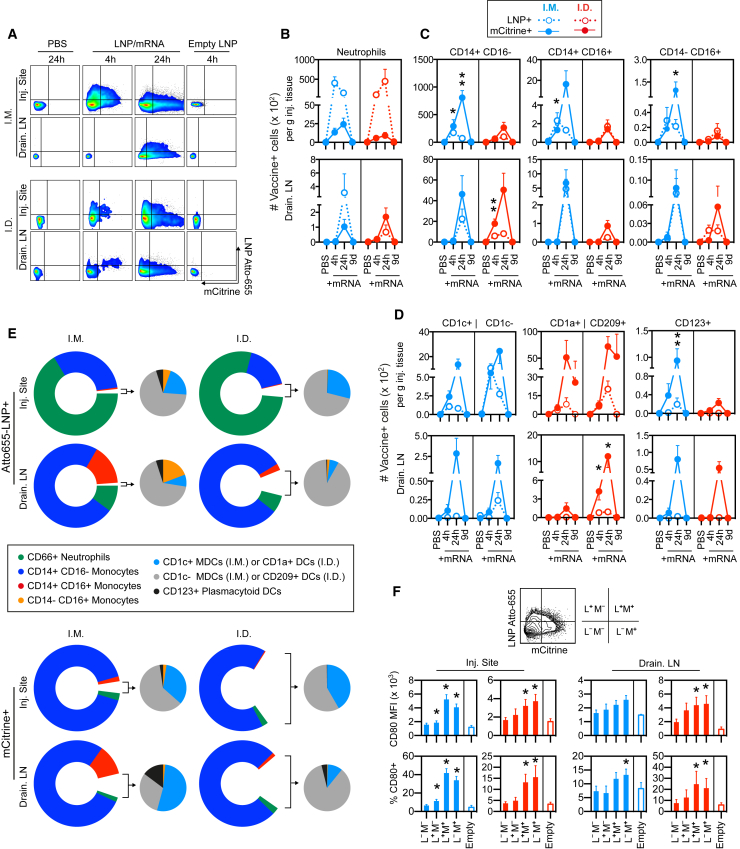
At 4 and 24 hr as well as 9 days, biopsies from the site of injection (skin or muscle) were analyzed for cell infiltration. Compared to the PBS-injection site, there was rapid recruitment of CD45+ immune cells to the LNP/mRNA-injected sites (Figures 2A and 2B). The level of cell infiltration was higher at 24 hr compared to 4 hr. Cell infiltration was found regardless of whether the LNP contained mRNA or not, indicating that LNP itself was capable of inducing recruitment. Multiple cell subsets were defined within the CD45+ population (Figure S2A-B). Injection of LNP alone resulted in a similar pattern of infiltrating cell subsets as LNP/mRNA (Figure S3A-B). At 24 hr, CD66abce+ neutrophils and classical CD14+ CD16− monocytes were the most frequent cell type infiltrating the skin and muscle after i.d. and i.m. administration, respectively (Figures 2C and 2D). Both skin and muscle also showed a noticeable increase in intermediate (CD14+ CD16+) monocytes as well as non-classical (CD14-/dim CD16+) monocytes reported to have efficient antigen uptake and/or produce inflammatory cytokines upon specific TLR stimulation.18, 19 The kinetics of cell infiltration was largely similar between i.m. and i.d. administration except that some monocyte and DC subsets remained at elevated levels nine days after i.d. delivery, although no other sign of inflammation was evident at the site.
Figure 2.
Rapid Immune Cell Infiltration in Response to LNP/mRNA Administration to Distinct Injection Sites
(A–F) Cell suspensions obtained from muscle and skin injected with PBS or indicated formulations at 4 and 24 hr or 9 days post-immunization were used for enumeration of immune cells per gram tissue from the injection sites. (A) Gating of infiltrating live CD45+ immune cells in muscle (i.m.) and skin (i.d.) injected with PBS, LNP without mRNA (Empty), or with mRNA content at 4 and 24 hr. (B) Comparison of CD45+ immune cell levels in muscle versus skin at 4 or 24 hr after injection of empty LNP (–) or LNP with mRNA cargo (+). (C and D) Characterization of the CD45+ mobilized immune cell subsets: (C) CD66abce+ neutrophils and (D) CD14+ CD16− classical monocytes, CD14+ CD16+ intermediate monocytes, and CD14− CD16+ non-classical monocytes. (E) CD11c+ myeloid dendritic cells (DCs) and CD123+ plasmacytoid DCs. (F) CD1c (BDCA-1) + or – DCs in muscle and CD1a+ or CD209 (DC-SIGN)+ APCs in skin at 4 and 24 hr (n = 5/group) and day 9 (n = 2/group). Filled and open circles show the mean and individual values, respectively. *p < 0.05, **p < 0.01; ns, not significant. Wilcoxon signed-rank test.
DCs consist of a heterogeneous group of cells; CD11c+ myeloid DCs (MDCs), including the CD1c+ MDC subset, are potent inducers of CD4+ T cell responses, and CD123+ plasmacytoid DCs (PDCs) are efficient producers of type I IFNs.20 The numbers of MDCs (CD1c+ and CD1c−) and PDCs were significantly elevated at both the muscle and skin vaccine injection sites (Figures 2E and 2F). The skin harbors epidermal CD1a+ Langerhans cells, CD209 (DC-SIGN)+ dermal DCs, CD1a+ dermal DCs, and CD209+ macrophages.21 The CD209+ APCs were found to be especially elevated after LNP/mRNA administration (Figure 2F). In conclusion, there was a rapid and in most cases transient infiltration of APCs to the LNP/mRNA-injected site.
