Abstract
Rationale: The etiology of sarcoidosis is unknown, but microbial agents are suspected as triggers.
Objectives: We sought to identify bacterial, fungal, or viral lineages in specimens from patients with sarcoidosis enriched relative to control subjects using metagenomic DNA sequencing. Because DNA from environmental contamination contributes disproportionately to samples with low authentic microbial content, we developed improved methods for filtering environmental contamination.
Methods: We analyzed specimens from subjects with sarcoidosis (n = 93), control subjects without sarcoidosis (n = 72), and various environmental controls (n = 150). Sarcoidosis specimens consisted of two independent sets of formalin-fixed, paraffin-embedded lymph node biopsies, BAL, Kveim reagent, and fresh granulomatous spleen from a patient with sarcoidosis. All specimens were analyzed by bacterial 16S and fungal internal transcribed spacer ribosomal RNA gene sequencing. In addition, BAL was analyzed by shotgun sequencing of fractions enriched for viral particles, and Kveim and spleen were subjected to whole-genome shotgun sequencing.
Measurements and Main Results: In one tissue set, fungi in the Cladosporiaceae family were enriched in sarcoidosis compared with nonsarcoidosis tissues; in the other tissue set, we detected enrichment of several bacterial lineages in sarcoidosis but not Cladosporiaceae. BAL showed limited enrichment of Aspergillus fungi. Several microbial lineages were detected in Kveim and spleen, including Cladosporium. No microbial lineage was enriched in more than one sample type after correction for multiple comparisons.
Conclusions: Metagenomic sequencing revealed enrichment of microbes in single types of sarcoidosis samples but limited concordance across sample types. Statistical analysis accounting for environmental contamination was essential to avoiding false positives.
Keywords: sarcoidosis, metagenomic, microbiome, bacterial 16S ribosomal RNA, fungal internal transcribed spacer ribosomal RNA
At a Glance Summary
Scientific Knowledge on the Subject
Sarcoidosis is believed to be an aberrant immune response to an unknown antigenic trigger in a genetically susceptible host. Microbial agents are suspected as triggers, although evidence is inconclusive. Metagenomic deep sequencing can quantitatively detect microbes that are unsuspected and/or unculturable, but it has not been systematically applied to sarcoidosis.
What This Study Adds to the Field
Targeted identification of all bacteria and fungi using 16S and internal transcribed spacer sequence tags, shotgun sequencing of all nucleic acids in viral preparations, and whole-genome sequencing was applied to sarcoidosis and control sample sets of lymphoid tissue, BAL, sarcoidosis spleen, and the Kveim reagent. Because environmentally derived sequences can confound detection in specimens with low authentic microbial content, extensive environmental sequence controls were incorporated. This analysis revealed enrichment of specific taxa in sarcoidosis compared with nonsarcoidosis specimens within individual sample sets, such as the fungus Cladosporium, although no taxa were consistently sarcoidosis enriched across sample sets. This study identifies agents as potential candidates for further validation. Analysis without accounting for environmentally derived sequences would have yielded multiple spurious hits, demonstrating the need for rigorous attention to environmental microbial sequences in metagenomic study of lung diseases. We provide a robust model to account for environmental contamination that is broadly applicable to other metagenomic studies.
Sarcoidosis is a multisystem disease characterized by an aberrant immune response that results in inflammation and granuloma formation. Sarcoidosis is believed to have an antigenic or inflammatory trigger that initiates the immune reaction in a susceptible host (1–3). Several susceptibility genes have been identified (4, 5), but the trigger remains obscure. Granulomatous inflammation is commonly seen in responses to microbial agents, as are other features of sarcoidosis immunopathology, such as oligoclonal CD4 T-cell expansion and T-helper cell type 1 polarization (1). No microbial cause has been definitively established for sarcoidosis, but candidates include species of Mycobacterium (6–9), as well as fungi (10) and Propionibacterium acnes (11, 12), a common skin bacteria.
The ability to detect rare or unculturable microbes has improved dramatically using deep DNA sequencing (13–15). Several studies have applied bacterial 16S ribosomal RNA (rRNA) gene sequencing to sarcoidosis, with differing results (6, 16, 17). No prior studies have interrogated fungal lineages with tag sequencing or used shotgun metagenomic sequencing for comprehensive studies of total DNA or purified viral particles.
We performed an intensive metagenomic investigation of multiple sarcoidosis sample sets using 16S rRNA gene sequencing to capture bacteria, internal transcribed spacer (ITS) sequencing for fungi, and whole-genome shotgun sequencing to characterize all microbes. Samples (Table 1) include two independent sets of formalin-fixed, paraffin-embedded (FFPE) granulomatous tissue biopsies from patients with newly identified sarcoidosis and control patients (sets A and B), and BAL from patients with newly diagnosed untreated stage II/III sarcoidosis and healthy control subjects (set C). We also interrogated a sample of the Kveim reagent (set D) (which is made from sarcoidosis-affected spleen and was used historically for sarcoidosis diagnosis by intradermal injection and monitoring for granuloma formation [18–20]), along with fresh granulomatous spleen from a patient with sarcoidosis (set E).
Table 1.
