Summary
ATOH1 is a master transcription factor for the secretory lineage differentiation of intestinal epithelial cells (IECs). However, the comprehensive contribution of ATOH1+ secretory lineage IECs to the homeostasis, repair, and tumorigenesis of the intestinal epithelium remains uncertain. Through our ATOH1+ cell-lineage tracing, we show here that a definite number of ATOH1+ IECs retain stem cell properties and can form ATOH1+IEC-derived clonal ribbons (ATOH1+ICRs) under completely homeostatic conditions. Interestingly, colonic ATOH1+ IECs appeared to exhibit their stem cell function more frequently compared with those of the small intestine. Consistently, the formation of ATOH1+ICRs was significantly enhanced upon dextran sodium sulfate colitis-induced mucosal damage. In addition, colonic ATOH1+ IECs acquired tumor stem cell-like properties in the azoxymethane-DSS tumor model. Our results reveal an unexpected contribution of colonic ATOH1+ IECs to maintaining the stem cell population under both homeostatic and pathologic conditions and further illustrate the high plasticity of the crypt-intrinsic stem cell hierarchy.
Keywords: Atoh1, intestinal secretory cell, plasticity, de-differentiation, intestinal stem cell, colonic inflammation, tumorigenesis, DSS, AOM-DSS
Graphical Abstract

Highlights
-
•
Intestinal ATOH1+ cells can exhibit stem cell properties under homeostatic conditions
-
•
Recruitment of ATOH1+ cell-derived stem cells is enhanced by inflammation
-
•
Cell-intrinsic NF-kB signaling promotes generation of ATOH1+ cell-derived stem cells
-
•
ATOH1+ tumor stem cells contribute to the development of colitis-associated tumors
Ishibashi et al. report the contribution of ATOH1+ intestinal epithelial cells to the maintenance, regeneration, and tumorigenesis in the colon. They find that a definite number of ATOH1+ intestinal epithelial cells retain stem cell properties under homeostatic conditions. Also, generation of ATOH1+ cell-derived stem cells is significantly enhanced by the inflammatory environment and contributes to the development of colitis-associated tumors.
Introduction
The intestinal epithelium is maintained by intestinal stem cells (ISCs), which reside at the bottom of the crypt (Okamoto and Watanabe, 2016). ISCs are identified by the expression of specific genes, such as leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5) (Barker et al., 2007). The differentiation of intestinal epithelial cells (IECs) predominantly proceeds in a unidirectional manner, starting from the ISCs to the terminally differentiated cells. In the earliest phase of this process, progenitor cells are committed either to a secretory or absorptive lineage based on the expression of key transcription factors such as atonal homolog-1 (ATOH1) or hairy and enhancer of split-1 (HES1) (Jensen et al., 2000, Yang et al., 2001).
However, a recent study showed that small-intestinal IECs committed to the absorptive lineage can exhibit ISC-oriented de-differentiation upon severe epithelial damage (Tetteh et al., 2016). Other studies have further highlighted the plasticity and ISC-oriented de-differentiation of secretory lineage-committed IECs in the small intestine, which are identified by the expression of NGN3 (Schonhoff et al., 2004), Delta-like 1 (DLL1) (van Es et al., 2012), and doublecortin-like kinase 1 (DCLK1) (Westphalen et al., 2014). ATOH1+ label-retaining cells (LRCs) reside at the +4 position and serve as secretory progenitor cells, but they can also exhibit ISC properties upon severe tissue injury (Buczacki et al., 2013). These studies indicated that a collective proportion of ATOH1+ secretory lineage-committed IECs retain their potential to revert to ISCs and can exhibit ISC-specific properties upon severe tissue injury to compensate for the massive loss of genuine ISCs (Mills and Sansom, 2015). Consistent with this finding, a recent single-cell analysis of LGR5+ IECs identified the rare presence of ATOH1+LGR5+ double-positive cells in normal small-intestinal crypts (Kim et al., 2016). However, the comprehensive and dynamic contributions of these ATOH1+ potential ISCs under normal and pathologic conditions have yet to be described. In addition, the key factors that can recruit a specific subset of ATOH1+ IECs back to the ISC pool upon severe epithelial damage remain mostly uncertain.
In intestinal tumorigenesis, several studies have shown that LGR5+ ISCs are the cells of origin of sporadic adenomatous polyposis coli (Apc)-deficient tumors (bottom-up model) (Barker et al., 2009). Other studies have shown that villus cells can initiate tumor development through overexpression of GREM1 or constitutive activation of the nuclear factor kappa-B (NF-κB) pathway (Davis et al., 2015, Schwitalla et al., 2013). These studies raise the possibility that de-differentiation of lineage-committed IECs may constitute another pathway of intestinal tumorigenesis (top-down model). Accordingly, inflammatory bowel disease (IBD) patients develop colitis-associated cancers (CACs) via a pathway distinct from that of sporadic colon cancers. Established CACs are abundant in Atoh1+ cells and, thus, retain features of secretory lineage-committed cells, such as mucin production (Park et al., 2006). The robust expression of ATOH1 in CACs may be supported by environmental tumor necrosis factor alpha (TNF-α) and contributes to maintaining their highly malignant properties (Fukushima et al., 2015). However, the potential contribution of ATOH1+ secretory lineage-committed IECs as the origin of CAC tumor stem cells has yet to be described.
In this study, using the ATOH1+ cell-lineage-tracing model, we showed that colonic ATOH1+ IECs could give rise to functional ISCs under both homeostatic and pathologic conditions. The results highlight the unexpectedly broad contribution of ATOH1+ IEC-derived ISCs to the maintenance, regeneration, and progression of colitis-associated tumorigenesis in the colonic epithelium.
Results
Lineage Tracing of ATOH1+ IECs Labels Secretory Lineage IECs
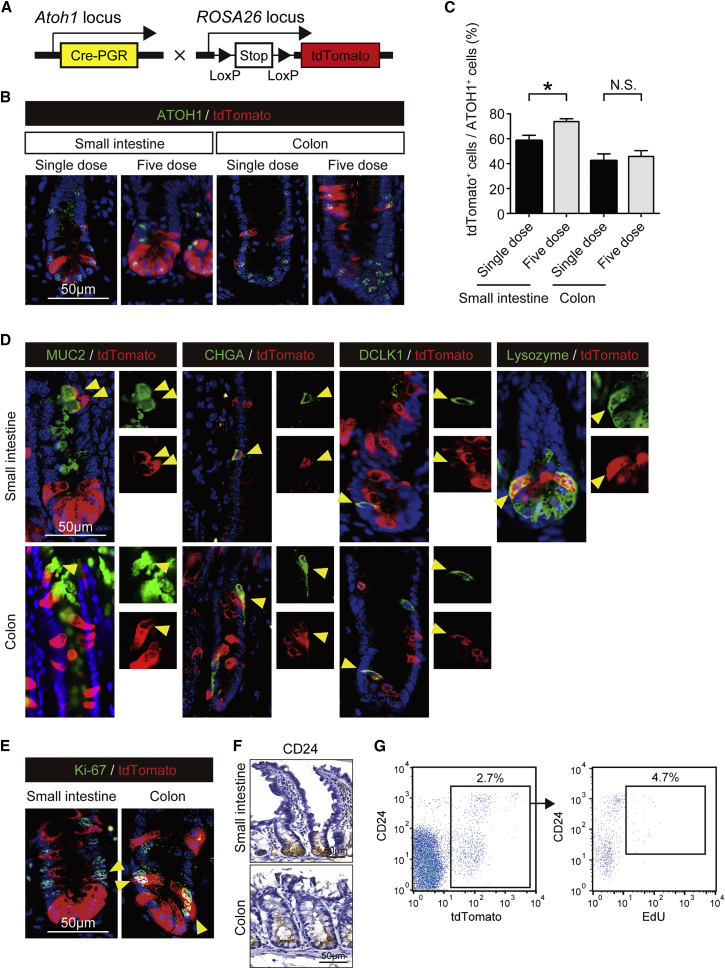
To elucidate the dynamic contribution of ATOH1+ IECs to the maintenance of the intestinal epithelium, we planned a lineage-tracing experiment. Here, we crossed Atoh1Cre-PGR mice (Rose et al., 2009) with Rosa26-LSL-tdTomato reporter mice to generate Atoh1Cre-PGR; Rosa26-LSL-tdTomato mice (Atoh1tdTomato mice, Figure 1A). In these mice, the effect of haploinsufficiency due to the knockin allele could not be observed, as confirmed through the analysis of Atoh1 mRNA and protein expression in the small intestine and colon (Figures S1A–S1C). To optimize the RU486-mediated tdTomato labeling of ATOH1+ IECs, we compared the labeling efficiency between a single dose of RU486 and the injection of RU486 for 5 consecutive days. Both protocols successfully labeled ATOH1+ IECs in the crypts of the small intestine and colon (Figure 1B). The 5-dose protocol resulted in a higher labeling efficiency (Figure 1C) and was therefore employed in the majority of the following experiments.