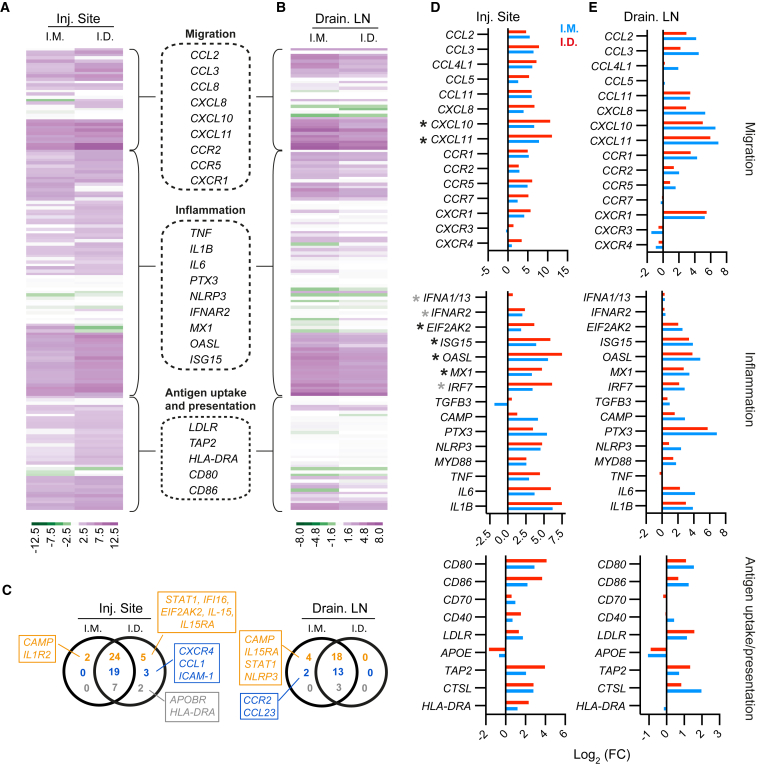
Upregulation of Genes Associated with IFN Responses at Injection Sites and in the Draining LNs
We analyzed whether the robust cell infiltration associated with LNP/mRNA administration was accompanied by modulation of genes associated with innate immune activation. Transcriptomic analyses were performed on biopsies from the injection sites as well as draining LNs collected at 24 hr post-immunization. A panel of 154 pre-selected genes were grouped by their involvement in inflammation, migration or antigen uptake and presentation (Figures 3A and 3B). From this panel, the i.d. and i.m. groups modulated a common set of 50 and 34 genes with log2 (fold change) ≥2 at the LNP/mRNA injection sites and in the draining LNs, respectively (Figure 3C). This suggests that i.m. and i.d. delivery of LNP/mRNA to a large extent induces similar innate immune activation at 24 hr. This may help explain the similar H10-induced adaptive responses in the groups after the prime-boost immunization (Figures 1A–1D).
Figure 3.
Gene Expression Analysis of the LNP/mRNA Injection Sites and the Draining LNs
(A–E) Gene regulation at the LNP/mRNA vaccine injection sites and in the draining LNs relative to corresponding PBS controls at 24 hr. log2 fold change (FC) represents log2 (ratio of the mean expressions at vaccine and PBS sites) (n = 4/group). (A and B) Heatmaps of selected genes at the injection sites (A) and draining LNs (B) that are primarily involved in inflammation, migration, antigen uptake, and presentation. Examples of these genes are in the dashed boxes. (C) The number of selected genes with log2 (FC) ≥ 2 exclusively or mutually expressed in i.m. and i.d. groups and involved in inflammation (orange), migration (blue), or antigen uptake and presentation (gray). Boxes show upregulated genes only in i.m. or i.d. group. (D and E) log2 (FC) of specific genes of interest at the vaccine injection sites (D) and in the draining LNs (E) related to the indicated pathways. Gray asterisks are genes involved in IFN signaling, and black asterisks are IFN-inducible genes.
Genes associated with inflammatory mediators, e.g., IL1B, MYD88, PTX3, and NLRP3, were upregulated in the LNP/mRNA-injected sites and respective draining LNs (Figures 3D and 3E). As reported recently, mice immunized with mRNA upregulated genes specifically connected to the IFN response, including type I IFN-inducible MX1 (MxA).22 In addition to upregulation of MX1, IFN-inducible chemokines CXCL10 (IP-10) and CXCL11 (I-TAC) were expressed at high levels especially in the skin of the i.d. group and at similar levels in the draining LNs of both groups. In fact, CXCL10 and CXCL11 were the highest upregulated IFN-inducible genes expressed at about 7- to 11-fold higher levels than the PBS control sites. While upregulation of the IFNα receptor 2 (IFNAR2) gene was detected after both i.d. and i.m. administration, expression of the IFNα 1/13 was mainly observed in the skin 24 hr post-immunization. Genes for LNP uptake (LDLR)23 plus antigen processing (CTSL) and antigen loading (TAP2) were also upregulated along with the T cell co-stimulatory molecules CD80 and CD86 at both the injection sites and in the draining LNs.
LNP/mRNA Exposure Results in Type I IFN Production and Phenotypic Differentiation of APCs
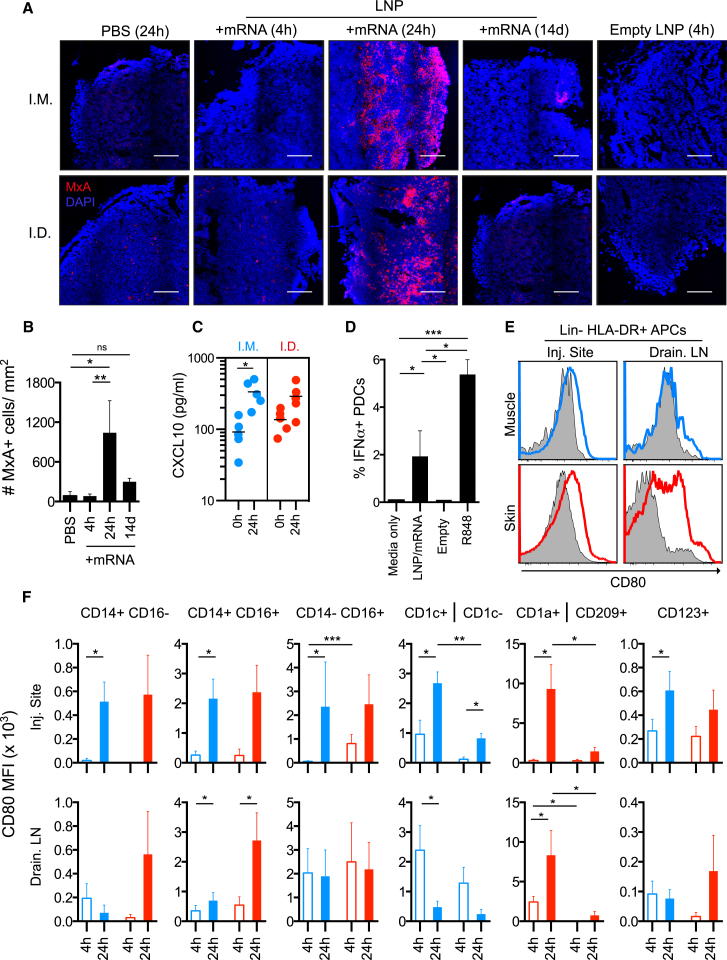
The gene expression pattern correlated with a transient type I IFN response on the protein level observed by upregulation of MxA in the draining LNs (Figure 4A) after LNP/mRNA administration. While MxA was not found at 4 hr, it was readily detected at 24 hr and returned to much lower levels after 14 days. As expected, MxA expression was not detected in control sites receiving PBS and empty LNP (Figures 4A and 4B). In line with the strong upregulation of the IFN-inducible CXCL10 gene at 24 hr after LNP/mRNA administration, the plasma levels of CXCL10 (IP-10) were also elevated (Figure 4C). Protamine-complexed mRNA and self-replicating RNA have been shown to induce IFNα in vitro14, 24 and RNA formulated in liposomes stimulated IFNα in vivo in mice in a TLR7-dependent manner.2, 22, 25 Since PDCs express high levels of TLR7 and are unique in their secretion of high levels of type I IFNs, we exposed them to LNP/mRNA in vitro. LNP/mRNA, but not empty LNP, induced detectable IFNα production in PDCs indicating a contribution by the mRNA content. However, the responses to LNP/mRNA were lower compared to the synthetic TLR7/8 agonist R848 (Figure 4D).
