Pneumonia
Pneumonia, defined as an acute infection of the lung parenchyma by microbial pathogens, affects more than 3 million people per year and is responsible for more than 50,000 deaths annually in the United States (1). Globally, pneumonia is an even bigger healthcare burden, affecting more than 150 million and responsible for more than 2 million deaths annually, primarily in children younger than 5 years of age (2). Pneumonia is the eighth most common cause of death in the United States, responsible for 53,282 deaths in the United States alone in 2013 (1). The World Health Organization has declared pneumonia as the single largest cause of childhood deaths worldwide, accounting for the deaths of 920,136 children (16% of all deaths) younger than 5 years of age (2). The elderly and those with comorbidities are also at increased risk of developing pneumonia. Given the aging population, this burden of pneumonia is likely to increase. Pneumonia can be classified as either hospital acquired or community acquired. The hospital-acquired varieties pose a more severe threat, as they are often caused by multidrug-resistant microbes (“superbugs”), and drug resistance in community-acquired pneumonia is an increasingly recognized problem. Consequently, novel therapies are urgently needed, and understanding and bolstering the host’s immune response to these microbial pathogens represents an exciting approach to combat this global threat.
The mucosal surface of the lung is constantly exposed to invasive microbial pathogens that have the potential to cause pneumonia in susceptible hosts. When these agents overwhelm host defense, invasion of microbes results in pneumonia. Some common bacterial agents that cause pneumonia are Streptococcus pneumoniae, group A Streptococcus, Klebsiella pneumoniae, Staphylococcus aureus, Mycoplasma pneumoniae, and Pseudomonas aeruginosa. Viral agents that cause pneumonia are influenza virus, respiratory syncytial virus (RSV), and severe acute respiratory syndrome corona virus (SARS-CoV). Fungi, such as Aspergillus fumigatus, Cryptococcus neoformans, and Paracoccidioides brasiliensis, can also cause pneumonia, particularly in immunocompromised individuals. An effective early immune response in the lower respiratory tract is crucial for a successful elimination of microbes. Cells of the innate immune system possess germline-encoded pattern-recognition receptors that can sense conserved microbial molecules referred to as pathogen-associated molecular patterns (PAMPs) and set off a cascade of immune responses (3). Among pattern-recognition receptors, nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) are unique cytosolic receptors, which constantly patrol for invading pathogens in cytoplasm. The review has been divided to describe inflammasome assembly and activation and their role in acute pneumonia.
Inflammasomes
An inflammasome consists of a self-oligomerizing scaffold of proteins that includes NLRs, typically with leucine-rich repeats (LRRs), and the adaptor molecule known as apoptosis-associated speck-like protein (ASC), which contains a caspase activation and recruitment domain (CARD) and recruits procaspase-1, leading to its activation. NLRs have a tripartite structure consisting of a conserved central domain called the nucleotide-binding and oligomerization domain (NBD or NOD), a COOH-terminal LRR domain, and an N-terminal protein–protein interaction domain. Activation of certain NLR family containing CARD domain (NLRC) and NLR family containing pyrin domain (NLRP) family members, such as the NLRC4, NLRP3, NLRP1, and possibly NLRP12, leads to formation of caspase-1–containing inflammasomes. The inflammasomes involved in pulmonary host defense against infections are listed in Table 1. On sensing a stimulus, the NLR member (depending on the inflammasome type) and ASC undergo multimerization followed by recruitment of procaspase-1 to ASC through a CARD–CARD interaction. This process leads to autocleavage of caspase-1, which is subsequently responsible for the release of the active (mature or cleaved) form of proinflammatory cytokines, such as IL-1β and IL-18. Inflammasome complexes (>700 kD) can be homogeneous (contain a single type of NLR protein) or heterogeneous (contain different NLR proteins).
Table 1.
Inflammasomes Activated during Pulmonary Diseases
| NLR/Non-NLR Member | Alternate Name | Stimuli | Salient Observations | References |
|---|---|---|---|---|
| NLRP3 | CIAS1, PYPAF1, Cryopyrin, NALP3, CLR1.1 | Bacterial | ||
| Bacteria and toxins: Staphylococcus aureus, Streptococcus pneumoniae, Klebsiella pneumoniae, and intracellular bacteria (Francisella tularensis) | Caspase-1 activation, prevention of lung permeability | 8, 18, 19, 30 | ||
| Moraxella catarrhalis | RNA: IL-1β and IL-18 secretion in macrophages | 10 | ||
| LPS | Phagosomal membrane lysis, bacterial escape to cytosol for caspase-1 activation | 30 | ||
| Microbial nucleic acids | Extracellular ATP opens pore mediated by pannexin-1, delivering MDP from intracellular vesicle to cytosol, caspase-1 and IL-1β activation | 8 | ||
| Cytosolic delivery, K+ efflux | 30 | |||
| Viral | ||||
| Sendai virus | NLRP3 inflammasome is activated via MAVS, which are intact mitochondrial outer membrane proteins | 50 | ||
| Influenza virus, PB1-F2 and M2 proteins | Mitofusin 2 (Mfn 2) leads to increased interaction between MAVS and NLRP3. RNA of the virus, PB1-F2 protein and M2 proteins are probable ligands. Possible mechanisms for activation include lysosomal rupture and ROS models | 51, 52 | ||
| Human rhinovirus, viroporin 2B | Activation by IFN pathway via RIG-I | 73 | ||
| Viroporin 2B causes influx of Ca2+ from Golgi and ER into cytosol, activating NLRP3 inflammasome along with NLRC5 inflammasome | 58 | |||
