Summary
The intestinal epithelium in the Drosophila midgut is maintained by intestinal stem cells (ISCs), which are capable of generating both enterocytes and enteroendocrine cells (EEs) via alternative cell fate specification. Activation of Delta-Notch signaling directs ISCs for enterocyte generation, but how EEs are generated from ISCs remains poorly understood. Here, we identified Phyllopod (Phyl) as a key regulator that drives EE generation from ISCs. Phyl, which is normally suppressed by Notch, functions as an adaptor protein that bridges Tramtrack 69 (Ttk69) and E3 ubiquitin ligase Sina for degradation. Degradation of Ttk69 allows the activation of the Achaete-Scute Complex (AS-C)-Pros regulatory axis, which promotes EE specification. Interestingly, expression of AS-C genes in turn further induces Phyl expression, thereby establishing a positive feedback loop for continuous EE fate specification and commitment. This positive feedback circuit-driven regulatory mechanism could represent a common strategy for reliable and irreversible cell fate determination from progenitor cells.
Keywords: Drosophila midgut, enteroendocrine cell, Notch, Ttk69, Phyl, Sina, Scute, cell specification, cell commitment
Graphical Abstract
Highlights
-
•
Phyl acts as a limiting factor for EE specification in Drosophila
-
•
Sina-Phyl degrades Ttk69 to promote EE specification
-
•
Sina-Phyl-Ttk69 and Scute form a positive feedback loop to drive EE commitment
-
•
Phyl is transcriptionally suppressed by Notch
In this article, Rongwen Xi and Chang Yin show that, in adult Drosophila midgut, the Sina-Phyl-Ttk69 complex and Scute form a positive feedback regulatory loop to drive cell fate commitment from enteroendorine cell progenitor cells. The findings provide important insights into faithful cell fate commitment from multipotent stem cells.
Introduction
A fundamental question in developmental biology is how cells acquire their fates. Specification of cell fate occurs during animal development, as well as in renewable adult tissues in which new cells are constantly generated by resident stem cells. Although transcription factors are commonly involved in determining cellular identities (Graf and Enver, 2009, Zernicka-Goetz et al., 2009), how their expression and activity are regulated to control progressive and reliable cell fate determination is in general poorly understood and requires detailed analysis in each individual developmental context.
Intestinal epithelium in Drosophila midgut provides a relatively simple and genetically tractable experimental system for studies of cell fate specification from stem cells (Biteau et al., 2011, Jiang and Edgar, 2011). Intestinal stem cells (ISCs) in Drosophila posterior midgut periodically produce committed progenitor cells termed enteroblasts (EBs) that differentiate further into either absorptive enterocytes (ECs) or secretory enteroendocrine cells (EEs) (Micchelli and Perrimon, 2006, Ohlstein and Spradling, 2006). The exit of ISC self-renewal and control of the binary fate decision of EBs is primarily controlled by Delta (Dl)-Notch signaling (Ohlstein and Spradling, 2007, Perdigoto et al., 2011). EBs with high Notch activation will adopt an EC fate, whereas EBs with low Notch activity will adopt an EE fate (Ohlstein and Spradling, 2007). Notch activation induces expression of the genes of the enhancer of split complex (E(spl)-C), which functions to promote ISC differentiation by antagonizing the bHLH transcription factor Daughterless (Bardin et al., 2010). A number of genes or pathways have been implicated in regulating EE specification, including the transcriptional repressor Tramtrack 69 (Ttk69) (Wang et al., 2015), the acheate-scute complex (AS-C) genes (Amcheslavsky et al., 2014, Bardin et al., 2010) that encode several basic-helix-loop-helix (bHLH) transcriptional factors, and the EE-determination transcription factor Prospero (Pros) (Biteau and Jasper, 2014, Wang et al., 2015, Zeng and Hou, 2015), among others (Beebe et al., 2010, Biteau and Jasper, 2014, Jiang et al., 2009, Kapuria et al., 2012, Lin et al., 2010, Quan et al., 2013). We previously presented evidence to support the essential role of a Ttk69-AS-C-Pros regulatory axis that controls EE specification from ISCs. AS-C gene expression is normally repressed by Ttk69, and a Ttk69-null mutation forced all progenitor cells to adopt an EE fate. While it is thus clear that the regulation of AS-C and its attendant activation of Pros by Ttk69 controls EE specification, it remains unclear how Ttk69 is itself regulated (Wang et al., 2015).
In addition to recent work showing the role of Ttk in EE specification in the midgut, decades of studies have demonstrated that Ttk regulates cell fate specification in the development of other organs such as the eye and external sensory organs (Badenhorst, 2001, Badenhorst et al., 2002, Giesen et al., 1997, Guo et al., 1995, Li et al., 1997, Okabe et al., 2001, Tang et al., 1997, Xiong and Montell, 1993). Post-translational modification of Ttk has been shown to promote R7 photoreceptor and sensory organ precursor (SOP) specification: the E3 ubiquitin ligase Seven in absentia (Sina) and an adaptor protein Phyllopod (Phyl) that ubiquitinate Ttk, which is subsequently degraded by the proteasome (Li et al., 1997, Li et al., 2002, Pi et al., 2001, Tang et al., 1997). Here, we investigated the function of sina and phyl in the adult Drosophila midgut, and this led us to reveal a positive feedback loop that drives EE commitment from ISCs.
Results
sina and phyl Are Both Required for EE Specification in the Adult Drosophila Midgut
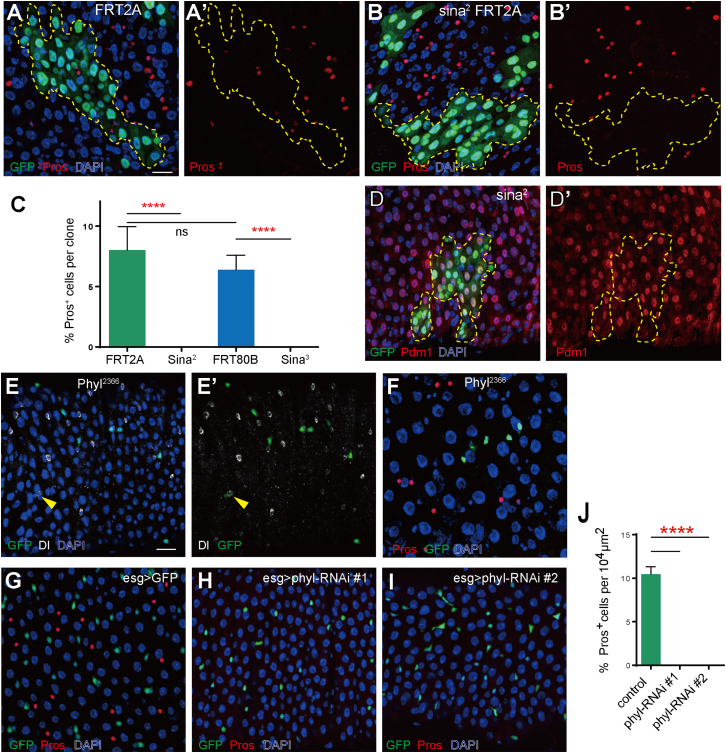
To determine whether sina has a role in the ISC lineages in the adult midgut, we used the MARCM system to generate sina homozygous mutant ISC clones in heterozygous animals by induced mitotic recombination, and then analyzed the cell composition of GFP-marked clones originated from ISCs 1–2 weeks after clone induction (ACI) (Lee and Luo, 1999, Lin et al., 2008, Wang et al., 2015, Xu et al., 2011). Normally, during progenitor cell differentiation, about 10%–20% of EBs adopt the EE fate; the rest of the EBs adopt the EC fate. As a consequence, EE cells only represent a small fraction of ISC progeny in the midgut epithelium (Biteau and Jasper, 2014, Ohlstein and Spradling, 2007). Quantitative analysis of wild-type ISC clones at day 10 ACI revealed that EEs, which can be specifically identified using Pros as a marker, constituted approximately 6%–8% of the total cell population within the clones. In contrast, virtually no Pros-expressing cells could be detected in the GFP-marked sina2 mutant clones (Figures 1A, 1B, and 1C). The sina2 mutant allele encodes in a truncated protein that lacks 105 amino acids of the C terminus of the Sina protein (Carthew and Rubin, 1990). GFP-marked clones of sina3, another loss-of-function allele of sina, exhibited an identical EE loss phenotype to that of sina2 mutant clones (Figures S1A and S1B). We also stained these sina2 mutant clones with Drosophila Tachykinin (dTK), a neuropeptide that is secreted by EEs. Virtually no dTK+ cells could be found in sina2 mutant clones (Figure S1C). It is noteworthy that the size (cell number) of the clones was largely comparable between wild-type and sina mutant ISC clones, indicating that loss of sina does not affect ISC proliferation. Staining with antibodies against Pdm1, an EC marker, revealed that ECs were properly differentiated in sina mutant clones (Figure 1D). Taken together, these observations suggest that sina is specifically required for EE specification from ISCs.
