Summary
The identification of cell surface proteins on stem cells or stem cell derivatives is a key strategy for the functional characterization, isolation, and understanding of stem cell population dynamics. Here, using an integrated mass spectrometry- and microarray-based approach, we analyzed the surface proteome and transcriptome of cardiac progenitor cells (CPCs) generated from the stage-specific differentiation of mouse and human pluripotent stem cells. Through bioinformatics analysis, we have identified and characterized FZD4 as a marker for lateral plate mesoderm. Additionally, we utilized FZD4, in conjunction with FLK1 and PDGFRA, to further purify CPCs and increase cardiomyocyte (CM) enrichment in both mouse and human systems. Moreover, we have shown that NORRIN presented to FZD4 further increases CM output via proliferation through the canonical WNT pathway. Taken together, these findings demonstrate a role for FZD4 in mammalian cardiac development.
Keywords: Frizzled 4, cell surface capture, mass spectrometry, microarray, Norrin, cardiac progenitor cells, cardiomyocytes, cardiac development, regenerative medicine
Highlights
-
•
Identified and characterized FZD4 as a new marker for lateral plate mesoderm
-
•
FZD4, in conjunction with FLK1 and PDGFRA, increases cardiomyocyte enrichment
-
•
FZD4 is expressed in the human system and shows enrichment in cardiomyocytes
-
•
NORRIN addition shows increase in cardiomyocyte output from FZD4 progenitor cells
In this article, Zandstra and colleagues have identified FZD4 as a new marker for lateral plate mesoderm. Using FZD4, they showed an increase in cardiac progenitor cell purity and a subsequent increase in cardiomyocyte induction in both the mouse and human systems. These findings demonstrate a role for FZD4 in mammalian cardiac development.
Introduction
The identification of cell surface proteins on stem cells or stem cell derivatives is a key strategy for the functional characterization, isolation, and understanding of stem cell population dynamics (Nunomura, 2005). Extensive analysis of the identified stem cell types can enhance our understanding of the signaling events that stimulate fate decisions during branching points in the differentiation process. In mouse cardiac development, the heart is derived from cardiac progenitor cells (CPCs), which emerge from the lateral plate mesoderm (LPM) of the embryo (Kinder et al., 1999, Rana et al., 2013). Reports on the enrichment of CPCs (Cai et al., 2003, Passier et al., 2005) show that it is possible to use in vitro cardiomyocyte (CM) differentiation of mouse pluripotent stem cells (mPSCs) (Amit et al., 2000, Cameron et al., 2006, Gerecht-Nir et al., 2004, Sachinidis et al., 2003) as a model system to study cardiac development. A brachyury (BRY)+, fetal liver kinase 1 (FLK1)+ population has been shown to mark two separate mesodermal progenitor populations that arise sequentially; the first gives rise to the hemangioblast and subsequent endothelial and hematopoietic lineages, and the second is enriched for early cardiovascular progenitors that display CM, smooth muscle cell (SMC), and endothelial cell (EC) lineage potential (Kattman et al., 2006). Another type of progenitor cell expressing NK2 transcription factor related, locus 5 (NKX2.5), and kit oncogene (C-KIT) is capable of clonal expansion and differentiation to CMs, conduction system cells, and SMCs in vitro (Wu et al., 2006). However, these cells do not give rise to cells of the endothelial lineage, suggesting that they represent a more differentiated progenitor population that has already segregated from the endothelial fate. Finally, isolated islet 1 (ISL1)+ cells have been shown to be enriched for CPCs (Moretti et al., 2006). More specifically, ISL1+NKX2.5+FLK1+ cells give rise to CM, SMC, and EC lineages, while ISL1+FLK1+NKX2.5− cells produce SMCs and ECs, and ISL1+FLK1−NKX2.5+ cells produce CM and SMC lineages. It is clear that a number of genes have been implicated in early cardiogenesis; however, only a small fraction of these genes represent surface markers, including FLK1, platelet-derived growth factor α (PDGFRA), (Bondue et al., 2011), and, more recently, C-X-C chemokine receptor type 4 (CXCR4) (Nelson et al., 2008). Accordingly, there is a need for additional surface markers that can identify additional cardiogenic subpopulations, which would allow for the enrichment of pure of CPCs and thus enable a better understanding of cell dynamics during heart development.
Cell surface analysis of mPSCs and human pluripotent stem cells (hPSCs) have previously revealed a large set of diverse markers and signaling molecules associated with mPSC maintenance and development (Bausch-Fluck et al., 2015, Boheler et al., 2014, Zhang et al., 2009). We have extended this analysis to encompass mPSC differentiation to cardiac mesoderm and ultimately to CMs. Using mass spectrometry (MS), we identified 246 surface markers during key stages of mesoderm specification and early cardiac development in vitro. We also performed microarray analysis on the CPC subpopulations isolated by surface markers and cross-referenced the proteomic data to identify candidate proteins specific to CPCs. These proteins were further validated using qRT-PCR and flow cytometry, leading to the selection of five promising CPC marker candidates. In this context, we focused on the surface receptor Frizzled 4 (FZD4) for further analysis. Supporting our technical validation, FZD4 is known to be involved in the Wingless (WNT) signaling pathway, which plays an essential role in cardiac development (Cadigan and Nusse, 1997, Cohen et al., 2008, Gessert and Kühl, 2010).
Here, we demonstrated that day 3.75 (d3.75) mPSC-derived FZD4+ cells are enriched for LPM, which gives rise to cardiomyocytes, while the FZD4− population is enriched for paraxial mesoderm, which gives rise to somites. We also have shown that within the FLK1+PDGFRA+ CPC population are FZD4+ and FZD4− subpopulations, in both the mPSC and hPSC systems. Furthermore, we have provided evidence that activation of the WNT signaling pathway with the addition of the ligand NORRIN further increases the cardiogenic output of the FZD4+ cell population. In conclusion, FZD4 is a marker that can be used to segregate pre-cardiac mesoderm and FZD4-NORRIN signaling increases CM output through canonical WNT activation, which further validates the role of FZD4 in cardiac induction.