Figure 1.
Establishment of ATOH1+ Cell-Lineage Tracing
(A) Schematic representation of the alleles used to establish the Atoh1tdTomato mice.
(B) Co-staining of ATOH1 (green) and tdTomato (red) in small-intestinal and colonic tissues. Atoh1tdTomato mice were administered RU486 in either a single dose (single dose) or for 5 consecutive days (five-dose) and were then analyzed on the day following the final treatment. Note that all of the tdTomato+ IECs co-expressed ATOH1.
(C) Quantification of ATOH1+ IEC labeling efficiency based on the analysis shown in (B). Data are expressed as the mean ± SEM of biological replicates (n = 3). ∗p < 0.05, N.S., not significant.
(D) Co-staining of secretory IEC markers (green) and tdTomato (red). tdTomato-labeled MUC2+ goblet cells, CHGA+ enteroendocrine cells, DCLK1+ tuft cells, and Lysozyme+ Paneth cells are shown (yellow arrowheads).
(E) Co-staining for Ki-67 (green) and tdTomato (red) revealed tdTomato+ Ki-67+ double-positive IECs (yellow arrowheads).
(F) Immunostaining of CD24 using small-intestinal and colonic tissue of a wild-type mice.
(G) Representative flow plots of the small-intestinal IECs recovered from the Atoh1tdTomato mice on the day after completion of the five-dose RU486 treatment and EdU labeling. The CD24high/mid tdTomato+ fraction (combined population of CD24high and CD24mid cells) was further analyzed based on EdU labeling (right).
See also Figure S1.
The analysis performed 24 hr after a single dose of RU486 showed that all secretory lineage IECs and some Ki-67+ IECs were initially labeled by tdTomato (Figures 1D and 1E). Conversely, all of the tdTomato+ IECs were completely negative for HES1 (Figure S1D) and for other absorptive lineage markers (Figure S1E). To further confirm the labeling of mitotic IECs, the uptake of 5-ethynyl-2′-deoxyuridine (EdU) was examined in ATOH1+ IECs. Using CD24 as a marker for lower crypt IECs (Figure 1F) (Sato et al., 2012), we found that 4.7% of the CD24high/mid tdTomato+ IECs were also positive for EdU (Figure 1G).
These results collectively confirmed that our ATOH1+ IEC lineage-tracing system initially labeled both post-mitotic and mitotic secretory lineage-committed IECs in a highly specific manner.
Atoh1+IECs that Retain an ISC-like Phenotype Exist within Normal Intestinal Crypts
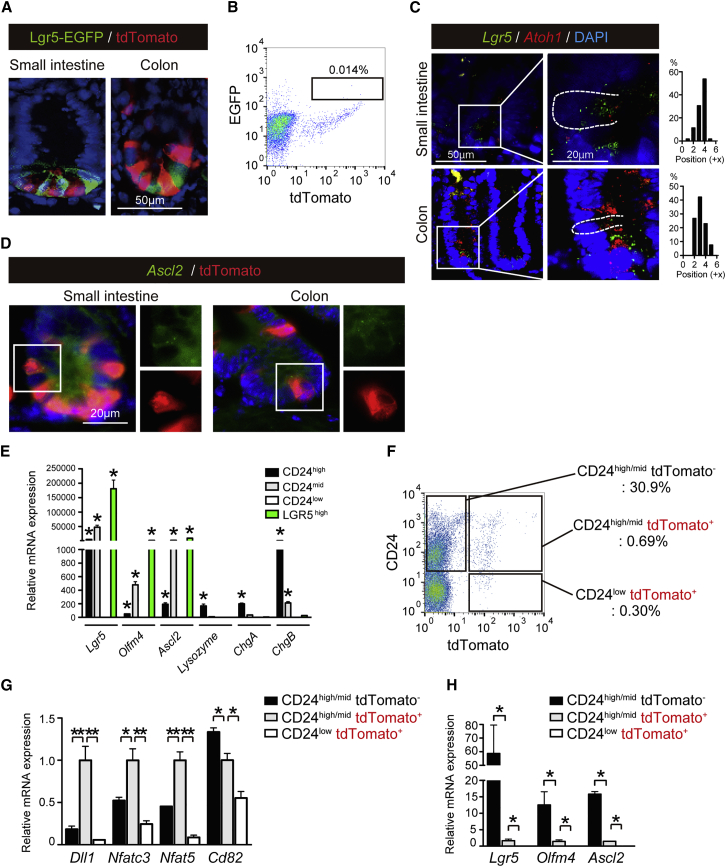
LGR5+ ISCs are located at the bottom of the crypt between Paneth cells (Barker et al., 2007). To determine whether any LGR5+ ISCs were labeled by our lineage-tracing system, we crossed our Atoh1tdTomato mice with Lgr5-EGFP-IRES-creERT2 mice to generate Lgr5-EGFP-IRES-creERT2; Atoh1Cre-PGR; Rosa26-LSL-tdTomato mice (Lgr5EGFPAtoh1tdTomato mice). The induction of Atoh1Cre-PGR allele-dependent tdTomato labeling in Lgr5EGFPAtoh1tdTomato mice showed that the tdTomato+ IECs were clearly distinct from LGR5+ ISCs (Figure 2A). However, flow cytometric analysis of ATOH1+ IECs revealed a rare population of LGR5-EGFP+ ATOH1+ double-positive IECs in the small intestine of Lgr5EGFPAtoh1tdTomato mice (Figure 2B). Consistently, RNAscope in situ hybridization (RNAscope ISH) clearly exhibited Lgr5+Atoh1+ double-positive cells, most frequently at the +4 position or +3 position of the small intestine and colon, respectively (Figure 2C). The integrity of our RNAscope analysis was validated by using positive control tissues (Figures S2A and S2B). Also, tdTomato+ crypt cells co-expressing Ascl2 were found in both regions (Figure 2D).
Figure 2.
ATOH1+ IECs Include a Cell Population that Retains the Expression of Stem Cell-Specific Genes
(A) Co-staining of Lgr5-EGFP (green) and tdTomato (red) in the small-intestinal and colonic crypts of Lgr5EGFPAtoh1tdTomato mice on the day following the completion of the five-dose RU486 treatment.
(B) Representative flow plots of the small-intestinal IECs recovered from the Lgr5EGFPAtoh1tdTomato mice on the day following the completion of the five-dose RU486 treatment.
(C) RNAscope in situ hybridization (RNAscope ISH) for Lgr5 (green) and Atoh1 (red) in the small-intestinal and colonic crypts of wild-type mice. The white dotted line shows the cell margin of a Lgr5+Atoh1+ double-positive cell. Position number of Lgr5+Atoh1+ double-positive cells (n = 30 of three independent experiments for each analysis) within a crypt was determined based on its relative position from the bottom of the crypt. Images of the colon are re-used in Figure 4C.
(D) Co-analysis of RNAscope ISH for Ascl2 (green), with immunostaining for tdTomato (red). Ascl2+tdTomato+ double-positive cells are identified in the small-intestinal and colonic crypt. A magnified view of the area identified by the white square is shown in the right side.
