Abstract
Unambiguous cell line authentication is essential to avoid loss of association between data and cells. The risk for loss of references increases with the rapidity that new human pluripotent stem cell (hPSC) lines are generated, exchanged, and implemented. Ideally, a single name should be used as a generally applied reference for each cell line to access and unify cell-related information across publications, cell banks, cell registries, and databases and to ensure scientific reproducibility. We discuss the needs and requirements for such a unique identifier and implement a standard nomenclature for hPSCs, which can be automatically generated and registered by the human pluripotent stem cell registry (hPSCreg). To avoid ambiguities in PSC-line referencing, we strongly urge publishers to demand registration and use of the standard name when publishing research based on hPSC lines.
Keywords: human pluripotent stem cells, cell line nomenclature, cell data referencing, cell line authentication, cell databases, stem cell registry
In this article Kurtz, Seltmann and colleagues propose a standard nomenclature and registry for human pluripotent stem cells. The nomenclature is based on fixed name elements and provides a unique identifier for PSC lines suitable for referencing over diverse data sources, registries, publications, and cell banks. Rigorous application is needed to reduce risks from misidentification and false referencing of lines and data.
Main Text
Risks Associated with Lack of a Unique Identifier for hPSC Lines
Although a nomenclature was previously proposed for human pluripotent stem cell (hPSC) lines (Luong et al., 2011), this was only adopted by a few cell banks. Its widespread acceptance was hindered by lack of a central registry, which would be necessary to avoid name duplications, and thus utility of the nomenclature was limited. In addition, it was not possible to encode in the name relationships between lines, for example whether different lines derive from the same donor; moreover, the proposed nomenclature contained modules for optional information, such as disease or patient number, which were poorly defined, thereby hindering standardization. The low community penetration of this first nomenclature proposal resulted in reality in the continuing lack of a standard unique identifier or name. This coincides with an accelerating growth of research and application areas for hPSCs, leading to the establishment of many new lines, which are independently named by an increasing number of autonomous generators. The consequences of this development are the generation of identical names for different lines and, conversely, the accumulation of synonyms. For example, generic names such as iPS-1, iPS-WT, or ALS-1 are accumulating (Luong et al., 2011), making cross-referencing impossible. Conversely, CRL2429 fibroblasts from the American Type Culture Collection or line BJ provided by Stemgent have been used by many laboratories to generate iPSC lines, each assigned with a different name suggestive of unrelatedness. Furthermore, renaming of lines is common and occurs when hPSC banks and registries store cells from many providers, when names are altered with ownership changes, or when lines with isogenic or other genetic modifications are established. We found that 10.2% of approximately 1,900 hPSC lines registered in the hPSC registry (hPSCreg) (https://hpscreg.eu/https://hpscreg.eu) have multiple synonyms, and 84.4% have at least one synonym. In the Cellosaurus database (http://web.expasy.org/cellosaurus/), 15.1% of the 6,252 listed hPSC lines have multiple synonyms and 49.5% have at least one additional synonym. For example, the hESC line WA09 has 10 synonyms used in literature: H9, WA 09, WA9, H9.hESCs, H9 ESC, H9 hES, H9ES, GE09, WAe003-A, WICELLe003-A, and WAe009-A. Consequently, cross-referencing becomes progressively ambiguous. Moreover, the relationships between lines are not traceable as lines from the same donor, such as different clones derived from source cells or genetically modified sublines, become disconnected from their parental lines (Table 1).
Table 1.