| Human respiratory syncytial virus, viroporin | Acts via its small hydrophobic viroporin. Synthesis of pro–IL-1β by TLR2/MyD88/NF-κB pathways, ROS generation, K+ efflux resulting in inflammasome and caspase-1 activation and IL-1β release | 57, 59 | ||
| Adenovirus | ATP-mediated signaling leads to NLRP3 activation via purinergic receptor P2X7. DAMPs released by cells such as HMGB-1 and ATP can also trigger inflammasome activation | 56, 63 | ||
| Fungal | ||||
| Aspergillus fumigatus | Activates Syk tyrosine kinase, caspase-1 maturation, IL-1β secretion | 76 | ||
| Cryptococcus neoformans, Paracoccidioides brasiliensis | Caspase-1 processing, IL-1β secretion, and neutrophil recruitment | 74, 79 | ||
| IL-18 and IL-1β secretion | 80, 81 | |||
| NLRP12 | NALP12, PYPAF7, Monarch1, NLRP RNO2, PAN6, CLR19.3 | Bacterial | ||
| Yersinia pestis (ligand unknown but associated with T3SS) | TLR-4 recognizes LPS, upregulation of NLRP12, pro IL-1β, IL-18. NLRP12 forms inflammasome in response to Y. pestis, IL-1β, IL-18 secretion, inducing IFN-γ | 24 | ||
| K. pneumoniae (ligand unknown) | NLRP12 upregulation in pneumonia, augmented NF-κB, MAPK, and expression of histone deacetylases, improved host survival, bacterial clearance, neutrophil recruitment and accumulation in lungs | 25 | ||
| F. tularensis, S. aureus, Pseudomonas aeruginosa | Higher bacterial burden and defect in transendothelial migration of neutrophils to the lung parenchyma from circulation in Nlrp12−/− mice, but no role in assembly of inflammasome (caspase-1 independent). NLRP12 important for CXCL1 secretion by macrophages | 26 | ||
| NLRC4 | CARD12, CLAN, CLR2.1, IPAF | Bacterial | ||
| Legionella pneumophila | Bacterial flagellin via T4SS activates caspase-1, requires NAIP5, macrophage cell death | 30, 35, 40 | ||
| P. aeruginosa | Bacterial flagellin/rod activate caspase-1 via T3SS, non–flagellin-expressing strains also activate caspase-1 via NLRC4, ligand unknown | 30, 35, 37, 38, 41 | ||
| Burkholderia pseudomallei | TLR-5 important for protection; NLRC4 detects rod protein of T3SS, followed by caspase-1 activation and section of IL-1β | 35, 37, 39 | ||
| Escherichia coli | NAIP5 acts as inflammasome receptor for T3SS rod proteins. T3SS rod proteins activate NLRC4 with proinflammatory cytokine secretion via caspase-1 activation | 37 | ||
| K. pneumoniae | Activation of NLRC4, host protection via IL-1β, IL-17A, KC, MIP-2, LIX, ICAM-1, VCAM-1, NF-κB and MAPK activity, TNF-α, ligand unknown | 42 | ||
| NLRC5 | NOD27, NOD4, CLR16.1 | Viral | ||
| Human rhinovirus | NLRC5 and viroporin 2 B colocalize in Golgi. NLRC5 interacts with NLRP3 and leads to caspase-1 maturation and IL-1β release | 57, 58 | ||
| Sendai virus and poly(I:C) | Inflammasome activation associated with type I IFN pathway via IRF-3 and RANTES (CCL5) | 65 | ||
| Non-NLR inflammasome | ||||
| AIM2 | — | Bacterial | ||
| Double-stranded DNA | Oligonucleotide/oligosaccharide-binding domain of AIM2 senses DNA, interacts with ASC via its PYD domain to activate caspase-1; caspase-1–associated pyroptosis also observed | 14, 56, 63 | ||
| F. tularensis | IL-1β production by AIM2 inflammasome on bacterial lysis | 3 | ||
| S. pneumoniae | Caspase-1 activation, production of IL-1β and IL-18 and pyroptosis | 46 | ||
| Fungal | ||||
| A. fumigatus | IL-1β and IL-18 processing in conjunction with NLRP3 inflammasome | 77 |
Definition of abbreviations: AIM2 = absent in melanoma 2; ASC = apoptosis-associated speck-like protein; CARD = caspase activation and recruitment domain; CXCL1 = C-X-C motif chemokine ligand 1; DAMPs = damaged-associated molecular patterns; ER = endoplasmic reticulum; HMGB = high-mobility group box 1; ICAM = intercellular adhesion molecule; IRF = IFN regulatory factor; KC = keratinocyte-derived chemokine; LIX = LPS-induced CXC chemokine; MAPK = mitogen-activated protein kinase; MAVS = mitochondrial antiviral signaling; MDP = muramyl dipeptide; MIP-2 = macrophage inflammatory protein 2; NAIP = NLR family apoptosis inhibitory protein; NF-κB = nuclear factor κB; NOD = nucleotide-binding and oligomerization domain; NLR = NOD–like receptor; PYD = pyrin domain; RANTES = regulated upon activation, normal T-cell expressed and secreted; RIG-1 = retinoic acid-inducible gene-I; ROS = reactive oxygen species; TNF = tumor necrosis factor; TLR = Toll-like receptor; VCAM = vascular cell adhesion molecule.
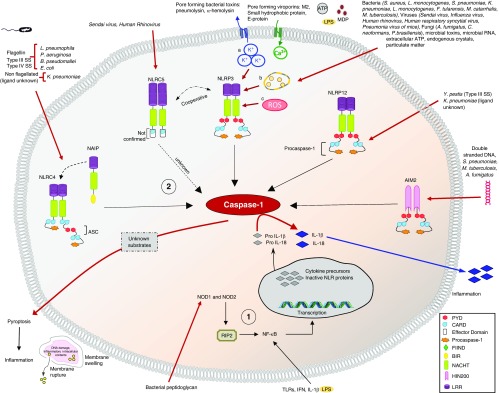
Successful inflammasome activation and IL-1β production depends on two signals, the first of which is a priming step that involves recognition of PAMPs by Toll-like receptors (TLRs) as well as recognition of microbial peptidoglycan by NOD1 and NOD2 in the cytosol (4). This triggers a signaling cascade that results in the transcription of pro–IL-1β and pro–IL-18. The second signal involves the activation of inflammasomes by intracellular stimuli (for example, the T3 and T4 secretion systems) and results in the activation of caspase-1, which cleaves the precursor forms of IL-1β and IL-18 into their mature and secreted forms. The different inflammasomes known to play a role in acute pneumonia and their mechanism of action are described in Figure 1. This two-step sequence for activation of mature proinflammatory cytokines serves as a safety mechanism preventing uncontrolled activation and secretion of cytokines, which could lead to severe tissue and organ injury in response to microbial infection or to a noninfectious stimulus, such as damage-associated molecular patterns. Although numerous stimuli can induce inflammasome activation, the downstream signaling cascades are generally conserved, although the precise molecular and cellular mechanisms involved in inflammasome activation remain elusive.
Figure 1.
