Summary
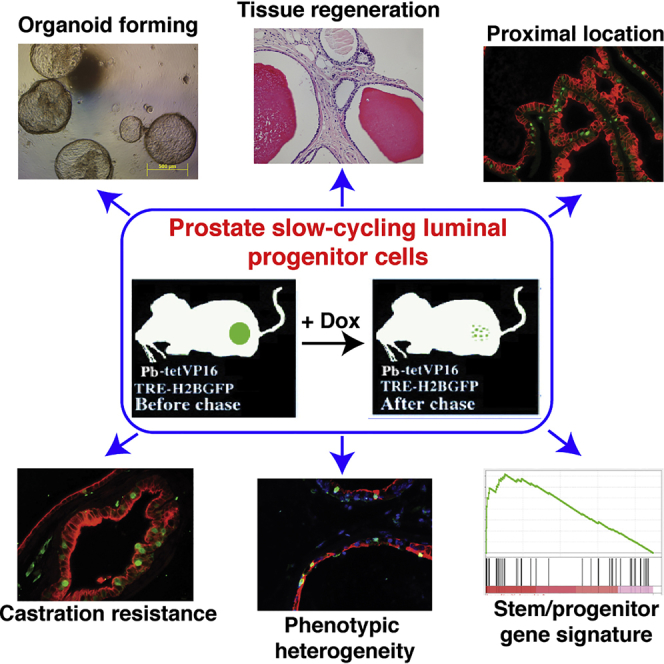
The existence of slow-cycling luminal cells in the prostate has been suggested, but their identity and functional properties remain unknown. Using a bigenic mouse model to earmark, isolate, and characterize the quiescent stem-like cells, we identify a label-retaining cell (LRC) population in the luminal cell layer as luminal progenitors. Molecular and biological characterizations show that these luminal LRCs are significantly enriched in the mouse proximal prostate, exhibit relative dormancy, display bipotency in both in vitro and in vivo assays, and express a stem/progenitor gene signature with resemblance to aggressive prostate cancer. Importantly, these LRCs, compared with bulk luminal cells, maintain a lower level of androgen receptor (AR) expression and are less androgen dependent and also castration resistant in vivo. Finally, analysis of phenotypic markers reveals heterogeneity within the luminal progenitor cell pool. Our study establishes luminal LRCs as progenitors that may serve as a cellular origin for castration-resistant prostate cancer.
Keywords: prostate stem cells, luminal progenitors, label-retaining cells, quiescence, prostate cancer, prostate, differentiation, cancer stem cells
Graphical Abstract
Highlights
-
•
A bigenic mouse model to study prostatic slow-cycling luminal epithelial cells
-
•
Prostate label-retaining cells (LRCs) exhibit stem/progenitor cell activities
-
•
Luminal LRCs are developmentally bipotent and display a progenitor gene signature
-
•
Luminal LRCs resist castration and molecularly resemble aggressive prostate cancer
In this article, Tang and colleagues, by using an unbiased bigenic mouse model to specifically earmark, prospectively isolate, and functionally characterize the quiescent stem-like cells, report a label-retaining cell (LRC) population in the mouse prostate luminal cell layer that possesses many stem/progenitor cell activities. These luminal LRCs are developmentally bipotent and intrinsically castration resistant.
Introduction
The prostatic gland contains basal and luminal epithelial cells, together with rare neuroendocrine cells. Luminal cells express cytokeratin (CK) 18 and androgen receptor (AR) and are androgen dependent, whereas basal cells express CK5 and stem cell (SC) transcription factor p63 and are androgen independent (Shen and Abate-Shen, 2010). Developmentally, the murine prostate originates from an ancestral p63+AR− basal SC population (Pignon et al., 2013). Studies using prostate regeneration assays demonstrate that multipotent SCs capable of differentiating into all prostatic cell types are localized in the basal layer (Kwon and Xin, 2014). Therefore, basal cells are thought to represent the main pool of prostate stem cells (PSCs) (Wang et al., 2013). However, lineage-tracing studies indicate that basal cells rarely generate luminal cells during adult tissue homeostasis but display plasticity under the inductive influence of embryonic urogenital mesenchyme (UGM) cells in tissue regeneration assays, acquiring facultative stem/progenitor cell properties and generating luminal cells (Choi et al., 2012, Wang et al., 2013). Consequently, both cell layers in adult murine prostate are self-sustained by lineage-restricted stem/progenitor cells (Choi et al., 2012), although more primitive SCs reside in the basal layer (Ousset et al., 2012). An in vitro organoid assay recently identified a small fraction (<1%) of luminal cells functionally defined as multipotent luminal progenitors in that they were able to generate organoids containing both basal and luminal cells (Karthaus et al., 2014). Beyond homeostasis, several rare luminal progenitor populations have been reported in regressed mouse prostates, including castration-resistant NKX3.1-expressing (CARN) (Wang et al., 2009), SCA-1+ (Kwon et al., 2016), and castration-resistant BMI1-expressing (CARB) (Yoo et al., 2016) cells. The precise relationship between these luminal progenitor cell populations remains unclear.
The prostate has been a model for studying tissue SCs, because it undergoes atrophy upon castration and regeneration upon re-administration of androgen, and this regression-regeneration cycle can be repeated multiple times. Somatic SCs are generally dormant and this cardinal slow-cycling feature is frequently utilized to identify putative SCs by labels that become diluted as a result of cell division (Tang, 2012). Studies have shown that label-retaining cells (LRCs) in many organs are enriched for SCs (dos Santos et al., 2013, Foudi et al., 2009, Szotek et al., 2008, Tsujimura et al., 2002, Tumbar et al., 2004, Wang et al., 2012). Previously, 5-bromodeoxyuridine (BrdU) was employed to perform pulse-chase experiments to identify candidate SCs. In the prostate, a long-term chased BrdU+ cell population, encompassing both basal and luminal cells, which resides in the proximal region of mouse prostatic ducts and exhibits attributes of epithelial SCs was proposed as PSCs (Tsujimura et al., 2002). Whether these dormant cells truly represent SCs has not been answered mainly due to the technical infeasibility of purifying out live BrdU+ cells for functional studies. More recently, cell surface markers coupled with fluorescence-activated cell sorting (FACS) have been used to dissect the subsets of cells within a bulk population. These assays depend on known SC markers, and, notably, the majority of widely used markers (e.g., SCA-1, CD49f) preferentially identify prostate basal stem-like cells (Lawson et al., 2007, Lukacs et al., 2010a, Stoyanova et al., 2012, Xin et al., 2005), leaving the luminal cell compartment under-studied. Lineage-tracing technology has greatly enhanced our understanding of SC development; however, lineage-tracing studies only suggest that a certain cell population harbors SCs, but could not pinpoint which precise cell(s) within the population is SC (Rycaj and Tang, 2015).
In this study, we employed a bigenic mouse model to identify, isolate, and characterize the stem-like properties and gene expression profiles of quiescent LRCs from mouse prostates expressing a tunable H2B-GFP driven by the promoter of a luminal lineage-preferential gene probasin (Suraneni et al., 2010). Biological and molecular studies show that long-term chased luminal LRCs are inherently resistant to castration and can generate organoids in vitro and prostatic glands in vivo. Notably, the LRC-derived organoids and prostatic glands contain both basal and luminal cells, suggesting the bipotency of LRCs. RNA sequencing (RNA-seq) analysis of this rare population reveals a progenitor-like transcriptome that resembles therapy-resistant prostate cancer (PCa). Analysis of a spectrum of phenotypic markers previously linked to epithelial cell stemness reveals heterogeneity in luminal progenitor populations. Collectively, our study augments our understanding of prostate luminal progenitors and provides valuable insights into their potential roles in prostate development and, possibly, cancer initiation and progression.