Sample Sets Studied
| Sample Set | Sample Type | Study Group | No. of Subjects | Collection Site | Bacteria | Fungi | RNA Virus | DNA Virus |
|---|---|---|---|---|---|---|---|---|
| A | Tissue | Sarcoid | 45 | Gdansk | 643 | 1,180 | N/A | N/A |
| Tissue | Control | 37 | Gdansk | 207 | 236 | N/A | N/A | |
| Paraffin only (paired) | Environmental control | 82 | Gdansk | 465 | 1,081 | N/A | N/A | |
| Blanks | Reagent control | 27 | Gdansk | 74 | 84 | N/A | N/A | |
| B | Tissue | Sarcoid | 30 | Philadelphia | 5,548 | 2,136 | N/A | N/A |
| Tissue | Control | 19 | Philadelphia | 2,813 | 2,703 | N/A | N/A | |
| Blanks | Reagent control | 5 | Philadelphia | 285 | 55 | N/A | N/A | |
| C | BAL | Sarcoid | 16 | Philadelphia | 3,105 | 25 | 1 | 85 |
| BAL | Healthy subjects | 12 | Philadelphia | 1,604 | 13 | 1 | 40 | |
| Prewash (paired) | Environmental control | 24 | Philadelphia | 823 | 28 | 4 | 99 | |
| Blanks | Reagent control | 4 | Philadelphia | 157 | 22 | 0 | 38 | |
| D | Kveim reagent | Sarcoid | 1 | New York | 1,725 | 20 | N/A | 4 |
| Water | Environmental control | 1 | Philadelphia | 1,035 | 3 | N/A | 26 | |
| E | Spleen | Sarcoid | 1 | Philadelphia | 1,156 | 19 | N/A | 3 |
| Saline wash of instruments | Environmental control | 2 | Philadelphia | 408 | 2 | N/A | 31 | |
| Water | Reagent control | 1 | Philadelphia | 1,035 | 3 | N/A | 26 |
Definition of abbreviation: N/A = not available.
An often-underappreciated feature of sequence-based microbial detection is that at low levels of true signal, sequences can be dominated by microbial DNA from environmental sources introduced during sample collection, storage, DNA extraction, or other steps (21, 22). This particularly confounds analysis of samples in which the authentic content of microbial DNA is low, such as lung bronchoscopies and tissue biopsies (21, 23–25). Even with the most careful preparation, however, there is often no way to eliminate environmental sequences completely, so further computational and statistical methods must be used to identify contamination. We thus used extensive environmental sampling and applied novel statistical modeling to minimize false-positive calls. By investigating several independent sample sets and tissue types, we were able to interrogate whether sarcoidosis-enriched sequences appeared consistently across sample sets. Some of the results of these studies have been previously reported in the form of an abstract (26).
Methods
Archived Tissue Samples
Two sets of FFPE sarcoidosis and control tissues were analyzed. Set A (from Gdańsk) were mediastinal lymph nodes showing noncaseating granulomas typical of sarcoidosis and negative by staining for acid-fast or fungal elements. Controls were mediastinal lymph nodes with normal or nonspecific reactive histology. Set B (from Philadelphia) consisted of mediastinal nodes containing granulomas typical of sarcoidosis and negative by fungal and acid-fast stain. Controls were histologically normal nodes from cancer staging procedures. Stored specimens were retrieved and 10-μm cuts taken under aseptic conditions. Paraffin block environmental controls were cut concurrently with tissue specimens. For set A, these were matched from the same block as tissue; for set B, they were not from the same block.
BAL
BAL fluid (set C) was obtained from subjects undergoing diagnostic bronchoscopy (from Philadelphia) for suspected new diagnosis of pulmonary sarcoidosis who had chest X-rays consistent with parenchymal (Scadding stage II/III) involvement. Subjects included here had sarcoidosis confirmed by standard criteria and exclusion of alternative diagnoses. BAL was performed using standard clinical protocols. Control BAL was obtained from healthy volunteers who underwent research bronchoscopy (23). Before bronchoscopy, an environmental control (bronchoscope prewash) was obtained as previously described (23). BAL and prewash were placed immediately on ice and stored at −80°C until analysis.
Kveim and Spleen Tissue
An aliquot of Kveim reagent ([19]; set D) was analyzed that was prepared at Mt. Sinai Hospital (New York) for clinical diagnostic use as described (27) and stored under sterile conditions.
Sarcoidosis-involved spleen (set E) was obtained from an individual with long-standing disease (from Philadelphia), previously but not currently treated, who underwent splenectomy for symptomatic splenomegaly. Tissue was freshly dissected from the organ and frozen at −80°C. An aliquot of the saline used for tissue homogenization served as a matched environmental control.
Human Subjects
Tissue samples were obtained from anonymized tissue archives. Bronchoscopy and spleen donor subjects provided written informed consent under institutional review board–approved protocols.
Sequence Analysis of 16S and ITS rRNA Gene Segments
Details of extraction, amplification, Illumina sequencing, and taxonomic assignment are in the online supplement. The bacterial 16S ribosomal RNA gene was amplified using V1V2 primers (see Table E2 in the online supplement); this relatively short amplicon was chosen to maximize amplification efficiency for rare sequences from low–microbial biomass samples (23, 28). The fungal ribosomal RNA internal transcribed spacer ITS1 region was amplified using ITS1F/ITS2 primers (25, 28, 29) (Table E2). Sequences were organized into operational taxonomic units (OTUs) at 97% identity. Statistical analysis was performed at the individual OTU level and at genus and family levels.
Virome Analysis
Virome analysis was performed on BAL and matched prewash specimens (30, 31). To enrich for viruses, fluid was pelleted and acellular material subject to size-exclusion concentration, followed by nuclease treatment to digest nonencapsulated nucleic acids. Nucleic acids were then extracted, and DNA was subjected to whole-genome amplification using GenomiPhi. RNA was reverse transcribed to cDNA and polymerase chain reaction amplified. Resulting libraries were shotgun sequenced; reads were quality filtered and then annotated using a custom database we constructed that included all complete bacterial, fungal, archaeal, and viral genomes in RefSeq release 79 (32). All nonviral reads were removed from consideration. We found many reads annotated to viruses later determined to be either from reagents or misannotation of human reads (31), which were therefore excluded. Details are in the online supplement.
Whole-Genome Sequencing
DNA from sarcoidosis spleen tissue and Kveim reagent was subjected to whole-genome sequencing (WGS) on an Illumina HiSeq. Reads were quality filtered, processed, and classified using Kraken (33) with our custom database (described above), with low-complexity regions masked before querying. Details are in the online supplement. The analytic pipeline is available at https://github.com/eclarke/sunbeam.
Accessing Sequence Data
Sequence data are available in the National Center for Biotechnology Information Sequence Read Archive under BioProject ID PRJNA392272.