(E) qRT-PCR analysis of CD24high, CD24mid, and CD24low IECs in the small intestine of the wild-type mice (n = 3). Small-intestinal LGR5high IEC population of the Lgr5EGFP mice (n = 3) served as a positive control for ISCs. Data are expressed as the mean ± SEM. ∗p < 0.01.
(F) Representative flow plots of the small-intestinal IECs recovered from the Atoh1tdTomato mice on the day following the completion of the five-dose RU486 treatment.
(G and H) qRT-PCR analysis of cell fractions that were acquired in (F).
Data are expressed as the mean ± SEM of values normalized by the expression of β-actin (n = 3 of biological replicates). ∗p < 0.01, ∗∗p < 0.05.
See also Figure S2.
CD24 is commonly expressed by small-intestinal and colonic crypt IECs (Figure 1F) and therefore used to identify and isolate LGR5+ ISCs or Paneth cells (von Furstenberg et al., 2011). Our analysis of the CD24high, CD24mid, and CD24low fraction of small-intestinal IECs confirmed that the CD24high and CD24mid population includes LGR5+ ISCs, Paneth cells, and enteroendocrine cells (Figure 2E). In contrast, the CD24low population did not include these cell populations. Therefore, the combined population of CD24high and CD24mid small-intestinal IECs (CD24high/mid) was collected from the Atoh1tdTomato mice on the day after the administration of a single dose of RU486 and sorted into tdTomato+ and tdTomato− cell fractions (Figure 2F). qRT-PCR analysis of these cell populations showed that gene markers for secretory cell progenitors (Dll1) and genes recently identified as markers of LRCs (Nfatc3, Nfat5) (Buczacki et al., 2013) were highly expressed in the CD24high/mid tdTomato+ IECs (Figure 2G). Exceptionally, CD82 showed highest expression in CD24high/midtdTomato− IECs, indicating its preferential expression in the Atoh1− IEC population. In addition, ISC markers such as Lgr5, Olfm4, and Ascl2 presented the highest expression in the CD24high/midtdTomato− population (Figure 2H). However, expression of these genes was also detected in the CD24high/midtdTomato+ IEC population, although at a much lower level. These results sufficiently indicated that ATOH1+ IECs in the normal intestinal epithelium consist of a heterogeneous cell population, including secretory lineage progenitor cells, and a small population of IECs retaining ISC-specific gene expression.
Atoh1+ IECs Constitutively Exhibit ISC Properties under Homeostatic Conditions
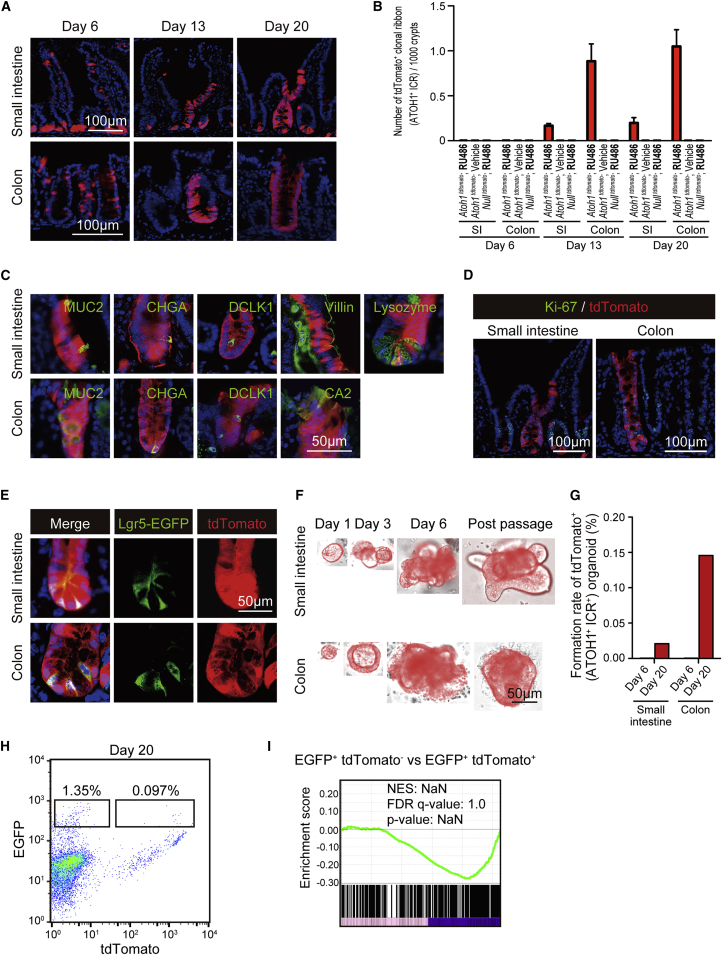
To identify the possible contribution of ATOH1+ IECs as potential ISCs, the dynamic changes in the tdTomato+ IEC distribution were traced for up to 20 days. On day 6, tdTomato+ IECs were found both in the crypts and in the villi, showing mostly a scattered distribution pattern (Figure 3A). At a later phase, the overall number of tdTomato+ IECs showed a clear decrease, indicating that most of these cells exhibit only a short lifetime. However, we found clusters of tdTomato+ IECs in both the small intestine and the colon on day 13 and day 20 (Figure 3A). These tdTomato+ IEC clusters clearly formed a continuous array of IECs along the crypt-villus axis, indicating that they represent clones of IECs arising from a common ATOH1+ IEC-derived ISC origin. Thus, we would like to designate these clusters of IECs as ATOH1+ IEC-derived clonal ribbons (ATOH1+ICRs), representing crypts dominated by ATOH1+ IEC-derived ISCs. The incidence of ATOH1+ICR generation was higher in the colon than in the small intestine, reaching up to 1.0 ribbon per 1,000 crypts (Figure 3B). No ATOH1+ICR formation was found by RU486 treatment in mice carrying Rosa26-LSL-tdTomato allele alone (Nulltdtomato) or by vehicle-alone treatment in Atoh1tdTomato mice. Compared with the previous lineage tracing of small-intestinal DLL1+ IECs (van Es et al., 2012) or DCLK1+ IECs (Westphalen et al., 2014), colonic ATOH1+ IECs formed clonal ribbons at a frequency that was at least 10-fold higher under homeostatic conditions. Such a regional difference may simply represent the preferential labeling efficiency of our system or may represent the difference in the population size of ATOH1+ IECs that retain ISC-like properties. Also, as we did not use a multi-color reporter, it remains possible that the ATOH1+ICRs observed in vivo may not necessarily represent a single-clone origin but may be composed of multi-cell origin.
Figure 3.
ATOH1+ IECs Give Rise to LGR5+ ISCs that Can Form Clonal Ribbons under Homeostatic Conditions
(A) Lineage tracing in Atoh1tdTomato mice showing the formation of tdTomato+ clonal ribbons (ATOH1+ICRs) in the small intestine and colon. Mice were treated by the five-dose protocol of RU486 and killed for analysis on day 6, day 13, and day 20.
(B) Quantification of ATOH1+ICR formation frequency in the small intestine and colon, using the tissue sections of Atoh1tdTomato mice or mice carrying Rosa26-LSL-tdTomato allele alone (Nulltdtomato). Atoh1tdTomato mice treated with vehicle alone (vehicle, n = 3), or Nulltdtomato mice treated with RU486 (n = 3) served as negative control. Atoh1+ICR was defined as those clusters of tdTomato+ cells completely occupying the whole crypt-villus unit, as assessed by the examination of at least three serial sections. The number of ATOH1+ICR was normalized against the total number of crypts. Data are expressed as the mean ± SEM.
(C) Co-staining of cell-lineage markers, such as MUC2, CHGA, DCLK1, Villin, CA2, and Lysozyme (green) with tdTomato (red) on day 20.
(D) Co-staining of Ki-67 (green) and tdTomato (red) in the Atoh1+ICR observed on day 20.
(E) Fluorescence images of tdTomato+ cells (red) co-expressing Lgr5-EGFP (green) in the ATOH1+ICR of Lgr5EGFPAtoh1tdTomato mice on day 20. Mice were treated as described in (A).