Most Common Risks Leading to Ambivalent and Wrong Cell Line Names due to Absence of a Standard hPSC Nomenclature and Unique Identifier
| Reason | Risk | Risk Mitigation |
|---|---|---|
| Independent use of identical primary source cell for reprogramming (e.g., commercial provider of primary cells, time lapses between donations [e.g., healthy young and diseased old donor, donation in different labs by same donor]) | high | genetic profiling, preferentially by short tandem repeats (STR); use of acronym identifying donor in unique identifier; link unique identifier to STR profile |
| Distribution of line between labs and other resources such as banks and registries, which rename according to internal nomenclatures | high | establish unchangeable unique identifier; link other names as synonym |
| Change of ownership with renaming | very high | establish unchangeable unique identifier; link different owner's name as synonym |
| Renaming after genetic modification of line (subclones, isogenic lines, reporter lines) | high | establish link to unique identifier in the name |
| Using generic lab-style names (iPSC-1, ALS-4) | very high | establish unchangeable unique identifier; link generic name as synonym |
| Cell culture contamination with other lines and subsequent mix-up and misauthentication | high | genetic profiling, preferentially by STR |
The resulting equivocal hPSC-line names throughout different resources poses a cumulative long-term risk of dissociation between lines and annotated data and their further erroneous inheritance. Specifically, loss of association between an hPSC line and its donor data poses risks in phenotype-genotype association studies and further reduces reproducibility of experiments with already phenotypically variable hPSCs. An observable tendency is the ad hoc establishment of ever more hPSC lines tailored for a specific application in many individual laboratories (McKernan and Watt, 2013, Kobold et al., 2015). This is at least partially due to the difficulties of searching and directly comparing existing cell lines across the many resources and because of the intrinsic uncertainty with regard to authenticity and identity of lines established elsewhere. This trend is likely supported by the variable standards applied for quality assessment, cell and donor characterization, and consent and data formats applied by different labs, which further diminish utility and comparability of hPSC lines and, thus, scientific validation of results. The current efforts to develop common characterization and data standards are driven primarily by hPSC databases and hPSC banking initiatives (Seltmann et al., 2016, French et al., 2015). Success of these efforts will be essential to increase comparability and trust in the quality of the available hPSC lines.
Measures to Identify and Authenticate hPSC Lines
A minimal set of measures will considerably improve identification and authentication of hPSC lines. These are (1) the provision of a mandatory genetic identity profile as annotation to each hPSC line and (2) the application of a standardized name, which serves as a unique identifier (UI). Genetic authenticity profiles such as short tandem repeats (STR) or SNP profiles authenticate donor and all derived hPSC lines. However, it should be emphasized that genotype information including STR, SNP, or human leukocyte antigen patterns could be used to reveal donor identity if combined with other personal information (Isasi et al., 2014, Morrison et al., 2017). Hence, data access policies that comply with the data protection provisions stipulated by the donor's informed consent are required. These measures limit public access to genetic data. In addition, the application of different genetic profiling standards by different labs reduces comparability. Therefore, utilization of standardized names must be enforced as the second essential measure to ensure hPSC-line identity. Such a UI, or name, will unmistakably connect cell line-specific annotations from multiple datasets, including genetic profiles, the literature, and reference databases. To implement an UI, we propose to modify the previously proposed nomenclature for hPSCs (Luong et al., 2011) and adapt the recommended nomenclature system of the International Cell Line Authentication Committee (ICLAC) (http://iclac.org/resources/cell-line-names/). The modified nomenclature uses the previously proposed identifier for cell source, cell line, and cell type and complements this with codification of relatedness of a given hPSC line to its origin and its genetically modified derivatives (Table 2). An automated tool was realized to create these standardized hPSC names, which are available from a central registry.
Table 2.
Nomenclature: Elements and Scheme
| Name of Element | No. of Digits | Example | Comments | Maximal Possibilities | |
|---|---|---|---|---|---|
| 1 | generator acronym | 2–6 | XXXXXX | self-assigned code provided by the generator; capital letters | 321,272,380 |
| 2 | cell line type | 1 | “e” or “i” | e: ESC line | >2 |
| i: induced PSC line | |||||
| 3 | donor ID | 3 | 001 | alphanumerical 3-digit code | 46,655 |
| 4 | clone number | 2–3 | A | alphabetic capital letter preceded by hyphen | 702 |
| 5 | subclone or subline | 0–3 | −1 | alphanumerical 2-character code preceded by hyphen | 1,330 |
| total characters | 8–16 | >87 × 109 cell lines per generator acronym | |||
| >28 × 1018 unique names | |||||
Structure and elements of the hPSC nomenclature. Five descriptive elements compose a cell line name. (1) An acronym, for example of the generator institution, laboratory, a registry, or a bank. (2) The cell type—other symbols are, for example, (p) for parthenogenic or (s) for somatic cell nuclear transfer PSC lines. (3) A unique alphanumerical identifier. Depending on future needs, this could be extended to 4 digits (1,679,615 possible permutations). These first three elements together form a unique identifier linking the line to a single origin or donor, or blastocyst in case of ESC. Element (4) identifies each cell line, or clone, which was generated from the donor, and element (5) identifies genetically modified subclones or sublines derived from a given clone. The fixed string of elements helps to detect and avoid syntactic naming errors. The nomenclature does not include any personal information that might compromise the privacy and confidentiality of the donor, as recommended in ICLAC's guidelines for cell line naming. Automated generation of an autonomous donor identifier module as part of the name safeguards privacy even further. An example is provided in Table S1.