Statistical Analysis
Code and a complete description are in the online supplement. For sample sets A and C, which had paired environmental controls, we used the R package lme4 (34) to build a generalized linear mixed effects model (GLMM) to regress the number of reads of a taxon against the study group (sarcoid/healthy) and sample type (tissue/environmental control) (Figure E1). Environmental levels of the taxa in each sample/control pair were captured as a random effect. Enrichment was determined by the significance and directionality of the coefficient for the study group/sample type interaction term after fitting the model. For sample set B, which did not have matched environmental controls, we used the R package DESeq2 (35) to determine enrichment.
Because one could not predict a priori whether a putative sarcoidosis-associated microbial trigger would be a specific family, genus, species, or even OTU, lineages were tested at the individual OTU level and then aggregated and tested at the species, genus, and family levels. False discovery rate (FDR) correction was applied at each taxonomic level, and an FDR P value cutoff of 0.1 was considered significant. Although interrogating the data at each taxonomic level increased the risk of type I (false positive) errors, we considered this justified due to uncertainty over which taxonomic level might be linked to sarcoidosis and the exploratory nature of the study and mitigated by the multiple independent sample sets. Conversely, because requiring a lineage to reach FDR-corrected significance in multiple independent sample sets would increase the likelihood of type II errors, we also considered lineages that were significant after FDR correction in one sample set but only significant before FDR correction in other sample sets.
Results
Sample Sets Studied
We studied five sets of sarcoidosis samples and controls (Table 1). Two (sets A and B) were archival lymph node tissue from patients undergoing diagnostic biopsy, where the sarcoidosis tissue studied was histologically confirmed to show granulomas. Set A included environmental control paraffin blanks matched to the individual tissue block and analyzed in parallel. BAL (set C) was from patients with untreated pulmonary sarcoidosis and healthy volunteers. Reasoning that BAL would most likely reveal a microbial trigger early in the disease course with parenchymal lung involvement, we studied individuals newly presenting with radiological Scadding stage II/III. Environmental controls matched to each sample were prewashes of the bronchoscope used to collect the BAL. We analyzed an aliquot of the Kveim reagent (set D), which is derived from sarcoidosis-affected human spleen and used diagnostically by intradermal injection and monitoring for granuloma formation. Because this suggests an immunological response to a triggering antigen (20), we hypothesized that Kveim reagent may contain DNA traces of an etiological microbe. Finally, we tested fresh sarcoidosis-involved spleen (set E) paired with blank controls processed in parallel to model reagent contamination.
Set A: Lymph Node Tissue
Microbial lineages detected in set A by bacterial 16S and fungal ITS rRNA gene sequencing are shown as stacked bar graphs (Figure 1), with dominant taxa summarized in Figure E2. Each tissue was paired with a control paraffin shaving from the same sample block. Lymph node and environmental control samples are thus plotted side by side. In many cases, samples and paraffin controls appear similar.
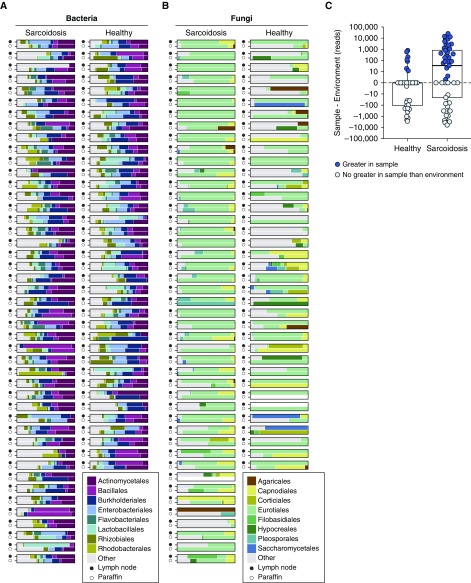
Figure 1.
Dominant bacterial and fungal orders in lymph node tissue samples and matched controls (set A). The major bacterial (A) and fungal (B) orders identified by 16S and internal transcribed spacer ribosomal RNA gene sequencing, respectively, are shown as proportions of total reads. Less-common lineages are aggregated under “Other.” For each pair, the solid circle indicates the formalin-fixed, paraffin-embedded lymph node sample, and the open circle indicates a slice of blank paraffin cut from the same block to serve as an environmental control. Empty (white) bar charts indicate that the sample was either not available or had no detectable lineages. The difference in Cladosporiaceae reads between a sample and its environmental control are shown in C. Solid circles represent samples with more Cladosporiaceae reads in the sample than the matched environmental control, and open circles represent samples in which the number of Cladosporiaceae reads were not greater than in the environment control. The abundances are shown as reads to more accurately reflect the input to the test, which used raw read counts as input. Normalization between differing sequencing depths was accounted for by modeling library size as a random effect for each sample (see Methods).
To investigate community structures in sarcoidosis and healthy lymph node samples, we calculated the UniFrac distance between each pair of samples and tested for clustering using permutational multivariate analysis of variance (PERMANOVA) (Figure E3). Bacterial communities were not significantly different between sarcoidosis and nonsarcoidosis tissues (Figure E3A), but fungal communities were different (Figure E3B; P = 0.027, R2 = 0.037). We then asked whether community differences might be attributed to differential contamination. We performed the same PERMANOVA test on paraffin controls from sarcoidosis and nonsarcoidosis samples. No significant difference was detected in bacterial 16S data (Figure E3C), but we did detect a difference in fungal ITS data (Figure E3D; P < 0.002, R2 = 0.091). Review of the specimen processing pipeline revealed that most sarcoidosis samples (31 of 45) were stored in a different building from nonsarcoidosis controls. A PERMANOVA test of the effects of storage site on the paraffin environmental controls revealed a significant effect on fungi (P < 0.00001) but not on bacteria. The environmental fungi responsible for site-specific differences were mostly of the Aspergillaceae family (negative binomial test, FDR P value = 0.019; Figure E2B).