Figure 4.
LNP/mRNA Induce Cellular Activation with Type I IFN Response Signature
(A–F) Analysis of cellular activation in situ post-immunization and in vitro. (A and B) Type I IFN-inducible MxA expression (red) in LNs draining PBS, LNP/mRNA, or empty LNP injection sites at the indicated time points (A) and the number of MxA+ cells per mm2 LN area in i.m. and i.d. group combined (B). n = 4/group. Mean ± SEM is shown. DAPI+ cell nuclei in blue. Scale bars, 200 μm. (C) Serum CXCL10 (IP-10) prior and 24 hr after immunization (n = 4/group). Bar represents the mean. (D) IFNα production in vitro by rhesus CD123+ PDCs stimulated for 16 hr with the indicated conditions. n = 8. Mean ± SEM is shown. (E) Expression of co-stimulatory CD80 at 24 hr on lineage (CD3/CD8/CD20)− HLA-DR+ APCs at the injection sites and in the draining LNs. Gray-filled histogram are PBS control sites, and blue (i.m.) and red (i.d.) lines denote LNP/mRNA sites from the same animal. (F) Compiled background (i.e., PBS site) subtracted from the mean fluorescence intensity (MFI) of CD80 on APC subsets at 4 and 24 hr. n = 4/group. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, ns (not significant). Wilcoxon signed-rank test.
LNP/mRNA may induce cellular activation directly but also in a bystander manner via type I IFNs as previously described.26 Consistent with the upregulated CD80 gene expression in the injection sites and LNs, we found that APCs at these sites upregulated CD80 surface expression compared to APCs from the donor-matched PBS injection sites (Figure 4E). The CD14+ CD16+ and the CD14− CD16+ monocyte subsets plus CD1a+ DCs infiltrating the injection sites and the skin-draining LNs showed the highest upregulation of CD80 (Figure 4F). In the i.m. group, CD1c+ MDCs were more efficient at upregulating CD80 than CD1c− DCs. CD1a+ APCs responded more strongly than CD209+ APCs both in the LNP/mRNA injected skin and its draining LNs. PDCs in both groups showed increased CD80 expression.
Interestingly, LNP alone, which was sufficient to induce immune cell infiltration to the injection sites (Figures 2A and 2B), did not show marked CD80 upregulation (Figure S4A). This suggests that the mRNA cargo is the main inducer of cellular activation of APCs. We confirmed that immune activation was not related to the specific protein encoded by the mRNA, as APCs upregulated CD80 or CD86 with similar efficiency in vitro when LNP contained mRNA encoding H10 or the fluorescent mCitrine protein (Figure S4B).
Multiple APC Subsets Translate the Vaccine mRNA In Vivo
Previous mouse studies showed translation of administered RNA in organs or tissues by in vivo bioluminescence2, 27, 28, 29 or by histological staining.4, 30 Although translated mRNA is detected by these approaches, they do not specify the infiltrating or tissue resident immune cells capable of vaccine mRNA translation. We used Atto655-labeled LNP with mRNA encoding the fluorescent mCitrine protein for separate detection of LNP uptake and mCitrine translation in specific target cells following administration.
A noticeable number of total APCs, as defined by expression of HLA-DR, were found to contain LNP and/or mCitrine already at 4 hr but were increased at 24 hr at the injection sites and in the draining LNs (Figure 5A). Distribution of the vaccine at the injection sites was also verified in situ (Figure S5A). The i.d. group showed a slightly more rapid appearance of mCitrine+ APCs in the draining LNs. No LNP+ or mCitrine+ cells were detected at the PBS or unlabeled empty LNP control sites demonstrating specificity of the signal and that the uptake and translation of LNP/mRNA are restricted to the vaccination sites and their draining LNs.
Figure 5.
Efficient LNP Uptake and Vaccine mRNA Translation Coincide with Cellular Activation
(A–F) Animals received mRNA-encoding fluorescent mCitrine protein encapsulated in Atto-655-labeled LNP at the indicated time points for analysis of LNP uptake and mCitrine mRNA translation per gram tissue from the injection sites and the draining LNs. (A) Uptake of LNP and translation of the delivered mRNA by lineage− HLA-DR+ APCs in suspensions of muscle (i.m.) and skin (i.d.) injection sites and the draining LNs at 4 versus 24 hr post-injection. Empty LNPs indicate unlabeled LNPs without mRNA. (B–D) Compiled data of LNP uptake and mRNA translation by neutrophils (B) and monocyte (C) or DC (D) subsets at 4 or 24 hr (n = 4/group) and day 9 (n = 2/group). Open and closed circles denote mean ± SEM of LNP+ and mCitrine+ cells, respectively. Asterisks indicate significantly higher mCitrine+ subsets compared to the opposing group or cell subset at the same time point. Mann-Whitney test. (E) Proportions of LNP+ or mCitrine+ cell subsets. Doughnut and pie charts represent the more versus less frequent cell subsets, respectively. (F) CD80 expression on HLA-DR+ Lin− APCs according to their LNP uptake (L) and mCitrine mRNA translation (M) as indicted by the quadrant gate. CD80 MFI and the percentage of CD80+ of these populations are compiled, and asterisks indicate significant differences compared to the L– M– population. n = 4/group. Mean ± SEM is shown. Wilcoxon signed-rank test. *p < 0.05, **p < 0.01.