Results
Integrated Mass Spectrometry and Microarray Analysis Identifies Early Mesoderm Surface Markers
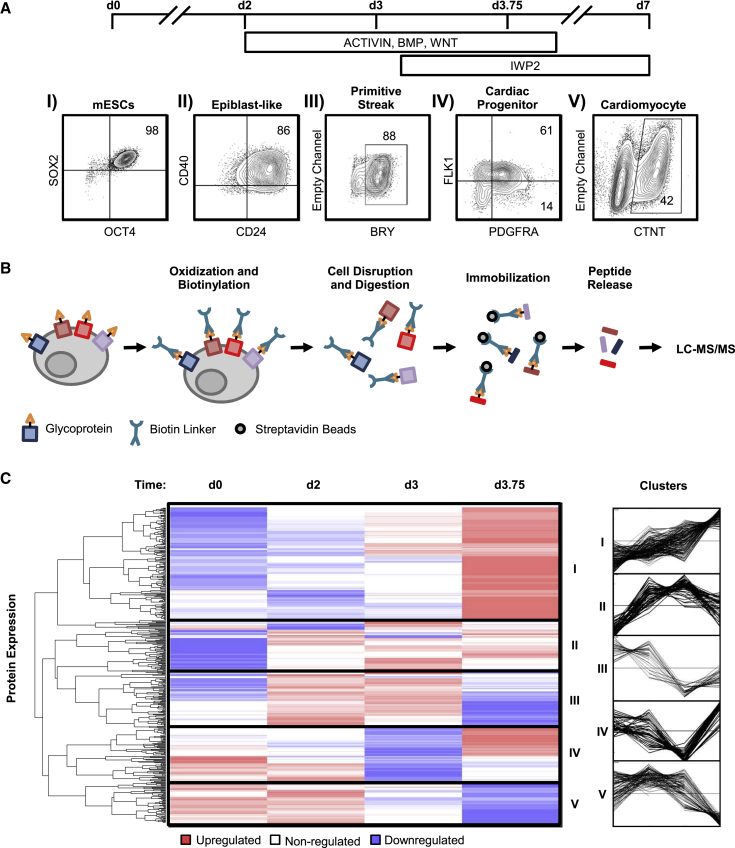
A staged protocol was applied in bioreactors to differentiate mPSCs to cardiomyocytes (Figure 1A). The stages were tracked by flow cytometry to characterize and assess purity starting with mPSCs (OCT4+SOX2+) followed by epiblast-like cells (CD24+CD40+), then primitive streak cells (BRY+), then early mesoderm containing CPCs (FLK1+PDGFRA+), and finally CMs (cardiac troponin T; CTNT+) (Figure 1AI–V). We then used MS to identify and characterize surface protein abundance profiles corresponding to the CPC stage of development (Figure 1AIV). MS analysis of surface proteins has been generally challenging due to the limited abundance of surface proteins relative to intracellular proteins, and difficulty in isolating hydrophobic membrane-bound proteins (Josic and Clifton, 2007, Macher and Yen, 2007). To address these concerns, we utilized a tag-based method (Wollscheid et al., 2009) to specifically label and exclusively isolate surface proteins for analysis (Figure 1B). Protein abundance at the cell surface was compared between time points using label-free quantification. Unsupervised k-means clustering was performed, revealing five distinct temporal patterns (Figure 1C). Cluster 1 contained 246 proteins whose abundance increased at the CPC stage and included known CPC markers, such as FLK1 and PDGFRA. Cluster 1 was also enriched for receptors relevant to mesoderm differentiation based on gene ontology analysis. However, the d3.75 time point, associated with the emergence of CPCs, is a mixed population, and thus it was difficult to determine which of the highly expressed surface proteins are associated with the FLK1+PDGFRA+ subpopulation. To better understand the underlying heterogeneity, we next performed a microarray time course focusing on the CPC and CM stages.
Figure 1.
Cell Surface Mass Spectrometry Yields Protein Abundance Profiles
(A) Cell production strategy involving embryoid body differentiation with cytokine addition at day 2 (d2) and d3 of differentiation. Cell types were assessed using flow cytometry with stage-specific markers (I–V). The number in the box represents the percent positive value.
(B) Surface proteins were labeled and processed into peptides using a cell surface capture procedure before LC-MS/MS analysis.
(C) Unsupervised k-means clustering of cell surface MS data was separated into 5 main clusters (red, upregulated; white, neutral; blue, downregulated; n = 4–5 in each group, pooled from 6 independent experiments). Line graph representations of each cluster show distinct profiles of up- and downregulation.
The CPC-stage cell population consists mostly of the cardiogenic FLK1+PDGFRA+ and hemogenic FLK1+PDGFRA− subpopulations (Kattman et al., 2006). We sorted the two subpopulations from the CPC stage and compared their transcriptome with that of the CTNT+ CMs (Figure 2A). Hierarchal clustering of the gene expression data showed distinct expression patterns unique to each of the subpopulations. When both of these subpopulations were compared with CM, both showed similar pathway enrichment; however, the FLK1+PDGFRA+ subpopulation showed 10-fold greater significance in cardiovascular pathway enrichment compared with the FLK1−PDGFRA+ population (Figure S1). Overall, 871 of 16,755 genes were differentially expressed (either up- or downregulated) when the FLK1+PDGFRA+ subpopulation was compared with the FLK1+PDGFRA− subpopulation. While these two subpopulations were relatively similar in gene expression, and were enriched for similar cardiac pathways, the upregulated pathways specific to cardiogenesis showed a higher enrichment, indicated by a larger −log(p value) (Figure 2B), suggesting that the CPCs reside in the FLK1+PDGFRA+ subpopulation. As stated previously, MS analysis at the CPC stage was performed on a heterogeneous population. To determine which subpopulation is associated with the identified surface markers, we applied gene-level segregation between hemogenic and cardiogenic subpopulations with respect to surface protein abundance by cross-referencing the genes with the identified proteins based on common patterns. As a result, the 246 surface proteins identified in cluster 1 were divided into three categories: 47 proteins uniquely expressed in the FLK1+PDGFRA+ subpopulation, 34 proteins in the FLK1+PDGFRA− subpopulation, and 165 proteins common to both (Figure 2C). Thus, by integrating the specificity of the MS-based protein identification with the purity of microarray analysis of sorted subpopulations, we have identified a list of 47 surface markers that are uniquely enriched in the FLK1+PDGFRA+ CPC subpopulation. We next set out to independently analyze and validate this dataset.
Figure 2.
Microarray Analysis Yields Subpopulation-Specific Surface Markers
(A) Gene analysis was performed on the sorted FLK1+PDGFRA−, FLK1+PDGFRA+, and CTNT+ populations in triplicate. Unsupervised clustering yielded distinct expression patterns unique to each cell type (red, upregulated, white, neutral, blue, downregulated; n = 3 in each group, pooled from 4 independent experiments).
(B) In a comparison between the FLK1+PDGFRA+ and FLK1+PDGFRA− populations, pathway enrichment analysis of the significantly upregulated and downregulated genes revealed pathways relevant to CM development.
(C) Proteins identified by MS from cluster 1 were further segregated as either uniquely or commonly expressed on FLK1+PDGFRA+ and FLK1+PDGFRA− cells using microarray data.
FZD4 Identified as a Potential Marker of a Subpopulation Enriched for Cardiac Progenitors
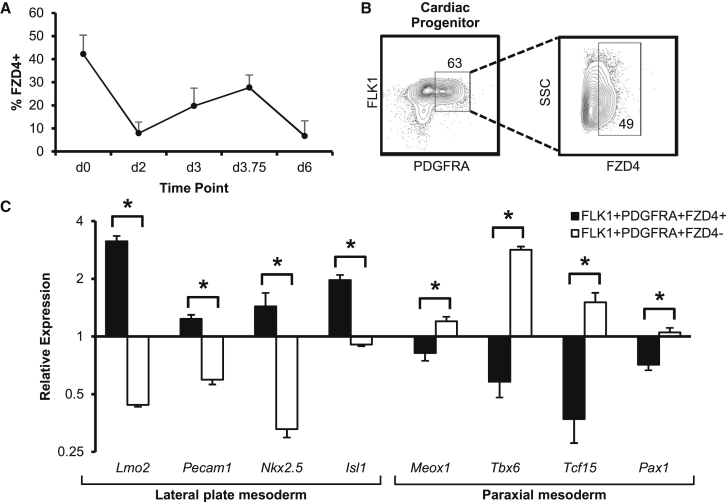
The 47 proteins that were enriched in the cardiogenic subpopulation were assessed and filtered based on literature search, mRNA expression and protein abundance correlation (Figure S2A), and antibody availability and efficacy (Figure S2B) to identify FZD4, ITGB5, LPAR4, and PLEXINB1 as the top candidates for further validation. Co-staining with these four candidate markers resulted in further segregation of cardiac progenitors within the heterogeneous FLK1+PDGFRA+ population (Figure S2C). Since FZD4 is involved in the WNT signaling pathway, which is developmentally relevant during early cardiac differentiation (Cohen et al., 2008, Gessert and Kühl, 2010, Ueno et al., 2007), FZD4 protein abundance was further evaluated by flow cytometry (Figure 3A). FZD4 abundance frequency decreased during primitive streak formation, increased at the cardiac progenitor stage, and decreased again when the CM population began to emerge. These observations suggested a potential role for FZD4 during CPC development, and we chose to further investigate FZD4 as a potential marker of a subpopulation with cardiac potential.