To account for environmental admixture statistically, we designed a GLMM that incorporated each sample’s matched environmental control (see Methods). In short, this approach uses the matched control to model the background levels of each taxon. Then, when testing for differential abundance of that taxon, the background levels are accounted for by a separate term in the regression rather than the study group term. We used this approach to test for differential abundance between sarcoidosis and healthy lymph node at the OTU, species, genus, and family level.
Among fungi, at the family level, Cladosporiaceae (within the Capnodiales order; Figure 1B) was significantly enriched in sarcoidosis (FDR P value = 0.049). At the OTU level, no individual taxa were significantly enriched after FDR correction, but two Cladosporium OTUs were significant before FDR correction (P < 0.05, FDR P = 1). The Cladosporiaceae fungal lineage is present in both tissue samples and paraffin blank controls but is most abundant in sarcoidosis tissue (Figure 1C). No bacterial lineages were significantly enriched in sarcoidosis after accounting for environmental contamination.
Set B: Lymph Node Tissue
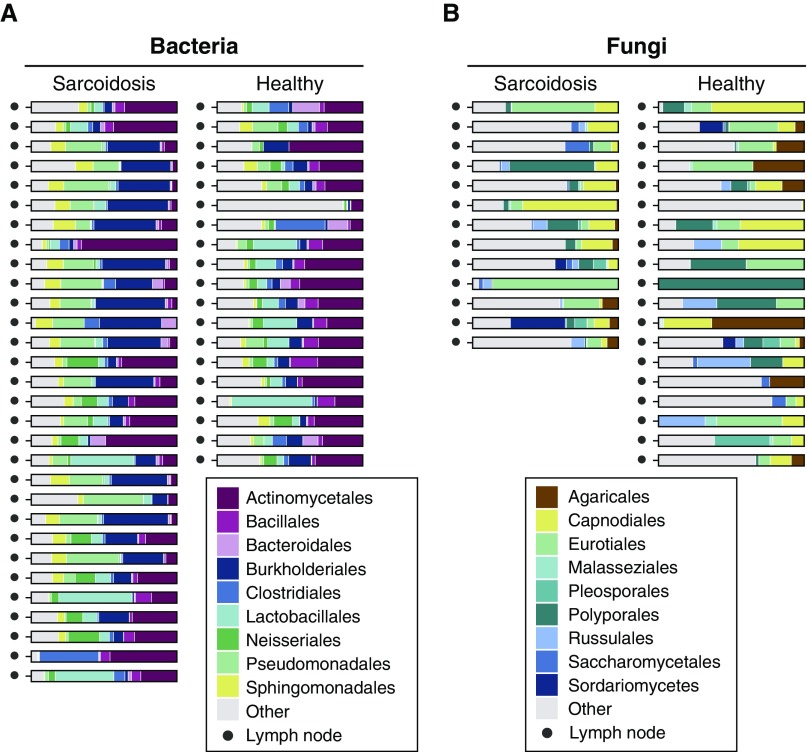
The dominant bacterial and fungal lineages in tissue set B are shown in Figure 2, with rank abundance plots in Figure E4. We compared community structure using UniFrac and PERMANOVA (Figure E5) and found differences in bacterial (but not fungal) populations between the sarcoidosis samples and healthy controls (P < 0.05). We then tested for differentially abundant taxa at the OTU, species, genus, and family levels. Numerous bacterial taxa were significantly enriched in sarcoidosis compared with control tissues (Table E3), including three OTUs in the Corynebacterium (order Actinomycetales) genus (FDR P = 1 × 10−5, 0.064, and 0.067, respectively) and four OTUs in the Rhodocyclaceae family (order Rhodocyclales; FDR P = 0.076, 0.003, 2 × 10−5, and 0.005, respectively). Other sarcoidosis-enriched bacteria were from the Sphinogomonadaceae family (order Sphingomonadales), the Comamonadaceaea and Oxalobacteraceaea families (order Burkholderiales), and the Moraxellaceae and Pseudomonadaceae families (order Pseudomonadales). No fungal lineages were sarcoidosis enriched in tissue set B after FDR correction, including Cladosporium (although fungi of this family do appear to be present in higher levels in the sarcoidosis samples; Figure E4B). Given the absence of paired environmental controls, these results are limited in isolation and serve mainly for comparison with other sample sets.
Figure 2.
Bacterial and fungal lineages in lymph node tissue samples (set B). The major bacterial (A) and fungal (B) orders identified by 16S ribosomal RNA and internal transcribed spacer (ITS) gene sequencing are shown as proportions of total reads. Less-common lineages are aggregated under “Other.” Seventeen samples failed to amplify any usable ITS sequences in B and are omitted. Blank paraffin controls matched to each tissue specimen were not available for these samples.
Set C: BAL
We analyzed DNA from whole BAL for bacteria and fungi using 16S and ITS gene sequencing (Figure 3, Figure E6), along with matched bronchoscope prewashes. Analysis using UniFrac and PERMANOVA (Figure E7) showed no significant differences between sarcoidosis and control bacterial communities. To identify taxa enriched in sarcoidosis while accounting for environmental input, we used the GLMM described above. We found that the bacterial family Corynebacteriaceae (order Actinomycetales) was enriched in sarcoidosis before FDR correction, but no taxa were enriched after FDR correction.
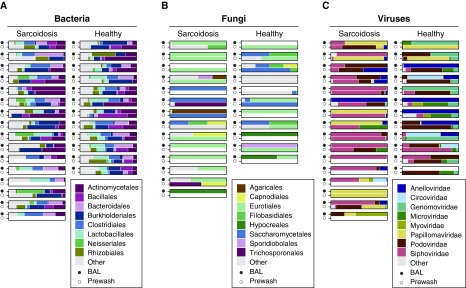
Figure 3.
Bacterial, fungal, and viral lineages in BAL (sample set C). The major bacterial (A) and fungal (B) lineages identified by 16S ribosomal RNA (rRNA) and internal transcribed spacer (ITS) rRNA sequencing and viral (C) lineages identified by shotgun sequencing of all nucleic acids in virus particle preparations are shown as proportions of the total reads. Data are shown at the order level for A and B and the family level for C. Less-common lineages are aggregated under “Other.” For each pair, the solid circle indicates the BAL fluid, and the open circle represents the prewash fluid for that scope. Empty (white) bar charts indicate that the sample was either not collected or had no detectable lineages. Three sample pairs failed to amplify any ITS sequences and are omitted from B.