Overview of the mechanisms of activation of various nucleotide-binding and oligomerization domain (NOD)-like receptors (NLR) and non-NLR inflammasomes in response to microbes, resulting in an inflammatory response via secretion of activated proinflammatory cytokines and pyroptosis. Steps in inflammasome activation: 1) Priming step involving factors such as Toll-like receptors (TLRs), NOD1, and NOD2; 2) inflammasome activation step. Mechanisms for activation of NLRP3 inflammasome: a) K+ efflux, b) lysosomal rupture, and c) mitochondrial reactive oxygen species (ROS) generation. A. fumigatus = Aspergillus fumigatus; AIM2 = absent in melanoma 2; ASC = apoptosis-associated speck-like protein; BIR = baculovirus inhibitor of apoptosis protein; B. pseudomallei = Burkholderia pseudomallei; CARD = caspase recruitment domain; C. neoformans = Cryptococcus neoformans; E. coli = Escherichia coli; FIIND = function to find domain; F. tularensis = Francisella tularensis; HIN200 = hematopoietic expression, interferon-inducible nature, and nuclear localization domain 200; K. pneumoniae = Klebsiella pneumoniae; L. monocytogenes = Listeria monocytogenes; L. pneumophila = Legionella pneumophila; LRR = leucine-rich repeat; M. catarrhalis = Moraxella catarrhalis; MDP = muramyl dipeptide; M. tuberculosis = Mycobacterium tuberculosis; NACHT = NAIP, CIITA (class II major histocompatibility complex transactivator), HET-E (incompatibility locus protein from Podospora anserine), and TP1 (mammalian telomerase-associated protein 1); NAIP = NLR family apoptosis inhibitory protein; NF-κB = nuclear factor-κB; P. aeruginosa = Pseudomonas aeruginosa; P. brasiliensis = Paracoccidioides brasiliensis; PYD = pyrin domain; RIP2 = receptor-interacting serine/threonine-protein kinase 2; S. aureus = Staphylococcus aureus; S. pneumoniae = Streptococcus pneumoniae; SS = secretion system; Y. pestis = Yersinia pestis.
Role of Inflammasomes in Bacterial Pneumonia
Secondary bacterial infections commonly complicate viral pneumonias and can be caused by diverse bacteria, such as S. pneumoniae, K. pneumoniae, S. aureus, M. pneumoniae, and P. aeruginosa. Bacterial pneumonia can also be manifest subsequent to viral pneumonia. Several inflammasomes are activated during different bacterial infections as part of the innate host immune response. The best characterized inflammasome is NLRP3 (5), which is primarily upregulated in immune and inflammatory cells after infection or inflammatory insult (6). The NLRP3 inflammasome is known to be activated by three diverse classes of stimuli: 1) invading microbial pathogens and their products, including lipopolysaccharide, muramyl dipeptide, nucleic acids, and pore-forming toxins; 2) endogenous danger signals like extracellular ATP, urate crystals, hyaluronan, and fibrillar amyloid-β; and 3) crystalline environmental pollutants, such as asbestos, silica, alum adjuvant, and ultraviolet irradiation (3, 5, 7–10).
The mechanism of NLRP3 inflammasome activation is widely accepted to be a two-step process, with regulation at both the transcriptional and post-translational levels (11, 12). The first signal is a priming event induced by TLR/nuclear factor (NF)-κB signaling that induces redistribution of ASC in the cytoplasm and expression of IL-1β and NLRP3 itself (5). The second signal is triggered by stimuli and leads to the activation of functional NLRP3 through recruitment of ASC and pro–caspase-1 to a cytoplasmic multimeric complex. Given the wide range of stimuli to which this inflammasome responds, multiple mechanisms of NLRP3 activation have been postulated and include K+ efflux via the purinergic receptor P2X, ligand-gated ion channel, 7 (P2X7), generation of mitochondrial reactive oxygen species (ROS), and release of mitochondrial DNA or cardiolipin (9, 13, 14). Recently, Ca2+ mobilization from the endoplasmic reticulum and extracellular spaces was shown to play an important role in NLRP3 inflammasome activation by orchestrating mitochondrial damage, ROS generation, and release of mtDNA into the cytosol (15). Furthermore, never in mitosis gene A (NIMA)-related kinase-7 (NEK7), was identified as an NLRP3-binding protein, functioning downstream of K+ efflux to regulate NLRP3 oligomerization and activation (16). These emerging mechanisms suggest that NLRP3 activation involves complex molecular processes with multiple activators and regulators in addition to assembly of the core inflammasome machinery.
The NLRP3 inflammasome contributes to host defense against numerous microbial infections (10, 17–19), and several pathogenic bacteria in the lung can activate NLRP3. Pneumolysin-activated NLRP3 confers protective immunity in pneumococcal pneumonia by maintaining lung barrier integrity (19). Similarly, NLRP3 limits bacterial growth in K. pneumoniae pneumonia by enhancing neutrophil recruitment and promoting macrophage necrosis via high-mobility group box 1 (HMGB-1) release (18). In a murine model of melioidosis, IL-1β and IL-18 processed by NLRP3 were found to provide host protection via regulation of inflammatory cell recruitment into the alveolar spaces (17). In a subsequent study, NLRP3 deficiency was found to promote bacterial clearance and result in the development of less severe necrotic pneumonia in S. aureus–induced infection (20). Furthermore, NLRP3 inhibition via P2X7 receptor blockade improved the outcome of lipopolysaccharide-induced acute lung injury (21).
The NLRP12 protein is predominantly expressed in monocytes/macrophages and granulocytes (22). NLRP12 is downregulated by TLR agonists like lipopolysaccharide and by microorganisms, including Mycobacterium tuberculosis, resulting in increased secretion of IL-6 (23). NLRP12 is believed to have a protective role in Yersinia pestis infection by inducing IL-18–mediated IFN-γ secretion (24). Similarly, NLRP12 signaling is important for host survival and bacterial clearance during K. pneumoniae infection, an effect that is mediated through the IL-17A/CXCL-1 (C-X-C motif chemokine ligand 1) axis (25). Furthermore, NF-κB signaling, cytokine and chemokine expression, and neutrophil recruitment were all reduced in Nlrp12−/− mice (25). Similarly, Nlrp12−/− mice infected with Francisella tularensis, S. aureus, and P. aeruginosa showed higher bacterial burden than wild-type/control counterparts. The authors demonstrated that on infection with these gram-positive and gram-negative bacteria, circulating neutrophils were similar between the control and Nlrp12−/− mice, whereas there was a defect in their transendothelial migration into the lung parenchyma from the circulation, as indicated by significantly fewer neutrophils in the lung parenchyma of Nlrp12−/− mice (26). Furthermore, they demonstrated that NLRP12 is important for CXCL1 secretion by macrophages infected with these bacteria, in vivo and in vitro. This was further confirmed by the decreased Cxcl1 expression in Nlrp12−/− macrophages. In addition, they suggested that NLRP12 does not play a role in the assembly of an inflammasome in response to F. tularensis, S. aureus, and P. aeruginosa, as IL-1β and caspase-1 production was not defective in Nlrp12−/− mice. Thus, the increased severity of infection in Nlrp12−/− mice is at least in part due to defective neutrophil migration and independent of caspase-1 activation (26).