Figure 3.
Flow Cytometry and qPCR Validation of Proteins
(A) FZD4 protein abundance time course was assessed using flow cytometry (n = 4 independent experiments, mean ± SEM).
(B) FLK1+PDGFRA+ subpopulation concomitantly stained with FZD4 show FZD4+ and FZD4− subpopulations. The number in the box represents the percent positive value.
(C) qPCR interrogation of lateral plate and paraxial mesoderm genes on sorted FLK1+PDGFRA+FZD4+ and FLK1+PDGFRA+FZD4− populations. FLK1+PDGFRA+FZD4+ cells showed an enrichment of lateral plate mesoderm genes while FLK1+PDGFRA+FZD4− cells showed an enrichment of paraxial mesoderm genes (mean ± SEM, n = 4 independent experiments, ∗p < 0.05).
Gene Expression Analysis on Sorted CPC Subpopulations Indicates that FZD4 Marks Pre-cardiac Mesoderm in CPCs
We next set out to examine the role of FZD4 in CPC development. We first used a sorting strategy to determine whether the FZD4+ and FZD4− populations captured a bifurcation point in early cardiac differentiation (Figure 3B). Cardiac progenitors were generated (as described in Figure 1A) and the gated FLK1+PDGFRA+ population was sorted into FZD4+ and FZD4− subpopulations (Figure S4A). Enrichment of FLK1, PDGFRA, and FZD4 abundance relative to the mock sorted sample within each compartment was confirmed using both flow cytometry (Figure S4B) and qPCR analysis (Figure S4C), which demonstrated high purities and minimal contamination from adjacent subpopulations.
We analyzed the FZD4+ and FZD4− populations directly after being sorted for genes that are expressed as the cells migrate from the primitive streak toward the anterior lateral region of the embryo, specifically looking at the bifurcation point segregating lateral plate and paraxial mesoderm. Cardiomyocytes are derived from the LPM and are marked by LIM domain only 2 (Lmo2), platelet endothelial cell adhesion molecule 1 (Pecam1), NK2 homeobox 5 (Nkx2.5), and Isl1, while paraxial mesoderm are marked by mesenchyme homeobox 1 (Meox1), T-box 6 (Tbx6), transcription factor 15 (Tcf15), and paired box 1 (Pax1) (Cheung et al., 2012). We observed that the FZD4+ population expressed higher levels of LPM markers while the FZD4− population expressed higher levels of paraxial mesoderm markers (Figure 3C). These results indicated that FZD4 may distinguish between lateral plate and paraxial mesoderm and may be used to further purify the early mesoderm population for CPCs.
FZD4-Expressing CPCs Yield Higher CM Outputs
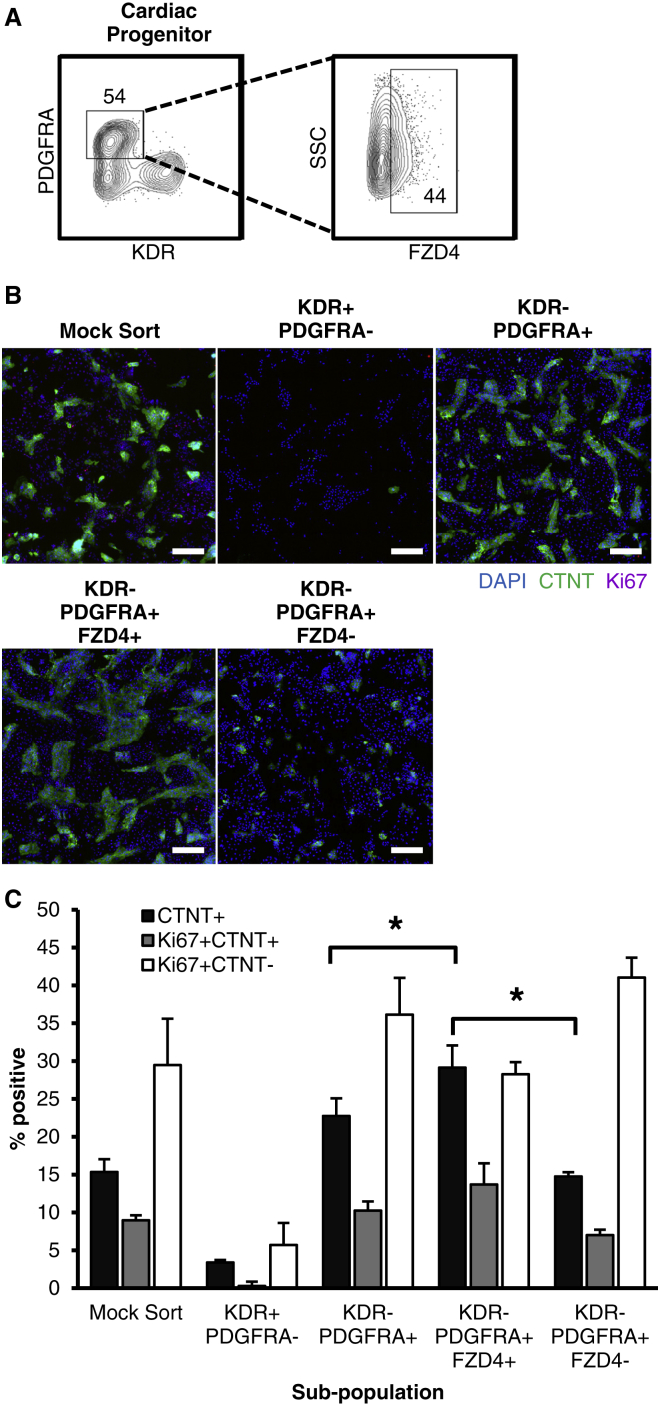
Given that our gene expression analysis showed that the FZD4+ CPC subpopulation was enriched for markers of LPM, which contains the CPC population during embryogenesis, we examined whether our in vitro FZD4+ expressing population was enriched for CPCs. Sorted FZD4+ and FZD4− populations were seeded onto a 384-well plate and cultured for 3 days until beating CMs were observed (Figure S5A). Bright-field imaging revealed that the FLK1+PDGFRA+FZD4+-derived fraction maintained robust beating and web-like networks, while beating was generally not observed in the FLK1+PDGFRA+FZD4−-derived fraction, which displayed static cell monolayers (Figure S5B). This observation was consistent with gene expression (Ctnt, α-myosin heavy chain [α-mhc], Isl1, and Pecam1) (Figure S5C), flow cytometry (CTNT) (Figure 4A), and immunofluorescence (CTNT) (Figure 4B) analyses of the sorted populations.