Fungal sequences in BAL were sparse (Figure 3B), concordant with previous reports on BAL fungal detections (25). However, the genus Aspergillus (within the Eurotiales order; Figure 3B) was enriched in sarcoidosis (FDR P = 0.042).
Virome Analysis of Sarcoidosis BAL
We investigated the lung virome in sarcoidosis by generating virus particle preparations from acellular BAL and matched prewashes and deep sequencing both RNA and DNA. Initial inspection revealed abundant reads annotated as HHV6/HHV7. These reads matched human simple sequence repeats (31) and were therefore removed. We also purged sequences that were present in blank controls and attributable to viral enzymes used as reagents and thus likely reagent derived. Our approach was designed to detect both DNA and RNA viruses, but we did not recover any RNA viruses that did not likely originate from reagent contamination.
The majority of remaining viral sequences were phages of the Siphoviridae and Iridoviridae lineages (Figure 3C). The data initially suggested a much richer population of viruses in sarcoidosis BAL than that of healthy control subjects. However, viral sequences in the sarcoidosis cohort’s prewash controls were also richer than the control cohort’s prewash (Figure E8). This difference is likely because subjects with sarcoidosis underwent bronchoscopy in a clinical endoscopy suite, whereas healthy volunteers were sampled in a different facility used for research studies. This suggests that each location contributed a different environmental background of virus sequences, likely originating in lavage saline or water used to rinse bronchoscopes after cleaning.
We therefore used the same GLMM approach to account for environmental differences when testing for enriched viral species. No viruses were sarcoidosis enriched at any taxonomic levels tested. Without accounting for environmental input, the enrichment analysis would have been confounded by differences resulting from bronchoscopy locations.
Sets D and E: Kveim Reagent and Sarcoidosis Spleen
We analyzed Kveim reagent and fresh spleen from a patient with sarcoidosis. Three separate pieces of spleen were tested, along with controls to capture sequences from the environment. Kveim, spleen and controls were subject to 16S and ITS sequence analysis (Figures 4A and 4B) and also shotgun WGS (Figure 4C). WGS yielded mostly human sequences, which were removed; the remaining sequences were queried for microbial annotations.
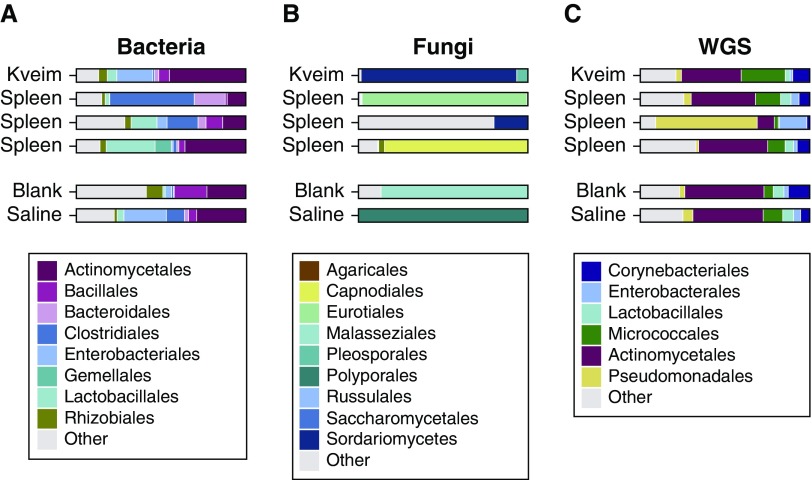
Figure 4.
Microbial lineages in the Kveim reagent (set D) and a granulomatous sarcoid-involved spleen (set E). The major lineages in sample sets D (Kveim) and E (sarcoidosis spleen) shown by sequencing. (A) Results from 16S sequencing, (B) internal transcribed spacer sequencing, and (C) results from whole-genome shotgun sequencing (WGS), after filtering, as described in the online supplement. Less-common lineages are aggregated under “Other,” including fungal detections in C.
The predominant bacteria found by both 16S and WGS were in the Propionibacteriaceae family (within the Actinomycetales order) and were detected across all samples, including controls. Other ubiquitous taxa included Corynebacteriaceae and Pseudomonadaceae (of the Actinomycetales and Pseudomonadales orders, respectively). Some differences were seen between 16S and WGS analysis for other taxa, likely resulting from the relative representation of sequences within 16S and WGS databases. No taxa were present only in sarcoidosis samples and not environmental controls.
Fungal detections were sparse in both ITS sequencing and WGS and inconsistent between methods. Cladosporiaceae (order Capnodiales) was detected by ITS in one spleen sample, but not by WGS. This may be due to a paucity of database genomic sequences for Cladosporiaceae, limiting detection in WGS annotation. There was no consistent fungal detection in sarcoidosis samples versus controls.
In the WGS data, we initially detected alignments annotated as Toxoplasma gondii in the Kveim and spleen samples. We also detected reads annotating to an unfinished Mycobacteria genome. However, these sequences were found to match human microsatellite simple repeats and so were judged to be false positives and removed (detailed in the online supplement). This is an issue for WGS data but not for 16S or ITS analysis, as the untargeted approach allowed capture of low-complexity repeat DNA. The only viral reads detected were from bacteriophages and were also found in the controls; they were thus inferred to be environmentally derived.
Shared Lineages
No bacterial or fungal lineages were significantly enriched after FDR correction in more than one sample set. To broaden our search, we examined lineages that were significantly enriched in one sample set after FDR correction and queried their abundance in the other sets. Enriched lineages are summarized in Table 2.
Table 2.