Infiltrating neutrophils were efficient at internalizing LNP but showed low capacity to translate the mRNA (Figure 5B). In contrast, classical CD14+ CD16− monocytes were found to be most abundant mCitrine+ immune cells at 24 hr after LNP/mRNA delivery. In addition, APC populations such as the CD16+ monocyte subsets, CD1c+ and CD1c− DCs as well as CD123+ PDCs and CD1a+ and CD209+ APCs at the LNP/mRNA injection sites also showed clear mCitrine translation especially at 24 hr (Figures 5C and 5D). Among the less abundant APCs, CD14+ CD16+ and CD14− CD16+ monocytes in the LNP/mRNA-draining LNs of the i.m. group were proportionally the most efficient cells at LNP uptake at 24 hr, while CD14+ CD16+ monocytes and PDCs translated more mCitrine (Figure 5E). Overall, there was more mCitrine+ and less LNP+ monocytes and DCs at 24 hr compared to 4 hr. The inverse relation may indicate that a large proportion of the acquired LNP has been degraded intracellularly at 24 hr and the mRNA cargo has advanced to translation. Detachment or quenching of the Atto-655 dye of the LNPs is unlikely since LNP was readily detectable in neutrophils at both time points (Figure 5B). While CD1a+ and CD209+ APCs in skin showed some low mCitrine signal at day 9, no other mCitrine+ immune cells were detected at the injection sites or draining LNs (Figures 5B–5D).
Uptake of LNPs was reported to be facilitated in the presence of ApoE.27 Monocytes and DCs highly express LDL- and LDL-like receptors.23 B cells also express LDL receptors,31 but were few in comparison to monocytes and DCs at the injection sites (Figures S2A and S2B), but LNP+ mCitrine + B cells and T cells existed in the draining LNs in both the i.d. and i.m. groups (Figure S5B).
mRNA Translation in APCs Leads to Phenotypic Differentiation
Since monocytes and DCs at the injection sites and in the draining LNs showed efficient LNP uptake and mRNA translation in addition to phenotypic differentiation, we assessed whether these features co-existed in the same cell. We found higher expression of the T cell co-stimulatory marker, CD80, in the APCs that showed mRNA translation (mCitrine+) compared to mCitrine- APCs, which suggests that cellular activation during mRNA translation ultimately result antigen-loaded APCs optimized for antigen presentation (Figure 5F). As expected, there was no CD80 upregulation on cells exposed to empty LNP without mRNA. We observed the same pattern in isolated rhesus and human APCs exposed in vitro to LNP/mRNA (Figure S5C).
Priming of T Cell Responses Is Restricted to the mRNA Vaccine-Draining Lymph Nodes
We have earlier shown that the location of vaccine+ APCs after protein/adjuvant vaccine administration is exclusively in the vaccine-draining LNs leading to priming of CD4+ T cells at this site.32 We investigated if H10-specific CD4+ T cells were initially generated by the LNP H10 mRNA vaccine in the LNs draining vaccine administration versus irrelevant distally located LNs at day 9 post-immunization. Carboxyfluorescein succinimidyl ester (CFSE)-labeled LN suspensions were cultured with H10 peptides to stimulate proliferation of H10-specific CD4+ T cells identified by CFSE-dilution. CD4+ T cell proliferation was strictly found in the vaccine-draining LNs and not present in the irrelevant control gut-draining LNs (Figure 6A). This demonstrates that priming of responses occurs exclusively in the draining LNs of mRNA vaccine administration. The location of the priming was confirmed by additional experiments where IFNγ production by H10-specific CD4+ T cells was detected only in the vaccine-draining LNs after expansion for 3 days followed by restimulation with H10 peptides (Figure 6B). Thus, LNP/mRNA-induced immunity, as represented by the priming of H10-specific CD4 T cells, was restricted to the LNs draining vaccine administration sites. Collectively, this study shows that mRNA vaccine delivery results in rapid cell infiltration and type I IFN-polarized activation coinciding with effective translation of the mRNA in APCs, which result in induction of robust vaccine-specific immunity.
Figure 6.
Priming of H10-Specific CD4+ T Cells Is Restricted to Draining LNs
(A and B) H10-naive animals received i.m. or i.d. immunizations with LNP with H10 mRNA upon which LNs draining vaccine injection or irrelevant non-vaccine draining LNs were harvested 9 days later for evaluation of H10-specific of CD4+ T cell responses. (A) CFSE-labeled suspensions of LNs draining i.m. or i.d. LNP/H10 mRNA injection sites or irrelevant gut draining control LNs at day 9 post-immunization were cultured for 3 days with H10 peptides for CFSE dilution by responding and proliferating H10-specific CD4+ T cells. (B) IFNγ production after 6 hr H10 peptide stimulation by H10-specific CD4+ T cells in the indicated LN cell suspensions that were cultured for 3 days with H10-peptides (i.e., to allow expansion of H10-specific CD4+ T cells) or with media only. n = 2/group. Mean ± SD is shown.
Discussion
mRNA and modified mRNA vaccines have emerged as a promising vaccine platform and are being evaluated in preclinical studies as well as human trials for infectious diseases.3, 4, 5, 6, 16 Despite this, several fundamental aspects of the mechanisms of action of mRNA vaccines are largely unknown. In this study, we show that two immunizations of LNP/H10 mRNA without adjuvant induced antibody titers that persisted above the protective level for seasonal influenza transmission during the study period of 25 weeks. We found that APCs were targeted by the vaccine both at the site of administration and in the draining LNs, leading to rapid translation of the vaccine antigen and phenotypic differentiation particularly in the APCs that produce the antigen. The innate immune cell activation was characterized by mRNA-mediated type I IFN responses, which were transient and local. Generation of vaccine-specific immunity developed within 9 days after immunization and exclusively occurred in the vaccine-draining LNs regardless of which administration route (i.d. or i.m.) was used.