The expression of NLRC4, a CARD domain–containing protein, is predominantly confined to the hematopoietic compartment (27). The NLRC4 inflammasome includes NLRC4, ASC, caspase-1, and NLR family apoptosis inhibitory protein (NAIP). The requirement of ASC in this inflammasome is pathogen specific (28, 29), because the NLRC4 inflammasome has its own CARD domain and therefore can activate caspase-1 without ASC recruitment (30). ASC may instead modulate NLRC4 activity, possibly through an indirect interaction with NLRC4, as this protein lacks a pyrin domain (PYD) (31). On stimulation, the central NACHT (NAIP, CIITA [class II major histocompatibility complex transactivator], HET-E [incompatibility locus protein from Podospora anserine], and TP1 [mammalian telomerase-associated protein 1]) domain of the NLRC4 oligomerizes with the NACHT domain of NAIP, resulting in the formation of the NLRC4 inflammasome (32). The NLRC4–NAIP proteins recognize pathogen-derived motifs in a very specific manner, with NLRC4 and NAIP performing complementary roles. NLRC4 recruits caspase-1 via a CARD–CARD interaction, whereas NAIP senses the PAMPs (33). The human genome encodes for only one functional NAIP protein, whereas there are four paralogs in mice (34). Mouse NAIP1 and NAIP2 bind to needle and rod proteins of the type III secretion system, whereas NAIP5 and NAIP6 bind to flagellin. The human NAIP recognizes type III secretion system needles as well as type IV secretion system–associated flagellin; however, it does not bind to type III secretion system rod proteins (35).
Several studies have documented the activation of NLRC4 in the presence of type III and type IV secretions involving the injection of needles and formation of pores in host cell membranes causing translocation of virulence factors such as flagellin and rods. These effects are observed in P. aeruginosa, Legionella pneumophila, Burkholderia pseudomallei, and Escherichia coli infections in mice (28, 35–39). Furthermore, the mechanism of recognition and activation appears to be different for different pathogens. For example, flagellin from Pseudomonas is recognized by NLRC4, whereas flagellin from L. pneumophila is sensed by NAIP5 (29, 31, 36, 40). However, recent studies have shown that nonflagellated strains of P. aeruginosa can also activate NLRC4 (41). We have also demonstrated the activation of NLRC4 by nonflagellated, gram-negative K. pneumoniae (42). These studies indicate that NLRC4 can recognize unidentified ligands other than flagellin. Furthermore, NLRC4 has been shown to provide protective immunity in murine models of lung infection with K. pneumoniae, B. pseudomallei, and L. pneumophila by limiting bacterial colonization in the lungs and dissemination to extrapulmonary organs (17, 35, 40, 42). However, in a separate study, mice deficient for Nlrc4 exhibited improved survival compared with wild-type mice after P. aeruginosa infection due to attenuated lung injury (43).
The IFN-inducible, HIN200 (hematopoietic expression, interferon-inducible nature, and nuclear localization domain 200) and PYD domain–containing protein, AIM2, is localized in the cytoplasm. Although not a member of the NLR protein family, AIM2 forms an inflammasome in response to intracytoplasmic, double-stranded DNA from a variety of microbial species as well as mammalian cells or even the host DNA itself (8, 44, 45). S. pneumoniae genomic DNA activated AIM2 inflammasome and resulted in activation of caspase-1 and maturation of IL-1β, and IL-18 in macrophages (46). AIM2 binds directly to DNA and can engage ASC by means of its PYD domain, thereby mediating caspase activation (5). Unlike NLR-based inflammasomes where the central NACHT domain is responsible for oligomerization and scaffold formation, the AIM2 inflammasome binds ligand (double-stranded DNA) via its C-terminal HIN domain, causing clustering of the inflammasome components to the many available binding sites on the DNA. This results in an activated inflammasome that can activate caspase-1 and promote maturation and secretion of proinflammatory cytokines (45, 47). Hence, AIM2 may play an important role in host defense during F. tularensis infection, which involves pathogen release to cytoplasm after evasion of the phagosome (5, 47). In another study, it was found that the cytosolic F. tularensis induced a type I IFN response by IRF (interferon regulatory factor)-3–dependent pathway. Furthermore, IFN is critical for inflammasome activation and caspase-1–dependent release of IL-1β and IL-18 (48).
Role of Inflammasomes in Viral Pneumonia
The CDC has declared pneumonia (including influenza infections) as the eighth leading cause of deaths in the U.S. population in 2014 (49). Next to bacteria, viruses are the most common causative agents of pneumonia. In viral infections, outer mitochondrial membrane proteins, known as mitochondrial antiviral signaling or IPS-1 (interferon-β promoter stimulator)/cardif/VISA (virus-induced signaling adapter) proteins, can activate the NLRP3 inflammasome, resulting in its oligomerization and subsequent activation and downstream signaling. Sendai virus infection, for example, is known to induce NLRP3 inflammasome activation via mitochondrial antiviral signaling proteins (50). In addition, the influenza virus can trigger the innate immune response via Mitofusin 2, promoting the association of NLRP3 and mitochondrial antiviral signaling proteins. Moreover, the influenza virus infection is associated with an increase in expression of all components of the NLRP3 inflammasome (51). In addition, viral RNA and its nonstructural protein PB1-F2 (translated from an alternative open reading frame in the PB1 gene) are speculated to be involved in inflammasome activation (51, 52). Furthermore, PB1-F2 can further activate the release of IL-1β by aggregating in phagosomes (52). In another study, expression of the influenza virus proton-specific ion channel M2 protein in the Golgi activated the NLRP3 inflammasome.