Figure 1.
sina and phyl Are both Required for EE Specification in the Adult Drosophila Midgut
Wild-type, sina, or phyl homozygous mutant MARCM clones (GFP, green) examined on day 10 after clone induction (ACI).
(A–B′) Clones co-stained with anti-Pros (red). (A and A′) A wild-type FRT2A clone. (B and B′) A sina2FRT2A clone. Note the absence of Pros+ cells in sina mutant clones (dashed lines and the separated red channels).
(C) The proportion of Pros+ cells per clone in wild-type and sina mutant clones on days 7–10 ACI. Mean ± SEM. n = 10 for FRT 2A clones, n = 24 for sina2 clones, n = 16 for FRT 80B clones, and n = 22 for sina3 clones. ∗∗∗∗p < 0.0001 (Student's t test).
(D and D′) A sina2 clone co-stained with anti-Pdm1 (dashed lines and the separated red channels).
(E and E′) phyl2366FRT42D mutant clones co-stained with anti-Dl (white). The yellow arrowhead indicates a phyl mutant cell positive for Dl expression.
(F) phyl2366FRT42D mutant clones co-stained with anti-Pros (red). There were no Pros+ cells within the mutant clones.
(G–I) Knockdown of phyl in esg+ cells causes the loss of Pros+ cells (red) in the midgut. (G) control midgut. (H and I) phyl-RNAi midgut. Crosses were made at room temperature and midguts were dissected 10 days after eclosion.
(J) The percentages of Pros+ cells in esg>GFP and esg>phyl-RNAi midguts. Mean ± SEM. n = 14 for wild-type control midgut, n = 12 for phyl-RNAi#1 midgut, n = 15 for phyl-RNAi#2 midgut. ∗∗∗∗p < 0.0001 (Student's t test).
Scale bars, 20 μm. See also Figures S1 and S2.
Previous studies in Drosophila eye and external sensory organs have demonstrated that Phyl is an essential adaptor protein that bridges Sina and Ttk to enable Ttk polyubiquitination and degradation. Direct interaction between Sina and Ttk is rather weak, but the presence of Phyl allows the formation of a strong Sina-Phyl-Ttk protein complex for subsequent protein modification and degradation, which is essential for proper photoreceptor differentiation (Li et al., 2002, Ou et al., 2003). To explore the function of phyl in EE fate specification, we generated GFP-marked MARCM clones homozygous for the phyl (phyl2366) mutant allele. Strikingly, at day 10 ACI, when the wild-type ISC clones had typically grow into patches of 10–30 cells comprising both ISCs and their derived progeny (polyploid ECs and diploid EEs) (Figure S2A), the phyl mutant ISC clones comprised only 1–3 cells (Figures 1E and 1F). Clones homozygous for the phyl2245 mutant allele, a genetic null, showed a similar growth defect (Figure S2C). The growth retardation of these mutant clones persisted over time; even at 3 weeks ACI, they still had only 1–3 cells (Figure S2B). Some cells of the mutant clones developed into large polyploid cells. Co-staining with the ISC marker Dl and the EE marker Pros revealed that some phyl mutant cells were positive for Dl (Figure 1E) and that none of the mutant clones (>500 clones examined) contained any Pros+ cells (Figure 1F). Staining with antibodies against phosphor-histone H3 (PH3) showed that none of the mutant clones (>300 clones examined) contained any mitotic cells, although PH3+ cells were present in wild-type clones and in the wild-type cells surrounding the mutant clones (Figures S2D and S2E). We also generated MARCM clones with phyl-RNAi expression, these clones were also devoid of EEs, and their clone sizes were also smaller than the wild-type clones, but this size phenotype was much milder than the phyl mutant clones, and many properly differentiated ECs were found in each clone (Figure S3). Because RNAi often reduces gene products but does not eliminate them, the presence of large EE-less clones suggests that the EE specification function of phyl is more sensitive to the gene dosage than the ISC proliferation function of phyl. Collectively, these results demonstrate that the loss of phyl causes decreased proliferation of ISCs and failed EE differentiation without affecting EC differentiation.
To further evaluate the role of phyl in regulating EE specification from ISCs, we used a GAL4-UAS binary expression system to knock down phyl expression (Brand and Perrimon, 1993). esg-GAL4 was used to drive the ISC- and EB-specific expression of RNAi targeting phyl in female esg-Gal4,UAS-GFP; UAS-phyl-RNAi (esg>phyl-RNAi) flies. While Pros+ EEs were scattered throughout wild-type intestinal epithelia (Figure 1G), the epithelia of esg>phyl-RNAi midguts were completely devoid of Pros+ cells (Figures 1H and 1I). There was no apparent difference in the esg>GFP+ cell populations between wild-type and esg>phyl-RNAi midguts (Figures 1G–1J). These results further support the notion that phyl is required for EE specification from ISCs. Therefore, similar to sina, phyl is indispensable for EE specification in the midgut epithelium. But unlike sina, phyl has additional roles in promoting ISC proliferation. It is noteworthy that although only posterior midgut regions were shown for the functional studies of sina and phyl, both genes are essential for EE generation along the length of the midgut, including anterior, middle, and posterior midgut regions.
Phyl, but Not Sina, Acts as a Limiting Factor for EE Cell Fate Specification
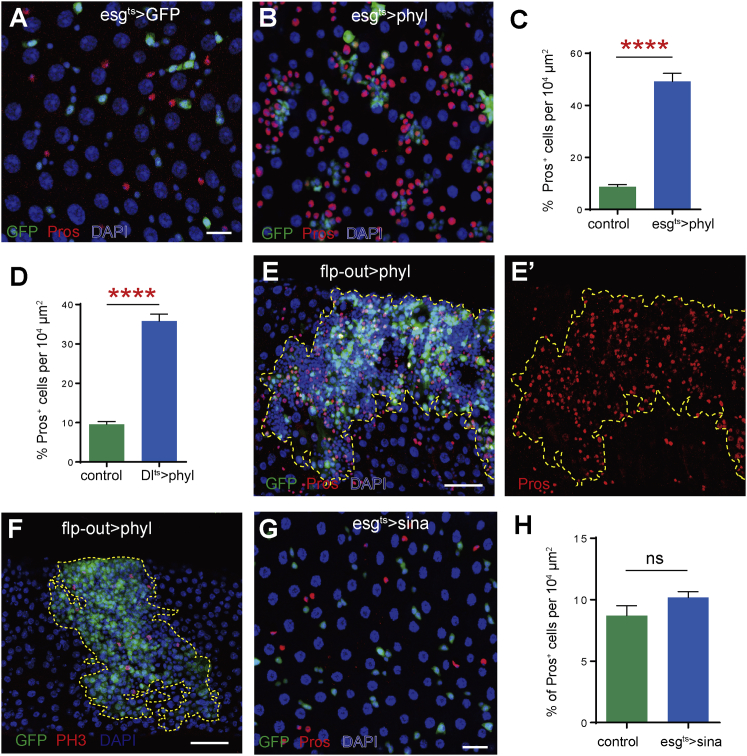
To evaluate whether phyl is sufficient to induce EE fate specification, we overexpressed phyl using the temperature-inducible GAL4-UAS expression system (McGuire et al., 2004). We found that conditional overexpression of phyl for 7 days in adult both ISCs and EBs using the esg-GAL4ts (esg-GAL4, Tub-GAL80ts) driver was sufficient to induce excessive production of EEs (Figures 2A–2C). Further, the continuous accumulation of EEs in the midgut epithelium eventually led to the development of multilayered EE-like tumors by around 2 weeks (not shown). Conditional overexpression of phyl using an ISC-specific driver, Dl-GAL4ts, also induces extra production of EEs, albeit to a less pronounced extent than in the esg-GAL4; UAS-phyl flies (Figure 2D). Interestingly, conditional overexpression of phyl in Notch-activated EBs using Su(H)Gbe-GAL4ts blocked EB differentiation, induced re-entry into mitosis, and caused some of these accumulated EBs to differentiate into EEs (Figure S4), although normally the Notch-activated EBs are post-mitotic and only differentiate into ECs. This suggests that Phyl expression is able to force EC-committed EBs to re-enter the cell cycle and to promote differentiation into EE fate instead. To determine whether this EE-specification-promoting effect occurs in a lineage-autonomous or a non-lineage-autonomous manner, we overexpressed phyl in a clonal fashion using a flp-out cassette technique (Struhl and Basler, 1993). We generated clones comprised exclusively of phyl-overexpressing cells (marked by GFP). Strikingly, there was a massive accumulation of Pros+ EE cells and Dl+ ISC-like cells within these clones, but the population and distribution of EE cells outside the clones remained normal (Figures 2E and S5). It is thus clear that the overexpression of phyl was sufficient to induce lineage-autonomous specification of EE cell fate. Consistent with our finding that phyl is required for ISC proliferation (Figures 1E and 1F), the overexpression of phyl also significantly increased ISC proliferation in a lineage-autonomous manner, as revealed by the significantly increased proportion of mitotic cells in these clones (Figure 2F). The ISC cell proliferation and EE cell specification effects of phyl together caused the rapid development of EE-like tumors in all the phyl overexpression clones (Figures 2E and 2F).