Efficient targeting of professional APCs by mRNA vaccines may be one mechanism by which vaccine-specific responses are rapidly and efficiently generated.2 Using multicolor flow cytometry of cell suspensions from the site of injection and the draining LNs, we found that the predominant cell populations that were mobilized after LNP/mRNA vaccine delivery were neutrophils, monocytes and DCs. This infiltration pattern is reminiscent of what was reported on mouse muscle receiving the SAM vaccine in nanoemulsion4 and by our previous observations with administration of an adenoviral vector33 or protein antigen with different adjuvants in the NHP model.32 However, here we found that although these cell populations efficiently internalized LNP according to their high endocytic activity, only monocytes and MDCs showed high translation of the mRNA-encoding protein. We found that H10 responses were specifically generated in the vaccine-draining LNs suggesting that APCs at this location must have been either directly targeted by the vaccine or internalized antigen sequestered from adjacent cells. Also, our data indicate that the APCs that translated protein also specifically underwent maturation, characterized by upregulation of co-stimulatory molecules required in the antigen presentation process. Furthermore, type I IFNs have been reported to support upregulation of molecules involved in antigen presentation and processing in vitro,34 which is in line with our data on increased gene expression of CD80, TAP2, CTSL and HLA-DR along with an IFN response.
Both by i.d. and by i.m. delivery, multiple monocyte and DC subsets at the injection sites and in the draining LNs showed translation of the mRNA. Interestingly, apart from classical monocytes, our LNP/mRNA vaccine also targeted the CD14+ CD16+ intermediate monocytes, which have been shown to support antibody responses by stimulating plasmablast differentiation35 and secrete pro-inflammatory cytokines upon TLR7 stimulation.18 By i.m. administration CD11c+ MDCs including the CD1c+ DCs superior at stimulating strong CD4+ T cell responses36 and the CD1c− population containing the CD141+ DCs with cross-presenting abilities37 showed efficient production of the antigen. In contrast, by i.d. administration skin DCs, i.e., CD1a+ DCs and CD209+ DC populations38 were targeted. Thus, antigen production and presentation to CD4+ T cells are likely performed by different APC subsets depending on the delivery route of the LNP/mRNA vaccine. Rapid targeting and activation of skin DCs and efficient transport to draining LNs could explain why the i.d. group showed stronger initial responses. In addition, only skin monocytes and DCs showed evidence of antigen translation at day 9, indicating prolonged antigen availability. In vivo imaging of mice receiving LNP/mRNA by different routes showed that i.d. delivery resulted in the most durable expression of mRNA-encoded protein.27
PDCs infiltrated the injection sites by either delivery route but showed low translation of the mRNA. Since PDCs exhibit low antigen presentation capacity36 they likely contribute to the responses induced by mRNA vaccines by other functions, e.g., production of type I IFNs. Moreover, as the size of the LNPs (approximately 100 nm) enables both passive or cell-associated transport to the draining LNs, this allows for delivery of LNP/mRNA to a range of cells and may explain the rather strong signals in B cells and T cells in draining LNs. Because B cells express TLR739 and present antigens, this may facilitate interactions with cognate T helper cells during the process of B cell differentiation into vaccine-specific antibody secreting cells.
The lack of antigen+ cells at the PBS or non-injected control sites demonstrates a restricted distribution to the injection site and the draining LNs of the LNP/mRNA vaccine after administration, which is in line with earlier studies on mRNA delivery in mice4, 29 as well as in NHPs immunized with protein antigen in adjuvant.32 Therefore, the presence of professional APCs with translated mRNA and with upregulated co-stimulatory molecules in vaccine-draining LNs provides an ample milieu for priming of naive CD4+ T cells. Not surprisingly, we found ongoing generation of vaccine-specific CD4+ T cells only in the vaccine-draining LNs. Detection of mRNA-encoded antigens in the LNs peaked at 24 hr whereas the antibody responses were sustained for weeks. This is in line with the short detection time for vaccine protein antigen+ cells32 that also induces durable vaccine immunity.40 It is possible that vaccine antigen levels below the detection limit on follicular DCs in the LNs sufficiently provide antigens to maintain vaccine responses, however vaccine-induced plasma cells in bone marrow secrete antibodies independent of cognate antigens.41
A recurrent finding in mouse reports on mRNA delivery is the induction of type I IFN responses.2, 22, 25, 42, 43 We show that LNP/mRNA vaccine delivered to distinctly different clinically relevant immunization sites induced significant type I IFN responses both at the gene and protein level in the draining LNs. The mRNA construct used in this study was modified to increase translational efficiency while avoiding excess innate activation.8 This type of mRNA constructs have shown promise for regenerative11, 12 and cancer therapies.2, 13 Type I IFNs have been shown to be critical for inducing anti-tumor responses both in mice and in humans in response to intravenously administered RNA aimed at cancer immunotherapy.2, 13 However, other studies utilizing mice devoid of the IFNα receptor showed that IFNα reduces the antigen expression and consequently the induction of antigen-specific immunity,25 while we recently found that the level of immunity was not affected.8 Whether type I IFNs are beneficial or detrimental for the generation of vaccine-induced immunity is an intense topic of debate.2, 8, 25, 42 Contradictory results may relate to differences in IFNα amounts stimulated by the specific mRNA formulations and/or the route of administration. The timing of transfection, cellular activation and initiation of translation of the vaccine mRNA in vivo may also be critical to circumvent any type I IFN-mediated block in mRNA translation.
Furthermore, IFNα is known to support CD8+ T cell immunity,44, 45 but we observed low H10-specific CD8+ T cells in the rhesus macaque model. mRNA vaccines have been shown to induce CD8+ T cell responses in mice29 but remains to be investigated more thoroughly in NHPs and humans. The modest CD8+ T cell responses in our study may be explained by the limited CD8+ T cell epitopes present in influenza HA for rhesus macaques as shown for humans.46
Comparison of trivalent versus monovalent seasonal influenza vaccines showed that the more potent trivalent vaccine also induced stronger type I IFN responses in vaccinated individuals.47 While HA-specific antibodies confer protection for influenza, other infections may require a stronger polyfunctional Th1 CD4+ T cell response than observed in this study. Whether this can be achieved by co-formulating the mRNA vaccine with a strong Th1 skewing adjuvant like poly I:C remains to be explored.48, 49, 50 We have previously reported that high-magnitude vaccine responses obtained by protein antigen together with a TLR7 agonist and alum was associated with a potent type I IFN response in NHPs,32 similar to what we observed in the current study.
To conclude, our data obtained from a rhesus macaque model, appreciated for its high translational value for human vaccine responses, provide mechanistic insights by which modified mRNA vaccines initiate innate immune activities at the injection site and in draining LNs that culminate in the priming of CD4+ T cells. A much better understanding of how strong vaccine responses can be elicited, tailored and sustained over time with modified mRNA construct would have significant impact on improving the design of this new vaccine platform.
