Abstract
Rationale: Deep learning is a powerful tool that may allow for improved outcome prediction.
Objectives: To determine if deep learning, specifically convolutional neural network (CNN) analysis, could detect and stage chronic obstructive pulmonary disease (COPD) and predict acute respiratory disease (ARD) events and mortality in smokers.
Methods: A CNN was trained using computed tomography scans from 7,983 COPDGene participants and evaluated using 1,000 nonoverlapping COPDGene participants and 1,672 ECLIPSE participants. Logistic regression (C statistic and the Hosmer-Lemeshow test) was used to assess COPD diagnosis and ARD prediction. Cox regression (C index and the Greenwood-Nam-D’Agnostino test) was used to assess mortality.
Measurements and Main Results: In COPDGene, the C statistic for the detection of COPD was 0.856. A total of 51.1% of participants in COPDGene were accurately staged and 74.95% were within one stage. In ECLIPSE, 29.4% were accurately staged and 74.6% were within one stage. In COPDGene and ECLIPSE, the C statistics for ARD events were 0.64 and 0.55, respectively, and the Hosmer-Lemeshow P values were 0.502 and 0.380, respectively, suggesting no evidence of poor calibration. In COPDGene and ECLIPSE, CNN predicted mortality with fair discrimination (C indices, 0.72 and 0.60, respectively), and without evidence of poor calibration (Greenwood-Nam-D’Agnostino P values, 0.307 and 0.331, respectively).
Conclusions: A deep-learning approach that uses only computed tomography imaging data can identify those smokers who have COPD and predict who are most likely to have ARD events and those with the highest mortality. At a population level CNN analysis may be a powerful tool for risk assessment.
Keywords: artificial intelligence (computer vision systems), neural networks, chronic obstructive pulmonary disease, X-ray computed tomography
At a Glance Commentary
Scientific Knowledge on the Subject
Deep learning has been used to analyze and categorize a variety of medical data including imaging. However, little work has been done on the use of deep learning to directly predict outcomes, and few studies have validated deep-learning approaches in cohorts entirely different than those in which they were developed.
What This Study Adds to the Field
In this study we show that a deep learning–based analysis of computed tomography scans of the chest can categorize smokers as having chronic obstructive pulmonary disease or not, and can directly predict outcomes including acute respiratory disease events and mortality. In addition, we have found that this deep-learning approach can be developed in one cohort and applied to a separate cohort without any additional training. This approach may provide a useful tool for identifying high-risk subgroups in large populations that can be applied to multiple different cohorts and may enable the identification of specific clusters of patients with chronic obstructive pulmonary disease who share unique imaging and clinical features.
Quantitative image analysis has become a cornerstone of clinical investigation. For such conditions as chronic obstructive pulmonary disease (COPD), objective computed tomographic (CT) measures of the lung parenchyma, airways, pulmonary vasculature, and the chest wall have all been shown to be useful for disease diagnosis, stratification, and risk prediction (1–5). Although objective CT analysis has provided clinically relevant insights into COPD it is essentially a radiographic method of anatomic and physiologic analysis that relies on the prespecification of radiographic features thought likely to be associated with certain clinical outcomes. New techniques in computer vision, natural image analysis, and machine learning have enabled the direct interpretation of imaging data, going directly from the raw image data to clinical outcome without relying on the specification of radiographic features of interest (6, 7). One such machine learning approach is deep learning, a term that includes convolutional neural network (CNN) analysis (7–20). CNN and other deep learning–based models are trained using large amounts of data from individuals with known outcomes, such as known disease diagnoses or clinical events like death. Once trained, the CNN model can then use data from other individuals to determine their probability for that event, and can rapidly assess risk across large populations without the need for the manual extraction or review of specific clinical or radiographic features (21). We hypothesized that deep learning analyses of imaging data could predict clinically relevant outcomes in smokers without the prespecification of features of interest.
Methods
Data Acquisition
Cohorts
Details regarding the cohorts, including cohort design and methods regarding acute respiratory disease (ARD) event reporting and mortality assessment, are available in the online supplement.
Briefly, COPDGene is an observational longitudinal study funded by the NHLBI of 10,300 smokers whose goal is to define the epidemiologic associations and genetic risk factors for the development of COPD (22). Participants with active lung diseases other than COPD and asthma were excluded from participation and all participants underwent baseline testing including an extensive interview, volumetric high-resolution CT scan of the chest, and spirometry. Smokers with and without COPD were enrolled and are now returning for their 5-year interval follow-up visit.
The ECLIPSE (Evaluations of COPD Longitudinally to Identify Predictive Surrogate End-points) study was a 3-year multicenter multinational longitudinal study of 2,164 subjects with Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage 2–4 COPD and 582 control subjects that was completed in 2011 (23). Participants were excluded if they had known respiratory diseases other than COPD or severe alpha-1 antitrypsin deficiency. Study procedures were performed at baseline, 3 months, 6 months, and then every 6 months for a total of 3 years. Spirometry was performed at baseline, and CT scans were performed at baseline, Year 1, and Year 3. Only the 2,164 ECLIPSE participants with COPD were included in this study, and of those 1,928 completed the 3-year follow-up (23).
ARD events
ARD events occur in smokers with and without COPD and are temporary increases of respiratory symptoms including cough, sputum production, and dyspnea warranting a change in therapy (24, 25). For this study, severe events (those requiring hospitalization) were not considered separately from mild and moderate events. Because of the high rate of ARD events in ECLIPSE, for this study a subject was considered to have had an ARD event if at least one occurred within the first year of follow-up. To ensure comparable results across cohorts, in COPDGene the primary outcome for this study was also an ARD event within the first year of follow-up. In COPDGene a secondary analysis was also performed in which a subject was considered to have had an ARD event if at least one occurred within the first 3 years of follow-up.