Results
Establishing a Bigenic Mouse Model to Label Slow-Cycling Cells in Prostatic Epithelium
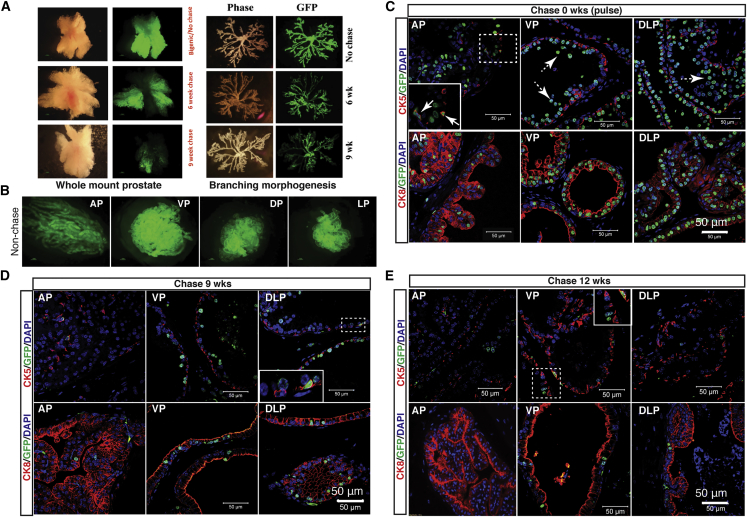
To study the dormant cell population in the luminal cell compartment, we adopted a Tet-Off system similar to that described previously (Tumbar et al., 2004) by engineering a Pb-tetVP16 transgenic line to express tetracycline repressor-VP16 controlled by probasin (Pbsn)-based ARR2Pb promoter (Zhang et al., 2000; Figure S1A). By crossing the Pb-tetVP16 mice with the tetracycline-responsive element-regulated mCMV/H2B-GFP reporter mice (Tumbar et al., 2004), we generated the bigenic mice, Pb-tetVP16-GFP, in which GFP expression is ultimately driven by Pbsn promoter (Figures S1A and S1B). In this way, without doxycycline (DOX) administration (pulse), the prostate tissues would be largely GFP+. Upon DOX administration (chase), the prostate will gradually lose the GFP signal due to cell division, while infrequently cycling and dormant cells would retain GFP for an extended period of time (Figure S1A). Indeed, the whole prostate or microdissected prostate branches from the unchased young adult (6 weeks) animals were green, and GFP intensity dropped accordingly at different intervals of chase (Figure 1A). These data demonstrate the successful establishment of a bigenic mouse model to fluorescently label slow-cycling cells in the prostatic epithelium.
Figure 1.
Identification of H2B-GFP LRCs
(A) Loss of GFP signals in DOX-chased prostates. Shown are gross GFP images in whole-mount prostates (left) and microdissected prostate branches (right) isolated from bigenic mice chased for 0 weeks (no chase), 6 weeks, and 9 weeks.
(B) Gross GFP images in different lobes of prostates dissected from unchased adult Pb-tetVP16-GFP bigenic mice.
(C–E) Double IF of CK5 or CK8 and GFP in different prostate lobes harvested from bigenic mice chased (on DOX diet) for 0 weeks (C), 9 weeks (D), and 12 weeks (E). Arrows and dashed arrows in (C) (top) indicate CK5+GFP+ basal cells and luminal cells shed into the lumen, respectively. AP, VP, DP, and LP refer to anterior, ventral, dorsal, and lateral prostate lobes, respectively. Dashed boxed regions are enlarged (solid boxes). Scale bars, 50 μm.
See also Figure S1.
H2B-GFP Primarily Labels Prostate Luminal Cells
We assessed the expression and distribution of GFP+ cells in the adult prostate glands. Consistent with the reported activity of endogenous Pbsn promoter (Zhang et al., 2000), GFP signal was higher in ventral prostate (VP) and dorsal and lateral prostate (DLP) than that in anterior prostate (AP) (Figure 1B). As a marker of differentiation and androgen action in the mouse prostate, Pbsn is primarily expressed in the luminal epithelial cells. Double immunofluorescence (IF) staining of CK5 or CK8 and GFP indicated that, as expected, GFP+ cells were mainly localized to the luminal compartment in unchased prostates (Figure 1C). Consistent with the varying gross GFP intensity in different prostate lobes (Figure 1B), the frequency of GFP+ cells was much higher in VP (79.4%) and DLP (79.4%) than that in AP (43.8%) (Figures 1C and S1C; Table S1). We also observed some GFP+CK5+ double-positive cells (Figure 1C, solid arrows) and non-epithelial GFP+ cells in the stromal compartment (Figure S1C), suggesting that the Pbsn promoter is also active in a small subset of basal and stromal cells, as reported previously (Valdez et al., 2012, Wang et al., 2006). Interestingly, during the course of this work and consistent with a prior study (Wang et al., 2014a), we frequently noticed many GFP+ luminal cells in the lumen (Figure 1C, dashed arrows), implying that luminal cells have a faster turnover than basal cells (see below).
Kinetics of H2B-GFP LRCs in Basal and Luminal Cell Layers
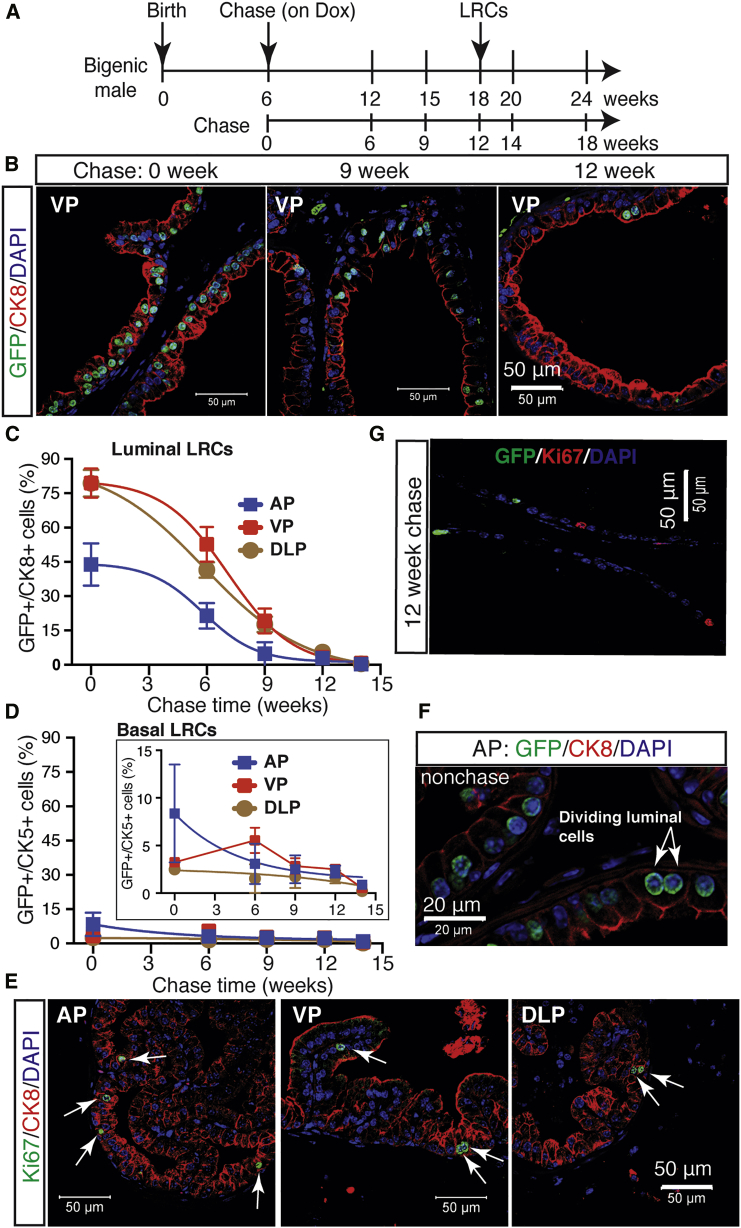
To characterize optimal chase time and kinetic changes in GFP+ cells, we quantified the percentage of GFP+ cells at different time points after chase according to their lineage identity (Figures 1C–1E and 2A). We started chasing the male mice at 6–8 weeks when the prostate is generally well developed. The initial luminal cell-labeling efficacy was 79.4%, 79.4%, and 43.8% for VP, DLP, and AP, respectively (Table S1). As expected, upon chase, prostatic cells gradually lost GFP signal (Figures 2B and S2A). By 9-week chase, the percentage of GFP+ luminal cells dropped from ∼80% to <20% in VP, and by 14-week chase and after, GFP+ cells were rarely seen (Figures 2C and S2A). After a 12-week chase, the luminal GFP+ cells remained at 2%–6% in different lobes (Figure 2C). Consequently, we referred to the GFP+ cells that persisted for at least a 12-week chase as LRCs. This time point is in line with studies in other organs using similar H2B-GFP mouse models (dos Santos et al., 2013, Szotek et al., 2008, Wang et al., 2012). Our model also labeled a small subset of CK5+ basal cells, with 2%–8% GFP+ cells in unchased lobes and 1%–3% GFP+ cells after a 12-week chase (Figure 2D; Table S1). Interestingly, compared with the relatively flat curve of basal GFP+ cell kinetics, luminal GFP+ cells decreased much faster as a function of chase (Figures 2C and 2D). IF analysis revealed Ki-67+ cells exclusively in luminal layer in all lobes (Figure 2E). Furthermore, we frequently and exclusively observed symmetrically dividing GFP+ cells in the luminal layer (Figure 2F). These observations validated faster proliferation and loss of GFP signal in luminal cells compared with basal cells. Quantitatively, unchased young adult bigenic mice displayed a small percentage of prostatic GFP+ cells that co-expressed the proliferation marker Ki-67 (4.03%, 0.42%, and 1.80% for AP, VP, and DLP, respectively); in contrast, after a 12-week chase no GFP+ cells were stained positive for Ki-67 (Figures 2G and S2B), confirming the quiescence of LRCs.