The influenza A virus M2 protein can directly trigger caspase-1 activation by promoting nucleotide transport into the cytosol (53). Several viruses are associated with ATP and ATP-sensitive K+ efflux, both of which can activate NLRP3 inflammasomes (5). For example, K+ efflux associated with the ATP-sensitive channel has been observed in RSV and influenza virus infections (53, 54). Moreover, it was suggested that cells dying as a result of influenza virus can cause NLRP3 activation by release of ATP (53). In another study, genomic influenza RNA failed to elicit an inflammasome response in bone marrow–derived macrophages in the absence of ATP (55). The importance of ATP-related NLRP3 activation in adenovirus infection was demonstrated in macrophage and epithelial cocultures (56). Furthermore, they also showed that ATP signaling via the P2X7 receptor is essential for NLRP3 activation in vivo (56).
Viroporin 2B of human rhinovirus causes activation of NLRP3 and CARD containing NLRC5, resulting in caspase-1 maturation and IL-1β release. Furthermore, this cytotoxic pore-forming protein is believed to control ion channel activity, causing an influx of cytosolic Ca2+ from Golgi and endoplasmic reticulum, resulting in inflammasome activation (57). Overlapping activation by the same pathogen and similar responses of NLRP3 and NLRC5 on human rhinovirus infection is indicative of a heterogeneous inflammasome or cooperative action between these two inflammasomes (58). The human RSV acts through its small hydrophobic viroporin, resulting in caspase-1 maturation and IL-1β production. The ion channel activity of the viroporin disrupts the intracellular ion balance, activating the NLRP3 inflammasome (59). After infection with RSV, pro–IL-1β synthesis is triggered by the TLR2/MyD88 (myeloid differentiation primary response 88)/NF-κB pathway, and this is accompanied by ROS generation and K+ efflux, leading to formation of the NLRP3 inflammasome (54). Both of these signals result in maturation and activation of caspase-1 and release of IL-1β. The small hydrophobic viroporin affects the ion gradient by forming a pore or channel in the plasma membrane, and the subsequent ionic changes cause inflammasome activation (54). This was confirmed by the failure of inflammasome activation by RSV mutants lacking the small hydrophobic viroporin, viral ion channel–inhibiting drugs, and by lipid raft disruptors (57).
The small hydrophobic protein of the metapneumovirus was found to act similarly to a viroporin and can affect membrane permeability (60). In another study in children, Il-1β, Il-18, Nlrp3, and IκBα genes were upregulated in metapneumovirus infection. Moreover, the severity of the disease was associated with upregulated Il-1β and Nlrp3 gene expressions indicating the deleterious effect of NLRP3 inflammasome activation (61). Adenovirus induces NLRP3 inflammasome activation in macrophages. This was confirmed by the decreased innate immune responses of macrophages from Asc knockout and Nlrp3 knockout mice (62). Intracellular damaged-associated molecular patterns such as HMGB-1 and ATP that are released by cells because of apoptosis, necrosis, or pyroptosis postinfection can also trigger NLRP3 activation (63). Adenovirus infection was found to trigger NLRP3 activation via the purinergic receptor P2X7 via ATP-mediated signaling (56), and endosomal rupture was also found to activate the NLRP3 inflammasome response (64). Moreover, deletion of the P2X7 gene resulted in higher survival rates in mice, suggesting that the excessive activation of the NLRP3 inflammasome can be damaging in adenovirus infection (56).
NLRC5 possesses a similar structure to other NLR family members with a central NACHT domain and also has the longest chain of LRRs at the COOH terminus among all human NLRs. However, opinion is divided with respect to the role of the effector domain at this molecule’s N-terminus. NLRC5 was reported to adopt a death domain fold without homology to CARD or PYD domains (65), and Kuenzel and colleagues (66) have reported that it does possess a CARD domain at the N-terminus. Moreover, Davis and colleagues (58) have shown the activation of an inflammasome with NLRP3 cooperation with procaspase-1, pro–IL-1β, and ASC in primary human monocytes. On infection with the human rhinovirus, there is colocalization of NLRC5 and the 2B viroporin in the Golgi. Furthermore, NLRC5 complexed with ASC interacts with NLRP3, resulting in inflammasome formation and innate immune response, such as elevated IL-1β release (57). NLRC5 was also shown to be activated by RNA signatures of Sendai virus and poly(I:C) in THP-1 (leukemic monocyte) cell lines and was found to regulate the type I IFN pathway via IRF-3 and the early-phase chemokine, RANTES (regulated upon activation, normal T-cell expressed and secreted) (CCL5) (65).
A novel coronavirus, SARS-CoV, causes severe acute respiratory syndrome (SARS), which is characterized by severe pneumonia. Although the mechanism of inflammasome activation by SARS-CoV infection is unclear, it is associated with the excessive release of proinflammatory cytokines, including IFN-γ, IL-6, MCP-1 (monocyte chemoattractant protein-1), IL-18, and IL-1β in the blood, lungs, and lymph nodes (67, 68). Pediatric cases of SARS were also associated with elevated IL-1β and IL-18, indicating inflammasome activation and caspase-1 maturation (69). A possible mechanism for inflammasome activation by SARS-CoV is through its E protein, which is classified as a viroporin because it forms ion-conductive pores in planar lipid bilayers (in vitro) and localizes in the endoplasmic reticulum–Golgi intermediate compartment (in vivo). The E protein of SARS-CoV acts as a cation channel and favors Na+ influx over K+ influx, in vitro (70). Another study showed that the E protein increases Ca2+ influx, which may result in activation of the NLRP3 inflammasome. The severe pathologies associated with SARS-CoV could be due to excessive activation of NLRP3 (71).
In addition to inflammasomes and associated cytokines such as IL-1β and IL-18, invading viruses can also induce IFNs other than IFN-γ, particularly type I IFN and type III IFN. Some studies have demonstrated activation of inflammasome by IFN; however, these results are not conclusive. It was suggested that type I IFN prevents de novo synthesis of IL-1β precursor and inhibits NLRP3 inflammasome and caspase-1 activation (72). However, another study in primary human lung epithelial cells revealed the IFN-mediated activation of NLRP3 inflammasome in influenza infection by RIG-I (retinoic acid-inducible gene-I) (73). Pothlichet and colleagues (73) have shown the activation of NLRP3 by IFN-1 pathway via RIG-I during influenza infection in vivo and in vitro. RIG-I binds to ASC and caspase-1, and the mRNA expression of TLR3 and NLRP3 was RIG-I–dependent in lung epithelial cells. Moreover, optimum IL-1β secretion required upregulation of TLR3 and NLRP3 mediated by RIG-I (73). Thus, further studies to discern the role of IFN in inflammasome activation are needed.