Deep-Learning Structure
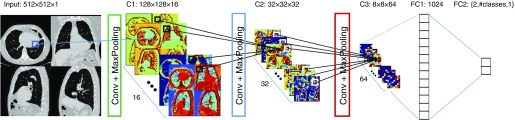
CT interpretation was enabled using the system shown in Figure 1. Because of constraints caused by the processing capabilities of existing graphical processing units the full high-resolution CT images from an individual cannot be used by the CNN. Therefore an object detector was used to automatically extract four canonical CT slices at preselected anatomic landmarks (26). This dimensionality reduction step “normalizes” the CT data using anatomic information. These images were joined into a single montage and included an axial slice centered at the heart at the level of the mitral valve, two sagittal reformatted slices centered at the left and right hilum, and a coronal reformatted slice centered in the ascending aorta. The CNN consisted of three convolutional layers alternating with rectified linear and max-pooling operations. The final two layers of the CNN were fully connected and the size of the last fully connected layer varied (two, number of classes and one, respectively) based on the task (binary classification, categorical classification, or regression). Further details regarding the deep learning structure are available in the online supplement.
Figure 1.
The input of the convolutional neural network is a composite image of four canonical views of the computed tomography scan: an axial slice at the level of the mitral valve, a coronal slice taken at the level of the ascending aorta, and two sagittal slices at the level of the right and left hila. The image is analyzed with a convolutional neural network consisting of three convolutional layers (Conv) followed by max-pooling operations, each reducing the image size fourfold in each direction. At the end of the convolutional layers are two fully connected networks, the first one of 1,024 neurons and the second one of variable size depending on the problem at hand: classification, multiclass classification, or regression.
Training Methodology
The COPDGene dataset was divided into two nonoverlapping subcohorts to be used for model development and testing. The model development cohort consisted of a group to be used for model training (Training) and a group used for optimization of the metaparameters of the CNN (Validation; n = 1,000). Further details regarding training and optimization including selection of cohort size and the sensitivity of our models to training set size and different image reconstruction characteristics are available in the online supplement. The testing set (Testing; n = 1,000) consisted of subjects whose data were not used for model training or optimization. The reported COPDGene results were obtained from those 1,000 Testing subjects. Finally, the CNN developed using COPDGene data was then applied to the CT images collected in ECLIPSE without additional training.
Statistical Analysis
Data are presented as means and SD where appropriate. For all binary categorical outcomes evaluated using the CNN-based probabilities (presence or absence of COPD, occurrence of an ARD event, and mortality), an individual was categorized as being predicted by the CNN to have that outcome if the CNN-derived probability was greater than 0.5 for that category or event. For GOLD staging, the per-subject stage with the highest probability was chosen. All analyses were replicated in the ECLIPSE cohort using the CNN models trained in the COPDGene cohort training set. Because the CNN-based approach is a global assessment of risk, no multivariable analyses were performed. Analyses were performed using MedCalc (MedCalc Software) and SAS 9.4 and JMP 12 (SAS Institute). P values less than 0.05 were considered to be statistically significant.
COPD detection and staging
COPD detection was limited to COPDGene because there were a limited number of smokers in ECLIPSE without expiratory airflow obstruction. Clinically, participants were defined as having COPD if the ratio of their FEV1 to their FVC was less than 0.7 (27). The ability of the CNN to identify participants with COPD was evaluated using logistic regression for the calculation of the C statistic and the Hosmer-Lemeshow calibration test using 10 risk categories. The C statistic is a measure of the model’s discrimination, or how well it performs with regard to assessing who does and does not have COPD, whereas the Hosmer-Lemeshow goodness-of-fit test assesses the model calibration, which is the agreement between the observed and predicted risk (28, 29). Note that for the Hosmer-Lemeshow test a significant P value (<0.05 in this study) indicates poor calibration. Therefore the desired outcome for a predictive model is for the Hosmer-Lemeshow test P value to be nonsignificant (28).
The GOLD stage was defined by spirometry only (stages 1–4), and the classification by the CNN into the appropriate stage was evaluated by the percentage of correctly classified cases and by the percentage of cases whose classification lay within one class of the clinical stage (27).
Univariate associations between CNN predicted FEV1 and actual FEV1 were assessed using Pearson correlation.
ARD events
CNN performance for the prediction of ARD was expressed using logistic regression for the calculation of odds ratios and C statistic, as well as with the Hosmer-Lemeshow calibration test using 10 risk categories. In addition, the number of events per CNN probability quartile was evaluated using the Cochran-Mantel-Haenszel (CMH) trend test, and the performance of the CNN for the prediction of ARD events was compared with the performance of a univariate logistic regression model that used low-attenuation area (LAA), a well-validated measure of emphysema severity (1, 2, 30, 31). LAA was defined as the percentage of lung with a density less than −950 Hounsfield units, and was dichotomized at the median for binary analyses.
Mortality
Three year, all-cause mortality prediction using the CNN was assessed using Cox proportional hazards, and all covariates were evaluated using the Martingale residuals method and found not to violate the proportional hazards assumption (32). Three-year mortality was selected to allow for comparison between the COPDGene and ECLIPSE cohorts because only 3 years of follow-up were available in the latter. Model discrimination was measured using the C index and model calibration was assessed using the Greenwood-Nam-D’Agnostino (GND) test. The C index and the GND test are the survival analysis analogs to the C statistic and Hosmer-Lemeshow test described previously. As with the Hosmer-Lemeshow test, a nonsignificant P value for the GND test suggests no evidence of poor calibration and is therefore the desired outcome for a predictive model (28, 33–35). The GND test becomes unstable when there are fewer than 5 events per group so it was performed with four risk categories (35). Kaplan-Meier analyses stratified by CNN-predicted probability quartile for death were performed to aid in the visualization of the results. As with the ARD event analyses, the performance of the CNN was compared with the performance of a univariate Cox regression model using LAA for the prediction of mortality. In addition, in the ECLIPSE cohort and in the subgroup of participants with COPD in the COPDGene cohort, the performance of the CNN was also compared with the performance of the body mass index, airflow obstruction, dyspnea, and exercise capacity (BODE) index, a multicomponent predictor of mortality (36). The BODE score was dichotomized at the median for binary analyses.