(F) Successful establishment of tdTomato+ organoids from the small-intestinal and colonic tissues recovered from Atoh1tdTomato mice on day 20.
(G) The efficiency of tdTomato+ organoid formation was quantified using tissues collected from the Atoh1tdTomato mice. Total cumulative number of tdTomato+ organoids acquired from four mice is normalized by the total number of organoids.
(H) Representative flow plots showing the increased presence of Lgr5-EGFP+tdTomato+ IECs in the small intestine of the Lgr5EGFP-CreERT2Atoh1tdTomato mice on day 20 compared with the data on day 6 shown in Figure 2B.
(I) Designated cell fractions in (H) were subjected to microarray analysis and then compared and analyzed for the adult stem cell-specific gene set (M1999) by GSEA.
See also Figure S3.
These ATOH1+ICRs contained all lineages of differentiated IECs and Ki-67+ IECs (Figures 3C and 3D), thereby indicating the possibility that the resident ATOH1+ IECs had acquired genuine ISC-specific function and thus took over the whole crypt-villus unit.
Accordingly, analysis of the ATOH1+ICRs in Lgr5EGFPAtoh1tdTomato mice showed the clear existence of LGR5-EGFP+tdTomato+ double-positive IECs in ATOH1+ICRs (Figure 3E). To further verify the acquisition of ISC properties by ATOH1+ IECs, we employed the organoid culture system. Small-intestinal or colonic tissues from Atoh1tdTomato mice were subjected to organoid culture at 20 days after RU486 induction. Among the established organoids, we found organoids that completely consisted of tdTomato+ IECs (Figure 3F). These tdTomato+ organoids could be re-organized across passages. The efficiency of forming these tdTomato+ organoids was highly consistent with the time-dependent appearance of ATOH1+ICRs in vivo (Figure 3G). Induction of ATOH1+ICR formation appeared to be driven by an IEC-intrinsic mechanism, as tdTomato+ organoids could be formed through RU486-mediated induction in vitro (Figure S3A) and maintained beyond passaging (Figure S3B). However, the efficiency of forming tdTomato+ organoids in vitro was lower in the organoids of Atoh1tdTomato mice than in those of Lgr5-EGFP-IRES-creERT2; Rosa26-LSL-tdTomato mice (Lgr5tdTomato mice) (Figures S3C and S3D). In addition, tdTomato+ organoids could be re-organized from a single tdTomato+ cell, also at a lower frequency compared with LGR5high cells (Figures S3E and S3F). Those single-cell-derived tdTomato+ organoids could be re-organized beyond passaging (Figure S3G) and give rise to both secretory and absorptive cells (Figure S3H). These results indicate the acquirement of multi-potency by ATOH1+ IECs in vitro.
To further identify the properties of ATOH1+ IEC-derived ISCs, LGR5-EGFP+tdTomato+ double-positive ISCs were collected from Lgr5EGFPAtoh1tdTomato mice on day 20 (Figure 3H), and global gene expression in these cells was compared with LGR5-EGFP+ single-positive ISCs. Gene set enrichment analysis (GSEA) showed that the expression of stem cell-signature genes in LGR5-EGFP+tdTomato+ double-positive ISCs was comparable with that in LGR5-EGFP+ single-positive ISCs (Figure 3I). These results confirmed that a specific population of ATOH1+ IECs is constitutively re-directed to exhibit genuine ISC-specific properties and participate in the maintenance of the normal intestinal epithelium.
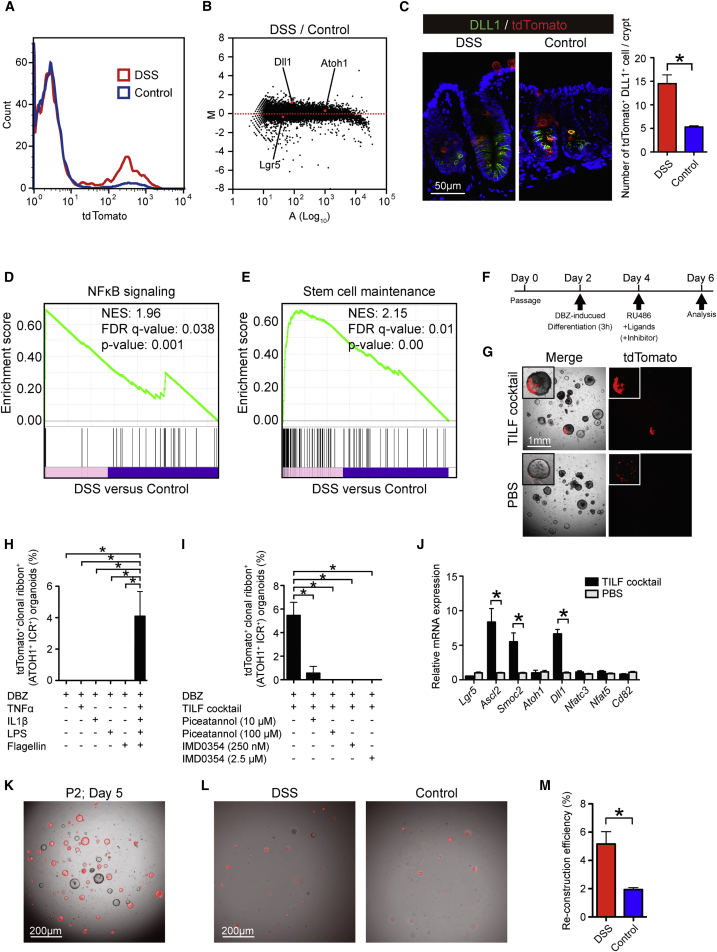
Colonic Inflammation Promotes the Generation of ATOH1+ IEC-Derived Clonal Ribbons
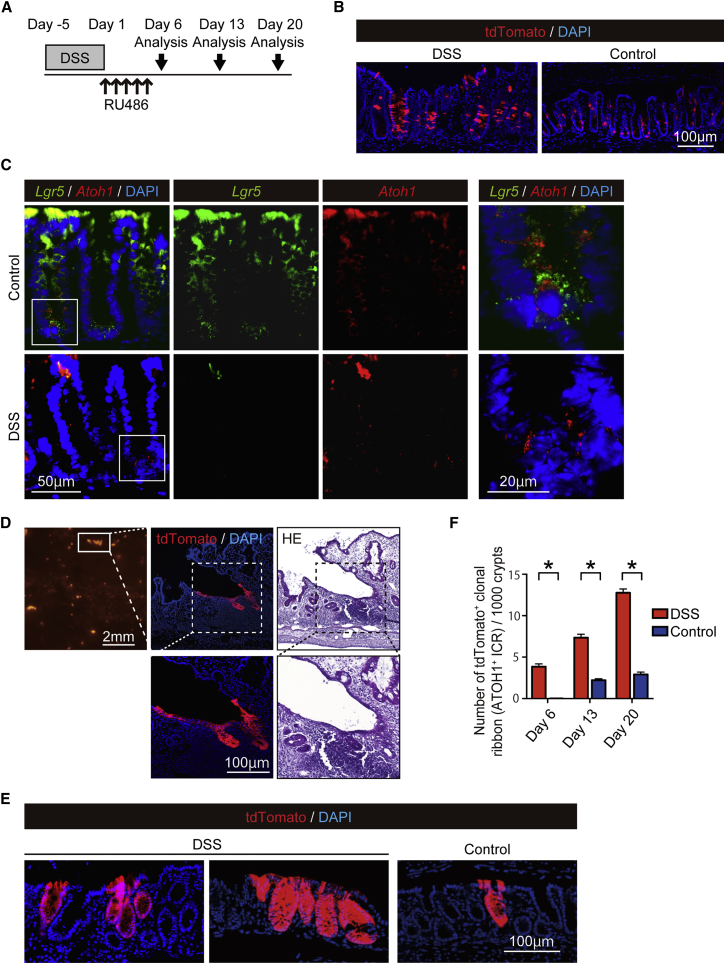
Following epithelial injury, various signaling pathways are activated within the intestinal epithelium to aid in optimization of the regeneration program (Okamoto et al., 2009). In the present study, only a minimal increase in the number of small-intestinal ATOH1+ICRs was observed following irradiation-induced damage (control group, 0.20 ribbons per 1,000 crypts; irradiation group, 0.63 ribbons per 1,000 crypts; data not shown). Thus, we examined whether the recruitment of colonic ATOH1+ IECs to the ISC pool and the subsequent formation of ATOH1+ICRs could be enhanced in a dextran sodium sulfate (DSS) colitis model. A significant reduction in body weight in the DSS-colitis mice indicated the successful induction of colitis (Figure S4A). At day 2, we found only a small number of tdTomato+ IECs in the rectum of DSS-colitis mice (Figure S4B). However, at day 6, we found clear increase in the number of tdTomato+ IECs in DSS-colitis mice (Figure 4B). At this time period, LGR5+ATOH1+ double-positive cells were not clearly identified in the rectal crypts of DSS-colitis mice (Figure 4C). However, a clear and persistent existence of ATOH1+ IECs was observed in the rectum of these DSS-colitis mice (Figures S4C and S4D). Accordingly, a significant increase in the mucosal area occupied by tdTomato+ IECs was observed during the recovery phase (day 6, day 13, and day 20) of the damaged mucosa (Figures S4E and S4F). Importantly, ATOH1+ICRs appeared in the rectum of DSS-colitis mice as early as day 6, a time period when they were never observed in the control mice. Some of the massive tdTomato+ area in the DSS-colitis mice at day 13 represented a cluster of tdTomato+ IECs extending from the ulcer-edge crypts, which covered the surface of the adjacent wound (Figure 4D). This observation indicated that ATOH1+ IEC progenies actively contribute to the early phase of tissue repair by forming the wound-associated epithelium (Seno et al., 2009). Consequently, we found an increase in the number of ATOH1+ICR formation and massive clusters of ATOH1+ICRs in the rectum of DSS-colitis mice at a later phase (day 20, Figures 4E and 4F). Such an increase in the number of ATOH1+ICR was never found by pre-labeling ATOH1+ cells before DSS treatment (Figures S4G and S4H), thus indicating that colitis-induced ATOH1+ICR formation is initiated by ATOH1+IEC-derived cells that are less sensitive to DSS-induced damage, or ATOH1+ cells that have newly appeared during the post-DSS labeling period.