Figure 2.
Dynamics and Characterization of LRCs
(A) Scheme for tracking the dynamics of prostatic GFP+ cells in hormonally intact mice, in which we started chasing the bigenic animals at 6 weeks of age and analyzed the prostate tissues at different time points post chase.
(B) IF images of CK8 and GFP in the VP isolated from bigenic mice chased for different time intervals.
(C and D) Quantification of GFP+ cells in luminal (C) and basal (D) cell populations as a function of chase time. At each time point, two to four mice were examined, and data are shown as the mean ± SE.
(E) IF staining of Ki67 and CK8 in different prostate lobes in wild-type adult mice. Arrows indicate Ki67+CK8+ cells.
(F) IF of GFP and CK8 in the AP of unchased adult bigenic mice showing symmetrical division of luminal cells.
(G) IF staining of Ki67 and GFP in the VP of bigenic mice chased for 12 weeks.
Luminal LRCs Are Enriched in the Proximal Prostatic Ducts and Mark Progenitors Capable of Regenerating Prostate Glands In Vivo
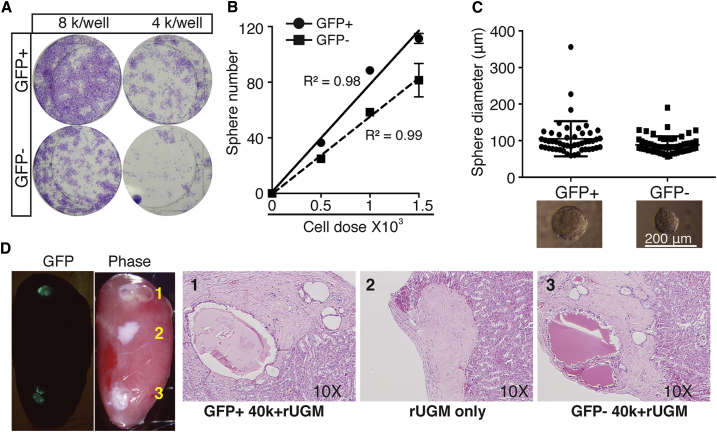
We next performed in vitro and in vivo SC-related assays, first in bulk GFP+ LRCs. The lineage-negative (Lin−) GFP+ cells freshly purified from the prostates chased for 12 weeks (Figure S3A) displayed higher capacity to form 2D colonies and 3D spheres with larger size (Figures 3A–3C) than matched GFP− cells. In vivo tissue regeneration assays demonstrated that both GFP+ and GFP− cells could readily regenerate prostate glandular structures with secretions in the lumen (Figure 3D). The origin of the recombinants was verified by GFP signal and staining (Figures 3D and S3B). It is worth noting that, after a 12-week chase, the LRCs were composed of both luminal and basal cells (ratio = 2:1; Table S1). As basal cells represent the main PSC pool, it is expected that GFP− population, which contained the majority of both luminal and basal cells, would also efficiently regenerate prostate tissues in vivo. Regardless, our data indicate that the long-term chased LRC population harbors stem/progenitor cells with tissue regeneration ability in vivo.
Figure 3.
The LRC Population Harbors Tissue Regeneration Ability In Vivo
Freshly purified lineage-depleted (Lin−) prostatic LRCs (i.e., GFP+ cells from 12-week-chased bigenic mice) exhibit higher stem/progenitor activities than matched non-LRCs (i.e., GFP− cells). Shown are colony formation (A), limiting-dilution sphere (B), sphere size measurement (C), and in vivo prostate tissue regeneration (D) assays. In (D), 1, 2, and 3 indicate the three representative recombinants. Results shown for each panel were representative data of at least two to three independent experiments showing consistent results. For (B) and (C), data are shown as the mean ± SD derived from technical replicates. See also Figure S3.
To “zoom in” on the biological differences between luminal and basal LRCs, we purified GFP+ cells from the two cell compartments using marker staining and FACS (Valdez et al., 2012) to identify basal (SCA-1+CD49fhi), luminal (SCA-1−CD49flo), and stromal (SCA-1+CD49f−/lo) cells (Figure S4A). Since the Pbsn promoter is active in only a small subset of basal cells (Valdez et al., 2012, Wang et al., 2006), we considered our Pb-tetVP16-GFP bigenic model non-optimal to characterize basal SCs. In support, basal LRCs exhibited limited advantages over basal non-LRCs in SC-related assays, exhibiting slightly increased 2D clonal capacity and nearly equivalent sphere-forming cell frequency (Figures S4B and S4C), although GFP+ basal cells did form larger 3D spheres than GFP− basal cells (Figure S4D). Overall, these results could be explained by low labeling efficiency in the basal cell layer and high SC activity of stochastic basal cells. Therefore, for the remainder of this study we focused on luminal LRCs.
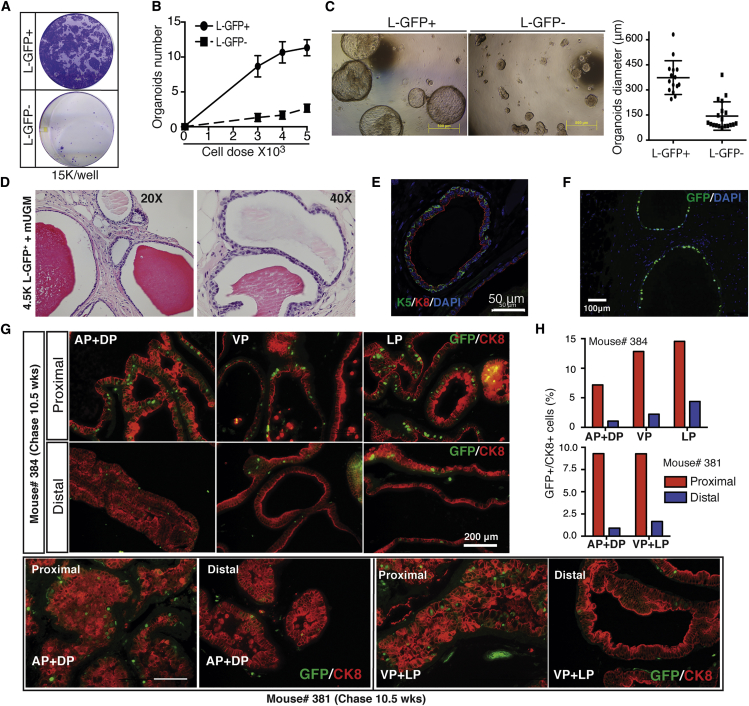
Since our initial luminal cell-labeling efficacy was ∼80% for VP and DLP, we first tested whether luminal progenitor cell activity was equally distributed between the labeled and unlabeled (∼20%) luminal populations in unchased adult animals. Both 2D clonal (Figure S4E) and 3D organoid (Figure S4F) assays revealed similar activities in unchased luminal (L)-GFP+ and L-GFP− cells, validating the suitability of our model in comparing stem/progenitor cell activities in chased L-GFP+ and L-GFP− cells. Freshly purified luminal LRCs from 12-week-chased prostates exhibited significantly higher clonal and organoid-forming (Figures 4A and 4B) capacities than matched luminal GFP− cells. Importantly, luminal GFP+ cells formed large hollow organoids, whereas only a limited number of luminal GFP− cells formed small compact spheres (Figure 4C). Importantly, luminal LRC-derived organoids had a glandular structure reminiscent of normal prostate, with p63+ and CK5+ basal cells mainly residing in the peripheral layer (Figure S4G). This result is in line with recent findings that only multipotent luminal progenitors can form organoids in vitro (Karthaus et al., 2014). When we mixed L-GFP+ or L-GFP− cells (4.5 × 103) with mouse UGM and implanted subcutaneously into NOD-SCID-IL2Rγnull (NSG) mice, only L-GFP+ cells generated prostatic glandular structures with overt secretions in the lumen (Figure 4D), whereas even higher numbers of luminal GFP− cells (up to 18 × 103) failed to give rise to prostatic tissue (not shown). These LRC-regenerated glands were normal-appearing with a layer of CK8+ luminal cells lining the lumen and a discontinuous layer of CK5+ basal cells encapsulating the structure (Figure 4E). The identity of injected cells and the LRC origin of recombinants were verified by GFP staining (Figure 4F), and the level of GFP was still detectable after several rounds of cell proliferation during prostate regeneration (not shown). These results together indicate that luminal LRCs mark luminal progenitors with bilineage differentiation capacity in vitro and in vivo.