Figure 2.
Phyl, but Not Sina, Acts as a Limiting Factor for EE Cell Fate Specification
(A and B) Conditional overexpression of phyl in esg+ cells for 7 days resulted in excessive EEs (anti-Pros, red) in the midgut.
(C) The percentages of Pros+ cells in the epithelia of esgts>GFP and esgts>phyl midguts. Mean ± SEM. n = 15 for esgts>GFP midguts, and n = 16 for esgts>phyl midguts. ∗∗∗∗p < 0.0001 (Student's t test).
(D) Conditional overexpression of phyl in Dl+ cells for 7 days also produced excessive EEs. The percentage of Pros+ cells in midguts of indicated genotypes. Mean ± SEM. n = 23 for Dlts>GFP midguts, and n = 22 for Dlts>phyl midguts. ∗∗∗∗p < 0.0001 (Student's t test).
(E and E′) A flp-out clone (marked by GFP, green) with phyl overexpression co-stained with anti-Pros (red). Dashed lines depict the clone margin.
(F) The phyl overexpression flp-out clones (green) co-stained with anti-PH3 (red).
(G) Epithelium of esg-GAL4ts>sina midgut stained with anti-Pros (red).
(H) Quantitative data on the percentages of Pros+ cells in the epithelia esgts>GFP and esgts>sina midguts. n = 15 for wild-type control midguts, n = 23 for u-sina midguts.
Scale bars: (A [also applies to B] and G) 20 μm; (E and F) 50 μm. See also Figure S3.
Using a similar approach, we also conditionally overexpressed sina in ISCs and EBs. sina overexpression did not cause any obvious abnormalities in either the ISC or EE population (Figures 2G and 2H), suggesting that sina only plays a permissive role for EE specification, and phyl is not only permissive but also instructive for EE specification from ISCs. Therefore, phyl seems to act as a limiting factor for EE fate specification from ISCs.
Similar to the phenotypes resulting from phyl overexpression, the loss of ttk69 also causes excessive ISC proliferation and unidirectional generation of EEs and is also able to induce EC-committed EBs to re-enter mitosis and induce their differentiation into EE fate instead. Conversely, similar to phyl ablation, the overexpression of ttk69 causes ISC quiescence and failure of EE differentiation (Wang et al., 2015). These phenotypic similarities support the idea that phyl and ttk69 function in a common regulatory pathway to control EE specification.
Epistatic Relationships among Sina, Phyl, Ttk69 and AS-C complex in EE Specification
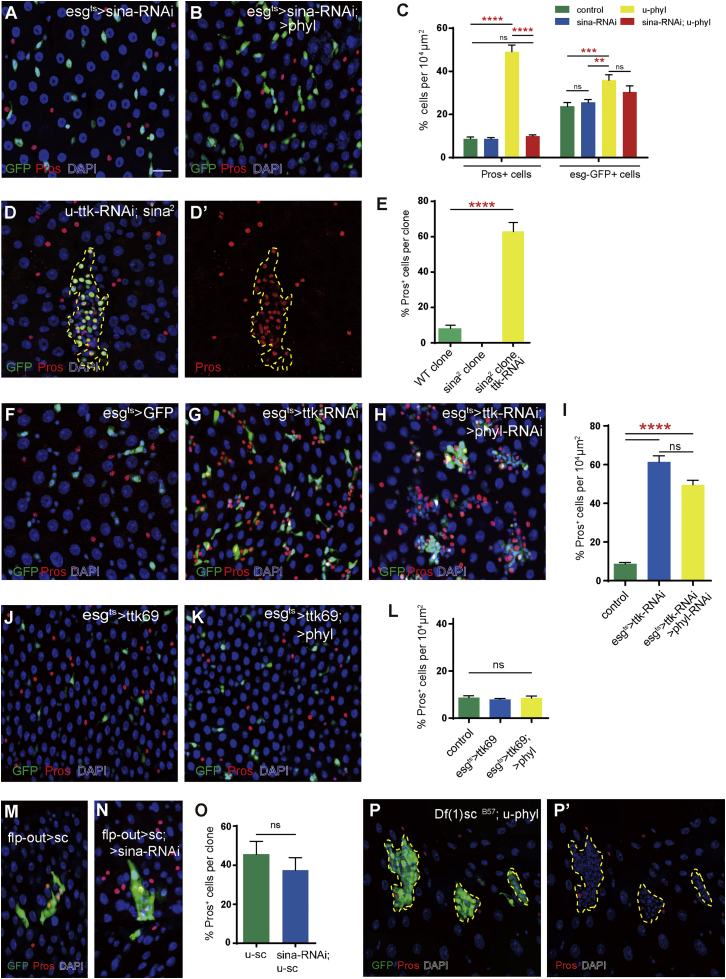
The similar requirement of Sina, Phyl, and Ttk69 for EE specification strongly suggests that the Sina-Phyl-Ttk69 protein complex, which is involved in eye and SOP development, may also be involved in the regulation of EE specification in the Drosophila midgut. If indeed Sina functions as an ubiquitin ligase and Phyl functions as an adaptor for recruitment, ubiquitination, and degradation of Ttk69, the excessive EE phenotype caused by phyl overexpression should be dependent on sina activity. Recall that we earlier showed that the conditional overexpression of phyl in adult esg+ cells for 7 days effectively induced the generation of excessive EEs in the epithelium (Figure 2B). However, this phenotype was completely suppressed when sina was depleted via RNAi (Figures 3A–3C). Therefore, the ability of phyl to induce EE specification is dependent on sina activity. As an extra note, we found that the slight increase in of esg>GFP+ cells following phyl overexpression was not effectively suppressed by sina-RNAi (Figures 3A–3C), indicating that the function of phyl in promoting ISC proliferation is likely independent of sina.
Figure 3.
Epistatic Relationships among Sina, Phyl, Ttk69 and AS-C complex in Regulating EE Specification
(A and B) Midgut epithelia of indicated genotypes stained with anti-Pros (red). Flies were shifted to the restrictive temperature for 7 days before analysis. GFP, green.
(C) The percentages of Pros+ and GFP+cells in the epithelia of indicated genotypes. Mean ± SEM. n = 15 for esgts>GFP control, n = 14 for esgts>sina-RNAi, n = 16 for esgts>phyl, and n = 12 for esgts>sina-RNAi; u-phyl midguts. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (Student's t test). ns, no significant difference.