Implementation of the hPSC Nomenclature in hPSCreg
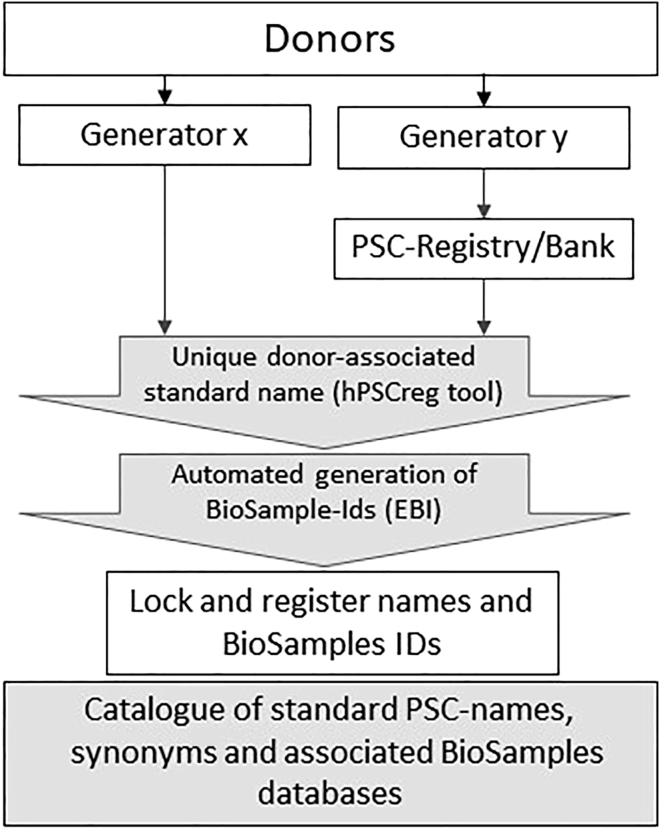
The automated tool is implemented in hPSCreg and is accessible online as a manual data entry form (https://hpscreg.eu/user/cell_line_name/create) or programmatically using the RESTFull application programming interface (API) (https://hpscreg.eu/about/naming-tool#api). Upon entering the essential set of name elements (Table 2 and Supplemental Experimental Procedures), a standardized name is automatically created and further manual editing blocked. The name can be generated at any time during or after hPSC-line generation. Once a standard name is generated, it is locked and a printable quick-response code containing the name is automatically provided. Pre-existing synonyms of established lines are linked to the new standard name to avoid any loss of linkage to familiar names such as WA09 or H9 (standard name WAe009-A). Standard cell line names are centrally registered in the hPSCreg catalog. In addition, the names are deposited in the cell line database Cellosaurus (http://web.expasy.org/cellosaurus) and a BioSamples identifier (http://www.ebi.ac.uk/biosamples/) is assigned to each cell line subsequent to naming (Figure 1). To demonstrate the utility of the naming tool, an application example has been successfully implemented at the European Bank of induced Pluripotent Stem Cells (EBiSC), which deposits iPSC lines from multiple laboratories and generates the standard name via hPSCreg (De Sousa et al., 2017).
Figure 1.