Figure 4.
Luminal LRCs Mark Progenitors Capable of Regenerating Prostate Glands In Vivo
(A–C) Freshly purified Lin− luminal GFP+ LRCs exhibit stem/progenitor activities. Shown are higher colony- (A) and organoid- (B) forming capabilities and larger sphere sizes (C) in L-GFP+ compared with L-GFP− cells. For (B) and (C), data are shown as the mean ± SD derived from technical replicates.
(D–F) Only luminal GFP+, but not GFP− cells, are capable of regenerating prostate tissues in vivo. Shown are H&E staining (D) and IF of CK5 and CK8 (E), and GFP (F) in prostate glands regenerated in vivo from sorted luminal GFP+ cells co-injected with mouse UGM.
(G and H) Enrichment of luminal LRCs in the proximal mouse prostate. Different prostate lobes dissected from bigenic mice at 10.5 weeks of DOX chase were divided longitudinally into two portions (distal and proximal) and subjected to IF of GFP and CK8. Shown are representative images (G) and quantification data (H) from two different mice. Results shown (A)–(F) are representative data of at least two to three independent experiments showing consistent results. For (G) and (H), two bigenic mice were analyzed and data represent the means from cell number counting of five to eight random high-magnification (×20) images of each indicated category.
Previous studies using a BrdU label-retention strategy suggested that putative PSCs (including both basal and luminal cells) are slow cycling and enriched in the proximal region of the prostate (Tsujimura et al., 2002). We sought to examine whether our luminal LRCs are also enriched in the proximal prostate. Each prostate lobe from mice chased for 10.5 weeks was dissected longitudinally into two portions, representing the distal and proximal prostatic ducts. IF analysis of GFP and CK8 indicated that the GFP+CK8+ luminal LRCs were significantly enriched in the proximal prostatic ducts adjacent to the urethra (Figures 4G and 4H).
Interestingly, when we examined the mRNA and protein level of PBSN in the pelvic urogenital sinus (UGS) of newborn and 1-week-old male mice, we observed that the Pbsn mRNA and PBSN protein were minimally expressed in newborn (d1) UGS but readily detectable in 1-week-old UGS (Figures S4H–S4J), suggesting that Pbsn is expressed early on in urogenital tissues and Pbsn promoter activity is not necessarily active only in mature fully differentiated luminal cells. Indeed, we detected GFP+ cells co-expressing CK8 and relatively high levels of PBSN protein in pelvic UGS of 1-week-old bigenic mice (Figure S4K). These results indicate that our LRC labeling system is also suitable in marking luminal progenitor cells at early developmental stages.
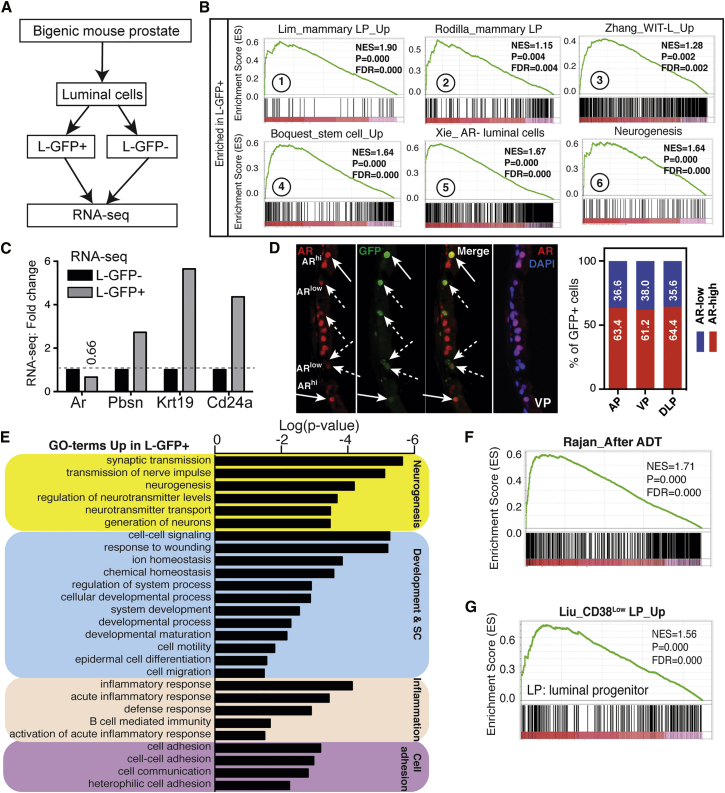
Luminal LRCs Display a Luminal Progenitor Gene Expression Signature
We next sought to determine the molecular features of L-GFP+ progenitors prepared from bigenic mice chased for 12 weeks. By performing genome-wide RNA-seq analysis using rRNA-depleted total RNAs (Figure 5A), we obtained an average of ∼77 million reads per sample with an average mapping rate of 93% to the reference mouse genome (UCSC version mm10; Figure S5A). MA plot indicated appropriate normalization of our RNA-seq data (Figure S5B). Gene set enrichment analysis (GSEA) of RNA-seq transcriptome revealed exclusive enrichment in L-GFP+ (over L-GFP−) cells of gene signatures related to luminal epithelial progenitors and SCs (Figure 5B, plots 1–4). Significantly, a recently reported signature of an AR-deleted luminal cell population with a stem-like phenotype in the mouse prostate (Xie et al., 2017) was significantly enriched in our luminal LRCs (Figure 5B, plot 5). Furthermore, we recently developed an in vitro 2D culture system that allows enrichment of luminal progenitors from primary human prostate tissues (Zhang et al., 2017). The gene signature of this cultured-enriched human luminal progenitors (WIT-L) was also enriched in luminal LRCs (Figure 5B, plot 3). In contrast, and, as expected, signatures associated with non-progenitor cells (luminal progenitor_down), AR signaling and steroid/androgen hormone metabolism were only enriched in L-GFP− cells (Figure S5C). It has been suggested that luminal progenitors are less dependent on AR signaling (Agarwal et al., 2015, Kwon et al., 2014). Consistent with an enrichment of AR/androgen signaling in L-GFP− cells (Figure S5C) and with AR− luminal cell signature (Xie et al., 2017) in L-GFP+ cells (Figure 5B, plot 5), Ar mRNA expression was decreased in L-GFP+ cells (Figure 5C). IF analysis of GFP and AR using titrated anti-AR antibody dilutions revealed significant heterogeneity of AR expression within luminal cell population and indicated that ∼37% ± 1.6% of GFP+ luminal cells expressed low AR protein (Figures 5D and S5D). Interestingly, CK19, a marker for luminal transit amplifying cells (Hudson et al., 2001, Korsten et al., 2009, Wang et al., 2001), and CD24a, a cell surface marker frequently used to enrich mammary luminal progenitors (dos Santos et al., 2013, Rodilla et al., 2015), were dramatically increased in L-GFP+ cells (Figure 5C). Intriguingly, we observed higher levels of Pbsn in L-GFP+ versus L-GFP− cells (Figure 5C), perhaps indicative of the luminal nature of LRCs and also due to LRC identification by virtue of Pbsn promoter activities. Real-time qPCR analysis in two independently sorted biological samples confirmed slightly reduced Ar mRNA levels in L-GFP+ cells and also validated the differentially expressed patterns of about a dozen genes in L-GFP+ versus L-GFP− cells (Figure S5E; data not shown). Intriguingly, L-GFP+ cells, compared with L-GFP− cells, expressed higher levels of luminal markers (Nkx3.1, Krt8, and Krt18) but lower levels of basal genes (Trp63 and Krt5) (Figure S5E).