Results
Image Preprocessing
Baseline imaging and patient characterization data were available for 9,983 COPDGene subjects and 1,672 ECLIPSE subjects (Table 1; see Table E1 in the online supplement). Automated slice selection correctly identified the appropriate axial, sagittal, and coronal structures in 9,408 (92.4%) of the COPDGene participants and 1,547 (92.5%) ECLIPSE participants.
Table 1.
Baseline Characteristics of the COPDGene and ECLIPSE Cohorts
| COPDGene | ECLIPSE | |
|---|---|---|
| Age, yr, mean (SD) | 59.5 (9.0) | 63.6 (7.1) |
| Sex, % female (n) | 46.7 (4,819) | 33.0 (582) |
| Race, % black (n) | 33.2 (3,420) | * |
| BMI, mean (SD) | 28.8 (6.3) | 26.7 (5.6) |
| Pack-years, mean (SD) | 44.2 (24.9) | 50.3 (27.4) |
| Current smoking, % (n) | 52.6 (5,417) | 35.5 (626) |
| FEV1 % predicted, mean (SD) | 76.6 (25.6) | 47.5 (15.9) |
| MMRC score, mean (SD) | 1.4 (1.4) | 1.5 (1.8) |
| Percent LAA, mean (SD) | 6.2 (9.6) | 13.4 (12.0) |
| BODE | 1.4 (1.8) | 4.8 (2.2) |
| GOLD stage | |
|
| 0 | 42.8 (4,387) | 0.0 (0) |
| 1 | 7.7 (791) | 0.0 (0) |
| 2 | 18.8 (1,926) | 42.0 (741) |
| 3 | 11.4 (1,164) | 43.7 (770) |
| 4 | 5.9 (607) | 13.95 (216) |
| ARD event, % (n) | |
|
| Reported at least one ARD event within 1 yr of enrollment | 8.8 (791)† | 54.7 (966) |
| Reported at least one ARD event within 3 yr of enrollment | 38.2 (3,426)† | ‡ |
| Death, % (n) | |
|
| Died within 3 yr of enrollment | 5.1 (458)§ | 9.8 (211) |
| Died during follow-up | 12.8 (1,160)§ | ‡ |
Definition of abbreviations: ARD = acute respiratory disease; BMI = body mass index; BODE = body mass index, airflow obstruction, dyspnea, and exercise capacity; COPD = chronic obstructive pulmonary disease; ECLIPSE = Evaluations of COPD Longitudinally to Identify Predictive Surrogate End-points; GOLD = Global Initiative for Chronic Obstructive Lung Disease; LAA = low-attenuation area; MMRC = Modified Medical Research Council Dyspnea Scale.
Detailed summary statistics for each of the COPDGene subgroups (training, validation, and testing) are available in the online supplement. Except where indicated percentages are percent of overall cohort.
No race data available.
Number with longitudinal ARD event data available = 8,966.
Not analyzed.
Number with mortality data available = 9,057.
COPD Detection and Staging
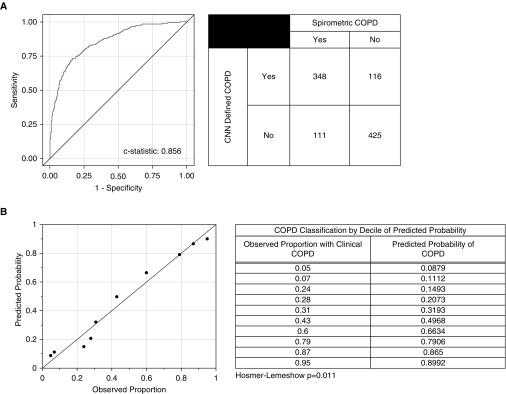
The CNN model correctly determined the presence or absence of COPD in 773 of the 1,000 subjects in the testing cohort with a C statistic of 0.856 (Figure 2A). Although the Hosmer Lemeshow test indicated poor calibration (P = 0.011), visual inspection of the model calibration by deciles demonstrated a reasonable fit between the CNN model predicted probabilities of COPD and the observed probability of COPD (Figure 2B).
Figure 2.
Detection of chronic obstructive pulmonary disease (COPD) by convolutional neural network (CNN) in the COPDGene cohort. (A) Receiver operating characteristic curve, C statistic, and summary table for the CNN prediction of COPD in the COPDGene testing cohort. Clinical COPD was defined based on FEV1/FVC less than 0.7. The CNN defined COPD based on CNN predicted probability of COPD greater than 0.5. (B) The predicted probabilities are the predicted probability of the outcome (COPD) by the CNN. The observed proportions are the observed proportions of participants in that decile who had the outcome. Reference lines indicate perfect correlation (slope = 1; intercept = 0). The Hosmer-Lemeshow test is a test for evidence of poor calibration. That is, a nonsignificant P value (>0.05) indicates no evidence for poor calibration.
With regard to CNN prediction of GOLD stage, accurate designation of the exact stage was achieved 51.1% of the time and was correct or off by one stage 74.9% of the time in the COPDGene cohort. The same model applied to the ECLIPSE cohort correctly predicted the GOLD stage in 29.4% of the cases and was correct or off by one stage in 74.6%. Finally, in the COPDGene testing cohort there was a strong correlation between CNN predicted FEV1 and actual FEV1 (r = 0.734; P < 0.001).