Figure 4.
FZD4 Enriches CPC in the FLK1+PDGFRA+ Population, Increasing Subsequent CM Yield
(A and B) Cardiac output of sorted progenitor subpopulations based on CTNT abundance measured by flow cytometry (A; the number in the box represents the percent positive value) and immunofluorescence staining CTNT-expressing cells (green) and DAPI nuclear stain (blue) (B). Scale bars represent 500 μm.
(C) Quantified from flow cytometry, d7 CTNT+ percentage from d3.75 input CPC populations sorted using CPC-associated markers FLK1, PDGFRA, and FZD4. CTNT abundance is significantly higher in the FLK1+PDGFRA+FZD4+ population relative to both the FLK1+PDGFRA+ and FLK1+PDGFRA+FZD4− population (mean ± SEM, n = 4 independent experiments, ∗p < 0.05).
(D) (I) d3.75 (input) CPC marker abundance was correlated to the cardiomyocyte percentage (CTNT+) of the d7 (output) populations. (II) In the mock sorted d7 output, the CTNT abundance has a higher correlation with the FLK1+PDGFRA+FZD4+ input marker than FLK1+PDGFRA+. (III) FZD4 alone also exhibited a higher correlation than FLK1+PDGFRA+ with the output CTNT abundance in the mock sorted sample (n = 4 independent experiments, ∗p < 0.05).
To compare the level of cardiac induction between the FZD4+ and FZD4− subpopulations, we quantified the frequency of CTNT abundance and used 5-ethynyl-2′-deoxyuridine (EdU) to estimate cell proliferation (Figure 4C). Compared with the mock sorted controls that showed a CTNT+ frequency of 31.1% ± 8.6%, the CPC (FLK1+PDGFRA+) population showed a significantly enriched CTNT+ output of 53.9% ± 4.3%, and the non-CPC (FLK1−PDGFRA−) showed a lower output of 5.0% ± 2.0%. Additionally, the FLK1+PDGFRA+FZD4+ (62.1% ± 6.0%) and FLK1+PDGFRA+FZD4− (38.7% ± 6.4%) condition showed a significant increase and decrease of CTNT+ abundance, respectively, relative to the FLK1+PDGFRA+ population. We also observed this trend in the CTNT+EdU+ subpopulation, indicating that there is an increased baseline proliferation of CTNT+ cells in the Flk1+PDGFRA+FZD4+ subpopulation. This suggests that the increased CTNT+ expression in FZD4+ sorted populations may be due to a proliferation-based mechanism.
To explore how FZD4 abundance correlates with output CTNT response, we compared the different d3.75 (input) CPC markers with cardiomyocyte percentage (CTNT+) of the d7 (output) populations (Figure 4DI). A high correlation between an input CPC marker and output CTNT abundance can be indicative of the predictive power of the CPC marker. In the mock sorted d7 output, CTNT abundance correlated more highly with the FLK1+PDGFRA+FZD4+ input marker than with FLK1+PDGFRA+ alone (Figure 4DII). Moreover, FZD4 alone also exhibited a higher correlation than FLK1+PDGFRA+ with the output CTNT abundance in the mock sorted sample (Figure 4DIII). The other output populations showed relatively no correlation with input markers, which can be expected since these populations are sorted, essentially negating any positive or negative biases the input markers may have had. Taken together, these results indicate that at the CPC stage, FZD4 can be used either alone or in combination with FLK1 and PDGFRA to predict the number of CMs after 3 days of differentiation, and that higher FZD4 abundance at the CPC stage will lead to greater CM output.
Higher CM Output from FZD4+ CPCs Is Also Observed during hPSC Differentiation
To determine whether FZD4 abundance patterns and signaling effects were conserved in hPSCs, we generated hPSC-derived CPCs in a similar BMP/ACTIVIN A-induced system (Yang et al., 2008). In the human system, CPCs are also identified using the surface markers kinase insert domain receptor (KDR; analog to FLK1 in mouse) and PDGFRA. However, in contrast to mouse CPCs, the KDR−PDGFRA+ subpopulation contains the CPC. Co-staining with FZD4 reveals a similar segregation of the cardiogenic KDR−PDGFRA+ population into a FZD4+ and FZD4− subpopulations, consistent with what we have observed in the mouse system (Figure 5A). The FZD4+ and FZD4− fractions were sorted and reseeded, and after 7 days the abundance of CTNT and the cell proliferation marker, Ki67, was analyzed using immunofluorescence (Figure 5B) and quantified using image analysis (Figure 5C). We observed a similar increase in the CTNT+ percentage in the PDGFRA+FZD4+ subpopulation relative to the FZD4− subpopulation. This trend is also seen in the Ki67+ percentage within the CMs, suggesting that the increase in CM frequency is due to increased proliferation. These results indicate that FZD4 enriches for CMs and suggests that the increase in CMs is due to increased proliferation, which is consistent with our observations in the mouse system.
Figure 5.
FZD4 Enriches CPC in hPSC-Derived Cardiomyocytes
(A) FZD4+ and FZD4− subpopulations were sorted from PDGFRA+ cardiac progenitor cells and differentiated into cardiomyocytes. The number in the box represents the percent positive value.
(B and C) Intracellular staining for CTNT (green), DAPI nuclear stain (blue), and Ki67 (purple) (B) shows that the FZD4+ compartment exhibited an increased number of cardiomyocytes as compared with the FZD4− population (scale bar represents 500 μm), quantified (C) using CellProfiler (mean ± SEM, n = 3 independent experiments, ∗p < 0.05).