Figure 5.
Luminal LRCs Display a Luminal Progenitor Gene Signature Associated with CRPC
(A) Scheme of RNA-seq experiments using freshly purified L-GFP+ and L-GFP− cells from bigenic mice chased for 12 weeks.
(B) Representative GSEA results in luminal GFP+ cells.
(C) Differential expression of the indicated genes in RNA-seq in L-GFP+ versus L-GFP− cells.
(D) IF of GFP and AR in 12-week-chased animals. Shown are representative images (left) and quantification data (right; a total of 41, 71, and 73 GFP+ cells for AP, VP, and DLP, respectively, were counted from four to six random high-magnification (×20) images of each lobe). In (D), solid arrows point to GFP+/ARhi cells and dashed arrows to GFP+/ARlow cells.
(E) Functional annotation by DAVID of genes preferentially upregulated in luminal GFP+ cells.
(F and G) GSEA results for the enrichment of indicated gene signatures in luminal GFP+ cells.
To further dissect the transcriptomic profiles in L-GFP+ cells (Table S2), we performed the pathway/network enrichment analysis by Ingenuity Pathway Analysis and DAVID gene ontology annotation. The two approaches generated similar results and revealed neurogenesis, SC and development, inflammation/immunity, and cell adhesion being the top gene categories overrepresented in L-GFP+ cells (Figures 5E and S5F). Interestingly, we have recently shown that normal human prostate basal/SCs (Zhang et al., 2016b) and PSA−/lo prostate cancer SCs (Qin et al., 2012) also preferentially express many neurogenesis-related genes. Further GSEA corroborated that, compared with L-GFP− cells, the slow-cycling L-GFP+ cells were highly enriched in gene networks involved in neural/neuronal development and functions (Figures 5B, plot 6, and S5G). Surprisingly, we observed no obvious enrichment of cell-cycle-related pathways in L-GFP+ cells, and a survey of key cell-cycle regulators revealed a generally decreased expression of positive cell-cycle genes, while cell-cycle-negative regulators remained unchanged (Figure S5H). These results implicate potential involvement of other mechanisms (e.g., transforming growth factor β signaling [Salm et al., 2005]) in driving the quiescence of LRCs.
Neurogenesis and Inflammation Gene Signatures Link the Gene Expression Profile in Luminal LRCs to Aggressive and Castration-Resistant PCa
Our recent studies (Zhang et al., 2017) linked the gene expression profile in WIT-L human prostate luminal progenitors to aggressive subtypes of PCa such as CRPC (castration-resistant PCa) and neuroendocrine PCa. Further, attenuated AR activity and androgen deprivation therapy (ADT) promote a stem-like cell phenotype in PCa (Schroeder et al., 2014) and inflammation has been shown to expand a progenitor-like luminal cell pool (Liu et al., 2016) and decrease AR signaling in human prostatic luminal cells (Zhang et al., 2016a). These discussions prompted us to determine whether the gene expression profile in luminal LRCs could be linked to clinical features of PCa. GSEA revealed a significant enrichment of gene signatures of WIT-L human luminal progenitors in L-GFP+ cells (Figure 5B, plot 3), suggesting that the luminal LRC gene profile may associate with aggressive PCa phenotypes. In support, gene signatures in patient CRPC after failure of ADT were enriched in L-GFP+ cells (Figures 5F and S5I). These latter observations may suggest a global progenitor-like gene expression profile in human CRPC.
Of interest, a gene signature of mouse prostate tumors originated from basal cells (Wang et al., 2013) was modestly but significantly enriched in L-GFP+ cells (Figure S5J), suggesting that prostate tumors with a basal cell origin may be maintained by luminal progenitor-like cells, consistent with an earlier study (Stoyanova et al., 2013). Importantly, a recently reported gene signature of a human CD38low prostatic luminal progenitor population was dramatically enriched in our L-GFP+ cells (Figure 5G). Of note, the CD38low human luminal progenitors are tightly associated with inflammation in PCa, express an inflammatory signature with reduced AR and androgen signaling, and associate with disease progression and poor outcome (Liu et al., 2016). Thus, our mouse luminal LRCs share molecular features with human prostatic luminal progenitors.
Luminal LRCs Resist Castration In Vivo
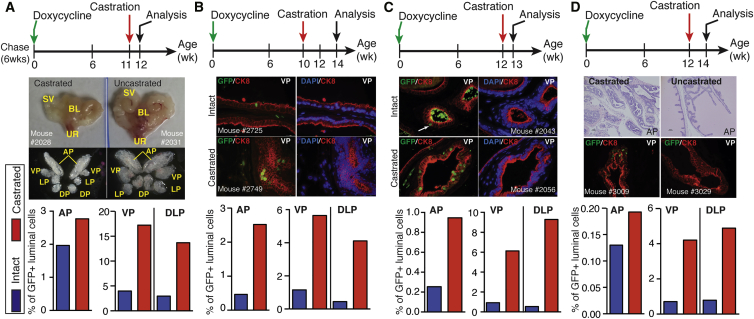
The cellular origin(s) of castration resistance is a central question in PCa research and a long-standing hypothesis is that stem/progenitor cells may preferentially survive ADT. Consequently, we evaluated the in vivo castration sensitivity of luminal LRCs. To first assess whether our model could label luminal cells intrinsically resistant to castration, we castrated the unchased Pb-tetVP16-GFP male mice at 6–8 weeks and then started DOX chase and examined the abundance of GFP+ cells in prostate lobes at different time points after castration. Numerous GFP+ cells (8.0%, 29.3%, and 20.1% for AP, VP, and DLP, respectively) were observed at even 12 weeks after castration (Figure S6A), suggesting that our model marks a proportion of luminal cells that are intrinsically castration resistant in physiological conditions. We next sought to evaluate whether long-term chased LRCs resist castration. We castrated 10- to 12-week-chased bigenic male mice and analyzed prostates 1–4 weeks later (Figures 6A–6D). As short as 1 week after castration, seminal vesicles and prostate glands shrank significantly (Figures 6A and 6C), suggesting successful androgen ablation. In all experimental settings, the frequency of luminal LRCs (GFP+CK8+) dramatically increased post castration, indicating that GFP+ cells possessed intrinsic survival advantages over GFP− cells (Figures 6A–6D). For example, in uncastrated bigenic mice chased for 14 weeks, very few GFP+ cells were seen (Figure S2A); in contrast, abundant GFP+ cells remained in the prostates of mice castrated at 10–12 weeks and harvested at 13- to 14-week chase (Figures 6B–6D). We also examined whether LRCs resist apoptosis after castration by co-IF analysis of GFP and cleaved caspase-3 in the prostates castrated at 12- to 13-week chase and harvested at 3 days, 1 week, and 2 weeks post castration (Figure S6B). Apoptosis in mouse prostatic luminal cells generally peaks at 3–4 days after castration and persists, at declining levels, in the following 1–3 weeks (Kato et al., 2013). Notably, we did not detect any GFP+ LRC cells expressing cleaved caspase-3 in all three experimental settings (Figure S6B), suggesting that castration-induced apoptosis in LRCs is a rare event. To further determine whether the increased number of GFP+ LRCs was due to proliferation besides resistance to apoptosis, we performed co-IF of Ki67 and GFP in the above experimental settings. A small number of Ki67+ epithelial cells were observed in regressed prostates (e.g., 1-week [Figure S6C] or 2-weeks [Figure S6D] post castration), but no GFP+ Ki67+ LRCs were found (Figure S6D), consistent with the dormancy of LRCs. These results collectively demonstrate that luminal LRCs preferentially survive androgen deprivation.