ARD Events
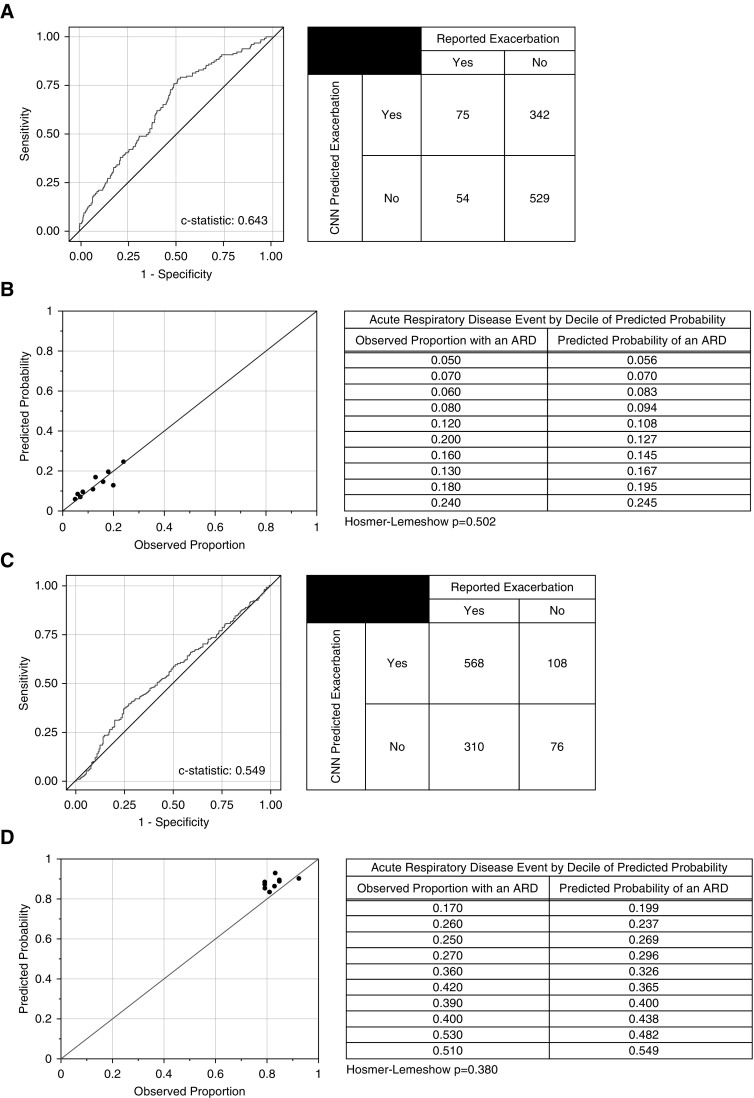
In the COPDGene testing cohort, the CNN model of ARD events had a C statistic of 0.64, and those subjects predicted by the model to be at risk for an ARD event had a 2.15 higher odds of having an event compared with those who were not (P < 0.001) (Figure 3A). As shown in Figure 3B, the CNN prediction model was well calibrated with regard to risk prediction with Hosmer-Lemeshow (P = 0.502), suggesting no evidence of poor calibration. This is further supported by the fact that the number of individuals with an event in the higher CNN probability quartiles was greater than in the lower quartiles (CMH, P < 0.001) (see Table E2A).
Figure 3.
Prediction of acute respiratory disease (ARD) events by convolutional neural network (CNN) in the first year of follow-up. (A) Receiver operating characteristic curve, C statistic, and summary table for the CNN prediction of ARD events in the COPDGene testing cohort. (B) The predicted probabilities are the predicted probability of the outcome (ARD) by the CNN in the COPDGene testing cohort. The observed proportions are the observed proportions of participants in that decile who had the outcome. Reference lines indicate perfect correlation (slope = 1; intercept = 0). The Hosmer-Lemeshow test is a test for evidence of poor calibration. That is, a nonsignificant P value (>0.05) indicates no evidence for poor calibration. (C) Receiver operating characteristic curve, C statistic, and summary table for the CNN prediction of ARD events in the ECLIPSE cohort. (D) The predicted probabilities are the predicted probability of the outcome (ARD) by the CNN in the ECLIPSE cohort. The observed proportions are the observed proportions of participants in that decile who had the outcome. Reference lines indicate perfect correlation (slope = 1; intercept = 0). The Hosmer-Lemeshow test is a test for evidence of poor calibration. That is, a nonsignificant P value (>0.05) indicates no evidence for poor calibration.
In the ECLIPSE cohort the CNN model of ARD events had a C statistic of 0.55 (Figure 3C). Those subjects predicted by the model were not at a significantly higher risk for an ARD event than those who were not (P = 0.125). As shown in Figure 3D, the CNN prediction model was well calibrated with regard to risk prediction with Hosmer-Lemeshow (P = 0.380), suggesting no evidence of poor calibration. Also, as shown in Table E2B, the number of individuals with an event in the higher CNN probability quartiles was slightly greater than in the lower quartiles (CMH, P = 0.049).
Of note, as shown in the online supplement, the CNN model had similar performance in the prediction of ARD events over 3 years of follow-up in the COPDGene cohort despite a broader range of event probabilities (see Table E3 and Figure E1).
By comparison, the model based on LAA for the prediction of ARD events within the first year of follow-up showed evidence of poor calibration (P = 0.032) (see Figure E2). Although the LAA-based model showed better calibration in the ECLIPSE cohort, its discrimination was quite poor (C statistic = 0.548), likely in part because of the limited range of event probabilities in that cohort (see Figure E3). Visual inspection of the calibration of the LAA-based model for the prediction of ARD events over 3 years of follow-up showed evidence of poor calibration, especially at low event probabilities (see Figure E4).