(D and D′) ISC clones of indicated genotypes co-stained with anti-Pros (red). Note the accumulation of Pros+ cells in sina2ttk-RNAi clones (dashed lines and the separated red channels).
(E) Quantitative data on the percentages of Pros+ cells in the clones shown in (D and D′). Mean ± SEM. n = 10 for wild-type FRT2A clones, n = 24 for sina2 mutant clones, n = 12 for ttk-RNAi; sina2 mutant clones. ∗∗∗∗p < 0.0001 (Student's t test).
(F–H) Midgut epithelia of (F) esgts>GFP, (G) esgts>ttk-RNAi, and (H) esgts>ttk-RNAi; phyl-RNAi stained with anti-Pros (red). Flies were shifted to restrictive temperature for 7 days before analysis.
(I) Quantitative data on the percentages of Pros+ cells in the epithelia shown in (G–H). Mean ± SEM. n = 15 for esgts>GFP, n = 16 for esgts>ttk-RNAi, and n = 10 for esgts>ttk-RNAi; phyl-RNAi midguts. ∗∗∗∗p < 0.0001 (Student's t test).
(J and K) Midgut epithelia of (J) esgts>UAS-ttk69 and (K) esgts>UAS-ttk69; UAS-phyl stained with anti-Pros (red). Flies were shifted to restrictive temperature for 7 days before analysis.
(L) Quantitative data on the percentages of Pros+ cells in the epithelia shown in (K and L). Mean ± SEM. n = 15 for esgts>GFP controls, n = 19 for esgts>UAS-ttk69, and n = 15 for esgts>UAS-ttk69; UAS-phyl midguts. ns, no significant difference.
(M) Overexpression of sc by using the flp-out system (green), and the clone was stained with anti-Pros (red).
(N) Overexpression of sc in sina-RNAi flp-out clones (green) were stained with anti-Pros (red).
(O) The proportion of Pros+ cells per clone in clones of indicated genotypes on day 7 ACI. Mean ± SEM. n = 7 for sc overexpression clones, and n=7 for sc overexpression, sina-RNAi clones. ns, no significant difference.
(P and P′) Overexpression of phyl in Df(1)scB57 mutant clones failed to induce excessive EE cells (anti-Pros, in red). Dashed lines depict the clone margin.
Scale bar: 20 μm.
We next tested the epistatic relationships between ttk69 and sina/phyl. If Sina/Phyl functions to degrade Ttk69, Ttk69 should be downstream of Sina/Phyl in regulating EE specification (therefore genetically epistatic to sina/phyl). Recall that sina2 mutant clones failed to generate any EEs (Figure 1B). However, the co-depletion of ttk in sina2 mutant clones caused EE tumor formation in a lineage-autonomous manner (Figures 3D and 3E), a phenotype virtually identical to that resulting from ttk depletion alone (Wang et al., 2015). Therefore, ttk69 is epistatic to sina in regulating EE specification. Similarly, we found that ttk is also epistatic to phyl. Depleting phyl via RNAi could not suppress the excessive EE phenotype caused by conditional depletion of ttk in esg+ cells (Figures 3F–3I), but overexpression of ttk69 was sufficient to prevent excessive EE phenotype caused by conditional overexpression of phyl (Figures 3J–3L). These results demonstrate that ttk69 is genetically downstream of sina/phyl in EE specification. The epistatic relationships among sina, phyl, and ttk69 indicate that, similar to what occurs during Drosophila eye and SOP development, Sina/Phyl likely function to promote EE generation through the proteolytic degradation of Ttk69.
We next tested the epistatic relationships between AS-C genes and sina/phyl. Although sina mutant clones are devoid of EEs, we found that overexpression of sc in sina-RNAi clones was still able to induce EE specification within the clones (Figures 3M–3O), supporting the notion that sc is epistatic to sina. Df(1)scB57 is a small chromosomal deficiency allele in which the entire AS-C genes are removed (Heitzler et al., 1996). Overexpression of phyl in induced Df(1)scB57 clones failed to induce the supernumerary EE phenotype (Figure 3P). These clones are composed of small progenitor cells that do not have Pros expression (Figure 3P′). The occasional Pros+ cells found in the clones are likely due to residual activity of gene products. Collectively, these data demonstrate that AS-C genes are genetically downstream of sina/phyl and ttk69, and ttk69 is genetically downstream of sina/phyl in EE specification. Therefore, a Sina-Phyl-Ttk69-AS-C regulatory axis controls EE specification in the Drosophila midgut.
Sina and Phyl Regulate Ttk69 Protein Stability
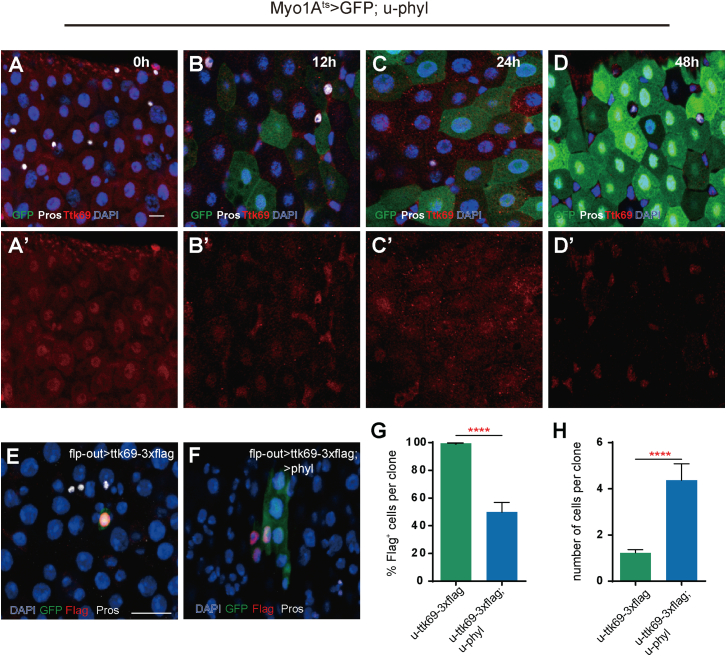
To test whether Sina/Phyl are able to regulate Ttk69 protein levels in the midgut, we used a highly specific antibody against Ttk69 (Wang et al., 2015) to monitor Ttk69 protein levels in the midgut following the manipulation of sina/phyl function. In wild-type midgut, Ttk69 was generally expressed in all epithelial cells, with the highest levels in ECs and the lowest levels in ISCs and EEs (Figure 4A) (Wang et al., 2015). Myo1A-Gal4ts (Myo1A-Gal4;Tub-GAL80ts), an EC-specific driver was used to conditionally ectopically express phyl in ECs. The impact of phyl expression on Ttk69 protein levels was analyzed 12, 24, and 48 hr after including phyl expression (Figures 4B–4D). Interestingly, downregulation of Ttk69 protein levels in ECs was observed as early as 12 hr (Figure 4B). Hardly any Ttk69 protein could be detected in ECs by 48 hr (Figures 4C and 4D). Therefore, the ectopic expression of phyl in ECs is able to rapidly downregulate Ttk69 protein levels in ECs. We also expressed a 3xFlag-tagged ttk69 transgene in flp-out clones and examined the effect of phyl expression on Ttk69-Flag protein levels. Because ttk69 overexpression inhibits ISC proliferation, Ttk69-Flag-overexpression clones hardly grew at all; many remained as single or double cell clones, and staining with anti-Flag antibody showed strong Ttk69-Flag protein expression in all of these clones (Figures 4E–4H). The addition of Phyl overexpression in Ttk69-Flag-overexpression clones caused clones to grow slightly bigger (Figures 4E–4H), indicating that the proliferation-inhibitory effect of Ttk69 is partially suppressed by Phyl overexpression. Importantly, the Ttk69-Flag protein level was significantly downregulated in the Phyl/Ttk69-Flag co-overexpression clones, and no Ttk69-Flag protein could be detected in the majority of the cells of these clones (Figures 4F and 4G). Collectively, these results demonstrate that Phyl overexpression is sufficient to downregulate the Ttk69 protein level in a lineage-autonomous manner. This conclusion is consistent with the idea that Sina and Phyl function together to promote EE specification by inhibiting Ttk69 through proteolytic degradation.