Figure 4.
Formation of ATOH1+ IEC-Derived Clonal Ribbon Is Significantly Promoted during Regeneration from Colitis-Induced Epithelial Damage
Formation of ATOH1+ IEC-derived clonal ribbon (ATOH1+ICR) was compared between the DSS-colitis mice (DSS) and control mice (Control).
(A) Experimental design for the lineage-tracing analysis of Atoh1+ IECs in DSS-colitis-induced Atoh1tdTomato mice.
(B) Labeling of ATOH1+ IECs-derived cells by tdTomato (red) at day 6 in the colon of DSS-colitis mice and control mice.
(C) Double RNAscope ISH showing expression of Atoh1 (red) and Lgr5 (green) in the colonic crypts of DSS-colitis mice and control mice. Magnified view of the designated area (white square) is shown in the right end column. Images of the colon are re-used in Figure 2C.
(D) An example of ATOH1+ICR forming the wound-associated epithelium in DSS-colitis mouse on day 13.
(E) Representative rectal sections showing ATOH1+ICRs (red) in the control mice (Control) and in the DSS-colitis mice (DSS) on day 20.
(F) Quantification of the ATOH1+ICR formation frequency in the rectum of DSS-colitis mice (n = 3) and control mice (n = 3). The number of ATOH1+ICR was normalized by the total number of crypts. Data are expressed as the mean ± SEM per 1,000 crypts. ∗p < 0.0001.
See also Figure S4.
Collectively, we found that generation of ATOH1+ICRs was significantly promoted at the earliest phase of the tissue repair process in DSS-colitis mice, and thereby partly contributed to the regeneration of colitis-associated wounds.
Inflammatory Cytokines and Bacterial Components Coordinately Promote the Formation of ATOH1+ICRs
To obtain insight into the mechanism underlying the promotion of ATOH1+ICR formation in the colitic environment, the tdTomato+ IEC population in the rectum of DSS-colitis-induced Atoh1tdTomato mice was collected (Figure 5A) and subjected to microarray analysis. MA plot analysis identified Dll1 as one of the highly upregulated genes in the tdTomato+ IEC population of the DSS-colitis mice (Figure 5B). Consistently, a clear increase in the number of DLL1+tdTomato+ double-positive IECs was confirmed in the rectal tissue of the DSS-colitis mice (Figure 5C).
Figure 5.
Inflammatory Cytokines and Bacterial Components Coordinately Promote the Formation of ATOH1+ICRs
(A) Representative histogram of rectal tdTomato+ IECs in the DSS-colitis mice (red) and control mice (blue) on day 6.
(B) MA plot demonstrating difference in global gene expression between tdTomato+ IECs of DSS-colitis mice and control mice.
(C) Double immunostaining of DLL1 (green) and tdTomato (red) using colonic tissues of DSS-colitis mice and control mice at day 6. The number of tdTomato+DLL1+ double-positive cells was quantified by analysis of rectal tissue obtained from DSS-colitis mice (n = 3) and control mice (n = 3). Data are expressed as the mean ± SEM. ∗p < 0.05.
(D) The global gene expression data from (B) were compared with the registered NF-κB signaling gene set (M1983) through GSEA.
(E) The global gene expression data from (B) were compared with the registered adult stem cell gene set (M1999) through GSEA.
(F) In vitro analysis of ATOH1+ICR formation using colonic organoids from Atoh1tdTomato mice. Organoids were cultured under DBZ-induced secretory lineage-differentiation conditions for 3 hr and subjected to in vitro lineage-tracing analysis following the addition of RU486 with cytokines or bacterial components (Ligands). The addition of PBS alone served as a control.
(G) Phase-contrast view and tdTomato fluorescence view (red) of the organoids treated as shown in (F). The data are from organoids on day 6. Note that the organoids in the control condition (PBS) showed a scattered distribution pattern of tdTomato+ IECs, whereas those treated with the combination of TNF-α, IL-1β, LPS, and flagellin (TILF cocktail) clearly showed a continuous stream of tdTomato+ IECs (upper left inset).
(H) Quantitative analysis of the in vitro lineage-tracing analysis. The number of organoids showing in vitro ATOH1+ICR was normalized by the total organoids that were established during the experimental period. The presented data are from three independent analyses including over 100 organoids from each experimental condition. Data are expressed as the mean ± SEM. ∗p < 0.05.
(I) Quantitative analysis of the in vitro lineage-tracing analysis. Data are expressed as the mean ± SEM (three independent analyses). ∗p < 0.05. Piceatannol or IMD0354 was added at the same time as the TILF cocktail.
(J) qPCR analysis of colonic organoids. Data are expressed as the mean ± SEM of values normalized against the expression of β-actin (three independent analyses). ∗p < 0.05.
(K) Single-cell organoid reconstruction assay of colonic organoids treated by TILF cocktail. Colonic organoids were treated as described in (F), and those TILF-treated tdTomato+ organoids were further subjected to single-cell passage. Picture shows organoids at day 5 after second passage from TILF addition (P2; Day5).
(L) Single-cell organoid reconstruction assay using colonic IECs isolated from DSS-colitis mice or control mice at day 3.
(M) Quantification of organoid reconstruction assay shown in (L), using DSS-colitis mice (n = 3) and control mice (n = 3). Data are expressed as the mean ± SEM. ∗p < 0.001.
See also Figure S5.
Pathway analysis of the microarray data additionally revealed that gene sets representing the general inflammatory response were most significantly upregulated. Furthermore, we found that the response to interleukin (IL)-1 and TNF-α was significantly upregulated in the ATOH1+ IECs of the DSS-colitis mice (Figure S5A). The NF-κB pathway represents the major downstream signaling pathway shared by TNF-α and IL-1β (Mercurio and Manning, 1999). A previous study highlighted the importance of constitutive NF-κB pathway activation in the de-differentiation of mouse small-intestinal villus IECs (Schwitalla et al., 2013). Our GSEA consistently revealed that the NF-κB pathway gene set was significantly upregulated in the tdTomato+ IECs of the DSS-colitis mice (Figure 5D), as were genes related to stem cell maintenance (Figure 5E). Thus, in DSS colitis, ATOH1+ IEC-derived cells appeared to gain stem cell-like gene expression under an increased activation level of the NF-κB pathway.