Mortality
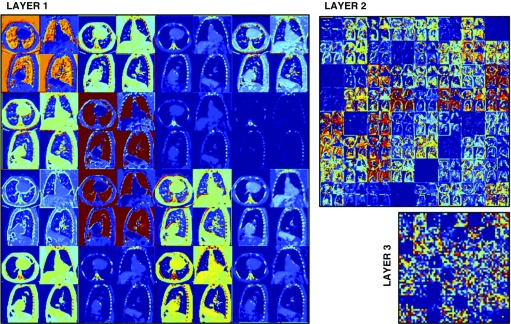
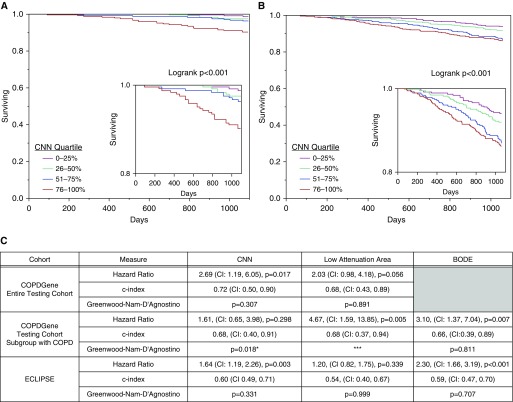
An example of the response of the network trained to predict mortality is shown in Figure 4 and a summary of the assessment of the CNN mortality model is shown in Figures 5A–5C. Kaplan-Meier survival analysis results by quartile of CNN predicted probability of death for the COPDGene testing cohort and the ECLIPSE cohort are shown in Figures 5A and 5B. In both the COPDGene testing cohort and ECLIPSE, the CNN model for mortality showed fair discrimination based on C indices of 0.72 (confidence interval, 0.50–0.90) and 0.60 (confidence interval, 0.49–0.71) respectively, and no evidence of poor calibration as indicated by nonsignificant GND P values (Figure 5C). In both ECLIPSE and the subgroup of individuals with COPD in the COPDGene testing cohort (n = 391), the CNN model showed similar or better discrimination for mortality as the BODE index (Figure 5C). However, in both of these groups, the confidence interval for the C index for both predictors included values less than 0.5, suggesting relatively poor discrimination overall. In addition, based on the GND test there was evidence for poor calibration of the CNN model in the COPD subgroup. However, it should be noted that because of a low overall mortality rate there were fewer than five deaths in two of the CNN COPD subgroup quartiles, which may have resulted in test instability. Finally, the LAA-based model performed slightly worse than the CNN-based model with regard to discrimination in both the COPDGene testing cohort and ECLIPSE, and similarly in the COPDGene testing cohort subgroup with COPD (Figure 5C).
Figure 4.
Response of the neural network trained to predict mortality for a testing case. The large, medium, and small subimages represent the first, second, and third convolutional layers, respectively. For each layer, the responses of the individual filters are used to generate a composite image. The image values have been limited between 0 (dark blue) and 0.5 (red) for display purposes. Different filters enhance different areas of the image, such as the lungs, the chest wall, or the bone structures. The interpretation of the second and third convolutional layers is impeded by the lack of resolution of the response images.
Figure 5.
Mortality predictions by convolutional neural network (CNN) in the COPDGene and ECLIPSE cohorts. (A and B) Kaplan-Meier survival analyses for the CNN prediction of all-cause mortality in the COPDGene testing cohort (A) and in the ECLIPSE cohort (B). Insets show survival probabilities of 0.8 and greater. (C) Summary table of the hazard ratios, C indices, and tests of calibration (Greenwood-Nam-D’Agnostino) comparing CNN-based risk prediction with low-attenuation area–based and BODE-based risk prediction. The hazard ratios for the CNN are expressed as the risk in the group predicted by the CNN to be at higher risk of death compared with those predicted to be at a lower risk of death. The hazard ratios for low-attenuation area and BODE are expressed as the risk of those with higher values compared with those with lower values, dichotomized at the median. The C index is analogous to the C statistic for logistic regression. The Greenwood-Nam-D’Agnostino test is a test for evidence of poor calibration. That is, a nonsignificant P value (>0.05) indicates no evidence for poor calibration. *Fewer than five events in two of the quartiles, which may make the estimate unstable. ***One subgroup had 0 events, which precludes the Greenwood-Nam-D’Agnostino test from being performed. BODE = body mass index, airflow obstruction, dyspnea, and exercise capacity; CI = confidence interval; COPD = chronic obstructive pulmonary disease; ECLIPSE = Evaluations of COPD Longitudinally to Identify Predictive Surrogate End-points.
Discussion
Our investigation suggests that a deep learning–based approach, CNN analysis, applied to the CT imaging data of current and former smokers, can identify those individuals with COPD, characterize disease severity, and predict clinical outcomes including ARD events and death.
A multitude of studies using both qualitative and quantitative imaging techniques have shown the utility of CT imaging in assessing lung function, categorizing disease severity, and predicting outcomes in patients with a variety of lung diseases (30). For instance, both the percentage of lung occupied by low-density emphysematous tissue and measurements of airway structure on CT have been shown to be highly associated with lung function (37–40). Other examples of CT-based metrics include the ratio of the diameters of the pulmonary artery to aorta, which is associated with acute respiratory exacerbations, and bronchiectasis, which is associated with a longer recovery from acute exacerbations and increased mortality (25, 41, 42). Although studies of these measures and many others have revealed a great deal about respiratory diseases, they require prior knowledge of the anatomic and physiologic implications of disease to prespecify which radiographic features are of interest. In addition, those studies that rely on qualitative analysis may suffer from a loss of data because of individuals with indeterminate findings (43).