Materials and Methods
Generation of Modified mRNA and Lipid Nanoparticles
Modified mRNA encoding the HA of H10N8 influenza A virus (A/Jiangxi-Donghu/346/2013) or the yellow fluorescent protein mCitrine were generated as previously described.8 The lipid mixture was combined with a 50 mM citrate buffer (pH 4.0) containing mRNA at a 3:1 ratio (aqueous:ethanol) using a microfluidic mixer (Precision Nanosystems, Vancouver, BC, Canada). For formulations containing GLA (Avanti Lipids, Alabaster, AL, USA), lipids were combined in a molar ratio of 50:9.83:38.5:1.5:0.17 (ionizable lipid:DSPC:cholesterol:PEG-lipid:GLA). All formulations were dialyzed against PBS, concentrated using Amicon ultra centrifugal filters (EMD Millipore, Billerica, MA, USA) and passed through a 0.22 μm filter. Particles were 80–100 nm in size with >95% RNA encapsulation.
Immunizations
This study was approved by the Local Ethical Committee on Animal Experiments. Twelve Chinese rhesus macaques were housed in the Astrid Fagraeus laboratory at Karolinska Institutet according to guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care. All procedures were performed abiding to the provisions and general guidelines of the Swedish Animal Welfare Agency. Animals were allocated to three groups (n = 4/group) receiving either i.m. or i.d. administration of LNP/H10 mRNA (50 μg) or LNP/H10 mRNA formulated with GLA adjuvant (5 μg) by the i.m. route. Animals were immunized at week 0 and 4 and the group receiving LNP/H10mRNA with GLA was boosted again at week 15. For innate immune response studies, animals received two i.m. or i.d. injections of Atto-655 labeled LNP/mCitrine mRNA (50 μg/site) at different sites at 4 and 24 hr respectively (n = 4/group). PBS (24 hr) and empty non-labeled LNP (4 or 24 hr) injections served as internal controls. For evaluation of priming of CD4+ T cells and vaccine tracking at day 9, contralateral sites received i.m. or i.d. injections with a 1:1 mixture of Atto-655 labeled LNPs containing either mCitrine (50 μg) or H10 mRNA (50 μg). The final injection volumes were 0.5 (i.m.) or 0.1 mL (i.d.) for immunogenicity study and 1 (i.m.) or 0.25 mL (i.d.) for innate response study. i.m. injections were administered as described33 and i.d. injections were split into three separate deliveries (< 10 mm apart) on a marked injection site.
Analyses of Vaccine-Specific Responses after Prime-Boost Immunizations
The HAI assay was performed with 0.5% turkey red blood cells (Rockland Antibodies and Assays, Limerick, PA, USA) diluted in PBS. Non-specific HAI was prevented by incubating serum overnight at 37°C with receptor destroying enzymes (Denka Seiken, San José, CA, USA). Serial dilutions (1:2) of serum samples were performed in V-bottom 96-well plates in duplicates, starting with a 1:10 dilution. Recombinant HA of H10N8 influenza A virus (four units), A/Jiangxi-Donghu/346/2013 (Medigen, Frederick, MD, USA) was added to diluted serum and incubated for 30 min at room temperature. The reciprocal of the last serum dilution resulting non-agglutinated red blood cells represented the HAI titer. Titers <10 were assigned as 1. Stimulation of H10-specific recall CD4+ T cell responses was performed on U-bottom 96-well plates, where 1.5 million peripheral blood mononuclear cells (PBMCs) per well were stimulated overnight with overlapping peptides of full-length H10 protein (2 μg) in presence of brefeldin A (5 μg/ml, Sigma, St. Louis, MO, USA). Production of IFNγ, IL-2, and TNF by restimulated H10-specific CD4 T cells was evaluated after intracellular staining (Table S1) as described.36 H10-specific CD4+ T cells were estimated by flow cytometry, and their polyfunctionality was analyzed using SPICE software (v.5.35).
Analyses of CD4+ T Cell Responses in Draining LNs after Priming
PBMCs and cell suspensions from LNs draining vaccine injection or gut-draining (mesenteric) control LNs after prime immunization were labeled with 1 μM CFSE (Invitrogen, Carlsbad, CA, USA) for 7 min at 37°C, and labeling was stopped by neat fetal calf serum (FCS) and washed with complete media (10% FCS, 1% penicillin/streptomycin/glutamine in RPMI [all from GIBCO, Stockholm, Sweden]). CFSE-labeled cells were cultured for three days in absence or presence of H10 peptides (3 μg, 15-mers overlapping by five amino acids, 95% purity) (Genscript, Piscataway, NJ, USA) to assess CFSE dilution by responding H10-specific CD4 T cells. To evaluate IFNγ production by H10-specific CD4 T cells after prime, LN cells or PBMCs were expanded in culture with H10 peptides (3 μg) for three days, washed, and restimulated (2 μg H10 peptides) for 6 hr in presence of brefeldin A prior to intracellular cytokine staining (ICS) as described above.
Tissue Single-Cell Suspensions
Injected muscle tissues were sampled during necropsy and separately stored in RPMI on ice as described.33 Injected skin (25 mm radius) was dissected for cell suspension. All tissues were separately processed without pooling 1 hr after harvesting. Muscle and skin were weighed, normalized to 1 g by removal of fat, connective tissue, and excess muscle or skin, respectively. Muscles were processed and digested with Liberase as described in detail.33 Skin was digested with Liberase TH (0.26WU/ml, Roche, Indianapolis, IN, USA) plus DNase (0.1 mg/ml, Sigma) at 37C° for 1 hr under agitation, and Liberase activity was quenched with complete media. Skin digestions were filtered through 70 μm cell strainers (BD, Stockholm, Sweden), washed with PBS, and immediately stained for flow cytometry. Lymph nodes (LNs) were minced with scissors and mechanically disrupted in 70 μm cell strainers using a plunger, washed, and stained immediately. PBMCs were obtained via standard protocols.
Cell Characterization by Flow Cytometry
Filtered and washed suspensions representing approximately 1 gram of tissue or 5 × 106 LN cells were stained with Live/Dead fixable Blue dead cell stain kit according to the manufacturer’s protocol (Invitrogen), blocked with FcR-blocking reagent (Miltenyi Biotec, San Diego, CA, USA), and stained with cocktail of fluorescent antibodies (Table S1). Stained samples were spiked with AccuCount beads (Spherotech, Lake Forest, IL, USA), and cell subset numbers were calculated according to the manufacturer’s protocol. A total of 1–2 million events per sample were acquired using LSR Fortessa flow cytometer (BD) and analyzed with FlowJo software (Tree Star).