Figure 4.
Sina and Phyl Regulate Ttk69 Protein Stability
(A–D) phyl was conditionally expressed in EC cells driven by Myo1Ats and stained with anti-Ttk69 (red). (A and A′) The midgut from Myo1A-GAL4ts; u-GFP control flies. (B–D′) The midguts from Myo1A-GAL4ts; u-GFP; u-phyl flies treated at restrictive temperature for indicated length of time. Note that the Ttk69 protein level in small cells remained unaltered in all images, and the seemingly increased level in shown in (B–D) is because of increased signal contrast.
(E and F) flp-out GFP clones (green) with u-ttk69-3xflag overexpression (E) or with u-ttk69-3xflag and u-phyl co-overexpression (F) stained with anti-Flag antibody (red). Anti-Pros, white.
(G and H) The proportion of Flag+ cells (G) and the number of total cells per clone (H) in clones of indicated genotypes on 7 day ACI. Mean ± SEM. n = 35 for ttk69-3xflag overexpression clones, and n=22 for ttk69-3xflag and phyl both overexpression clones. ∗∗∗∗p < 0.0001 (Student's t test).
Scale bars: (A) 10 μm (also applies to B–D); (E) 20 μm (also applies to F).
Phyl Is Transiently Upregulated in EE Progenitor Cells and Is Positively Regulated by Sc
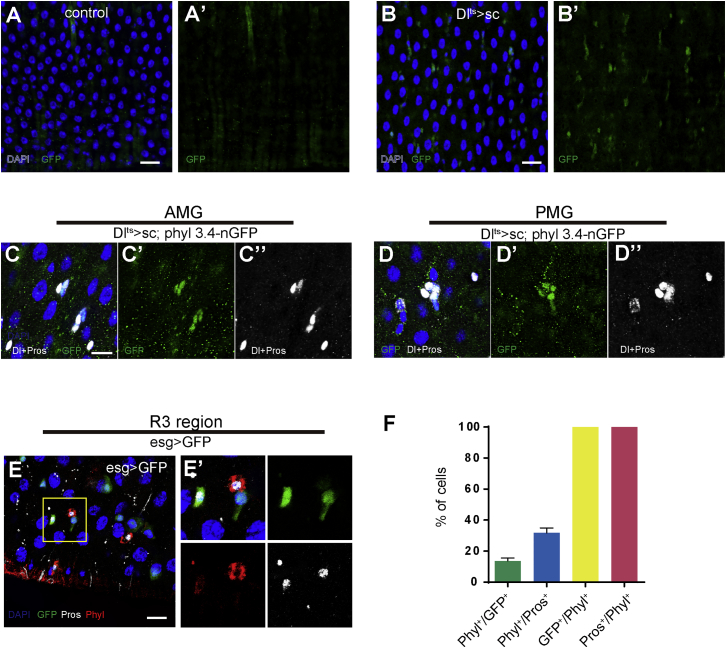
Given that phyl appears to be a limiting factor in EE specification, characterizing how phyl is regulated should deepen our understanding of the EE specification process. We therefore examined phyl expression using phyl3.4-GFP, an in vivo GFP reporter driven by a 3.4 kb phyl promoter fragment (Pi et al., 2004). Interestingly, the expression of phyl3.4-GFP was barely detectable in the midgut epithelium (Figure 5A). We also generated a polyclonal antibody against Phyl, and the anti-Phyl signal was also largely undetectable in the intestinal epithelium along the length of midgut, except that it was detected in the cytoplasm of some diploid cells in the copper cell (R3) region where gastric stem cells reside (Buchon et al., 2013, Strand and Micchelli, 2011, Wang et al., 2014). These Phyl+ diploid cells were co-stained with low levels of Pros and esg>GFP, indicating that they could be differentiating EE progenitor cells (Figures 5E and 5F). Regional differences in Phyl expression levels could be due to differential Dl-Notch signaling activities, as Notch activity is known to be relatively low in the R3 region (Marianes and Spradling, 2013, Strand and Micchelli, 2011, Wang et al., 2014), and Phyl expression is negatively regulated by Notch (described later).
Figure 5.
Phyl Is Transiently Upregulated in EE Progenitor Cells and Is Positively Regulated by Sc
(A–D′) Midguts of indicated genotypes stained with anti-GFP (green). (A) Flies of phyl3.4-nGFP. (B) Flies of Dl-GAL4ts;u-sc, phyl3.4-nGFP. Flies were shifted to restrictive temperature for 48 hr before analysis. Note that non-specific staining occurred on muscle fibers. Midguts of Dl-GAL4ts;u-sc, phyl3.4-nGFP stained with Dl and Pros antibody (white) in both the anterior (C) and the posterior (D) midgut regions.
(E) Flies of esg>GFP stained with anti-Pros (white) and anti-Phyl (red) in R3 region. Note that a high expression level of Phyl protein was observed in low-level GFP and Pros+ cells.
(F) Quantification of the percentage of indicated cells in R3 region of the midguts of esg>GFP flies. n = 4–6 midguts.
Scale bars: (A, B, and E) 20 μm; (C and D) 10 μm. See also Figure S4.
Despite failure to detect Phyl expression using both the GFP marker and the antibody in the anterior and posterior midgut, the functional requirement of phyl for EE specification along the length of the midgut suggests that phyl should be expressed there, but its expression level could be too low to be detected/reflected by antibody staining or by the transcriptional reporter. To test this hypothesis, we used a highly sensitive Tyramide Signal Amplification (TSA) method to amplify the reporter GFP signal and studied the phyl3.4-GFP expression pattern. This analysis revealed that the majority of diploid cells, including ISCs and EEs, at both the anterior and posterior regions of the midgut, had active phyl transcription (Figure S6). It thus seems clear that Phyl is generally expressed in ISCs at low levels in both anterior and posterior midgut regions. This baseline Phyl activity is likely important for ISC proliferation, as it was shown earlier that the proliferative ability of ISCs was strongly affected following the ablation of phyl.
We next examined whether phyl could be a transcriptional target of sc in the midgut, as phyl is known as a transcriptional target of Sc in external sensory organs (Pi et al., 2004). This is potentially interesting because sc is an essential AS-C gene known to be required for EE specification that functions downstream of ttk69 (Amcheslavsky et al., 2014, Bardin et al., 2010, Wang et al., 2015). As shown above, phyl expression was barely detectable in normal intestinal epithelium by either anti-Phyl staining or the phyl3.4-GFP reporter. However, the conditional overexpression of sc in ISCs for 48 hr using Dl-Gal4 rapidly induced phyl expression in progenitor cells, revealed by either anti-Phyl staining or the GFP reporter (Figure 5B compared with 5A, and data not shown). Co-staining with Pros and Dl markers revealed that, in both anterior and posterior midgut, the highest Phyl expression occurred in Dllow and Proslow cells, which are likely differentiating EEs, and its expression became diminished in Proshigh cells (Figures 5C and 5D). These observations suggest that Sc is able to transcriptionally activate phyl in the midgut progenitor cells. Taking this into consideration, the initial activation of Phyl could reinforce its expression via a positive feedback mechanism mediated by Sina-Phyl-Ttk69 and Sc, which may ensure rapid accumulation of the EE-determination factor Pros for EE specification and maturation. This positive feedback regulation of Phyl during EE differentiation is also consistent with the notion that Phyl seems to be accumulated at its highest levels in differentiating EEs.