Figure 6.
Luminal LRCs Resist Castration In Vivo
The Pb-tetVP16-GFP bigenic male mice were castrated at 11, 10, 12, and 12 weeks after chase and analyzed 1 week (A), 4 weeks (B), 1 week (C), and 2 weeks (D) later, respectively. Shown are representative images of urogenital organs and dissected prostate lobes (A), IF of GFP and CK8 in the VP (B–D) and H&E of AP (D) from bigenic mice with or without castration. Quantification of the percentage of GFP+ cells in CK8+ luminal cell population in different prostate lobes in above experimental castration settings is presented at the bottom. Arrow in (C) indicates a rare GFP+CK8+ luminal cell. See also Figure S6.
Phenotypic Analysis of LRCs Reveals Heterogeneity in Luminal Progenitor Cells
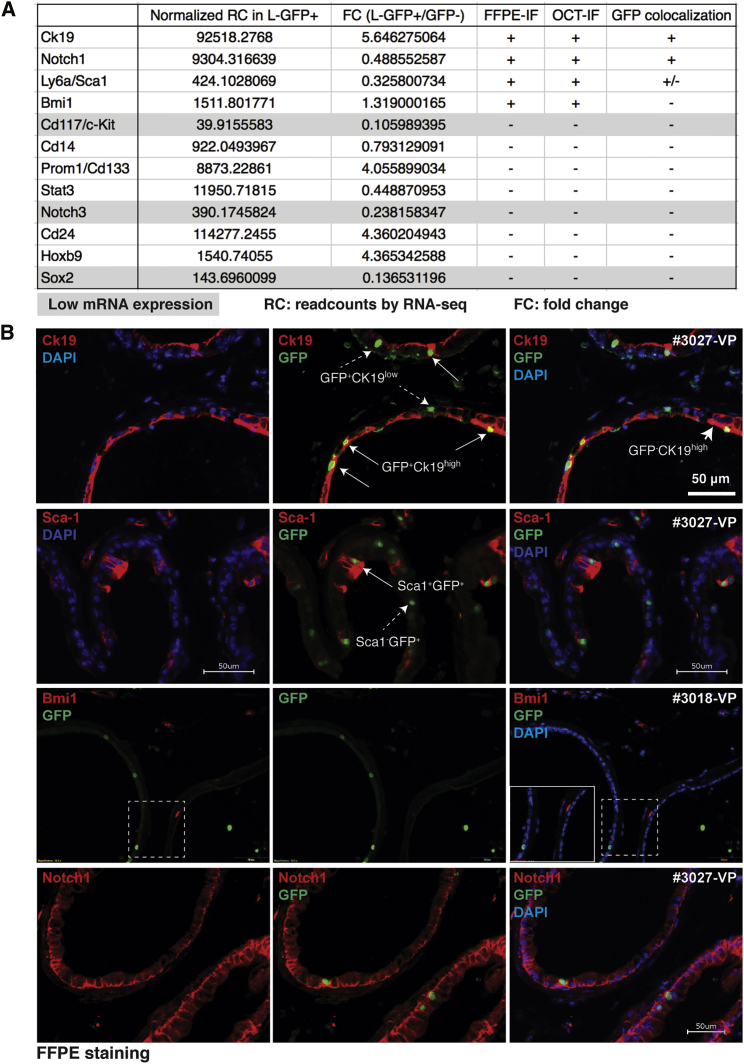
Several surface markers have been reported for mouse mammary luminal progenitors, including SCA-1 (or LY6A), CD133, CD117, CD14, CD24 (Shehata et al., 2012), NOTCH3 (Lafkas et al., 2013), and NOTCH1 (Rodilla et al., 2015). Other than SCA-1 (Burger et al., 2005), similar phenotypic studies in the mouse prostate under physiological (i.e., homeostatic) conditions are generally lacking. CK19 was proposed as a prostate luminal progenitor marker in several studies (Hudson et al., 2001, Korsten et al., 2009, Wang et al., 2001) and several SC factors such as BMI1 (Yoo et al., 2016), SOX2 (Kregel et al., 2013), STAT3 (Schroeder et al., 2014), and HOXB9 (de Pinieux et al., 2001) have been associated with stem-like prostate (cancer) cells under castration condition. We performed double IF staining of GFP and about a dozen of reported mouse prostate/mammary luminal progenitor markers on prostate tissues collected from bigenic mice chased for 12 weeks and correlated with the marker mRNA levels in RNA-seq (Figure 7A). Most of these markers were undetectable in normal uncastrated prostates in both formalin-fixed and paraffin-embedded cryosections (Figure 7A). Consistent with the RNA-seq data (Figures 5C and 7A), L-GFP+ cells expressed relatively abundant CK19 protein compared with L-GFP− cells, although not all CK19high cells were GFP+ (Figures 7B and S7A). A subset of L-GFP+ cells co-expressed SCA-1 at the proximal prostate, and not all SCA-1+ cells were GFP+ (Figure 7B), consistent with a recent report that SCA-1 identifies a distinct subset of androgen-independent luminal progenitors in the proximal prostate (Kwon et al., 2016). The 12-week-chased L-GFP+ cells did not co-localize with BMI1-expresing cells (Figures 7B and S7A), a rare population that mainly resides in the basal cell layer (Lukacs et al., 2010b), although Bmi1+ cells have recently been reported to reside, infrequently, in the luminal layer and mark rare castration-resistant luminal progenitors (Yoo et al., 2016). We detected a relatively low and homogeneous expression of NOTCH1 (Figure 7B), suggesting that, unlike in mammary tissues (Rodilla et al., 2015), NOTCH1 is not a preferential marker for prostate luminal progenitors. The positive staining of BMI1 and SCA-1 in stromal cells, and cleaved caspase-3 in apoptotic cells shed into the lumen after castration, served as positive control for IF analysis (Figure S7B). Together, these data suggest that prostate luminal progenitors are phenotypically, and, likely, functionally (as reflected by heterogeneous AR expression), heterogeneous.
Figure 7.
Phenotypic Heterogeneity of Luminal LRCs
(A) Summary of mRNA (measured in RNA-seq) and protein (measured by IF staining on both frozen and formalin-fixed paraffin-embedded [FFPE] sections) expression of the indicated phenotypic markers of the luminal progenitors.
(B) Double IF staining of GFP and the indicated markers in FFPE sections of prostate glands harvested from bigenic mice chased for 12 weeks. Boxed regions are enlarged.
See also Figure S7.
Discussion
Recent studies, employing 2D (Zhang et al., 2017) and 3D organoid (Karthaus et al., 2014) in vitro culture systems for human prostate and BrdU pulse-chase (Tsujimura et al., 2002) and lineage-tracing (Choi et al., 2012, Ousset et al., 2012) techniques for murine prostate, have indicated the existence of luminal progenitors within the luminal cell lineage. In this study, we established a bigenic mouse model to specifically label slow-cycling cells in the prostate epithelium, and further defined luminal LRCs as functional progenitors distinct from bulk luminal cells. These luminal LRCs have the following features: (1) they are relatively dormant and, like BrdU-retaining LRCs (Tsujimura et al., 2002), enriched in the proximal prostate; (2) they display bipotential differentiation ability in both in vitro organoid and in vivo prostate regeneration assays; (3) they express a progenitor gene signature with preferential expression of neurogenesis- and inflammation-associated genes, and this gene signature is dramatically enriched in treatment-failed PCa; (4) they survive androgen ablation by resisting castration-induced apoptosis and are further enriched in the prostate of castrated mice; and (5) they are phenotypically heterogeneous with only a subset expressing previously reported progenitor markers such as CK19 and SCA-1.