Summary of Taxa Enriched in Sarcoidosis over Controls in Lymph Node and BAL
| FDR P < 0.1 | non-FDR P < 0.05 | |
|---|---|---|
| Tissue set A | ||
| Bacteria | None | None |
| Fungi | Cladosporiaceae | OTU6408 (genus Cladosporium), Cladosporiaceae |
| Tissue set B | ||
| Bacteria | Many (113),* including Corynebacterium and Rhodocyclaceae | Many (252)* |
| Fungi | None | Many (149),† including one Cladosporium OTU (OTU7142) |
| BAL set C | ||
| Bacteria | None | OTU 104,987 (family Rhodocyclaceae), OTU 4,301,737 (genus Porphyromonas), Corynebacterium, Neisseria |
| Fungi | Aspergillus | Aspergillus |
| Viruses | None | None |
Definition of abbreviations: FDR = false discovery rate; OTU = operational taxonomic units.
See Table E3 for all enriched bacteria in set B.
See Table E4 for all enriched fungi in set B.
In tissue set A, fungi from the Cladosporiaceae family (order Capnodiales) were significantly enriched in sarcoidosis when tested as a group. A single Cladosporium OTU (OTU7142) was enriched in tissue set B before multiple testing correction (P = 0.042), although not the Cladosporiaceae family overall. Cladosporiaceae were detected but not enriched in sarcoidosis BAL. Finally, abundant Cladosporium reads were detected in one of three replicate spleen samples via ITS sequencing and not in the environmental controls, although it was not in WGS of spleen or Kveim.
In tissue set B, three OTUs belonging to the Corynebacterium bacterial genus (order Actinomycetales) were significantly enriched in sarcoidosis. Although no individual Corynebacterium OTUs were enriched in other sample sets, the Corynebacterium genus was enriched in sarcoidosis BAL before FDR correction (P = 0.02). Corynebacterium was detected but was not sarcoidosis enriched in tissue set A and was also detected in Kveim and spleen, as well as environmental controls.
Also in tissue set B, multiple OTUs in the Rhodocyclaceae (order Rhodocyclales) bacterial family were significantly enriched. A single Rhodocyclaceae OTU was enriched in BAL before FDR correction (OTU104987, genus Hydrogenophilus; P = 0.009). No Rhodocyclaceae lineages were detected in tissue set A, but they appeared in both Kveim and spleen as well as controls from sets D and E.
In BAL (set C), fungi in the Aspergillus genus (order Eurotiales) were significantly enriched in sarcoidosis. Aspergillus was detected in tissue set A but was not sarcoidosis enriched. Numerous Aspergillus lineages were also detected but not sarcoidosis enriched in tissue set B. Aspergillus species were not detected in Kveim or spleen in sets D and E but were found in the environmental and blank controls by WGS.
Discussion
This is the first study to interrogate microbial agents in sarcoidosis using a metagenomic approach combining bacterial and fungal sequence tag analysis, virome shotgun sequencing, and WGS. We anticipated that a causal microbe would be present in low abundance, so rigorous consideration of potential contamination would be critical for distinguishing authentic from environmentally derived sequences. Our findings were inconsistent across the five sample sets analyzed (Tables 1 and 2) but do provide candidates for further validation and strongly emphasize the importance of assessing environmental contamination.
Cladosporiaceae was significantly enriched in sarcoidosis specimens in tissue set A after adjustment for environmental admixture and multiple comparisons and also appeared in several other sample sets, although not with comparable statistical enrichment. Fungi in the Cladosporiaceae family are extremely common in the environment (36, 37), can trigger hypersensitivity pneumonitis and asthma (38, 39), and are capable of eliciting granulomatous inflammation (24, 40). This finding may warrant further investigation.
In tissue set B, we detected multiple sarcoidosis-enriched taxa, but interpretation is limited by the lack of matched environmental controls. Enriched taxa included several Corynebacterium OTUs, which were also sarcoidosis enriched before FDR correction in BAL (set C). Similarly, OTUs annotated as Rhodocyclaceae were significantly enriched in set B and enriched before FDR correction in set C. Corynebacterium are particularly interesting because they are known to elicit granulomatous responses in vivo (41, 42), although the association with sarcoidosis in this study was weak.
In addition to histopathological similarities, mycobacteria have been linked to sarcoidosis by immunological responses and/or sequence-based detection (6–9). However, we did not find enrichment of mycobacteria in sarcoidosis. Mycobacteria are difficult bacteria from which isolate DNA due to tough cell walls. To ensure our methods were robust, we confirmed detection via sequencing in known mycobacteria-infected tissue samples and biological specimens spiked with avirulent Mycobacterium tuberculosis (not shown). We also found low levels of mycobacteria in many samples and environmental controls. This suggests that our methods are not inherently insensitive to mycobacteria but that mycobacteria as a group were not enriched in these sarcoidosis specimens. However, the 16S variable region amplified, V1V2, cannot distinguish between mycobacterial species, which precludes detection of species-level differences. We also found that P. acnes was a ubiquitous environmental agent, concordant with other studies (43), with no evidence of enrichment in sarcoidosis.
Environmental admixture is an issue in any metagenomic survey and becomes increasingly important as the amount of authentic microbial content decreases (21). Such sequences can be introduced from specimen collection and storage, DNA extraction kits, the processing pipeline, or even “barcode error” inherent in Illumina deep-sequencing platforms that can allow low-level bleed-over in the sequencing process (22). Most importantly, in studies comparing subject groups, clinical samples collected at different times or locations may be contaminated with different environmental sequences. This is especially problematic when taxa of interest may also be environmentally present, so simple subtraction of background lineages is inappropriate. For example, sarcoidosis and healthy BALs were acquired in different locations and showed different background viromes. For tissue set A, specimen storage location differed between study groups, which led to enrichment of environmental fungi in one group and not the other.
A key component of our approach is the generalized linear mixed model, which enabled us to capture and control the effects of differential environmental admixture without losing the ability to test for differential abundance in environmental taxa. In tissue set A, without accounting for environmental input, a naive enrichment analysis would have identified fungal species within the Aspergillus and Penicillium genera as sarcoidosis enriched. The same is true for viruses in BAL (set C), which would have incorrectly identified greater phage populations in sarcoidosis. The GLMM approach presented here would enable handling of potential confounding effects of environmental admixture in microbiome studies generally and is particularly critical for specimens with low authentic microbial content when coupled with appropriate matched environmental controls for each clinical sample.