CXCL10 (IP-10) ELISA
Serum CXCL10 before and 24 hr after LNP/mCitrine mRNA injection was detected by Human CXCL10 Quantikine ELISA (R&D Systems, Minneapolis, MN, USA) using the manufacturer’s protocol.
In Situ Staining
Punch biopsies (4 mm) from the injection sites and the draining LNs were collected and snap-frozen in OCT (Sakura Finetek, Torrance, CA). Cryosections of muscle and skin from injection sites and LN biopsies (6 μm) were stained with anti-MxA antibody (Professors Haller and Kochs, University of Freiburg, Germany) as described.32 All, except LN, sections were also stained with wheat germ agglutinin-Alexa Fluor 594 (Invitrogen) to visualize plasma membrane. Sections were mounted with SlowFade Gold antifade regent with or without DAPI (Invitrogen). Images were acquired using a Nikon Eclipse Ti-E, Nikon A1 confocal microscope, 20× objective. MxA+ cells were quantified using Cell profiler software (Broad Institute).
In Vitro Stimulation
The Institutional Review Board approved the use of human blood. Rhesus macaque PBMCs or APCs isolated from human blood with RosettSep monocyte enrichment kit (Stemcell Technologies, Waterbeach, Cambridge, UK) were stimulated overnight with LNP/H10 mRNA (5 μg), LNP/mCitrine mRNA (5 μg), or unlabeled empty LNP (5 μg) where indicated. Detection of intracellular IFNα and cellular activation by CD80/CD86 upregulation was assessed by flow cytometry (Table S1).
Microarray and Data Analysis
Skin, muscle and LN tissues were stored in RNAlater (Invitrogen) at –20°C until use (n = 4/group). RNA was prepared using Trizol (Ambion, Austin, TX, USA) with Tissuelyser (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol. Cyanine-3 (Cy3)-labeled cRNA was prepared from 200 ng total RNA with Quick Amp Labeling Kit (Agilent, Stockholm, Sweden) according to the manufacturer’s protocol, followed by RNeasy column purification (QIAGEN). Cy3-cRNA was hybridized to Agilent Rhesus Macaque Gene Expression Microarrays v2 (part number G2519F-026806) for 17 hr at 65°C and processed according to the manufacturer’s protocol. Slides were scanned by Agilent DNA Microarray Scanner (G2505C), and images were processed in Agilent Feature Extraction Software. Local background-adjusted signals (gProcessedSignal) were additionally quantile normalized using the Bioconductor package to achieve consistency between samples. Changes in a pre-selected set of genes of interest, related to inflammation, migration, and antigen uptake/presentation were also considered using descriptive plots. Probesets corresponding to these genes were identified by manual curation of the array manufacturer’s annotation. Statistical analysis and generation of descriptive was carried out using customized scripts in R (v.3.3) and Python (v.3.5). Gene array data are deposited in GEO: GSE98211.
Statistical Analysis
Two-way ANOVA test with Tukey’s multiple comparisons was used for analysis of HAI titers and T cell cytokine responses, whereas the remaining compiled data were assessed by two-sided paired (Wilcoxon signed-rank test) or an unpaired (Mann-Whitney test) t test analysis. All analyses were performed using GraphPad Prism (v.5.0c) software and considered significant at *p < 0.05, **p < 0.01, and ***p < 0.001.
Author Contributions
F.L., G.L., and K.L. designed the research. F.L., G.L., A.L., E.A.T., S.O., and K.L. performed the experiments. F.L., G.L., J.R., H.S., and K.L. analyzed the data. G.C., L.A.B., K.B., K.H., S.J., and O.Y. designed the research and provided the LNP/mRNA vaccines. F.L. and K.L. wrote the paper. All authors reviewed the paper.
Conflicts of Interest
F.L., G.L., A.L., E.A.T., S.O., and K.L. declare no conflict of interest. G.C., K.B., K.H., S.J., and O.Y. are employees of Valera LLC, a Moderna Venture, which focuses on the development of mRNA therapeutics and vaccines for Infectious Diseases, and L.A.B., J.R., and H.S. are employees of Moderna Therapeutics.
Acknowledgments
We thank M. Spångberg and B. Eriksson at Astrid Fagraeus Laboratory. We also thank M. Vono, S. Domingo, and T. Sandberg for laboratory assistance. K.L. was supported by grants from the Swedish Research Council (Vetenskapsrådet) (grant no. 2015-02608). F.L. and G.L. are recipients of scholarships from the Fernström Foundation and the Swedish Society of Medicine, respectively. All vaccine formulations were supplied by Valera LLC, a Moderna Therapeutics Venture, under a Sponsored Research Agreement with Karolinska Institutet.
Footnotes
Supplemental Information includes five figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.08.006.