Phyl Is Negatively Regulated by Notch
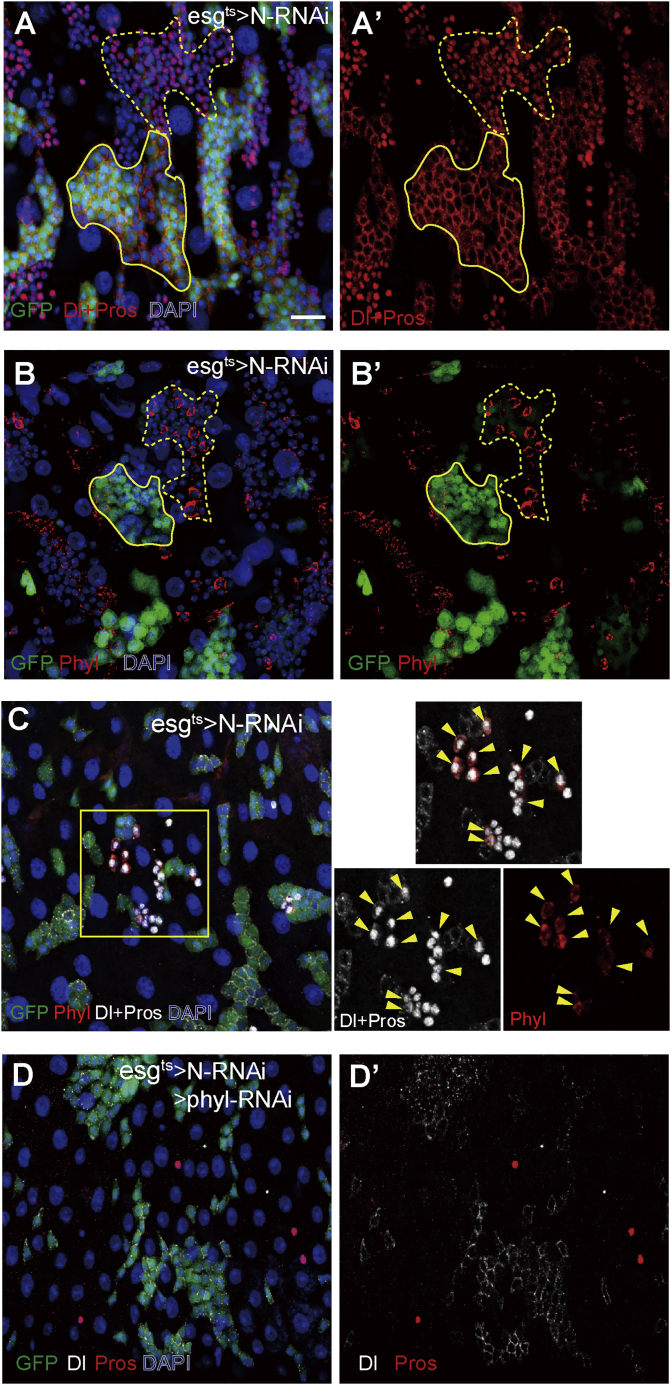
The binary fate choice of ISCs is primarily regulated by Dl-Notch signaling. As reported previously, loss of Notch in ISCs causes ISC-like and EE-like tumors (Figure 6A). Forced activation of Notch in ISCs, on the other hand, unidirectionally induces ISC differentiation into ECs (Micchelli and Perrimon, 2006, Ohlstein and Spradling, 2006, Ohlstein and Spradling, 2007), indicating that Notch is inactive or at low levels during EE generation. To test the potential regulatory relationships between Notch and Phyl, we examined Phyl expression in esg>Notch-RNAi epithelia. Although Phyl expression was hardly detectable in the anterior and posterior midgut, its expression was readily detectable in a subset of Pros+ tumor cells in esg>Notch-RNAi intestine (Figure 6B). Co-staining with Dl and Pros markers revealed that Phyl+ cells were mainly Pros+ and Dl+/low cells (Figure 6C and arrowheads in insets). Because differentiation into EE is accompanied by downregulation of Dl expression and initiation of Pros expression, these Pros+ and Dl+ cells are likely early EEs that are still in the process of differentiation toward maturation. It thus further reinforces the notion that Phyl is transiently upregulated in differentiating EEs. This transient upregulation of Phyl may lead to activation of the positive feedback loop for rapid accumulation of Pros in these cells and consequently EE commitment. To functionally test whether phyl expression is required for EE-like tumor cell generation, we co-depleted Notch and phyl in ISCs and studied the consequences. As a result, co-depletion of phyl caused the failure of EE-like tumor formation, although ISC-like tumors could still be formed (Figure 6D). The size of these phyl-RNAi N-RNAi ISC-like tumors appeared smaller than N-RNAi ISC-like tumors, but the difference was not statistically significant (Figure S7). It is thus possible that the cell proliferation activity of phyl could be dependent on Notch. We conclude that Phyl is normally suppressed by Notch, and loss of Notch causes derepression of Phyl, which drives continuous EE generation, leading to EE-like tumor formation. It is noteworthy that many Notch mutant cells, including all Dl+ ISC-like cells, do not show any detectable Phyl expression, suggesting that additional mechanisms are involved to regulate Phyl expression at the early stages of fate decision in ISCs.
Figure 6.
Phyl Is Negatively Regulated by Notch
(A and A′) Midguts of esgts>N-RNAi stained with anti-Dl and anti-Pros in red. Flies were shifted to restrictive temperature for 14 days before analysis. Note that ISC-like tumors (high GFP, solid line) and EE-like tumors (low GFP, dashed line) were induced.
(B and B′) Flies of esgts>N-RNAi stained with anti-Phyl. Note that a high expression level of cytoplasmic Phyl protein (red) was observed in a portion of cells within the EE-like tumor (low GFP, dashed line) but not in the ISC-like cells (high GFP, solid line).
(C) Flies of esgts>N- RNAi stained with anti-Phyl (red) and Dl, Pros (white). Note the Phyl+ cells were mainly Pros+ and Dl+/low cells (yellow arrowheads).
(D and D′) Flies of esgts>N-RNAi; phyl- RNAi stained with anti-Dl (white) and anti-Pros (red). Note that EE-like tumors disappeared when Notch and phyl were co-depleted.
Scale bar: 20 μm.
Discussion
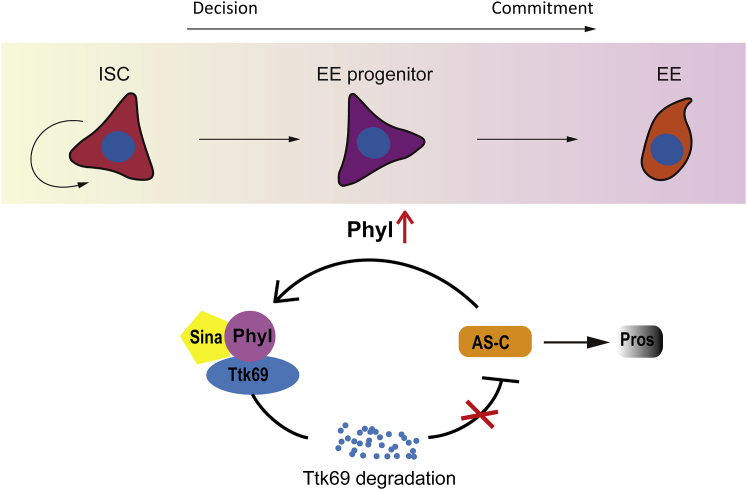
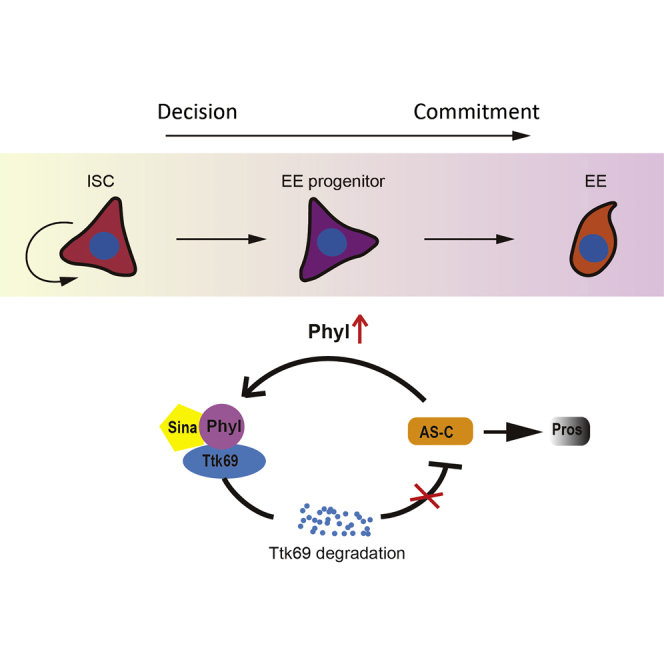
Our results collectively suggest a regulatory circuit in differentiating progenitors that drive EE fate commitment. Normally Phyl is suppressed by Notch. In EE progenitor cells that are Notchlow or inactive, Phyl is transiently upregulated, which acts as an adaptor protein to bring Ttk69 to the E3 ubiquitin ligase Sina for proteolytic degradation. The expression of the Ttk69 target gene sc is subsequently derepressed, which then induces expression of the EE fate determination factor Pros. Notably, expression of sc also induces phyl expression, thereby forming a positive feedback circuit that drives Ttk69 degradation and Pros accumulation, ultimately specifying EE fate (Figure 7).