Our study has several limitations. We investigated microbes that might be enriched in sarcoidosis at the time of diagnosis, which is the earliest time point feasible, but the time from actual disease onset is unknown, so an etiological trigger may no longer be present. Conversely, it is conceivable that microbial enrichment associated with sarcoidosis could be a consequence of the disease, rather than a cause. Use of samples from distinct geographic locations would reveal shared lineages, but sarcoidosis triggers may differ geographically. In addition, any triggers may not be enriched in subjects with sarcoidosis at all but may be ubiquitously present, with disease determined mainly by host susceptibility factors. Although we examined a total of 93 sarcoidosis and 72 nonsarcoidosis specimens, plus 150 environmental controls (for a total of 738 sequencing reactions), the number of samples in any one set was modest. For the FFPE samples, sensitivity may be lessened by damage DNA incurred by during the de-crosslinking step necessary to undo the formalin fixation (44). For our DNA virus methods, GenomiPhi amplification may introduce bias toward short circular DNA due to rolling-circle amplification, although this bias should be consistent across study groups. Finally, any primers chosen for tagged sequencing also have inherent biases and may be more sensitive to some microbes than others (such as with the V1V2 primers and Mycobacterium species, as discussed previously).
In summary, application of metagenomic sequencing and analytic approaches tailored to low–microbial biomass samples did not identify a single causative agent but identified several candidate agents as sarcoidosis enriched. These include the Cladosporiaceae fungal family and Corynebacterium bacterial taxa. The modest enrichment and limited concordance of these candidates in the sample sets precludes our ability to assert any causal relationship with sarcoidosis, but we believe these candidates may be of interest in future studies. More broadly, the model we present here increases the power of metagenomic studies in low–microbial biomass samples, such as lung and tissue specimens, by allowing researchers to account for and test environmental admixture, thus avoiding potential spurious identifications.
Acknowledgments
Acknowledgment
The authors thank the subjects who generously volunteered for this study, M. Padilla at Mount Sinai for the donation of Kveim reagent, A. Ziober for assistance with tissue specimens, W. Drake for avirulent mycobacteria, and members of the Collman and Bushman laboratories for help and suggestions.
Footnotes
Supported by National Institutes of Health grants U01HL112712 (Site-Specific Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis [GRADS] Study) and R01HL113252, and the Penn Center for AIDS Research (P30AI045008) and the Penn–Children’s Hospital of Philadelphia Microbiome Program.
Author Contributions: E.L.C., K.B., M.D.R., K.C.P., F.D.B., and R.G.C. designed research; A.S.F., I.I., W.B., B.R., H.M., A.D., L.A.L., M.D.F., M.D.R., and K.C.P. collected samples; E.L.C. performed research; E.L.C., F.D.B., and R.G.C. analyzed data; A.P.L., Y.H., C.E.H., and I.I. acquired data; E.L.C., F.D.B., and R.G.C. wrote the paper.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201705-0891OC on August 28, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Chen ES, Moller DR. Etiologic role of infectious agents. Semin Respir Crit Care Med. 2014;35:285–295. doi: 10.1055/s-0034-1376859. [DOI] [PubMed] [Google Scholar]
- 2.Chen ES, Moller DR. Etiologies of sarcoidosis. Clin Rev Allergy Immunol. 2015;49:6–18. doi: 10.1007/s12016-015-8481-z. [DOI] [PubMed] [Google Scholar]
- 3.Dubaniewicz A. Microbial and human heat shock proteins as ‘danger signals’ in sarcoidosis. Hum Immunol. 2013;74:1550–1558. doi: 10.1016/j.humimm.2013.08.275. [DOI] [PubMed] [Google Scholar]
- 4.Fischer A, Grunewald J, Spagnolo P, Nebel A, Schreiber S, Müller-Quernheim J. Genetics of sarcoidosis. Semin Respir Crit Care Med. 2014;35:296–306. doi: 10.1055/s-0034-1376860. [DOI] [PubMed] [Google Scholar]
- 5.Fingerlin TE, Hamzeh N, Maier LA. Genetics of sarcoidosis. Clin Chest Med. 2015;36:569–584. doi: 10.1016/j.ccm.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Drake WP, Pei Z, Pride DT, Collins RD, Cover TL, Blaser MJ. Molecular analysis of sarcoidosis tissues for mycobacterium species DNA. Emerg Infect Dis. 2002;8:1334–1341. doi: 10.3201/eid0811.020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ES, Wahlström J, Song Z, Willett MH, Wikén M, Yung RC, et al. T cell responses to mycobacterial catalase-peroxidase profile a pathogenic antigen in systemic sarcoidosis. J Immunol. 2008;181:8784–8796. doi: 10.4049/jimmunol.181.12.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Z, Marzilli L, Greenlee BM, Chen ES, Silver RF, Askin FB, et al. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J Exp Med. 2005;201:755–767. doi: 10.1084/jem.20040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubaniewicz A, Trzonkowski P, Dubaniewicz-Wybieralska M, Dubaniewicz A, Singh M, Myśliwski A. Mycobacterial heat shock protein-induced blood T lymphocytes subsets and cytokine pattern: comparison of sarcoidosis with tuberculosis and healthy controls. Respirology. 2007;12:346–354. doi: 10.1111/j.1440-1843.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 10.Suchankova M, Paulovicova E, Paulovicova L, Majer I, Tedlova E, Novosadova H, et al. Increased antifungal antibodies in bronchoalveolar lavage fluid and serum in pulmonary sarcoidosis. Scand J Immunol. 2015;81:259–264. doi: 10.1111/sji.12273. [DOI] [PubMed] [Google Scholar]