More recently, machine learning approaches, such as deep-learning and CNN-based analysis, have been used to establish a direct link between diagnostic images and disease categorization, bypassing the identification of features of interest. For example, Esteva and coworkers (21) recently showed that a CNN-based approach performed as well as dermatologists in the categorization of images of skin lesions as melanomatous or benign, and Lakhani and Sundaram (44) demonstrated that a similar method could categorize chest radiographs as having evidence of active tuberculosis or not. In this study we showed that deep learning using a CNN-based method was able to discriminate between smokers with and without COPD as well as previous image-based methods (45, 46).
Beyond disease categorization, we also developed CNN models for assessing the risk for ARD episodes and death, and showed that the models were well calibrated for those outcomes. Although a model’s ability to discriminate between those who are and are not likely to have an exacerbation is important, equally so is whether the predicted probability of an event based on the model is similar to the observed probability, a model characteristic known as calibration. Although the discriminatory ability of the deep-learning models for exacerbations and death was somewhat limited, they showed no evidence of poor calibration and they performed well across a wide range of event probabilities. In addition, visual comparison of the receiver operating characteristic and calibration curves suggested that the CNN models for ARD performed better than models based on a more standard objective CT measure, LAA. This was particularly true of model calibration at lower event probabilities where the LAA-based models showed evidence of poor calibration. Perhaps more importantly, the CNN-based method performed well not only in a separate testing component of the cohort in which it was developed, but also when applied to an entirely different cohort with much more severe disease. Together these findings suggest that this approach may be useful at a population level for identifying higher risk subgroups that should be targeted for new and existing interventions, and for assessing overall population level risk.
This study and approach do have significant limitations including high training computational cost and memory requirements, which limit the amount of data we can use to train our models. Ideally, one would use all of the CT images for training and prediction, but analytics on this scale are beyond the processing capabilities of existing graphical processing units. To address this, four representative images were used with the goal of obtaining the broadest sampling of features in the thorax. Thus this CNN in this study did not fully use all of the CT data available, and because the images used were preselected, the method was not entirely unguided. However, it should be noted that the four images chosen were selected based only on their inclusion of major thoracic structures, not based on any a priori hypotheses about the relationship between those structures and the outcomes evaluated. For example, images that would have allowed for the assessment of the ratio of the diameters of the pulmonary artery to aorta were not included in the montage. In addition, no specific features from the four image montage were selected and the CNN was unguided from that point.
A second challenge to this approach is the amount of data needed for training. As shown in the online supplement, our data suggest that several thousand subjects are needed to achieve stable model performance for select clinical outcomes.
From a clinical standpoint, the featureless nature of deep learning, or its ability to predict outcomes without specification of clinical or radiographic predictors of interest, is both a strength and a weakness. Understandably, this “black box” nature of deep learning, the fact that it does not tell the provider what from the images it is using to assign a probability, may result in discomfort with using the results, and may greatly limit its utility in the short term. In addition, there were clear decrements in the performance of the COPDGene-based deep-learning models in the ECLIPSE cohort. These decrements could be attributed to differences in cohort characteristics including disease severity and differences in the protocols used for image acquisition and reconstruction, but further work is needed to determine if this is the case.
With regard to other approaches for assessing disease-related risk, all of the models, including those based on both LAA and BODE, and with the notable exception of the CNN-based model in the COPDGene testing cohort, performed relatively poorly for the discrimination of mortality. Only the CNN-based model in the subgroup of patients with COPD in COPDGene showed evidence of poor calibration for the risk of mortality, although this latter finding may have been caused by the small number of events that may have made the GND test of calibration unstable. Together, these findings show the challenge of accurately predicting mortality in an individual patient with COPD, and suggest that this approach, at least in the near term, is better suited to population-based analyses.
That said, at a population level, the ability to use only one data source, such as CT scans, and not rely on the availability of multiple types of clinical data, such as is required for clinical models like the BODE index, is a particular strength of the deep-learning approach. This is especially true in systems where the volume or form of other clinical measures may be inadequate. For example, spirometry results may only be available in certain systems in unstructured text reports, paper form, or in scanned images of those paper results, which can be a barrier to their large-scale analysis. Although ongoing work using natural language processing and other approaches will likely eventually overcome this issue, it remains a challenge for the analysis of clinically acquired data in particular (47, 48). By contrast, CT image data is stored in a standard format, and the rapid growth in CT imaging for a wide range of indications means that it is increasingly available, even if its acquisition was not initially clinically indicated. Even when spirometry is available, several studies suggest that using spirometry alone, across a healthcare system COPD is frequently either misdiagnosed or missed as a diagnosis entirely, suggesting a role for other methods for diagnosis for epidemiologic studies and population-based research (49–52).
By using a method that relies less on the a priori specification of measures of interest on CT, deep learning may also allow for a more standardized approach to assessing disease risk across multiple populations, especially in future iterations when processing power enables the use of all of the CT images available. Finally, detailed inspection of the model response to the CNN layers, as provided in Figure 4, and detailed clinical evaluation of patients determined to be at risk for adverse outcomes by the CNN method may inform about imaging patterns and clinical characteristics that may provide insights about disease manifestations and etiology.
Deep learning, including CNN, can provide a fast and flexible method for the integration of imaging into biomedical research. In addition, it may allow for the assessment of population-wide disease. Unlike current reductionist methods that require the use of a summary statistic of a feature of interest, deep learning uses all of the data available in the image to predict clinically relevant outcomes. Although current processing power limits the number of images that this technique can be applied to at the moment, this exciting new field may ultimately enhance the ability to identify disease subtypes because it is not hindered by the ability to a priori specify what imaging data should be used for investigation. Therefore it may provide a more standardized approach to image analysis and overall risk assessment across research and clinical care networks.