To validate the contribution of NF-κB signaling to ATOH1+ICR formation, we performed in vitro analyses using organoid culture. Colonic organoids from Atoh1tdTomato mice were cultured under DBZ-induced differentiation conditions, which clearly promoted commitment to the ATOH1+ secretory cell lineage, and also promoted downregulation of ISC-specific genes (Figure S5B). To induce activation of the NF-κB pathway, two representative bacterial components, such as lipopolysaccharide (LPS) and flagellin, were employed in addition to TNF-α and IL-1β. The combined addition of these ligands (TILF cocktail) clearly upregulated the expression of IL-8 in DBZ-treated colonic organoids (Figure S5C) and induced the degradation of IκBα (Figure S5D), which confirmed the activation of the NF-κB pathway in secretory cell-committed organoids. Using these secretory cell-committed organoids, we proved that addition of the TILF cocktail could induce the formation of colonic ATOH1+ICRs in vitro (Figures 5F and 5G). This in vitro formation of ATOH1+ICRs in secretory cell-committed organoids clearly required the addition of all ligands (Figure 5H). Specific blockade of the NF-κB pathway using small-molecule compounds (Figure S5E) consistently inhibited the formation of ATOH1+ICRs in a dose-dependent manner (Figures 5I and S5F). Thus, we found that the NF-κB pathway functions as a key pathway in promoting the exhibition of ISC properties by ATOH1+ IEC-derived cells. Accordingly, the addition of TILF ligands clearly upregulated the expression of ISC markers Ascl2 and Smoc2, in addition to Dll1 (Figure 5J), suggesting that the expression of these genes may play a key role in the inflammation-induced formation of ATOH1+ICRs. Such a TILF-cocktail-induced upregulation of ISC markers was never observed in organoids treated without DBZ (Figure S5G). However, the expression level of Lgr5 remained unchanged both in DBZ-treated and untreated organoids (Figures 5I and S5G), possibly due to its low expression level in early reprogrammed ISCs.
To validate the ISC property of TILF-treated Atoh1+IEC-derived cells in vitro, we tested whether TILF-treated Atoh1tdTomoto mice organoids could newly reconstruct tdTomato+ organoids and expand their progeny beyond passaging. As expected, TILF-treated organoids could reconstruct tdTomato+ organoids beyond passaging from single isolated cells (Figure 5K). Also, an increased number of tdTomato+ organoids was re-constructed from single isolated IECs that were recovered from the DSS-colitis mice (Figures 5L and 5M), indicating enrichment of cells retaining ISC-like properties.
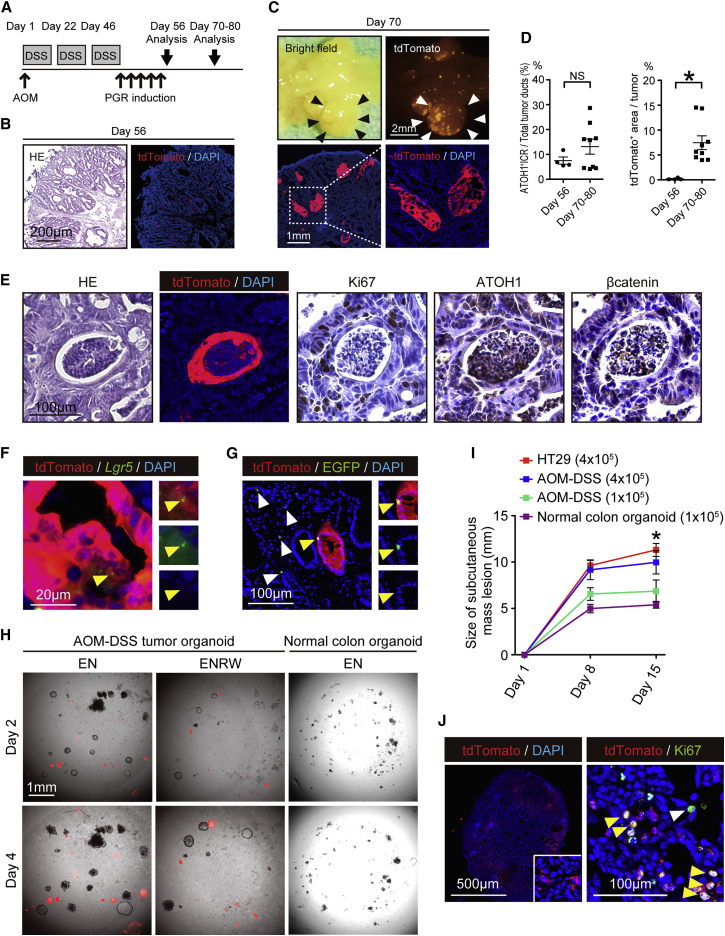
ATOH1+ IECs Comprise Colitis-Associated Tumors and Acquire Tumor Stem Cell Properties
CAC develops through a pathway distinct from that of sporadic colon cancer (Feagins et al., 2009, Thorsteinsdottir et al., 2011) and shows pathologic features such as high ATOH1 expression (Kano et al., 2013, Park et al., 2006). However, the precise role of ATOH1+ IECs in CACs remains uncertain. We have previously shown that TNF-α mediated activation of the NF-κB pathway in IECs plays an indispensable role in azoxymethane (AOM)-DSS tumor formation in mice (Onizawa et al., 2009). Hence, we asked whether the tumors of AOM-DSS model mice (Parang et al., 2016) harbor ATOH1+ tumor cells and exhibit features of CACs (Figure 6A). It was found that AOM-DSS tumors contained a substantial number of ATOH1+ tumor cells and expressed ATOH1 at a high level (Figures S6A–S6C). Furthermore, increased production of mucin was observed in these tumors (Figure S6D), suggesting that they faithfully phenocopy CACs.
Figure 6.
ATOH1+ IECs Contribute to the Formation of AOM-DSS-Induced Colitic Tumors and Acquire the Properties of Tumor Stem Cells
(A) Experimental design combining the AOM-DSS tumor model and ATOH1+ IEC lineage tracing.
(B) H&E staining and fluorescence images of a rectal tumor tissue section on day 56. A few isolated tdTomato+ cells (red) were found within the tumor tissue.
(C) Stereoscopic view (upper panels) and tissue section (lower panels) of a massive rectal tumor (black and white arrowheads) on day 70.
(D) Percentage ratio of tumor ATOH1+ICR or the area occupied by tdTomato+ tumor cells was analyzed in tumors at day 56 (n = 4) and day 70–80 (n = 8). Data are expressed as the mean ± SEM. ∗p < 0.05, N.S., not significant.
(E) Staining of serial sections showing ATOH1+, Ki-67+, and nuclear-localized β-catenin+ cells in the tumor ATOH1+ICR on day 70.
(F) RNAscope ISH analysis showing a Lgr5+ (green) tumor cell (yellow arrowhead) within the tdTomato+ (red) tumor area.
(G) Lgr5EGFPAtoh1tdTomato mice were treated as shown in (A). Lgr5-EGFP+ tdTomato+ double-positive tumor cells (yellow arrowhead) were found within the tumor ATOH1+ICRs of the rectal tumors on day 69, along with Lgr5-EGFP+ single-positive tumor cells in the surrounding tumor area (white arrowhead).
(H) Organoid culture using AOM-DSS tumor tissues from Atoh1tdTomato mice or normal colonic tissues from wild-type mice. Isolated cells were cultured in Wnt3a/R-Spondin-1-supplemented medium (ENRW) or Wnt3a/R-Spondin-1-depleted medium (EN). A merged view of the phase-contrast and tdTomato fluorescence (red) images is shown.