Studying prostatic luminal cells has been challenging due to their low stemness and the current infeasibility of efficient culturing in vitro. Prior studies have mainly employed known SC-related surface markers identified in other tissues to dissect the epithelial biology of the prostate. Currently, there is no well-accepted definition for prostate luminal progenitors (Zhang et al., 2017), and only a few studies have reported putative phenotypic markers of stem-like luminal cells in humans (i.e., CD38low; Liu et al., 2016) and in regressed mouse prostates (e.g., NKX3.1+ [Wang et al., 2009], SCA-1+ [Kwon et al., 2016], and BMI1+ [Yoo et al., 2016]). The slow-cycling luminal progenitor (i.e., L-GFP+) cells, identified here in a marker-independent manner, only partially overlap with a few of previously reported progenitor markers. In fact, most of the markers we analyzed, including CD117, CD14, CD133, STAT3, NOTCH3, CD24, HOXB9, and SOX2, seem to be expressed at too low levels to be reliably detected in the normal mouse prostate. Nevertheless, luminal LRCs appear to generally express low NOTCH1 and high CK19 levels, and a subset of LRCs stains positive for SCA-1. Consistent with a recent report (Yoo et al., 2016) showing that BMI1+ luminal cells are rarely seen in normal prostate, our LRCs do not co-localize with BMI1+ cells. These observations raise a question of whether the mouse prostate, under homeostatic conditions, might have distinct luminal progenitor subsets at varying proliferative states, like the hematopoietic system. For example, our LRCs mainly isolate slow-cycling luminal progenitor cells, while the SCA-1+LRCs− might represent fast-proliferating luminal progenitors. Regardless, our study, together with studies from others mentioned above, suggests the presence of multiple luminal progenitor cell populations in the unperturbed mouse prostate. Notably, our present study focuses on normal luminal progenitors; whether these LRCs change their phenotypic marker profiles and then overlap with some of the luminal progenitors (e.g., CARNs and CARBs) previously reported in regressed prostate remains to be determined.
Significantly, the slow-cycling LRCs, like NKX3.1-expressing CARNs (Wang et al., 2009), SCA-1+ (Kwon et al., 2016), and BMI1+ (Yoo et al., 2016) luminal progenitors in regressed prostates, are intrinsically resistant to castration, suggesting that the luminal cell layer in normal prostate harbors subsets of naturally androgen-insensitive cells that can survive androgen deprivation. Castration resistance in LRCs could be related to relatively low AR expression, as ∼37% LRCs express low levels of AR protein. Simultaneously or alternatively, castration resistance in LRCs may also be associated with their relative quiescence. In this regard, the L-GFP+ cells coordinately underexpress a cohort of positive cell-cycle regulator genes. Detailed dissection of LRCs in castrated prostates is needed to reveal mechanisms of androgen ablation resistance. One potential solution is to perform single-cell RNA-seq analysis to reveal cell-to-cell heterogeneity within the luminal cell compartment under both physiological and castration conditions.
The study of luminal progenitors in the prostate is of great significance to both basic and clinical research. First, luminal cells are the functional units mediating secretory activities and the luminal cell layer is self-sustaining (Choi et al., 2012), indicating the importance of progenitors in maintaining prostate homeostasis and glandular functions (Xie et al., 2017, Zhang et al., 2016a). Second, the luminal layer in situ seems to be overall more proliferative (Wang et al., 2013, Zhang et al., 2016b) and murine luminal cells are more susceptible to tumorigenic transformation to generate aggressive tumors (Wang et al., 2014b). This raises the possibility of luminal progenitors functioning as preferred cells-of-origin for CRPC. Our current findings that luminal LRCs preferentially survive androgen ablation provide support to this possibility. Third, stem-like luminal cells may function as tumor-propagating cells (Stoyanova et al., 2013), in line with mouse genetic studies (Abou-Kheir et al., 2010, Korsten et al., 2009) and with previous report showing that aggressive breast basal-like cancers actually originate from luminal progenitors rather than from basal SCs (Molyneux et al., 2010). Fourth, while this manuscript was under revision, a study identified a rare castration-resistant luminal progenitor cell population with a Lin−/SCA-1+/CD49fmed phenotype highly enriched in Pten-null prostate tumors (Sackmann Sala et al., 2017). Finally, the luminal progenitor gene expression profile can be linked to advanced and aggressive PCa subtypes (Liu et al., 2016, Sackmann Sala et al., 2017, Zhang et al., 2017; this study). This notion is further strengthened by the molecular resemblance of our luminal LRCs to recently reported human CD38low prostatic luminal progenitors (Liu et al., 2016), which has been shown to be associated with inflammation and PCa progression with poor outcome. In aggregate, our study presents a comprehensive biological and molecular characterization of slow-cycling prostate luminal progenitors. Further studies on the functional roles of luminal LRCs in tumorigenesis, and the molecular mechanisms that regulate their homeostasis and dormancy, may yield new predictive markers and therapeutic targets for aggressive CRPC.
Experimental Procedures
General procedures in producing and propagating transgenic animals have been described previously (Suraneni et al., 2010). All animal work was performed under the University of Texas MD Anderson Cancer Center and Roswell Park Cancer Institute IACUC approved protocols. Immunostaining was performed on either paraffin-embedded or OCT frozen sections. The mouse prostate tissues were enzymatically digested with collagenase IA and Dispase, followed by FACS analysis using the BD Aria (BD Biosciences).
Additional experimental procedures are detailed in the Supplemental Experimental Procedures.
Author Contributions
D.Z., C.J., and D.G.T. conceived and designed the study. D.Z., S.G., C.J., and A.T. performed the experiments; D.Z., Y.L., and J.S. performed RNA-seq and bioinformatics analysis. D.Z. and D.G.T. analyzed and interpreted the data. D.Z. and D.G.T. wrote the paper. All authors read and approved the final manuscript.
Acknowledgments
For the work conducted at the University of Texas MD Anderson Cancer Center (MDACC), we thank the Science Park Animal Core for animal maintenance and care, the Histology Core for IHC studies, P. Whitney for assistance in FACS, Y. Takata for technical help in RNA-seq, and the MDACC Department of Epigenetics and Molecular Carcinogenesis Flow Cytometry and Cell Imaging Core (FCCIC) headed by C.J. For work performed at Roswell Park Cancer Institute (RPCI), we acknowledge the support of several shared resources including flow and image cytometry (K. de Jong) and genomics resources. We thank Dr. J. Kirk for critically reading the manuscript and other Tang lab members for helpful discussions and suggestions. This project was supported by grants from the US NIH (R01-CA155693), Department of Defense (W81XWH-13-1-0352, W81XWH-14-1-0575, and W81XWH-16-1-0575), CPRIT (RP120380), and the Chinese Ministry of Science and Technology (MOST) grant 2016YFA0101203 (all to D.G.T.), and by RPCI and NCI center grant P30CA016056. This study also made use of the Science Park NGS Core, supported by CRPIT Core Facility Support Award RP120348 (to J.S.). We apologize to the colleagues whose work was not cited due to space constraint.
Published: December 21, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.016.
Contributor Information
Dingxiao Zhang, Email: dingxiao.zhang@roswellpark.org.
Dean G. Tang, Email: dean.tang@roswellpark.org.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE98760.