Acknowledgments
Acknowledgment
The authors gratefully acknowledge NVIDIA Corporation for the donation of the Titan Xp GPU used for this research.
Footnotes
Supported by NIH grants T32-HL007633 (S.Y.A.), R01-HL116931 (G.G., G.V.-S.-F., J.O.O., R.S.J.E., and G.R.W.), R01-HL116473 (R.S.J.E. and G.R.W.), K25-HL130637 (J.C.R.), and R01-HL089856 (J.C.R., R.S.J.E., and G.R.W.).
Author Contributions: The authors meet criteria for authorship as recommended by the International Committee of Medical Journal Editors. G.G., S.Y.A., R.S.J.E., and G.R.W. designed the study and wrote the initial manuscript. S.Y.A., F.N.R., and A.D. did the statistical analyses. G.V.-S.-F., J.O.O., J.C.R., and G.G. wrote the convolutional neural network algorithm and evaluated it in the available data. All authors contributed to the production of the final manuscript.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201705-0860OC on September 11, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hayhurst MD, MacNee W, Flenley DC, Wright D, McLean A, Lamb D, et al. Diagnosis of pulmonary emphysema by computerised tomography. Lancet. 1984;2:320–322. doi: 10.1016/s0140-6736(84)92689-8. [DOI] [PubMed] [Google Scholar]
- 2.Müller NL, Staples CA, Miller RR, Abboud RT. “Density mask”: an objective method to quantitate emphysema using computed tomography. Chest. 1988;94:782–787. doi: 10.1378/chest.94.4.782. [DOI] [PubMed] [Google Scholar]
- 3.Nakano Y, Muro S, Sakai H, Hirai T, Chin K, Tsukino M, et al. Computed tomographic measurements of airway dimensions and emphysema in smokers. Correlation with lung function. Am J Respir Crit Care Med. 2000;162:1102–1108. doi: 10.1164/ajrccm.162.3.9907120. [DOI] [PubMed] [Google Scholar]
- 4.Estépar RS, Kinney GL, Black-Shinn JL, Bowler RP, Kindlmann GL, Ross JC, et al. COPDGene Study. Computed tomographic measures of pulmonary vascular morphology in smokers and their clinical implications. Am J Respir Crit Care Med. 2013;188:231–239. doi: 10.1164/rccm.201301-0162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald ML, Diaz AA, Ross JC, San Jose Estepar R, Zhou L, Regan EA, et al. Quantitative computed tomography measures of pectoralis muscle area and disease severity in chronic obstructive pulmonary disease: a cross-sectional study. Ann Am Thorac Soc. 2014;11:326–334. doi: 10.1513/AnnalsATS.201307-229OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing systems 25 (NIPS 2012) New York: Curran Associates, Inc.; 2012. pp. 1097–1105. [Google Scholar]
- 7.Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. In: Cortes C, Lawrence ND, Lee DD, Sugiyama M, Garnett R, editors. Advances in neural information processing systems 28 (NIPS 2015) New York: Curran Associates, Inc.; 2015. pp. 91–99. [Google Scholar]
- 8.Lecun Y. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278–2324. [Google Scholar]
- 9.LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, et al. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989;1:541–551. [Google Scholar]
- 10.Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 11.Xu K, Ba J, Kiros R, Cho K, Courville AC, Salakhutdinov R, et al. Show, attend and tell: neural image caption generation with visual attention. arXiv:1502.03044. [Google Scholar]
- 12.Graves A, Mohamed A-r, Hinton GE. Speech recognition with deep recurrent neural networks. Presented at the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). May 26–31, 2013, Vancouver, BC, Canada. pp. 6645–6649. [Google Scholar]
- 13.Hinton GE, Deng L, Yu D, Dahl GE, Mohamed A-R, Jaitly N, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97. [Google Scholar]
- 14.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 15.Setio AA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging. 2016;35:1160–1169. doi: 10.1109/TMI.2016.2536809. [DOI] [PubMed] [Google Scholar]
- 16.Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, et al. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. 2016;35:1170–1181. doi: 10.1109/TMI.2015.2482920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 18.Ciresan D, Giusti A, Gambardella LM, Schmidhuber J.Deep neural networks segment neuronal membranes in electron microscopy images Pereira F, Burges CJC, Bottou L, Weinberger KQ.Advances in neural information processing systems 25 (NIPS 2012) New York: Curran Associates, Inc.2012. 2843–2851 [Google Scholar]
- 19.Miao S, Wang ZJ, Rui L. A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging. 2016;35:1352–1363. doi: 10.1109/TMI.2016.2521800. [DOI] [PubMed] [Google Scholar]
- 20.Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DR, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35:1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, et al. ECLIPSE investigators. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 24.Bowler RP, Kim V, Regan E, Williams AAA, Santorico SA, Make BJ, et al. Prediction of acute respiratory disease in current and former smokers with and without COPD. Chest. 2014;146:941–950. doi: 10.1378/chest.13-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells JM, Washko GR, Han MK, Abbas N, Nath H, Mamary AJ, et al. COPDGene Investigators. ECLIPSE Study Investigators. Pulmonary arterial enlargement and acute exacerbations of COPD. N Engl J Med. 2012;367:913–921. doi: 10.1056/NEJMoa1203830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez G, Washko G, San Jose Estepar R. Automated Agatston score computation in a large dataset of non ECG-gated chest computed tomography. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). Prague, Czech Republic: IEEE; 2016. pp. 53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 28.Lemeshow S, Hosmer DW., Jr A review of goodness of fit statistics for use in the development of logistic regression models. Am J Epidemiol. 1982;115:92–106. doi: 10.1093/oxfordjournals.aje.a113284. [DOI] [PubMed] [Google Scholar]
- 29.Holmberg L, Vickers A. Evaluation of prediction models for decision-making: beyond calibration and discrimination. PLoS Med. 2013;10:e1001491. doi: 10.1371/journal.pmed.1001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ash SY, Washko GR. The value of CT scanning. In: Anzueto A, Heijdra Y, Hurst JR, editors. Controversies in COPD. European Respiratory Society: 2015. pp. 121–133. [Google Scholar]
- 31.Washko GR, Parraga G, Coxson HO. Quantitative pulmonary imaging using computed tomography and magnetic resonance imaging. Respirology. 2012;17:432–444. doi: 10.1111/j.1440-1843.2011.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin DY, Wei LJ, Ying Z. Checking the Cox model with cumulative sums of martingale-based residuals. Biometrika. 1993;80:557–572. [Google Scholar]
- 33.Cook NR. C-statistics for survival data [accessed 2016 Sept 9]. Available from: http://ncook.bwh.harvard.edu/assets/survcmacs.v1.sas.