- 11.Ishige I, Usui Y, Takemura T, Eishi Y. Quantitative PCR of mycobacterial and propionibacterial DNA in lymph nodes of Japanese patients with sarcoidosis. Lancet. 1999;354:120–123. doi: 10.1016/S0140-6736(98)12310-3. [DOI] [PubMed] [Google Scholar]
- 12.Nishiwaki T, Yoneyama H, Eishi Y, Matsuo N, Tatsumi K, Kimura H, et al. Indigenous pulmonary Propionibacterium acnes primes the host in the development of sarcoid-like pulmonary granulomatosis in mice. Am J Pathol. 2004;165:631–639. doi: 10.1016/S0002-9440(10)63327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly BJ, Imai I, Bittinger K, Laughlin A, Fuchs BD, Bushman FD, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:7. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter E, Greinert U, Kirsten D, Rüsch-Gerdes S, Schlüter C, Duchrow M, et al. Assessment of mycobacterial DNA in cells and tissues of mycobacterial and sarcoid lesions. Am J Respir Crit Care Med. 1996;153:375–380. doi: 10.1164/ajrccm.153.1.8542146. [DOI] [PubMed] [Google Scholar]
- 17.Richter E, Kataria YP, Zissel G, Homolka J, Schlaak M, Müller-Quernheim J. Analysis of the Kveim-Siltzbach test reagent for bacterial DNA. Am J Respir Crit Care Med. 1999;159:1981–1984. doi: 10.1164/ajrccm.159.6.9701038. [DOI] [PubMed] [Google Scholar]
- 18.Siltzbach LE. The Kveim test in sarcoidosis: a study of 750 patients. JAMA. 1961;178:476–482. doi: 10.1001/jama.1961.03040440028006. [DOI] [PubMed] [Google Scholar]
- 19.Teirstein AS. Kveim antigen: what does it tell us about causation of sarcoidosis? Semin Respir Infect. 1998;13:206–211. [PubMed] [Google Scholar]
- 20.Klein JT, Horn TD, Forman JD, Silver RF, Teirstein AS, Moller DR. Selection of oligoclonal V beta-specific T cells in the intradermal response to Kveim-Siltzbach reagent in individuals with sarcoidosis. J Immunol. 1995;154:1450–1460. [PubMed] [Google Scholar]
- 21.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson LA, Smith P, Sengupta DJ, Prentice JL, Sandin RL. Molecular analysis of sarcoidosis lymph nodes for microorganisms: a case-control study with clinical correlates. BMJ Open. 2013;3:e004065. doi: 10.1136/bmjopen-2013-004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 2014;15:487. doi: 10.1186/s13059-014-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke EL, Rossman M, Patterson KC, Fitzgerald A, Feldman M, Litzky L, et al. Metagenomic analysis of microbial sequences in specimens from patients with sarcoidosis and controls [abstract] Am J Respir Crit Care Med. 2015;191:A6163. [Google Scholar]
- 27.Chase MW. The preparation and standardization of Kveim testing antigen. Am Rev Respir Dis. 1961;84:86–88. doi: 10.1164/arrd.1961.84.5P2.86. [DOI] [PubMed] [Google Scholar]
- 28.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant. 2015;15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbas AA, Diamond JM, Chehoud C, Chang B, Kotzin JJ, Young JC, et al. The perioperative lung transplant virome: torque teno viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant. 2017;17:1313–1324. doi: 10.1111/ajt.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peternel R, Culig J, Hrga I. Atmospheric concentrations of Cladosporium spp. and Alternaria spp. spores in Zagreb (Croatia) and effects of some meteorological factors. Ann Agric Environ Med. 2004;11:303–307. [PubMed] [Google Scholar]
- 37.Ezike DN, Nnamani CV, Ogundipe OT, Adekanmbi OH. Airborne pollen and fungal spores in Garki, Abuja (North-Central Nigeria) Aerobiologia (Bologna) 2016;32:697–707. doi: 10.1007/s10453-016-9443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tham R, Katelaris CH, Vicendese D, Dharmage SC, Lowe AJ, Bowatte G, et al. The role of outdoor fungi on asthma hospital admissions in children and adolescents: a 5-year time stratified case-crossover analysis. Environ Res. 2017;154:42–49. doi: 10.1016/j.envres.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 39.Chiba S, Okada S, Suzuki Y, Watanuki Z, Mitsuishi Y, Igusa R, et al. Cladosporium species-related hypersensitivity pneumonitis in household environments. Intern Med. 2009;48:363–367. doi: 10.2169/internalmedicine.48.1811. [DOI] [PubMed] [Google Scholar]
- 40.Silva CL, Ekizlerian SM. Granulomatous reactions induced by lipids extracted from Fonsecaea pedrosoi, Fonsecaea compactum, Cladosporium carrionii and Phialophora verrucosum. J Gen Microbiol. 1985;131:187–194. doi: 10.1099/00221287-131-1-187. [DOI] [PubMed] [Google Scholar]
- 41.Nureki S, Miyazaki E, Matsuno O, Takenaka R, Ando M, Kumamoto T, et al. Corynebacterium ulcerans infection of the lung mimicking the histology of Churg-Strauss syndrome. Chest. 2007;131:1237–1239. doi: 10.1378/chest.06-2346. [DOI] [PubMed] [Google Scholar]
- 42.Taylor GB, Paviour SD, Musaad S, Jones WO, Holland DJ. A clinicopathological review of 34 cases of inflammatory breast disease showing an association between corynebacteria infection and granulomatous mastitis. Pathology. 2003;35:109–119. [PubMed] [Google Scholar]
- 43.Mollerup S, Friis-Nielsen J, Vinner L, Hansen TA, Richter SR, Fridholm H, et al. Propionibacterium acnes: disease-causing agent or common contaminant? Detection in diverse patient samples by next-generation sequencing. J Clin Microbiol. 2016;54:980–987. doi: 10.1128/JCM.02723-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campos PF, Gilbert TMP. DNA extraction from formalin fixed material. Methods Mol Biol. 2012;840:81–85. doi: 10.1007/978-1-61779-516-9_11. [DOI] [PubMed] [Google Scholar]