Figure 7.
A Schematic Model for EE Specification Driven by the Positive Feedback Regulatory Circuit Composed of Sina/Phyl, Ttk69, and Sc
In EE progenitor cells derived from ISCs, Phyl is upregulated, which links EE-repressor Ttk69 to the E3 ubiquitin ligase Sina for degradation. Ttk69 degradation results in the derepression of AS-C genes, which subsequently induces the expression of EE-determination factor Pros to promote EE specification. The expression of Sc also induces Phyl expression, thereby forming a positive feedback regulatory circuit that continuously drives Phyl expression, Ttk69 degradation, and Pros accumulation, ultimately leading to EE commitment.
Regulatory circuits with feedback mechanisms are frequently used by multi-celled organisms to control the proportional generation of differentiated cell subtypes and for the homeostatic control of tissue maintenance and regeneration (Hsu and Fuchs, 2012, Tata and Rajagopal, 2016). In the Drosophila midgut, epithelial damage induces feedback regulation between ISCs and their progeny that promotes activation of ISCs for epithelial repair (Chen et al., 2016, Jiang et al., 2009). It has also been shown that differentiated EEs send feedback signals such as Slit molecules to ISCs to prevent excessive EE generation (Biteau and Jasper, 2014), but a recent study does not support the existence of such a feedback mechanism (Salle et al., 2017). Here, we identify a positive feedback circuit that functions in the process of EE specification. The engagement of this type of positive feedback circuit could conceivably serve several purposes. First, such a circuit could allow the rapid accumulation of fate regulators like Pros to a critical threshold for cell fate commitment, perhaps functioning to overcome the influence of proteolytic degradation of the fate regulators. Second, once induced, fate regulators may need to be present continuously to specify cell fate, and a positive feedback mechanism could ensure such an ongoing expression. Third, activation of this type of positive feedback circuit may also effectively prevent unwanted cell fate reversion. It is known that many committed progenitor cells are still able to change (revert) their fate under certain circumstances, as with the enteroendocrine progenitor cells in the mouse small intestine that can act as a reservoir for ISCs in response to ISC loss. The enterocyte-committed EBs in the fly ISC lineage also have the potential to revert their EE fate specification, such as following ttk69 depletion (Wang et al., 2015), or phyl overexpression reported in this study. In this context, the engagement of a positive feedback circuit at the start of EE specification could be an effective strategy to continuously ensure faithful commitment to the EE fate, especially under normal conditions.
How is the binary fate decision of ISCs, which directs the proportional generation of EC and EE from ISCs, regulated? A recent study from our group demonstrates that a transient activation of Sc in ISCs directs the generation of EEs from self-renewing ISCs (Chen et al., 2017). Although the current study primarily focuses on the role of Phyl in a positive feedback loop in differentiating EEs, Phyl could have an earlier function in ISCs to regulate Sc expression, and consequently cell proliferation and cell fate decisions. In addition, Phyl overexpression is able to induce many ISC-like cells, similar to that caused by the loss of Notch. A negative role for phyl in Notch signaling has been observed in developing eye imaginal discs, in which phyl is required for endocytic degradation of activated Notch (Nagaraj and Banerjee, 2009). Considering that Notch negatively regulates both sc and phyl transcription in the midgut, it is possible that the antagonistic activities of Phyl and Notch could participate in regulation of Sc and consequently cell fate decisions in ISCs.
Post-transcriptional regulation of Ttk by Sina and Phyl is known to determine neural cell fate versus non-neural cell fate in eye and sensory organ development, highlighting that Ttk-based mechanisms are used frequently in multiple developmental processes to regulate alternative cell fate decisions in Drosophila. Sina is an evolutionarily conserved E3 ubiquitin ligase. Similarly, BTB domain-containing proteins like Ttk are present in all eukaryotes (Perez-Torrado et al., 2006). Although mammals appear to have no Phyl homologs, this does not exclude the possibility that they may have functional counterpart(s). The loss of a mammalian sina gene, siah2, suppresses the neuroendocrine tumor phenotype in a mouse model of prostate cancer (Qi et al., 2010). It is therefore possible that protein complexes similar to Sina-Phyl-Ttk69 maybe function in diverse mammalian tissues, including for example in the epithelium inner lining of the digestive tract, to regulate cell fate decisions.
In short, this study identified a regulatory circuit composed of Sina-Phyl-Ttk69 and Sc that drives EE commitment from ISCs. The earliest event that initiates EE differentiation from ISCs, that is, the event that causes Notch inactivation and initial Phyl expression, is still unclear, but it is possibly linked to Numb-mediated symmetric cell division and other environmental cues (Salle et al., 2017). We propose that the engagement of a positive feedback circuit to drive cell fate specification, as revealed in the present study, may be a common mechanism employed to ensure faithful cell fate determination from progenitor cells in diverse organisms, including mammals.
Experimental Procedures
Fly Strains
The following fly stocks were used in this study: sina2 FRT2A (BDSC, #30724); sina3 FRT80B (BDSC, #26270); FRT42D phyl2366 (BDSC, #30723); phyl2245 (Kyoto Stock Center, #108363); FRT2A, FRT80B, FRT42D, Tub-Gal80ts, UAS-ttk69, Myo1A-Gal4, UAS-Notch-RNAi, and Act<stop<Gal4 (all obtained from BDSC); esg-Gal4 and UAS-GFP, (gift from Shigeo Hayashi, RIKEN Center for Development Biology, Japan); UAS-phyl-RNAi#1 (BDSC, #29433); UAS-phyl-RNAi#2 (VDRC, v35469); UAS-phyl (BDSC, #52015); UAS-sina.myc (BDSC, #30931); UAS-sina-RNAi (VDRC, v100691); UAS-sc (BDSC, #26687); phyl3.4-nGFP (a gift from Haiwei Pi, Department of Life Science, Chang-Gung University, Taiwan; Pi et al., 2004); Su(H)-Gal4 and Dl-Gal4 (a gift from Xiankun Zeng and Steven Hou, National Cancer Institute, USA; Zeng et al., 2010); UAS-Nicd (a gift from Ting Xie, Stowers Institute for Medical Research, USA); UAS-sc-3HA (Fly ORF, F000085) (Bischof et al., 2013). UAS-ttk69-3xflag was generated in the course of the present study. Briefly, the cDNA of ttk69 was cloned into the gateway attB-PUAST-3xflag vector (Drosophila Genomics Resource Center), and the plasmid was injected into embryos of attP40 flies.
Mosaic Analysis
GFP-marked clones in Drosophila midgut epithelium cells were generated using the MARCM system (Lee and Luo, 1999) and the flp-out technique (Struhl and Basler, 1993), as previously described (Lin et al., 2008, Lin et al., 2010, Xu et al., 2011). Female flies (between 3 and 5 days old) of a given genotype were exposed to heat-shock treatment for 1 hr at 37°C in a water bath. After the heat-shock treatment, flies were cultured at 25°C with regular food and were analyzed 4–14 days later.
Temperature Shift Assay
Flies carrying esgts (esg-Gal4,Tubulin-Gal80ts), Dlts, or Su(H)ts were crossed with appropriately matched transgenic flies contain UAS-transgenes at 18°C (Wang et al., 2015). Female flies (between 3 and 5 days old) of a given genotype were shifted from 18°C to 29°C and cultured with regular food that was refreshed every 2 days. Dissection and analysis were performed 7 days after the initial temperature shift or at other time points, as specified in the text.
Generation of Phyl Antisera
Polyclonal antibody directly against Phyl was generated in rabbit by using the synthetic peptide: TPAPIVYSKRRASRRSASVSC. The cysteine residue at the C terminus of the peptide was able to conjugate keyhole limpet hemocyanin. Serum obtained from immunized rabbit was purified by antigen affinity chromatography. Purified antiserum was used at a final dilution of 1:300.