Role of Inflammasomes in Fungal Pneumonia
Fungal infections can be a serious threat, especially to immunocompromised patients (74, 75). The role of the NLRP3 inflammasome in host defense against various fungal infections has been widely explored. However, the roles of other inflammasomes, especially NLRP6, NLRP12, and NLRC4, remain poorly understood. A. fumigatus, a common cause of fungal infections, leads to life-threatening conditions in immunocompromised individuals, notably organ transplant patients. The hyphal fragments of A. fumigatus cause NLRP3 inflammasome activation and subsequent IL-1β processing in human monocyte cell lines, both of which require Syk tyrosine kinase activity (76). Karki and colleagues (77) have demonstrated crucial collaborative roles of NLRP3 and AIM2 inflammasome in A. fumigatus infection using an immunocompromised mouse model. Mice lacking both NLRP3 and AIM2 were highly susceptible to pulmonary A. fumigatus infections when compared with wild-type mice, whereas mice deficient in either inflammasome exhibited a phenotype similar to wild-type mice. Moreover, NLRP3- and AIM2-mediated IL-1β and IL-18 secretion was found to be essential to confer protection against A. fumigatus in an immunocompromised mouse model (77). However, a separate study using Nlrp3-deficient mice demonstrated increased host protection and reduced fungal load in the lungs after infection. Intriguingly, when infected with a higher inoculum of the fungus, Nlrp3−/− mice were more susceptible than wild-type mice to A. fumigatus (78). This apparent discrepancy could be explained by differences in the doses of fungus used in the study.
C. neoformans is another opportunistic fungal pathogen that infects immunocompromised patients. Studies using mouse models and human macrophages have shown that the NLRP3 inflammasome is activated in response to acapsular C. neoformans infection. In mouse dendritic cells, secretion of IL-1β in response to C. neoformans requires NLRP3 activation and not NLRC4 or AIM2 inflammasomes. In an in vivo setting, adequate neutrophil recruitment and fungal clearance from lungs depend on NLRP3 activation (79). However, a separate study by the same group showed that internalized encapsulated C. neoformans can also activate both canonical caspase-1 and noncanonical caspase-8 inflammasome in mouse dendritic cells (74).
P. brasiliensis causes systemic granulomatous mycosis, which is endemic in certain countries of Latin America such as Brazil, Argentina, Venezuela, and Colombia (80, 81). The NLRP3 inflammasome contributes to host defense against P. brasiliensis by enhancing IL-18–mediated immune signaling (80) and by activating IL-1β for controlling infection (81).
Dysregulated Inflammasome Activation and Its Regulation
Inflammasome activation is critical for containment of pathogens. However, its uncontrolled activation, overriding the host regulatory mechanisms, can drive pathologic inflammatory responses leading to extensive host tissue damage. For example, S. aureus α-hemolysin–induced NLRP3 activation has been shown to have a deleterious role in pneumonia. Mice deficient in Nlrp3 displayed reduced lung pathologies that were associated with decreased neutrophil recruitment in the lungs (20). Although this study does not identify the precise mechanism of NLRP3-mediated lung pathologies, it implicates the ability of S. aureus–induced necroptosis to damage pulmonary architecture to facilitate phagocyte recruitment and subsequent bacterial dissemination. Similarly, mice deficient in Nlrc4 displayed enhanced bacterial clearance and decreased lung pathology (43). Moreover, functional type III secretion system from P. aeruginosa triggers NLRC4 inflammasome activation, and the subsequent IL-18 production was shown to drive excessive neutrophil recruitment and repress IL-17–mediated antimicrobial peptide expression. Furthermore, deletion of Nlrp3 or administration of anakinra (IL-1Ra) improved the outcomes of A. fumigatus or P. aeruginosa infections, as evidenced by attenuated inflammation and tissue damage in a mouse model (78). Deletion of the P2X7R gene was shown to improve survival rates of mice with adenovirus infection, suggesting that excessive activation of the inflammasome was responsible for the fatality observed in wild-type mice and acute respiratory distress syndrome associated with adenovirus infections (56). However, the histopathology of the lungs in P2X7R knockout mice was not different from those of the wild-type mice. These results suggest that their survival advantage could be related to attenuated host responses rather than the tissue damage caused by adenovirus. In another study in children with metapneumovirus infections, the severity of the disease was associated with upregulation of Il-1β and Nlrp3 mRNA transcripts, thus demonstrating the deleterious effect of overzealous inflammasome activation (61).
An efficient inflammasome-mediated host response would eventually subside. Therefore, the host regulatory mechanisms are critical to control the checkpoints of inflammasome activation. A classic example is the two-step activation of NLRP3 inflammasomes. The TLR-mediated first signal inducing NLRP3 transcription and distribution is critical before the second signal directly triggers their full activation. Similarly, several host CARD-only and PYD-only proteins regulate inflammasome activation as they decoy domain interactions to prevent recruitment of pro–caspase-1 and ASC to inflammasome complex, respectively (82). Different ASC variants generated through alternate splicing have distinct abilities to regulate inflammasome assembly and activation (83). Furthermore, NLRP10, an NLR lacking LRRs, negatively regulates inflammasome activation by inhibiting aggregation of ASC (84). In addition, mir-223 binds to the 3′ untranslated region of NLRP3 to suppress its expression at the translational level (85). Autophagy also limits NRLP3 inflammasome activation as it removes damaged mitochondria that can release excessive ROS and DNA into the cytoplasm (86). Similarly, deletion of autophagy-related protein 16–like 1, LC3B (structural protein of autophagosomal membranes), and beclin-1 promoted formation of NLRP3 assembly and subsequent IL-1β production to numerous stimuli (87, 88). Recently, identification of NEK7 as an NLRP3-binding protein that regulates NLRP3 oligomerization and activation (89) indicates that host regulatory mechanisms act at different signal transduction steps to limit uncontrolled inflammasome activation.