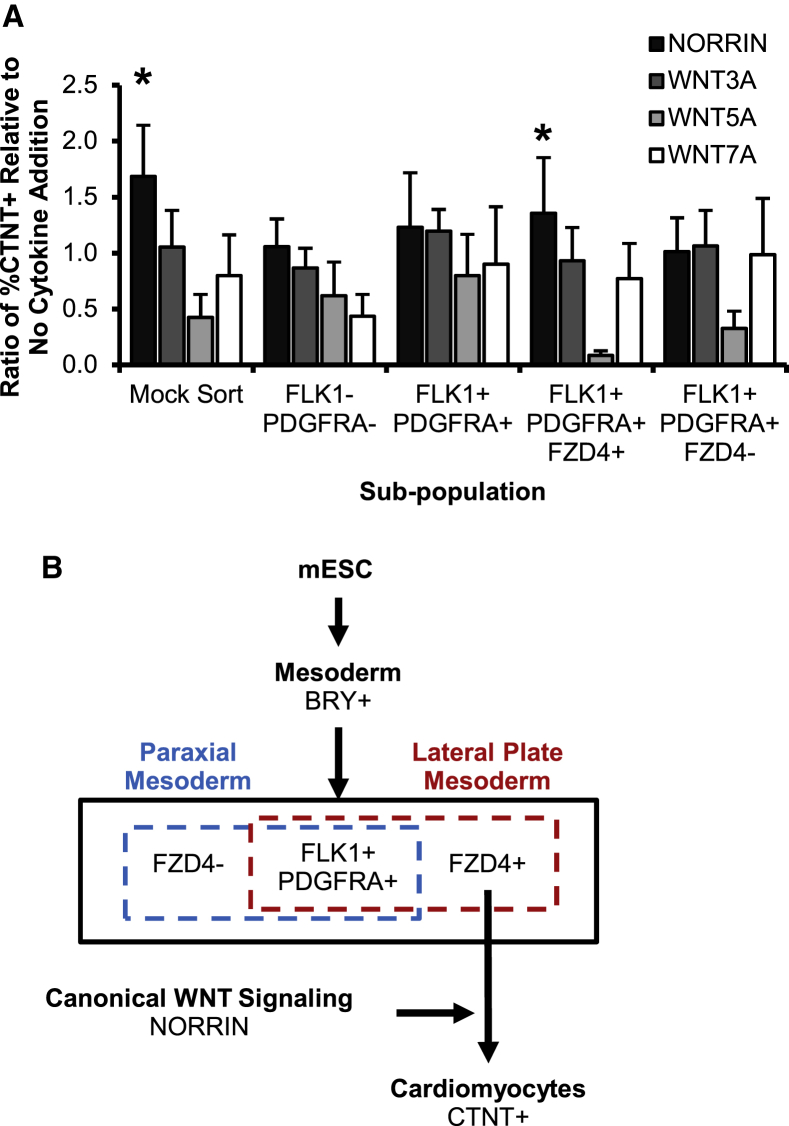
Canonical FZD4-NORRIN Signaling Enhances CM Output
We next examined whether the addition of ligands that bind FZD4 would have biological consequences on CM differentiation. We added ligands that bind with FZD4 (WNT3A, WNT5A, WNT7A, and NORRIN) to both mock sorted and sorted subpopulations at the CPC stage (d3.75) and measured the subsequent CTNT abundance after 3 days (Figure S6A). To determine optimal conditions for CM induction, we generated individual dose-response curves for each ligand (Figure S6B). The CM output was reported as a ratio relative to the base condition where no ligand was applied (Figure 6A). The ligands WNT3A, WNT5A, and WNT7A showed no significant difference with the ligand-free base condition. The addition of NORRIN, on the other hand, led to an increase in CTNT abundance in both mock sorted and FLK1+PDGFRA+FZD4+ subpopulations. These data show that the addition of the exogenous WNT ligand, NORRIN, further increases CTNT output in the FZD4+ cells relative to the absence of WNT stimulation, demonstrating a functional consequence for the abundance of FZD4 on CPCs. This provides evidence to suggest that NORRIN signals through the canonical WNT pathway to enhance cardiogenesis (Figure 6B). Given that our flow cytometry and qPCR results revealed a lack of Fzd4 expression at the CM stage compared with the CPC stage (Figure 3A), it is likely that FZD4-NORRIN signaling occurs during or immediately after the specification of FLK1+PDGFRA+ CPCs and acts to increase proliferation.
Figure 6.
Model of FZD4 Abundance in the Context of Early Cardiac Differentiation
(A) The addition of NORRIN further enhances CTNT response in both mock sorted and sorted FLK1+PDGFRA+FZD4+ populations (mean ± SEM, n = 3 independent experiments, ∗p < 0.05).
(B) Proposed model indicating the hierarchy of cells during differentiation and the relevant cell populations with their markers.
Discussion
In vitro analysis of LPM during differentiation has typically required the use of intracellular markers (such as Lmo2, Nkx2.5, and Isl1), which are not compatible for live cell sorting for further interrogation of enriched LPM subpopulations. Identification of membrane-expressed proteins that mark LPM would not only contribute to the functional analyses of in vitro mesoderm development but also enable live cell sorting of cardiac progenitors, which could then be enriched for further cell fate studies and the development of efficient cell manufacturing protocols. During embryogenesis, the surface protein FZD4 is expressed in LPM, from which cardiac mesoderm develops, defining one of the earlier specification points in cardiogenesis (Tan et al., 2013). We have shown evidence that receptor FZD4 can initiate the WNT signaling pathway, which has been shown in chick embryos to be required for LPM development during gastrulation (Sweetman et al., 2008). FZD4 has also been shown to map to a chromosomal region important for cardiac development (DeRossi et al., 2000). Additionally, Abdul-Ghani et al. (2011) have demonstrated that blocking FZD4 results in reduced cardiac induction, further implicating the importance of FZD4 in cardiac development. Here, we report that abundance of the surface marker FZD4 distinguishes lateral plate from paraxial mesoderm, and marks a pre-cardiac mesoderm population that can be used to more specifically select the CPC population during in vitro mPSC and hPSC differentiation.
While our results show that FZD4 abundance can be used to purify the CPC population and obtain subsequent CM enrichment, we also observed the presence of CM in the FZD4− subpopulation (38.2% ± 8.3% CTNT+ percentage). One possible cause may be the regeneration of the FZD4− population during culture post sorting. While the purities of the sorted FZD4+ and FZD4− populations are >95% as measured by both flow cytometry (Figure S4B) and qPCR (Figure S4C), the FZD4− population may generate a secondary FZD4+ subpopulation, albeit at a lower frequency, resulting in a lower CM population. In addition, it is also possible that the FZD4+ and FZD4− subpopulations are enriched for different types of progenitor cells, which could result in different types of CMs. Finally, this may indicate that another marker in addition to FZD4 may be required to fully purify the CPC subpopulation.
The FZD4 receptor is involved in WNT signaling, which is a key pathway during cardiogenesis (Cohen et al., 2008), consistent with our demonstration that NORRIN enhanced the CM output from our CPC subpopulation by increasing CM proliferation. It is known that the kinetics and type (i.e., canonical versus non-canonical) of WNT activation is critical in guiding cardiac induction and differentiation (Sumi et al., 2008). Early cardiac induction relies on canonical signaling to induce primitive streak formation and gastrulation. Subsequent canonical signaling must be inhibited in order for cardiac specification to occur, as the LPM moves laterally across the embryo. Canonical WNT activation is again required in later stages of cardiac development in a proliferative capacity to expand the cardiogenic population (Gessert and Kühl, 2010). In this study, we have shown that FZD4 is not only expressed on LPM but is also involved in increasing CM induction through increased proliferation. Unexpectedly, when our differentiation cultures were presented with WNT ligands at d3 (the equivalent of gastrulation in the embryo), there was no difference observed in CM output. However, upon addition of NORRIN at the CPC stage, there was an increase in CM enrichment due to an increase in proliferation. Further in-depth survival and proliferation studies are required to clearly distinguish between proliferation and selection mechanisms in CM development. Additionally, a move toward network-based WNT activation studies (Moon and Gough, 2016) would provide further evidence.