Immunostaining
Immunostaining of Drosophila midgut was performed as previously described (Lin et al., 2008). The following primary antibodies were used in this study: mouse anti-Dl (DSHB, C594.9B; 1:100); mouse anti-Pros (DSHB, MR1A; 1:100); rabbit anti-Tachykinin (a gift from Dick Nassel, Stockholm University, Sweden; 1:3,000); rabbit anti-Pdm1 (a gift from Xiaohang Yang, Zhejiang University, China; 1:1,000); mouse anti-phospho-Histone H3 (Cell Signaling Technology, #9706, 1:500); rabbit anti-GFP (Molecular Probes, A11122, 1:200); anti-Ttk69 (Wang et al., 2015); mouse anti-FLAG (Sigma, F1084; 1:300). Secondary antibodies were used in this study: goat anti-rabbit or anti-mouse IgGs conjugated to Alexa Fluor 488, 568, or Cy5 (Molecular Probes, A11034-A11036, A10524; 1:300). Signal amplification experiments were performed using a TSA kit (Invitrogen, TSA kit #22). Images were captured using a Nikon A1-R confocal microscope. All images were edited in Adobe Photoshop and were assembled in Adobe Illustrator.
Author Contributions
C.Y. and R.X. conceived and designed the experiments, analyzed the data, and wrote the manuscript. C.Y. performed the experiments.
Acknowledgments
We thank the members of the fly community, as cited in Experimental Procedures, for generously providing fly strains and antibodies; the Bloomington Drosophila Stock Center (BDSC), the Kyoto Stock Center, the Vienna Drosophila RNAi Center (VDRC), and Development Studies Hybridoma Bank (DSHB) for reagents; Xingting Guo for UAS-ttk69-Flag cloning, An-Ying Kang for microinjections, John Snyder for editing the manuscript, and the lab members for comments and discussions. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0103602) and the National Basic Research Program of China (2014CB850002) from the Chinese Ministry of Science and Technology.
Published: December 21, 2017
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.014.
Supplemental Information
References
- Amcheslavsky A., Song W., Li Q., Nie Y., Bragatto I., Ferrandon D., Perrimon N., Ip Y.T. Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Rep. 2014;9:32–39. doi: 10.1016/j.celrep.2014.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development. 2001;128:4093–4101. doi: 10.1242/dev.128.20.4093. [DOI] [PubMed] [Google Scholar]
- Badenhorst P., Finch J.T., Travers A.A. Tramtrack co-operates to prevent inappropriate neural development in Drosophila. Mech. Dev. 2002;117:87–101. doi: 10.1016/s0925-4773(02)00183-1. [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Perdigoto C.N., Southall T.D., Brand A.H., Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K., Lee W.C., Micchelli C.A. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J., Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development. 2013;140:2434–2442. doi: 10.1242/dev.088757. [DOI] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C.E., Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Jasper H. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7:1867–1875. doi: 10.1016/j.celrep.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Buchon N., Osman D., David F.P., Fang H.Y., Boquete J.P., Deplancke B., Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Rubin G.M. Seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- Chen J., Xu N., Huang H., Cai T., Xi R. A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. Elife. 2016;5:e14330. doi: 10.7554/eLife.14330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu N., Wang C., Huang P., Huang H., Jin Z., Yu Z., Cai T., Jiao R., Xi R. Transient Scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat. Cell Biol. 2017 doi: 10.1038/s41556-017-0020-0. [DOI] [PubMed] [Google Scholar]
- Giesen K., Hummel T., Stollewerk A., Harrison S., Travers A., Klambt C. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development. 1997;124:2307–2316. doi: 10.1242/dev.124.12.2307. [DOI] [PubMed] [Google Scholar]
- Graf T., Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Guo M., Bier E., Jan L.Y., Jan Y.N. Tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron. 1995;14:913–925. doi: 10.1016/0896-6273(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Bourouis M., Ruel L., Carteret C., Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hsu Y.C., Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Edgar B.A. Intestinal stem cells in the adult Drosophila midgut. Exp. Cell Res. 2011;317:2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria S., Karpac J., Biteau B., Hwangbo D., Jasper H. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet. 2012;8:e1003045. doi: 10.1371/journal.pgen.1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li S., Li Y., Carthew R.W., Lai Z.C. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Li S., Xu C., Carthew R.W. Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack. Mol. Cell. Biol. 2002;22:6854–6865. doi: 10.1128/MCB.22.19.6854-6865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Xu N., Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- Lin G., Xu N., Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of Drosophila intestinal stem cells. J. Mol. Cell Biol. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- Marianes A., Spradling A.C. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Nagaraj R., Banerjee U. Regulation of Notch and Wingless signalling by phyllopod, a transcriptional target of the EGFR pathway. EMBO J. 2009;28:337–346. doi: 10.1038/emboj.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Okabe M., Imai T., Kurusu M., Hiromi Y., Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- Ou C.Y., Pi H., Chien C.T. Control of protein degradation by E3 ubiquitin ligases in Drosophila eye development. Trends Genet. 2003;19:382–389. doi: 10.1016/S0168-9525(03)00146-X. [DOI] [PubMed] [Google Scholar]
- Perdigoto C.N., Schweisguth F., Bardin A.J. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585–4595. doi: 10.1242/dev.065292. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R., Yamada D., Defossez P.A. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Pi H., Huang S.K., Tang C.Y., Sun Y.H., Chien C.T. Phyllopod is a target gene of proneural proteins in Drosophila external sensory organ development. Proc. Natl. Acad. Sci. USA. 2004;101:8378–8383. doi: 10.1073/pnas.0306010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Wu H.J., Chien C.T. A dual function of phyllopod in Drosophila external sensory organ development: cell fate specification of sensory organ precursor and its progeny. Development. 2001;128:2699–2710. doi: 10.1242/dev.128.14.2699. [DOI] [PubMed] [Google Scholar]
- Qi J., Nakayama K., Cardiff R.D., Borowsky A.D., Kaul K., Williams R., Krajewski S., Mercola D., Carpenter P.M., Bowtell D. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Z., Sun P., Lin G., Xi R. TSC1/2 regulates intestinal stem cell maintenance and lineage differentiation through Rheb-TORC1-S6K but independently of nutritional status or Notch regulation. J. Cell Sci. 2013;126:3884–3892. doi: 10.1242/jcs.125294. [DOI] [PubMed] [Google Scholar]
- Salle J., Gervais L., Boumard B., Stefanutti M., Siudeja K., Bardin A.J. Intrinsic regulation of enteroendocrine fate by Numb. EMBO J. 2017;36:1928–1945. doi: 10.15252/embj.201695622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M., Micchelli C.A. Quiescent gastric stem cells maintain the adult Drosophila stomach. Proc. Natl. Acad. Sci. USA. 2011;108:17696–17701. doi: 10.1073/pnas.1109794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Tang A.H., Neufeld T.P., Kwan E., Rubin G.M. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Tata P.R., Rajagopal J. Regulatory circuits and bi-directional signaling between stem cells and their progeny. Cell Stem Cell. 2016;19:686–689. doi: 10.1016/j.stem.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Guo X., Dou K., Chen H., Xi R. Ttk69 acts as a master repressor of enteroendocrine cell specification in Drosophila intestinal stem cell lineages. Development. 2015;142:3321–3331. doi: 10.1242/dev.123208. [DOI] [PubMed] [Google Scholar]
- Wang C., Guo X., Xi R. EGFR and Notch signaling respectively regulate proliferative activity and multiple cell lineage differentiation of Drosophila gastric stem cells. Cell Res. 2014;24:610–627. doi: 10.1038/cr.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W.C., Montell C. Tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- Xu N., Wang S.Q., Tan D., Gao Y., Lin G., Xi R. EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev. Biol. 2011;354:31–43. doi: 10.1016/j.ydbio.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Zeng X., Chauhan C., Hou S.X. Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis. 2010;48:607–611. doi: 10.1002/dvg.20661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Hou S.X. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142:644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Morris S.A., Bruce A.W. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat. Rev. Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.