Conclusions
Pneumonia has emerged as a major cause of mortality globally, particularly of young children. Although the importance of inflammasomes in bacterial, viral, and fungal pneumonia is evident, much still remains to be learned, as new inflammasomes, their ligands, and downstream signaling cascades are constantly being discovered. It is important to note that although there is variation in the activation of inflammasomes by either bacterial or viral or fungal pathogens, the subsequent downstream signaling pathways are similar. Although the roles of several inflammasomes, including NLRP3, NLRP12, and NLRC4, as well as AIM2, have been established in the context of lung, including pneumonia, the exact role of other inflammasomes, such as NLRC5, NLRP6, and NLRP10, in pulmonary diseases remain elusive. Moreover, there is little or no information regarding the role of inflammasomes in the pathogenesis of disease by respiratory pathogens, such as Moraxella catarrhalis, SARS-CoV, metapneumovirus, hantavirus, parainfluenza virus, and Pneumocystis jirovecii. Understanding the interactions between different inflammasomes during the innate immune response is essential for identifying how immune sensors are stimulated by ligands and, ultimately, for development of therapies to attenuate excessive tissue damage. Molecular and cellular studies investigating crosstalk between TLRs and inflammasomes, and the spatial association of inflammasomes with intracellular adaptors, are needed for a more complete understanding of how host responses are integrated between immune sensors and how these sensors bridge innate and adaptive responses. The translational relevance of inflammasome research is most evident in the novel therapeutic targets it has identified. In this context, the importance of molecules downstream of inflammasomes, including caspase-1, IL-1β, and IL-1 receptor antagonists have been assessed in animal models to determine their potential as targets for combating deleterious inflammasome-mediated tissue damage. The challenge for the immediate future will be to apply our current understanding of inflammasomes to reduce tissue damage caused by excessive and unregulated inflammation using specific therapies in response to infections in both acute and chronic settings.
Footnotes
Supported by NIH grants F31HL137287 (S.P.), R01AI113720 and R01HL091958 (S.J.), and R01HL133336 (S.C.) and a Young Clinical Scientist Award from the Flight Attendant Medical Research Institute (S.C.).
Originally Published in Press as DOI: 10.1164/rccm.201707-1391PP on September 20, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Centers for Disease Control and Prevention. National Center for Health Statistics: pneumonia [accessed 2017 Dec 12]. Available from: http://www.cdc.gov/nchs/fastats/pneumonia.htm.
- 2.World Health Organization. Pneumonia fact sheet [updated 2016 Sep; accessed 2017 Dec 12]. Available from: http://www.who.int/mediacentre/factsheets/fs331/en/
- 3.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 4.Baral P, Batra S, Zemans RL, Downey GP, Jeyaseelan S. Divergent functions of Toll-like receptors during bacterial lung infections. Am J Respir Crit Care Med. 2014;190:722–732. doi: 10.1164/rccm.201406-1101PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Feldmeyer L, Keller M, Niklaus G, Hohl D, Werner S, Beer HD. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 8.Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha W, Mitoma H, Hanabuchi S, Bao M, Weng L, Sugimoto N, et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc Natl Acad Sci USA. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pétrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 15.Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci USA. 2012;109:11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 17.Ceballos-Olvera I, Sahoo M, Miller MA, Del Barrio L, Re F. Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1β is deleterious. Plos Pathog. 2011;7:e1002452. doi: 10.1371/journal.ppat.1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willingham SB, Allen IC, Bergstralh DT, Brickey WJ, Huang MT, Taxman DJ, et al. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol. 2009;183:2008–2015. doi: 10.4049/jimmunol.0900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witzenrath M, Pache F, Lorenz D, Koppe U, Gutbier B, Tabeling C, et al. The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. J Immunol. 2011;187:434–440. doi: 10.4049/jimmunol.1003143. [DOI] [PubMed] [Google Scholar]
- 20.Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, et al. Staphylococcus aureus α-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis. 2012;205:807–817. doi: 10.1093/infdis/jir846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Zhao J, Wang H, Liang Y, Yang N, Huang Y. Blockage of P2X7 attenuates acute lung injury in mice by inhibiting NLRP3 inflammasome. Int Immunopharmacol. 2015;27:38–45. doi: 10.1016/j.intimp.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lich JD, Williams KL, Moore CB, Arthur JC, Davis BK, Taxman DJ, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 23.Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–39924. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai S, Batra S, Del Piero F, Jeyaseelan S. NLRP12 modulates host defense through IL-17A-CXCL1 axis. Mucosal Immunol. 2016;9:503–514. doi: 10.1038/mi.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulland TK, Jain N, Hornick EE, Elliott EI, Clay GM, Sadler JJ, et al. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat Commun. 2016;7:13180. doi: 10.1038/ncomms13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 28.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozören N, Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 29.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 30.Brodsky IE, Monack D. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin Immunol. 2009;21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Case CL, Shin S, Roy CR. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun. 2009;77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kofoed EM, Vance RE. NAIPs: building an innate immune barrier against bacterial pathogens. NAIPs function as sensors that initiate innate immunity by detection of bacterial proteins in the host cell cytosol. BioEssays. 2012;34:589–598. doi: 10.1002/bies.201200013. [DOI] [PubMed] [Google Scholar]
- 33.Leissinger M, Kulkarni R, Zemans RL, Downey GP, Jeyaseelan S. Investigating the role of nucleotide-binding oligomerization domain-like receptors in bacterial lung infection. Am J Respir Crit Care Med. 2014;189:1461–1468. doi: 10.1164/rccm.201311-2103PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endrizzi MG, Hadinoto V, Growney JD, Miller W, Dietrich WF. Genomic sequence analysis of the mouse Naip gene array. Genome Res. 2000;10:1095–1102. doi: 10.1101/gr.10.8.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aachoui Y, Miao EA. Down with doublespeak: NAIP/NLRC4 inflammasomes get specific. J Exp Med. 2016;213:646. doi: 10.1084/jem.2135insight1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolle L, Yu FS, Kovach MA, Ballinger MN, Newstead MW, Zeng X, et al. Redundant and cooperative interactions between TLR5 and NLRC4 in protective lung mucosal immunity against Pseudomonas aeruginosa. J Innate Immun. 2015;7:177–186. doi: 10.1159/000367790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 40.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozören N, Brady G, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 41.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S, Batra S, Wakamatsu N, Pacher P, Jeyaseelan S. NLRC4 inflammasome-mediated production of IL-1β modulates mucosal immunity in the lung against gram-negative bacterial infection. J Immunol. 2012;188:5623–5635. doi: 10.4049/jimmunol.1200195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faure E, Mear JB, Faure K, Normand S, Couturier-Maillard A, Grandjean T, et al. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am J Respir Crit Care Med. 2014;189:799–811. doi: 10.1164/rccm.201307-1358OC. [DOI] [PubMed] [Google Scholar]