Supplemental Information
References
- Abou-Kheir W.G., Hynes P.G., Martin P.L., Pierce R., Kelly K. Characterizing the contribution of stem/progenitor cells to tumorigenesis in the Pten–/–TP53–/– prostate cancer model. Stem Cells. 2010;28:2129–2140. doi: 10.1002/stem.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S., Hynes P.G., Tillman H.S., Lake R., Abou-Kheir W.G., Fang L., Casey O.M., Ameri A.H., Martin P.L., Yin J.J. Identification of different classes of luminal progenitor cells within prostate tumors. Cell Rep. 2015;13:2147–2158. doi: 10.1016/j.celrep.2015.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger P.E., Xiong X., Coetzee S., Salm S.N., Moscatelli D., Goto K., Wilson E.L. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc. Natl. Acad. Sci. USA. 2005;102:7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinieux G., Legrier M.E., Poirson-Bichat F., Courty Y., Bras-Goncalves R., Dutrillaux A.M., Némati F., Oudard S., Lidereau R., Broqua P. Clinical and experimental progression of a new model of human prostate cancer and therapeutic approach. Am. J. Pathol. 2001;159:753–764. doi: 10.1016/S0002-9440(10)61746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos C.O., Rebbeck C., Rozhkova E., Valentine A., Samuels A., Kadiri L.R., Osten P., Harris E.Y., Uren P.J., Smith A.D., Hannon G.J. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:7123–7130. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D.L., Guy A.T., Fry P., O'Hare M.J., Watt F.M., Masters J.R. Epithelial cell differentiation pathways in the human prostate: identification of intermediate phenotypes by keratin expression. J. Histochem. Cytochem. 2001;49:271–278. doi: 10.1177/002215540104900214. [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Iaquinta P.J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C.M., Gao D., Begthel H., Sachs N. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Ishii K., Iwamoto Y., Sasaki T., Kanda H., Yamada Y., Arima K., Shiraishi T., Sugimura Y. Activation of FGF2-FGFR signaling in the castrated mouse prostate stimulates the proliferation of basal epithelial cells. Biol. Reprod. 2013;89:81. doi: 10.1095/biolreprod.112.107516. [DOI] [PubMed] [Google Scholar]
- Korsten H., Ziel-van der Made A., Ma X., van der Kwast T., Trapman J. Accumulating progenitor cells in the luminal epithelial cell layer are candidate tumor initiating cells in a Pten knockout mouse prostate cancer model. PLoS One. 2009;4:e5662. doi: 10.1371/journal.pone.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kregel S., Kiriluk K.J., Rosen A.M., Cai Y., Reyes E.E., Otto K.B., Tom W., Paner G.P., Szmulewitz R.Z., Vander Griend D.J. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS One. 2013;8:e53701. doi: 10.1371/journal.pone.0053701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O.J., Xin L. Prostate epithelial stem and progenitor cells. Am. J. Clin. Exp. Urol. 2014;2:209–218. [PMC free article] [PubMed] [Google Scholar]
- Kwon O.J., Valdez J.M., Zhang L., Zhang B., Wei X., Su Q., Ittmann M.M., Creighton C.J., Xin L. Increased Notch signalling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nat. Commun. 2014;5:4416. doi: 10.1038/ncomms5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O.J., Zhang L., Xin L. Stem cell antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential. Stem Cells. 2016;34:191–202. doi: 10.1002/stem.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafkas D., Rodilla V., Huyghe M., Mourao L., Kiaris H., Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D.A., Xin L., Lukacs R.U., Cheng D., Witte O.N. Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Grogan T.R., Hieronymus H., Hashimoto T., Mottahedeh J., Cheng D., Zhang L., Huang K., Stoyanova T., Park J.W. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 2016;17:2596–2606. doi: 10.1016/j.celrep.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs R.U., Goldstein A.S., Lawson D.A., Cheng D., Witte O.N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs R.U., Memarzadeh S., Wu H., Witte O.N. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7:682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B.D., Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Pignon J.C., Grisanzio C., Geng Y., Song J., Shivdasani R.A., Signoretti S. p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. USA. 2013;110:8105–8110. doi: 10.1073/pnas.1221216110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Liu X., Laffin B., Chen X., Choy G., Jeter C.R., Calhoun-Davis T., Li H., Palapattu G.S., Pang S. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V., Dasti A., Huyghe M., Lafkas D., Laurent C., Reyal F., Fre S. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycaj K., Tang D.G. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Res. 2015;75:4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann Sala L., Boutillon F., Menara G., De Goyon-Pelard A., Leprevost M., Codzamanian J., Lister N., Pencik J., Clark A., Cagnard N. A rare castration-resistant progenitor cell population is highly enriched in Pten-null prostate tumours. J. Pathol. 2017;243:51–64. doi: 10.1002/path.4924. [DOI] [PubMed] [Google Scholar]
- Salm S.N., Burger P.E., Coetzee S., Goto K., Moscatelli D., Wilson E.L. TGF-{beta} maintains dormancy of prostatic stem cells in the proximal region of ducts. J. Cell Biol. 2005;170:81–90. doi: 10.1083/jcb.200412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A., Herrmann A., Cherryholmes G., Kowolik C., Buettner R., Pal S., Yu H., Muller-Newen G., Jove R. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res. 2014;74:1227–1237. doi: 10.1158/0008-5472.CAN-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J., Stingl J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova T., Goldstein A.S., Cai H., Drake J.M., Huang J., Witte O.N. Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via beta-catenin signaling. Genes Dev. 2012;26:2271–2285. doi: 10.1101/gad.196451.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova T., Cooper A.R., Drake J.M., Liu X., Armstrong A.J., Pienta K.J., Zhang H., Kohn D.B., Huang J., Witte O.N., Goldstein A.S. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc. Natl. Acad. Sci. USA. 2013;110:20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni M.V., Schneider-Broussard R., Moore J.R., Davis T.C., Maldonado C.J., Li H., Newman R.A., Kusewitt D., Hu J., Yang P., Tang D.G. Transgenic expression of 15-lipoxygenase 2 (15-LOX2) in mouse prostate leads to hyperplasia and cell senescence. Oncogene. 2010;29:4261–4275. doi: 10.1038/onc.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szotek P.P., Chang H.L., Brennand K., Fujino A., Pieretti-Vanmarcke R., Lo Celso C., Dombkowski D., Preffer F., Cohen K.S., Teixeira J., Donahoe P.K. Normal ovarian surface epithelial label-retaining cells exhibit stem/progenitor cell characteristics. Proc. Natl. Acad. Sci. USA. 2008;105:12469–12473. doi: 10.1073/pnas.0805012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura A., Koikawa Y., Salm S., Takao T., Coetzee S., Moscatelli D., Shapiro E., Lepor H., Sun T.T., Wilson E.L. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J. Cell Biol. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez J.M., Zhang L., Su Q., Dakhova O., Zhang Y., Shahi P., Spencer D.M., Creighton C.J., Ittmann M.M., Xin L. Notch and TGFbeta form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell Stem Cell. 2012;11:676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hayward S., Cao M., Thayer K., Cunha G. Cell differentiation lineage in the prostate. Differentiation. 2001;68:270–279. doi: 10.1046/j.1432-0436.2001.680414.x. [DOI] [PubMed] [Google Scholar]
- Wang S., Garcia A.J., Wu M., Lawson D.A., Witte O.N., Wu H. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl. Acad. Sci. USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H., Halili M.V., Hu Y.P., Price S.M., Abate-Shen C., Shen M.M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sacchetti A., van Dijk M.R., van der Zee M., van der Horst P.H., Joosten R., Burger C.W., Grootegoed J.A., Blok L.J., Fodde R. Identification of quiescent, stem-like cells in the distal female reproductive tract. PLoS One. 2012;7:e40691. doi: 10.1371/journal.pone.0040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhu H.H., Chu M., Liu Y., Zhang C., Liu G., Yang X., Yang R., Gao W.Q. Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat. Commun. 2014;5:4758. doi: 10.1038/ncomms5758. [DOI] [PubMed] [Google Scholar]
- Wang Z.A., Toivanen R., Bergren S.K., Chambon P., Shen M.M. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8:1339–1346. doi: 10.1016/j.celrep.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Liu Y., Cai T., Horton C., Stefanson J., Wang Z.A. Dissecting cell-type-specific roles of androgen receptor in prostate homeostasis and regeneration through lineage tracing. Nat. Commun. 2017;8:14284. doi: 10.1038/ncomms14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lawson D.A., Witte O.N. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc. Natl. Acad. Sci. USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y.A., Roh M., Naseem A.F., Lysy B., Desouki M.M., Unno K., Abdulkadir S.A. Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation. Nat. Commun. 2016;7:12943. doi: 10.1038/ncomms12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Thomas T.Z., Kasper S., Matusik R.J. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141:4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- Zhang B., Kwon O.J., Henry G., Malewska A., Wei X., Zhang L., Brinkley W., Zhang Y., Castro P.D., Titus M. Non-cell-autonomous regulation of prostate epithelial homeostasis by androgen receptor. Mol. Cell. 2016;63:976–989. doi: 10.1016/j.molcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Park D., Zhong Y., Lu Y., Rycaj K., Gong S., Chen X., Liu X., Chao H.P., Whitney P. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat. Commun. 2016;7:10798. doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Lin K., Lu Y., Rycaj K., Zhong Y., Chao H.P., Calhoun-Davis T., Shen J., Tang D.G. Developing a novel two-dimensional culture system to enrich human prostate luminal progenitors that can function as a cell of origin for prostate cancer. Stem Cells Transl. Med. 2017;6:748–760. doi: 10.5966/sctm.2016-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.