Cardiac development is fairly conserved across multiple organisms, especially between the mouse and human system (Brade et al., 2006). We have demonstrated that FZD4 signaling promotes increased CM differentiation efficiency in the human system. During the CPC stage of hPSC differentiation, FZD4-NORRIN signaling increased CM output through increased proliferation. A previous study has also confirmed the increase in FZD4 abundance during the LPM stage in hPSCs and also implicated the role of FZD4 through the non-canonical WNT signaling pathway (Mazzotta et al., 2016). While our work indicates a canonical WNT signaling mechanism, both pathways may play a sequential role whereby immediately after progenitor specification canonical WNT signaling is required, with subsequent non-canonical signaling dominating to further promote CM specification. Further studies are required to tease out the intricate timing required for these signaling pathways to occur. In both cases, not only can FZD4 be used as a marker for enrichment analysis, but the FZD4 pathway can also be exploited to increase CM yield from hPSCs. Furthermore, several studies to develop clinical protocols for hPSC-derived CPC and CM therapies (Fernandes et al., 2015, Trounson and DeWitt, 2016) have highlighted the need for good-quality markers to identify and isolate the cell types required at high purity. To this end, our findings suggest that FZD4 is a highly promising candidate marker that can be used to increase CPC purity and be incorporated into the development of cardiac cell-based therapeutic applications.
Our sorting studies confirmed that FZD4 can be used in conjunction with previously known CPC markers, FLK1 and PDGFRA, to further resolve the CPC population and increase CM output. In addition to FZD4, our MS analysis identified other markers based on similar abundance patterns, which may also be important in the CPC stage of heart development. We already have initial validation of the abundance of three markers (LPAR4, ITGB5, and PLEXINB1) in terms of alignment with the proteomic data, and while further validation in terms of antibody specificity is required, these markers, including FZD4, can be used in combination to further resolve the different subpopulations present in the CPC population. For example, LPAR4 has been shown to be present in all four chambers of the developing rat heart (Wang et al., 2012), which may indicate a similar function in mouse cardiac development. In addition, ITGB5 contributes to transforming growth factor β-mediated cell adhesion to extracellular matrix and cell movement in mouse and human epithelial cells (Bianchi-Smiraglia et al., 2013), which may be involved during ingression of the LPM. PLEXINB1 is largely implicated in mouse neuronal development (Fazzari et al., 2007), although it can still play a role as a negative marker in improving the purification of the CPC subpopulation from the general heterogeneous population. Applying this process broadly, our analysis generated potential surface markers for additional stages of development (stem cell, epiblast, and primitive streak) and may provide further insights into the multiple cell types involved during the entire process of stem cell maintenance, CPC differentiation, and CM specification.
Conclusion
We have identified and characterized FZD4 as a marker for LPM and were able to utilize FZD4 as a marker, in conjunction with FLK1 and PDGFRA, to further purify CPCs and increase CM enrichment. Additionally, we have shown that FZD4 is also expressed in the hPSC system and allows for a similar enrichment in CM. Finally, NORRIN can be presented to the FZD4 receptor to induce WNT signaling-mediated proliferation, which results in an increase in CM output from CPCs. Taken together, these findings demonstrate a role for FZD4 in contributing to our understanding of the biology of mouse and human cardiac development.
Experimental Procedures
PSC Culture and Bioreactor Differentiation
The mouse PSC line (E14.1, 129/Ola) that expresses GFP driven by Brachyury expression (Fehling, 2003) was generously provided by Dr. Gordon Keller. PSCs were maintained on 0.2% gelatin (Sigma) coated tissue culture polystyrene (Fischer) in DMEM/nutrient mixture F-12 (Thermo Fisher Scientific) and Neurobasal medium (Thermo Fisher Scientific) supplemented with 1× B-27 supplement (Thermo Fisher Scientific), 1× N-2 supplement (Thermo Fisher Scientific), 2 mM Glutamax (Thermo Fisher Scientific), 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific), 0.05% BSA (Wisent), 1.5 × 10−4 M monothioglycerol (MTG; Sigma), 107 units/mL leukemia inhibitory factor (Millipore), and 10 ng/mL bone morphogenic protein 4 (BMP4; R&D Systems).
For the generation of embryoid bodies (EBs), PSCs were dissociated into single cells with TrypLE Express Enzyme (1×) (Thermo Fisher Scientific) and plated at 750,000 cells per 10 mL in a 100-mm Petri dish (BD Biosciences) and rotated at 60 rpm on a shaker plate. The differentiation medium used consisted of Iscove's modified Dulbecco's medium (Thermo Fisher Scientific) and Ham's F-12 nutrient mix (Thermo Fisher Scientific) supplemented with 1× B-27 supplement minus ascorbic acid (Thermo Fisher Scientific), 1× N-2 supplement, 2 mM Glutamax, 100 U/mL penicillin-streptomycin, 0.05% BSA, 1.5 × 10−4 M MTG, and 0.5 mM ascorbic acid (Sigma).
At d2, EBs were harvested and dissociated into single cells using TrypLE (Thermo Fisher Scientific) and reseeded into 100-mm Petri dishes (BD Biosciences) with differentiation medium further supplemented with 1 ng/mL BMP4, 2 ng/mL ACTIVIN A (R&D Systems), and 3 ng/mL WNT3A (R&D Systems). At d3, 2 μM inhibitor of WNT production (IWP2; Reagents Direct) was added to each Petri dish. At d3.75, cells were dissociated into single cells using TrypLE or 1 mM EDTA (Sigma) and seeded onto 0.2% gelatin-coated 96-well (Corning) or 384-well (Greiner) tissue culture plates at 200,000 cells and 50,000 cells, respectively, in StemPro-34 (Thermo Fisher Scientific), 2 mM Glutamax, 0.5 mM ascorbic acid, 150 μg/mL transferrin (Roche), 100 U/mL penicillin-streptomycin, and 2 μM IWP2. At d7, cells were harvested and analyzed.
Cells were incubated in a humidified 5% (v/v) CO2 air environment at 37°C.
Flow Cytometry, Cell Sorting, and Immunocytochemistry
EBs and cardiac cultures were harvested and dissociated to single cells in 1 mM EDTA or TrypLE and stained with the appropriate markers listed in Table S1. For cell surface markers, cells were stained in 0.5% fetal bovine serum (FBS; Gibco) in Hank's balanced salt solution (Thermo Fisher Scientific), and 7-aminoactinomycin D (Thermo Fisher Scientific) was used to denote live/dead cells. For intracellular proteins, cells were stained with LIVE/DEAD fixable near-IR dead cell stain kit for 633 or 635 nm excitation (Thermo Fisher Scientific) to denote live/dead cells, then fixed with 0.37% formaldehyde (Sigma) and permeabilized with 0.5% saponin (Sigma). Cell proliferation assays were performed according to the manufacturer's protocol using the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific). Stained cells were analyzed using an LSRFortessa (BD Biosciences). A gating strategy to mark live/dead cells along with singlet cells was used to minimize noise in the output signal (Figure S5D).
Cells were sorted using the FACSAria II (BD Biosciences) or FACSAria III (BD Biosciences), and analyzed using LSRFortessa (BD Biosciences).
All flow-cytometry data were analyzed using FlowJo software (vX.0.7; Treestar).
For immunocytochemistry, cells were fixed in 0.37% formaldehyde and permeabilized using 0.1% Triton X-100 (Sigma) in PBS (Thermo Fisher Scientific).