- 44.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang R, Tsuchiya K, Kawamura I, Shen Y, Hara H, Sakai S, et al. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J Immunol. 2011;187:4890–4899. doi: 10.4049/jimmunol.1100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kochanek KD, Murphy SL, Xu J, Tejada-Vera B. Deaths: final data for 2014. Natl Vital Stat Rep. 2016;65:1–122. [PubMed] [Google Scholar]
- 50.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 51.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, et al. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. Plos Pathog. 2013;9:e1003392. doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segovia J, Sabbah A, Mgbemena V, Tsai SY, Chang TH, Berton MT, et al. TLR2/MyD88/NF-κB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. Plos One. 2012;7:e29695. doi: 10.1371/journal.pone.0029695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee BH, Hwang DM, Palaniyar N, Grinstein S, Philpott DJ, Hu J. Activation of P2X(7) receptor by ATP plays an important role in regulating inflammatory responses during acute viral infection. Plos One. 2012;7:e35812. doi: 10.1371/journal.pone.0035812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Triantafilou K, Kar S, van Kuppeveld FJ, Triantafilou M. Rhinovirus-induced calcium flux triggers NLRP3 and NLRC5 activation in bronchial cells. Am J Respir Cell Mol Biol. 2013;49:923–934. doi: 10.1165/rcmb.2013-0032OC. [DOI] [PubMed] [Google Scholar]
- 58.Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Brickey WJ, et al. Cutting edge: NLRC5-dependent activation of the inflammasome. J Immunol. 2011;186:1333–1337. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax. 2013;68:66–75. doi: 10.1136/thoraxjnl-2012-202182. [DOI] [PubMed] [Google Scholar]
- 60.Masante C, El Najjar F, Chang A, Jones A, Moncman CL, Dutch RE. The human metapneumovirus small hydrophobic protein has properties consistent with those of a viroporin and can modulate viral fusogenic activity. J Virol. 2014;88:6423–6433. doi: 10.1128/JVI.02848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malmo J, Moe N, Krokstad S, Ryan L, Loevenich S, Johnsen IB, et al. Cytokine profiles in human metapneumovirus infected children: identification of genes involved in the antiviral response and pathogenesis. Plos One. 2016;11:e0155484. doi: 10.1371/journal.pone.0155484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 63.Yoo JK, Kim TS, Hufford MM, Braciale TJ. Viral infection of the lung: host response and sequelae. J Allergy Clin Immunol. 2013;132:1263–1276, quiz 1277. doi: 10.1016/j.jaci.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barlan AU, Danthi P, Wiethoff CM. Lysosomal localization and mechanism of membrane penetration influence nonenveloped virus activation of the NLRP3 inflammasome. Virology. 2011;412:306–314. doi: 10.1016/j.virol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hösel M, et al. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuenzel S, Till A, Winkler M, Häsler R, Lipinski S, Jung S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 67.Huang KJ, Su IJ, Theron M, Wu YC, Lai SK, Liu CC, et al. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 69.Ng PC, Lam CW, Li AM, Wong CK, Cheng FW, Leung TF, et al. Inflammatory cytokine profile in children with severe acute respiratory syndrome. Pediatrics. 2004;113:e7–e14. doi: 10.1542/peds.113.1.e7. [DOI] [PubMed] [Google Scholar]
- 70.Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, DeDiego ML, Torres J, Aguilella VM, et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nieto-Torres JL, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Kempen TS, Wenink MH, Leijten EF, Radstake TR, Boes M. Perception of self: distinguishing autoimmunity from autoinflammation. Nat Rev Rheumatol. 2015;11:483–492. doi: 10.1038/nrrheum.2015.60. [DOI] [PubMed] [Google Scholar]
- 73.Pothlichet J, Meunier I, Davis BK, Ting JP, Skamene E, von Messling V, et al. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. Plos Pathog. 2013;9:e1003256. doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen M, Xing Y, Lu A, Fang W, Sun B, Chen C, et al. Internalized Cryptococcus neoformans activates the canonical caspase-1 and the noncanonical caspase-8 inflammasomes. J Immunol. 2015;195:4962–4972. doi: 10.4049/jimmunol.1500865. [DOI] [PubMed] [Google Scholar]
- 75.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 76.Saïd-Sadier N, Padilla E, Langsley G, Ojcius DM. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. Plos One. 2010;5:e10008. doi: 10.1371/journal.pone.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karki R, Man SM, Malireddi RK, Gurung P, Vogel P, Lamkanfi M, et al. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iannitti RG, Napolioni V, Oikonomou V, De Luca A, Galosi C, Pariano M, et al. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat Commun. 2016;7:10791. doi: 10.1038/ncomms10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo C, Chen M, Fa Z, Lu A, Fang W, Sun B, et al. Acapsular Cryptococcus neoformans activates the NLRP3 inflammasome. Microbes Infect. 2014;16:845–854. doi: 10.1016/j.micinf.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 80.Ketelut-Carneiro N, Silva GK, Rocha FA, Milanezi CM, Cavalcanti-Neto FF, Zamboni DS, et al. IL-18 triggered by the Nlrp3 inflammasome induces host innate resistance in a pulmonary model of fungal infection. J Immunol. 2015;194:4507–4517. doi: 10.4049/jimmunol.1402321. [DOI] [PubMed] [Google Scholar]
- 81.Tavares AH, Magalhães KG, Almeida RD, Correa R, Burgel PH, Bocca AL. NLRP3 inflammasome activation by Paracoccidioides brasiliensis. Plos Negl Trop Dis. 2013;7:e2595. doi: 10.1371/journal.pntd.0002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- 83.Bryan NB, Dorfleutner A, Kramer SJ, Yun C, Rojanasakul Y, Stehlik C. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J Inflamm (Lond) 2010;7:23. doi: 10.1186/1476-9255-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Hasegawa M, Imamura R, Kinoshita T, Kondo C, Konaka K, et al. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int Immunol. 2004;16:777–786. doi: 10.1093/intimm/dxh081. [DOI] [PubMed] [Google Scholar]
- 85.Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 86.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 87.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 89.He Y, Zeng MY, Yang D, Motro B, Núñez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]