- 34.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 35.Demler OV, Paynter NP, Cook NR. Tests of calibration and goodness-of-fit in the survival setting. Stat Med. 2015;34:1659–1680. doi: 10.1002/sim.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celli BR, Cote CG, Marin JM, Casanova C, Montes de Oca M, Mendez RA, et al. The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med. 2004;350:1005–1012. doi: 10.1056/NEJMoa021322. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa M, Nasuhara Y, Onodera Y, Makita H, Nagai K, Fuke S, et al. Airflow limitation and airway dimensions in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:1309–1315. doi: 10.1164/rccm.200601-037OC. [DOI] [PubMed] [Google Scholar]
- 38.Coxson HO, Quiney B, Sin DD, Xing L, McWilliams AM, Mayo JR, et al. Airway wall thickness assessed using computed tomography and optical coherence tomography. Am J Respir Crit Care Med. 2008;177:1201–1206. doi: 10.1164/rccm.200712-1776OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coxson HO, Dirksen A, Edwards LD, Yates JC, Agusti A, Bakke P, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Investigators. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1:129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 40.Coxson HO, Hogg JC, Mayo JR, Behzad H, Whittall KP, Schwartz DA, et al. Quantification of idiopathic pulmonary fibrosis using computed tomography and histology. Am J Respir Crit Care Med. 1997;155:1649–1656. doi: 10.1164/ajrccm.155.5.9154871. [DOI] [PubMed] [Google Scholar]
- 41.Patel IS, Vlahos I, Wilkinson TM, Lloyd-Owen SJ, Donaldson GC, Wilks M, et al. Bronchiectasis, exacerbation indices, and inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:400–407. doi: 10.1164/rccm.200305-648OC. [DOI] [PubMed] [Google Scholar]
- 42.Martínez-García MA, de la Rosa Carrillo D, Soler-Cataluña JJ, Donat-Sanz Y, Serra PC, Lerma MA, et al. Prognostic value of bronchiectasis in patients with moderate-to-severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;187:823–831. doi: 10.1164/rccm.201208-1518OC. [DOI] [PubMed] [Google Scholar]
- 43.Putman RK, Hatabu H, Araki T, Gudmundsson G, Gao W, Nishino M, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Investigators. COPDGene Investigators. Association between interstitial lung abnormalities and all-cause mortality. JAMA. 2016;315:672–681. doi: 10.1001/jama.2016.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017:162326. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 45.Sørensen L, Nielsen M, Lo P, Ashraf H, Pedersen JH, de Bruijne M. Texture-based analysis of COPD: a data-driven approach. IEEE Trans Med Imaging. 2012;31:70–78. doi: 10.1109/TMI.2011.2164931. [DOI] [PubMed] [Google Scholar]
- 46.Mets OM, Buckens CF, Zanen P, Isgum I, van Ginneken B, Prokop M, et al. Identification of chronic obstructive pulmonary disease in lung cancer screening computed tomographic scans. JAMA. 2011;306:1775–1781. doi: 10.1001/jama.2011.1531. [DOI] [PubMed] [Google Scholar]
- 47.Sauer BC, Jones BE, Globe G, Leng J, Lu CC, He T, et al. Performance of a natural language processing (NLP) tool to extract pulmonary function test (PFT) reports from structured and semistructured Veteran Affairs (VA) data. EGEMS (Wash DC) 2016;4:1217. doi: 10.13063/2327-9214.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinchcliff M, Just E, Podlusky S, Varga J, Chang RW, Kibbe WA. Text data extraction for a prospective, research-focused data mart: implementation and validation. BMC Med Inform Decis Mak. 2012;12:106. doi: 10.1186/1472-6947-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernd L, Joan BS, Michael S, Bernhard K, Lowie EV, Louisa G, et al. BOLD Collaborative Research Group. the EPI-SCAN Team, the PLATINO Team, and the PREPOCOL Study Group. Determinants of underdiagnosis of COPD in national and international surveys. Chest. 2015;148:971–985. doi: 10.1378/chest.14-2535. [DOI] [PubMed] [Google Scholar]
- 50.Miller MR, Levy ML. Chronic obstructive pulmonary disease: missed diagnosis versus misdiagnosis. BMJ. 2015;351:h3021. doi: 10.1136/bmj.h3021. [DOI] [PubMed] [Google Scholar]
- 51.Berrington de González A, Mahesh M, Kim KP, Bhargavan M, Lewis R, Mettler F, et al. Projected cancer risks from computed tomographic scans performed in the United States in 2007. Arch Intern Med. 2009;169:2071–2077. doi: 10.1001/archinternmed.2009.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma S, Lucas CD. Increasing use of CTPA for the investigation of suspected pulmonary embolism. Postgrad Med. 2017;129:193–197. doi: 10.1080/00325481.2017.1281084. [DOI] [PubMed] [Google Scholar]