Microscopy and Image Analysis
Bright-field images were captured using an Olympus inverted microscope using cellSens software (Olympus). High-throughput image capture of fluorescent images was acquired at 10× and stitched together using Cellomics Arrayscan (Thermo Fisher Scientific). Image analysis was performed using Cell Profiler (Carpenter et al., 2006).
qPCR Analysis
Total RNA was extracted from samples using RNeasy mini or micro kits (Qiagen), and cDNA was generated using Superscript III reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer's protocols. cDNA was mixed with primers and SYBR Green Master Mix (Roche) and run on QuantStudio real-time PCR software (Applied Biosystems). Relative quantitative expression of genes was determined by the delta-delta cycle threshold method with GAPDH as an internal reference control. Primer sequences are listed in Table S2.
Microarray Analysis
Samples of mRNA were submitted to The Center for Applied Genomics, and Affymetrix Mouse Exon 1.0 ST arrays were used for the hybridization. Arrays were scanned, and the expression data were obtained in the form of .CEL files. Data were analyzed using R (v3.3.1) Bioconductor suite to extract, normalize, and summarize gene expression data.
Cell Surface Capture
Cell surface capture was performed as described previously (Wollscheid et al., 2009). In brief, cells or EBs were dissociated using 1 mM EDTA (Thermo Fisher Scientific) and 10% FBS (Hyclone). Exposed extracellular aldehydes were then oxidized with 1.6 mM sodium meta-periodate (Thermo Fisher Scientific) and reacted for 1 hr with 5 mM biocytin hydrazide (Biotium). The cells were then washed and lysed by sonication in a hypotonic lysis buffer, and the nuclei were pelleted by centrifugation. The supernatant containing the membranes and the cytoplasm were subjected to ultra-centrifugation. The microsomal pellet was collected and solubilized by addition of 0.1% RapiGest (Waters) and sonication. After overnight trypsin (Thermo Fisher Scientific) digestion, the biotinylated peptides were coupled to streptavidin beads (Thermo Fisher Scientific), thoroughly washed, and enzymatically released by PNGaseF (NEB). Peptides were then cleaned up over C18 tips (The NestGroup) and subjected to liquid chromatography-tandem MS (LC-MS/MS).
Mass Spectrometry Analysis
Samples were analyzed on an EASY-nLC 1,000 nano LC system (Thermo Fisher Scientific) connected to an Orbitrap Velos (Thermo Fisher Scientific) mass spectrometer, which was equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Peptide separation was carried out on a reversed-phase high-performance liquid chromatography column (75 μm × 10 cm) packed in-house with C18 resin (Magic C18 AQ 3 μm, Michrom BioResources) using a linear gradient from 90% solvent A (water, 0.2% formic acid, 1% acetonitrile) and 10% solvent B (water, 0.2% formic acid, 80% acetonitrile) to 65% solvent A and 35% solvent B for 60 min at a flow rate of 0.2 μL/min. The data-acquisition mode was set to acquire one high-resolution first-stage scan (ms1) in the ion cyclotron resonance cell followed by 10 collision-induced dissociation mass scans in the linear ion trap. For a high-resolution mass scan, 106 ions were accumulated over a maximum time of 500 ms, and the full width at half-maximum resolution was set to 60,000 (at m/z 300). Only mass signals exceeding 500 ion counts triggered a second-stage (ms2) attempt, and 104 ions were acquired for an ms2 scan over a maximum time of 250 ms. The normalized collision energy was set to 35%, and one microscan was acquired for each spectrum. Singly charged ions were excluded from triggering ms2 scans.
All acquired mass spectra were searched against the International Protein Index database (Version 3.26) using the SEQUEST algorithm (Eng et al., 1994). The SEQUEST database search criteria included: 0.2 Da mass tolerance for the precursor ion, 0.5 Da mass tolerance for the fragment ions, variable modifications of 0.984016 Da for asparagines (representing formerly N-glycosylated asparagines after deamidation through the PNGaseF treatment) and 15.994915 Da for methionines (covering rapidly oxidizing methionines), carbamidomethylation as static modification for cysteines, at least one tryptic terminus, and two missed cleavage sites. Statistical analysis of the data, including peptide and protein identification, was performed using the Trans-Proteomic Pipeline v4.3 (TPP; Seattle Proteome Center) including PeptideProphet and ProteinProphet (Keller et al., 2005). Peptides and proteins were detected and quantified at a confidence score of >1.3 using the TPP in combination with Progenesis software. The ProteinProphet probability score was set such that the false discovery rate was less than 1% as determined by ProteinProphet.
Label-free Quantification
Protein quantification was performed using Progenesis (Nonlinear Dynamics). After manually improving the alignment, peptides were quantified based on ms1 intensity, and filtered for sequences that have a UniProt accession number. Overall protein abundance was estimated using a 10% trimmed mean of all identified peptide abundances. Significance of differential expression in protein abundance was assessed by ANOVA.
Technical Validation of Candidate Proteins
From literature, identified proteins consisted of a mixture of mesoderm, endoderm, and neural related markers in addition to previously unannotated markers relevant to the CPC cell system (Table S3). Next, temporal mRNA expression profiles were used to validate profiles generated using MS. Samples of mRNA were collected at each stage of cardiac development (outlined in Figure 1A) for qRT-PCR analysis of gene expression for the 47 proteins associated with CPC. The Pearson correlation coefficient was calculated from the normalized mRNA expression and protein expression values (Figure S2A). Thirty-two proteins with a significant Pearson correlation coefficient (p < 0.05) were selected for antibody validation using flow cytometry. Fourteen antibodies that were available and rated for flow cytometry were first titrated against their reported positive control cell type. Those that displayed a positive signal were then used in a differentiation time course (d0 to d3.75). The antibodies that displayed 0% or 100% positive staining across all time points (indicative of non-specific binding) were discarded. The final panel of 10 candidate proteins (ANTXR1, CXCR4, EPHB2, FZD4, ITGA4, ITGB5, LPAR4, NCAM1, NT5E, and PLEXINB1) was selected based on antibody availability and differential expression (Figure S2B). These markers were then co-stained with FLK1 and PDGFRA to determine co-expression and their capacity to further resolve the FLK1+PDGFRA+ population. Using supervised gating, we found that the FLK1+PDGFRA+ population could be separated based on four markers (ITGB5, LPAR4, FZD4, and PLEXINB1) (Figure S2C). After the selection of FZD4 as a candidate marker, further validation was performed to ensure optimal antibody binding (Figure S3A) using a staining index metric (Telford et al., 2009) and measured non-specific background abundance using an isotype control (Figure S3B).
Statistical Analysis
All statistical tests were performed in R (v3.3.1) using a two-sided Mann-Whitney U test or Kruskal-Wallis one-way ANOVA with significance level α = 0.05.
Author Contributions
C.Y. initiated the project with guidance from P.W.Z. All analyses were done by C.Y. mPSC experiments were designed and performed by C.Y. Sorting studies were aided by T.Y. Microarray was performed by C.Y., H.S., S.K., N.D., and A.D.W. Cell surface capture was performed by C.Y. with help from D.B.-F. MS was performed by C.Y. with help from J.A.H., H.G., and A.P.F. The hPSC experiment was performed by H.S. C.Y. prepared the figures and wrote the manuscript with input from A.E., B.W., G.K., and P.W.Z.
Acknowledgments
C.Y. is grateful to all co-authors and members of the P.W.Z. lab for helpful feedback on the manuscript. He thanks the following labs for technical support: A.E. (mass spectrometry), B.W. (cell surface capture), and G.K. (mPSC differentiation). This research was funded by the Heart and Stroke Foundation, CIHR, and Medicine by Design.