(I) Growth of subcutaneous tumors initiated by transplantation of AOM-DSS tumor-cell-derived organoids (1 × 105 cells, n = 3; 4 × 105 cells, n = 3; AOM-DSS), normal colon-derived organoids (1 × 105 cells, n = 3; normal colon organoid), or HT29 cells (4 × 105 cells, n = 3; HT29) into nude mice. For the transplantation of AOM-DSS tumor cell-derived organoids, a mixture of tdTomato+ and tdTomato− organoids was used. Data are expressed as the mean ± SEM. ∗ indicates p < 0.001 of HT-29 and AOM-DSS (4 × 105) compared with normal colon organoid.
(J) Tissue analysis of the tumors formed through transplantation of AOM-DSS tumor cell-derived organoids (4 × 105 cells). Macroscopic view (left panel) showing the distribution of tdTomato+ cells (red) in a mosaic pattern. Immunostaining for tdTomato (red) and Ki67 (green) revealing a cluster of Ki67+tdTomato+ double-positive cells (yellow arrowhead) within the tumor tissue. In addition, Ki67+tdTomato− single-positive cells were found within the surrounding tumor tissue (white arrowhead).
See also Figure S6.
At the early stage of AOM-DSS tumor labeling in Atoh1tdTomato mice, only a few isolated tdTomato+ tumor cells were found (day 56, Figure 6B). However, clear expansion of the tdTomato+ area within the tumor was observed after day 70, which represented a continuous cluster of tdTomato+ tumor cells (Figures 6C and 6D). The ratio of these tumor ATOH1+ICRs among tumor-forming ducts did not increase during the tumor development (Figure 6D), but they clearly included Ki-67+ cells and ATOH1+ cells and showed nuclear translocation of β-catenin (Figure 6E). In contrast, they lacked expression of secretory cell markers other than MUC2 (Figure S6E). Importantly, some of the tdTomato+ cells co-expressed tumor stem cell markers such as CD44 or CD133 (Zeilstra et al., 2008, Zhu et al., 2009) (Figure S6F). Moreover, Lgr5+ cells were present in the tdTomato+ tumor clonal ribbons, as shown by the RNAscope ISH analysis (Figure 6F). Consistent with this finding, LGR5-EGFP+tdTomato+ double-positive tumor cells were found in the AOM-DSS tumors of the Lgr5EGFPAtoh1tdTomato mice, in addition to LGR5-EGFP+ single-positive tumor cells (Figure 6G). Taken together, these results collectively indicated that ATOH1+ IECs contributed to the development of AOM-DSS tumors by acquiring tumor stem cell properties. However, the existence of LGR5-EGFP+ single-positive tumor cells indicated that these AOM-DSS tumors were mosaic and potentially consisted of a heterogeneous population of ATOH1+ and ATOH1− tumor stem cells.
As expected, we observed that both tdTomato+ and tdTomato− AOM-DSS tumor cell organoids could be established in vitro (Figure 6H). The tdTomato+ AOM-DSS tumor organoids consisted solely of tdTomato+ cells and shared the Wnt-independent growth ability of the tdTomato− tumor organoids. Furthermore, they could be maintained across several passages and form tumors via subcutaneous transplantation into nude mice in a dose-dependent manner (Figure 6I). The re-produced tumors of the nude mice consisted of both tdTomato+ and tdTomato− proliferating cells (Figure 6J), which conserved the mosaic features of the original AOM-DSS tumor. These results collectively indicated that AOM-DSS tumors harbored ATOH1+ IEC-derived tumor stem cells, in addition to tumor stem cells of other origins. Thus, colitis-associated tumorigenesis may represent another pathologic condition in which the acquisition of stem cell-like properties by ATOH1+ IECs can be promoted.
Discussion
In our lineage-tracing model, the frequency of forming colonic ATOH1+ICRs reached 250–300 ribbons per mouse under homeostatic conditions and 1,500–1,800 ribbons per mouse in DSS colitis, which certainly exceeded previous models (Tetteh et al., 2016, van Es et al., 2012, Westphalen et al., 2014). Thus, our results highlight the contribution of ATOH1+ IECs in the maintenance of the ISC pool, particularly in the colon. We also succeeded in clearly visualizing the dramatic and time-dependent increase in the frequency of colonic ATOH1+ICR formation in the DSS-colitis model. This result underpins the previous report of DCLK1+ IECs, which indicated the increased ISC conversion of colonic DCLK1+ IECs in DSS-colitis mice but did not successfully identify increased formation of DCLK1+ IEC-derived clonal ribbons (Westphalen et al., 2014).
Previous studies have identified at least three subpopulations of small-intestinal ATOH1+ IECs that may retain their potential to undergo conversion back to ISCs: ATOH1+LGR5+ double-positive IECs at the +1 to +3 position (Kim et al., 2016), ATOH1+LRCs at the +4 position (Buczacki et al., 2013), and ATOH1+DLL1+ IECs at the +5 position (van Es et al., 2012). However, the origin of cells that can form colonic ATOH1+ICRs may be heterogeneous and may differ from the small-intestinal counterparts. For example, the existence of colonic LRCs is not well confirmed, and small-intestinal LRCs have never been shown to form clonal ribbons under homeostatic conditions (Kim et al., 2016). The existence of ATOH1+LGR5+ double-positive IECs in the normal or colitic colon has never been identified in previous reports. In our current analysis, we successfully identified a rare population of ATOH1+LGR5+ double-positive cells both in the small intestine and colon (Figure 2C). However, most of these ATOH1+LGR5+ double-positive cells appeared to be lost under induction of DSS colitis (Figure 4C). Therefore, ATOH1+LGR5+ double-positive cells may be the dominant origin of ATOH1+ICRs under homeostatic conditions but may be less involved in ATOH1+ICR formation under DSS colitis.
We have previously shown that DLL1+ IECs reside at the lowest part of the crypts, both in the small intestine and in the colon (Shimizu et al., 2014). Furthermore, an increase of DLL1+ colonic IECs was clearly confirmed in DSS-colitis mice (Figure 5C) (Shimizu et al., 2014). Therefore, the DLL1+ population of ATOH1+ IEC-derived cells may be one of the candidate subpopulations that can form ATOH1+ICRs in the colon, especially under inflammatory conditions. However, it remains possible that the increase of ATOH1+ICR formation in DSS-colitis mice depends on the concerted contribution of other unknown subpopulations. A similar de-differentiation has been observed in the airway secretory lineage IECs, suggesting that re-acquiring stem cell properties is inversely related to their maturity (Tata et al., 2013).
Our analysis of ATOH1+ IECs in DSS-colitis mice successfully identified cytokine- or bacterial-component-induced NF-κB activation as a key event in promoting ATOH1+ICR formation (Figures 5D–5J), which is consistent with a previous study involving genetic NF-κB activation in villus IECs (Schwitalla et al., 2013). Because NF-κB pathway activation in secretory lineage-committed organoids appeared to upregulate the expression of Dll1 (Figure 5J), it may be possible that NF-κB signaling promotes ATOH1+ICR formation via generating an increased number of DLL1+ secretory progenitor cells.
In the AOM-DSS model, ATOH1+ IECs acquired the properties of tumor stem cells and appeared to contribute to the mosaic development of tumors (Figure 6C). At present, it remains uncertain to what extent these cells precisely contribute to the overall development and progression of these tumors. The contribution of ATOH1+ tumor cells to the overall tumor formation may be underestimated due to the limited labeling efficiency of the present lineage-tracing system. However, the clear mosaic pattern harboring both ATOH1+ and ATOH1− tumor stem cells may illustrate one of the reasons why colitis-associated tumors are highly resistant to chemotherapeutic approaches. Recently, it has been shown that human colorectal cancer cells can inter-convert between differentiated tumor cells and tumor stem cells, thereby exhibiting resistance to tumor stem cell-specific ablation (Shimokawa et al., 2017). Further studies will be needed to confirm the contribution of ATOH1+ tumor cells to the heterogeneous development and progression of CACs found in IBD patients. Nevertheless, our results newly indicate ATOH1+ tumor cells as an additional therapeutic target cell population for CACs.