Supplemental Information
References
- 1.Moingeon P., Lombardi V., Saint-Lu N., Tourdot S., Bodo V., Mascarell L. Adjuvants and vector systems for allergy vaccines. Immunol. Allergy Clin. North Am. 2011;31:407–419. doi: 10.1016/j.iac.2011.03.001. xii. [DOI] [PubMed] [Google Scholar]
- 2.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 3.Bogers W.M., Oostermeijer H., Mooij P., Koopman G., Verschoor E.J., Davis D., Ulmer J.B., Brito L.A., Cu Y., Banerjee K. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J. Infect. Dis. 2015;211:947–955. doi: 10.1093/infdis/jiu522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 6.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A. Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Su H.H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rettig L., Haen S.P., Bittermann A.G., von Boehmer L., Curioni A., Krämer S.D., Knuth A., Pascolo S. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115:4533–4541. doi: 10.1182/blood-2009-11-247817. [DOI] [PubMed] [Google Scholar]
- 15.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 16.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox R.J. Correlates of protection to influenza virus, where do we go from here? Hum. Vaccin. Immunother. 2013;9:405–408. doi: 10.4161/hv.22908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cros J., Cagnard N., Woollard K., Patey N., Zhang S.Y., Senechal B., Puel A., Biswas S.K., Moshous D., Picard C. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lund H., Boysen P., Åkesson C.P., Lewandowska-Sabat A.M., Storset A.K. Transient migration of large numbers of CD14(++) CD16(+) monocytes to the draining lymph node after onset of inflammation. Front. Immunol. 2016;7:322. doi: 10.3389/fimmu.2016.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loré K., Adams W.C., Havenga M.J., Precopio M.L., Holterman L., Goudsmit J., Koup R.A. Myeloid and plasmacytoid dendritic cells are susceptible to recombinant adenovirus vectors and stimulate polyfunctional memory T cell responses. J. Immunol. 2007;179:1721–1729. doi: 10.4049/jimmunol.179.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGovern N., Schlitzer A., Gunawan M., Jardine L., Shin A., Poyner E., Green K., Dickinson R., Wang X.N., Low D. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages. Immunity. 2014;41:465–477. doi: 10.1016/j.immuni.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards D.K., Jasny E., Yoon H., Horscroft N., Schanen B., Geter T., Fotin-Mleczek M., Petsch B., Wittman V. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Elzen P., Garg S., León L., Brigl M., Leadbetter E.A., Gumperz J.E., Dascher C.C., Cheng T.Y., Sacks F.M., Illarionov P.A. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- 24.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montoya M., Schiavoni G., Mattei F., Gresser I., Belardelli F., Borrow P., Tough D.F. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood. 2002;99:3263–3271. doi: 10.1182/blood.v99.9.3263. [DOI] [PubMed] [Google Scholar]
- 27.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallen K.J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum. Vaccin. Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 31.De Sanctis J.B., Blanca I., Rivera H., Bianco N.E. Expression of low-density lipoprotein receptors in peripheral blood and tonsil B lymphocytes. Clin. Exp. Immunol. 1998;113:206–212. doi: 10.1046/j.1365-2249.1998.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang F., Lindgren G., Sandgren K.J., Thompson E.A., Francica J.R., Seubert A., De Gregorio E., Barnett S., O’Hagan D.T., Sullivan N.J. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 2017;9:9. doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- 33.Liang F., Ploquin A., Hernández J.D., Fausther-Bovendo H., Lindgren G., Stanley D., Martinez A.S., Brenchley J.M., Koup R.A., Loré K., Sullivan N.J. Dissociation of skeletal muscle for flow cytometric characterization of immune cells in macaques. J. Immunol. Methods. 2015;425:69–78. doi: 10.1016/j.jim.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simmons D.P., Wearsch P.A., Canaday D.H., Meyerson H.J., Liu Y.C., Wang Y., Boom W.H., Harding C.V. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J. Immunol. 2012;188:3116–3126. doi: 10.4049/jimmunol.1101313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwissa M., Nakaya H.I., Onlamoon N., Wrammert J., Villinger F., Perng G.C., Yoksan S., Pattanapanyasat K., Chokephaibulkit K., Ahmed R., Pulendran B. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe. 2014;16:115–127. doi: 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vono M., Lin A., Norrby-Teglund A., Koup R.A., Liang F., Loré K. Neutrophils acquire the capacity for antigen presentation to memory CD4(+) T cells in vitro and ex vivo. Blood. 2017;129:1991–2001. doi: 10.1182/blood-2016-10-744441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nestle F.O., Di Meglio P., Qin J.Z., Nickoloff B.J. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gujer C., Sundling C., Seder R.A., Karlsson Hedestam G.B., Loré K. Human and rhesus plasmacytoid dendritic cell and B-cell responses to Toll-like receptor stimulation. Immunology. 2011;134:257–269. doi: 10.1111/j.1365-2567.2011.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francica J.R., Sheng Z., Zhang Z., Nishimura Y., Shingai M., Ramesh A., Keele B.F., Schmidt S.D., Flynn B.J., Darko S., NISC Comparative Sequencing Program Analysis of immunoglobulin transcripts and hypermutation following SHIV(AD8) infection and protein-plus-adjuvant immunization. Nat. Commun. 2015;6:6565. doi: 10.1038/ncomms7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halliley J.L., Tipton C.M., Liesveld J., Rosenberg A.F., Darce J., Gregoretti I.V., Popova L., Kaminiski D., Fucile C.F., Albizua I. Long-lived plasma cells are contained within the CD19(-)CD38(hi)CD138(+) subset in human bone marrow. Immunity. 2015;43:132–145. doi: 10.1016/j.immuni.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broos K., Van der Jeught K., Puttemans J., Goyvaerts C., Heirman C., Dewitte H. Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T-cell responses despite the induction of type I interferon. Mol. Ther. Nucleic Acids. 2016;5:e326. doi: 10.1038/mtna.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol. Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kastenmüller K., Wille-Reece U., Lindsay R.W., Trager L.R., Darrah P.A., Flynn B.J., Becker M.R., Udey M.C., Clausen B.E., Igyarto B.Z. Protective T cell immunity in mice following protein-TLR7/8 agonist-conjugate immunization requires aggregation, type I IFN, and multiple DC subsets. J. Clin. Invest. 2011;121:1782–1796. doi: 10.1172/JCI45416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsh R.M., Bahl K., Marshall H.D., Urban S.L. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 2012;8:e1002352. doi: 10.1371/journal.ppat.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bui H.H., Peters B., Assarsson E., Mbawuike I., Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc. Natl. Acad. Sci. USA. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Athale S., Banchereau R., Thompson-Snipes L., Wang Y., Palucka K., Pascual V., Banchereau J. Influenza vaccines differentially regulate the interferon response in human dendritic cell subsets. Sci. Transl. Med. 2017;9:9. doi: 10.1126/scitranslmed.aaf9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tewari K., Flynn B.J., Boscardin S.B., Kastenmueller K., Salazar A.M., Anderson C.A., Soundarapandian V., Ahumada A., Keler T., Hoffman S.L. Poly(I:C) is an effective adjuvant for antibody and multi-functional CD4+ T cell responses to Plasmodium falciparum circumsporozoite protein (CSP) and αDEC-CSP in non human primates. Vaccine. 2010;28:7256–7266. doi: 10.1016/j.vaccine.2010.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffman R.L., Sher A., Seder R.A. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trumpfheller C., Caskey M., Nchinda G., Longhi M.P., Mizenina O., Huang Y., Schlesinger S.J., Colonna M., Steinman R.M. The microbial mimic poly IC induces durable and protective CD4+ T cell immunity together with a dendritic cell targeted vaccine. Proc. Natl. Acad. Sci. USA. 2008;105:2574–2579. doi: 10.1073/pnas.0711976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.