Published: December 14, 2017
Footnotes
Supplemental Information includes six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.008.
Accession Numbers
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) (GEO: GSE103560). The proteomic data have been deposited in ProteomeCentral (PRIDE: PXD007684).
Supplemental Information
References
- Abdul-Ghani M., Dufort D., Stiles R., De Repentigny Y., Kothary R., Megeney L.A. Wnt11 promotes cardiomyocyte development by caspase-mediated suppression of canonical Wnt signals. Mol. Cell. Biol. 2011;31:163–178. doi: 10.1128/MCB.01539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M., Carpenter M.K., Inokuma M.S., Chiu C.P., Harris C.P., Waknitz M.A., Itskovitz-Eldor J., Thomson J.A. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015;10:e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi-Smiraglia A., Kunnev D., Limoge M., Lee A., Beckerle M.C., Bakin A.V. Integrin-β5 and zyxin mediate formation of ventral stress fibers in response to transforming growth factor β. Cell Cycle. 2013;12:3377–3389. doi: 10.4161/cc.26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boheler K.R., Bhattacharya S., Kropp E.M., Chuppa S., Riordon D.R., Bausch-Fluck D., Burridge P.W., Wu J.C., Wersto R.P., Chan G.C.F. A human pluripotent stem cell surface N-glycoproteome resource reveals markers, extracellular epitopes, and drug targets. Stem Cell Rep. 2014;3:185–203. doi: 10.1016/j.stemcr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A., Tännler S., Chiapparo G., Chabab S., Ramialison M., Paulissen C., Beck B., Harvey R., Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T., Männer J., Kühl M. The role of Wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovasc. Res. 2006;72:198–209. doi: 10.1016/j.cardiores.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C.M., Hu W.S., Kaufman D.S. Improved development of human embryonic stem cell-derived embryoid bodies by stirred vessel cultivation. Biotechnol. Bioeng. 2006;94:938–948. doi: 10.1002/bit.20919. [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C., Bernardo A.S., Trotter M.W.B., Pedersen R.A., Sinha S. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat. Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E.D., Tian Y., Morrisey E.E. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–798. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- DeRossi C., Laiosa M.D., Silverstone A.E., Holdener B.C. Mouse fzd4 maps within a region of chromosome 7 important for thymus and cardiac development. Genesis. 2000;27:64–75. doi: 10.1002/1526-968x(200006)27:2<64::aid-gene30>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fazzari P., Penachioni J., Gianola S., Rossi F., Eickholt B.J., Maina F., Alexopoulou L., Sottile A., Comoglio P.M., Flavell R.A. Plexin-B1 plays a redundant role during mouse development and in tumour angiogenesis. BMC Dev. Biol. 2007;7:55. doi: 10.1186/1471-213X-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling H.J. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Fernandes S., Chong J.J.H., Paige S.L., Iwata M., Torok-Storb B., Keller G., Reinecke H., Murry C.E. Comparison of human embryonic stem cell-derived cardiomyocytes, cardiovascular progenitors, and bone marrow mononuclear cells for cardiac repair. Stem Cell Rep. 2015;5:753–762. doi: 10.1016/j.stemcr.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecht-Nir S., Cohen S., Itskovitz-Eldor J. Bioreactor cultivation enhances the efficiency of human embryoid body (hEB) formation and differentiation. Biotechnol. Bioeng. 2004;86:493–502. doi: 10.1002/bit.20045. [DOI] [PubMed] [Google Scholar]
- Gessert S., Kühl M. The multiple phases and faces of Wnt signaling during cardiac differentiation and development. Circ. Res. 2010;107:186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- Josic D., Clifton J.G. Mammalian plasma membrane proteomics. Proteomics. 2007;7:3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- Kattman S., Huber T., Keller G. Multipotent Flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Keller A., Eng J., Zhang N., Li X., Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100024. 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder S.J., Tsang T.E., Quinlan G.A., Hadjantonakis A.K., Nagy A., Tam P.P. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- Macher B.A., Yen T.-Y. Proteins at membrane surfaces—a review of approaches. Mol. Biosyst. 2007;3:705–713. doi: 10.1039/b708581h. [DOI] [PubMed] [Google Scholar]
- Mazzotta S., Neves C., Bonner R.J., Bernardo A.S., Docherty K., Hoppler S. Distinctive roles of canonical and noncanonical wnt signaling in human embryonic cardiomyocyte development. Stem Cell Rep. 2016;7:764–776. doi: 10.1016/j.stemcr.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R.T., Gough N.R. Beyond canonical: the Wnt and β-catenin story. Sci. Signal. 2016;9:eg5. doi: 10.1126/scisignal.aaf6192. [DOI] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Nelson T., Faustino R., Chiriac A., Crespo-Diaz R., Behfar A., Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Nunomura K. Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol. Cell. Proteomics. 2005;4:1968–1976. doi: 10.1074/mcp.M500216-MCP200. [DOI] [PubMed] [Google Scholar]
- Passier R., Oostwaard D.W., Snapper J., Kloots J., Hassink R.J., Kuijk E., Roelen B., de la Riviere A.B., Mummery C. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- Rana M.S., Christoffels V.M., Moorman A.F.M. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol. 2013;207:588–615. doi: 10.1111/apha.12061. [DOI] [PubMed] [Google Scholar]
- Sachinidis A., Fleischmann B.K., Kolossov E., Wartenberg M., Sauer H., Hescheler J. Cardiac specific differentiation of mouse embryonic stem cells. Cardiovasc. Res. 2003;58:278–291. doi: 10.1016/s0008-6363(03)00248-7. [DOI] [PubMed] [Google Scholar]
- Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/Â -catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Sweetman D., Wagstaff L., Cooper O., Weijer C., Münsterberg A. The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals. BMC Dev. Biol. 2008;8:63. doi: 10.1186/1471-213X-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.Y., Sriram G., Rufaihah A.J., Neoh K.G., Cao T. Efficient derivation of lateral plate and paraxial mesoderm subtypes from human embryonic stem cells through GSKi-mediated differentiation. Stem Cells Dev. 2013;22:1893–1906. doi: 10.1089/scd.2012.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford W.G., Babin S.A., Khorev S.V., Rowe S.H. Green fiber lasers: an alternative to traditional DPSS green lasers for flow cytometry. Cytometry A. 2009;75:1031–1039. doi: 10.1002/cyto.a.20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A., DeWitt N.D. Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 2016;17:194–200. doi: 10.1038/nrm.2016.10. [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Hou J., Han B., Nie Y., Cong X., Hu S., Chen X. Developmental changes in lysophospholipid receptor expression in rodent heart from near-term fetus to adult. Mol. Biol. Rep. 2012;39:9075–9084. doi: 10.1007/s11033-012-1778-6. [DOI] [PubMed] [Google Scholar]
- Wollscheid B., Bausch-Fluck D., Henderson C., O’Brien R., Bibel M., Schiess R., Aebersold R., Watts J.D. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.M., Fujiwara Y., Cibulsky S.M., Clapham D.E., Lien C., Schultheiss T.M., Orkin S.H. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wilson G.F., Soerens A.G., Koonce C.H., Yu J., Palecek S.P., Thomson J.A., Kamp T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.