Experimental Procedures
Mice
C57BL/6J mice and BALB/cAJcl-nu/nu mice were purchased from Japan Clea.
Atoh1Cre-PGR (Rose et al., 2009), Lgr5-EGFP-IRES-creERT2 (Barker et al., 2007), Rosa26-LSL-tdTomato mice (Madisen et al., 2010), or ApcMin mice were purchased from the Jackson Laboratory. Apcfl/fl mice were provided by RIKEN BRC (Colnot et al., 2004). The primers used for genotyping are listed in Table S1. Male and female mice at 8–10 weeks of age were used. All animal experiments were approved by the Animal Care and Use Committee of Tokyo Medical and Dental University (0160326A and 0170276A). For the induction of Cre-mediated recombination, mifepristone (RU486, 2 mg/body, Sigma-Aldrich) or tamoxifen (TMX, 2 mg/body, Sigma-Aldrich) was injected intraperitoneally into mice carrying the Atoh1Cre-PGR allele or the Lgr5-EGFP-IRES-creERT2 allele, respectively.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Okamoto et al., 2009). The primary antibodies employed for these assays are listed in Table S2. Antigen retrieval using citrate buffer was required for the staining of ATOH1, HES1 (Ito et al., 2000), chromogranin A (CHGA), DCLK1, Ki-67, tdTomato, and β-CATENIN.
Author Contributions
F.I., H.S., R.K., T. Nakata, and G.I. performed the histology experiments. F.I., S.F., S.A., K.S., and A.K. performed the organoid culture experiments under the supervision of T. Murano and T. Mizutani. Mouse experiments were performed by F.I. and S.N. under the supervision of S.O. and K.T. T. Nakamura and M.W. supervised the entire project. F.I. and R.O. wrote the manuscript.
Acknowledgments
We thank Dr. Tetsuo Sudo (Toray Industry) and Prof. Jane E. Johnson (UT Southwestern) for providing antibodies. This work was supported by MEXT/JSPS KAKENHI (16H05284, 226221307) and the Research Center Network Program and Practical Research Project from AMED (16bm0304001h0004, 17bm0304001h0005, 17ek0109188h0002).
Published: December 7, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.006.
Accession Numbers
Microarray data have been deposited in the Gene Expression Omnibus under accession numbers GEO: GSE81315 and GSE81451.
Supplemental Information
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Buczacki S.J.A., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Colnot S., Niwa-Kawakita M., Hamard G., Godard C., Le Plenier S., Houbron C., Romagnolo B., Berrebi D., Giovannini M., Perret C. Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Lab. Invest. 2004;84:1619–1630. doi: 10.1038/labinvest.3700180. [DOI] [PubMed] [Google Scholar]
- Davis H., Irshad S., Bansal M., Rafferty H., Boitsova T., Bardella C., Jaeger E., Lewis A., Freeman-Mills L., Giner F.C. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 2015;21:62–70. doi: 10.1038/nm.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins L.A., Souza R.F., Spechler S.J. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2009;6:297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- Fukushima K., Tsuchiya K., Kano Y., Horita N., Hibiya S., Hayashi R., Kitagaki K., Negi M., Itoh E., Akashi T. Atonal homolog 1 protein stabilized by tumor necrosis factor α induces high malignant potential in colon cancer cell line. Cancer Sci. 2015;106:1000–1007. doi: 10.1111/cas.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Udaka N., Yazawa T., Okudela K., Hayashi H., Sudo T., Guillemot F., Kageyama R., Kitamura H. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Kano Y., Tsuchiya K., Zheng X., Horita N., Fukushima K., Hibiya S., Yamauchi Y., Nishimura T., Hinohara K., Gotoh N. The acquisition of malignant potential in colon cancer is regulated by the stabilization of Atonal homolog 1 protein. Biochem. Biophys. Res. Commun. 2013;432:175–181. doi: 10.1016/j.bbrc.2013.01.034. [DOI] [PubMed] [Google Scholar]
- Kim T.-H., Saadatpour A., Guo G., Saxena M., Cavazza A., Desai N., Jadhav U., Jiang L., Rivera M.N., Orkin S.H. Single-cell transcript profiles reveal multilineage priming in early progenitors derived from Lgr5(+) intestinal stem cells. Cell Rep. 2016;16:2053–2060. doi: 10.1016/j.celrep.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F., Manning A.M. Multiple signals converging on NF-kappaB. Curr. Opin. Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- Mills J.C., Sansom O.J. Reserve stem cells: differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci. Signal. 2015;8:re8. doi: 10.1126/scisignal.aaa7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto R., Watanabe M. Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. J. Gastroenterol. 2016;51:11–21. doi: 10.1007/s00535-015-1098-4. [DOI] [PubMed] [Google Scholar]
- Okamoto R., Tsuchiya K., Nemoto Y., Akiyama J., Nakamura T., Kanai T., Watanabe M. Requirement of Notch activation during regeneration of the intestinal epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G23–G35. doi: 10.1152/ajpgi.90225.2008. [DOI] [PubMed] [Google Scholar]
- Onizawa M., Nagaishi T., Kanai T., Nagano K.-I., Oshima S., Nemoto Y., Yoshioka A., Totsuka T., Okamoto R., Nakamura T. Signaling pathway via TNF-alpha/NF-kappaB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G850–G859. doi: 10.1152/ajpgi.00071.2008. [DOI] [PubMed] [Google Scholar]
- Parang B., Barrett C.W., Williams C.S. AOM/DSS model of colitis-associated cancer. Methods Mol. Biol. 2016;1422:297–307. doi: 10.1007/978-1-4939-3603-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.T., Oh H.K., Gum J.R., Crawley S.C., Kakar S., Engel J., Leow C.C., Gao W.Q., Kim Y.S. HATH1 expression in mucinous cancers of the colorectum and related lesions. Clin. Cancer Res. 2006;12:5403–5410. doi: 10.1158/1078-0432.CCR-06-0573. [DOI] [PubMed] [Google Scholar]
- Rose M.F., Ahmad K.A., Thaller C., Zoghbi H.Y. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl. Acad. Sci. USA. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2012;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Fingerle A.A., Cammareri P., Nebelsiek T., Göktuna S.I., Ziegler P.K., Canli O., Heijmans J., Huels D.J., Moreaux G. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Seno H., Miyoshi H., Brown S.L., Geske M.J., Colonna M., Stappenbeck T.S. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc. Natl. Acad. Sci. USA. 2009;106:256–261. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Okamoto R., Ito G., Fujii S., Nakata T., Suzuki K., Murano T., Mizutani T., Tsuchiya K., Nakamura T. Distinct expression patterns of Notch ligands, Dll1 and Dll4, in normal and inflamed mice intestine. PeerJ. 2014;2:e370. doi: 10.7717/peerj.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., Sato T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H. Replacement of Lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir S., Gudjonsson T., Nielsen O.H., Vainer B., Seidelin J.B. Pathogenesis and biomarkers of carcinogenesis in ulcerative colitis. Nat. Rev. Gastroenterol. Hepatol. 2011;8:395–404. doi: 10.1038/nrgastro.2011.96. [DOI] [PubMed] [Google Scholar]
- van Es J.H., Sato T., van de Wetering M., Lyubimova A., Nee A.N.Y., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Furstenberg R.J., Gulati A.S., Baxi A., Doherty J.M., Stappenbeck T.S., Gracz A.D., Magness S.T., Henning S.J. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D.J., Nuber A.H., Brandtner A., Setlik W., Remotti H., Muley A. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Invest. 2014;124:1283–1295. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Bermingham N.A., Finegold M.J., Zoghbi H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- Zeilstra J., Joosten S.P.J., Dokter M., Verwiel E., Spaargaren M., Pals S.T. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008;68:3655–3661. doi: 10.1158/0008-5472.CAN-07-2940. [DOI] [PubMed] [Google Scholar]
- Zhu L., Gibson P., Currle D.S., Tong Y., Richardson R.J., Bayazitov I.T., Poppleton H., Zakharenko S., Ellison D.W., Gilbertson R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.