Naming Scheme for hPSC Lines
A donor is recruited and hPSC lines generated by diverse generators x, y, … n or deposited in banks and registries, which submit the essential name elements to hPSCreg to automatically generate a standard name. A BioSamples ID for the line and the donor is automatically provided in parallel for each cell line. The names and BioSamples IDs are locked and registered in a catalog in the hPSCreg database as well as in the Cellosaurus cell line database. Historical lines with non-standard names are often used in hPSC banks and registries. Registration of these lines in hPSCreg will automatically provide a standard name and link it to the pre-existing names, which remain as synonyms.
For validation of names, a semantic uniform resource locator (URL) for every registered and validated cell line is provided. This simple yet semantically meaningful system for information retrieval improves cell line identification and reduces syntax ambiguities. The URLs are persistent and will not change over time, allowing their use as uniform resource identifiers in an application ontology (Seltmann et al., 2016) (Supplemental Experimental Procedures).
Referencing of hPSC Line-Associated Data
The amount and type of information that can be encoded in a name is limited by the rather practical argument of maintaining human readability and label compatibility. A limit of 16 characters for a practicable hPSC name is used here. Several useful but impractical proposals have previously been made for encoding of disease and genetic information in a standard name. Although these attributes are of high interest in hPSC research, their integration as elements of a standard nomenclature would pose multiple obstacles. A disease is a potentially changing and complex donor attribute as it may be a transient condition; e.g., it may develop only after an hPSC line has been derived and additionally, donors may have multiple diseases or phenotypes or are mere carriers of a genetic disease risk without expressing the disease phenotype. Moreover, disease codification is not harmonized despite the existence of disease and phenotype vocabularies in diverse nomenclatures, taxonomies, and ontologies. As a result, diagnosis and disease acronyms are often complex, which could make the name unreadable, error prone, vague, and inoperable for computation. Similar issues arise with respect to genetic modifications, which could for example include karyotypes, SNPs, and transgenic modifications in isogenic clones associated with complex gene nomenclatures.
Instead of overcrowding the name with complex information, a more flexible approach is required to associate attributes such as cell type of origin, disease, phenotype, genetic background, and others. These annotations should be linked by the unique cell line name and made accessible through directly connected databases such as hPSCreg and can be further expanded through linked resources such as BioSamples. The BioSamples database at the European Bioinformatics Institute (EBI) (http://www.ebi.ac.uk/biosamples/https://www.ebi.ac.uk/biosamples/) aggregates information for reference samples such as cell lines, donors, and projects for which data exist in one of the assay databases such as ArrayExpress, the European Nucleotide Archive, or the European Genotype-Phenome Archives (Faulconbridge et al., 2014, Barrett et al., 2012). Furthermore, the BioSample(s) databases at the EBI, the National Center for Biological Information, and the DNA DataBank of Japan are cross-referenced. Linking the standard hPSC-line name to BioSamples IDs thus provides the information available in the BioSamples database, such as donor-related clinical records, biological samples, genes, and genotypes; BioSamples also models relationships such as “derived from,” providing the option to widely expand the information associated with the cell line (Figure 1).
Open Tasks and Enforcement of the Nomenclature
Provision of a standard nomenclature to build a name and UI is an important first step toward reducing risks in hPSC research through misreferencing and misidentification. According to the ISO/AWI 20387, an international standard on biobanks and biobanking, “unique identifier” is defined as a code that is associated with a single entity within a given system and demands a database or reference of names or identifiers, which is provided in hPSCreg. The nomenclature tool also adheres to the FAIR data guiding principles, aimed at enhancing the reusability of data holdings with specific emphasis on enhancing the ability of machines to automatically find and use the data, in addition to supporting its reuse by individuals (Wilkinson et al., 2016). The proposed nomenclature and RESTful API web service for the generation of a standard name, together with a central name registry, promote identification and accurate referencing of cell lines and research data. Currently, more than 300 of the hPSCreg registered lines only have the standard name, indicating increasing acceptance, which is also promoted through the International Stem Cell Banking Initiative and EBiSC (De Sousa et al., 2017, Kim et al., 2017).
As a key reference for reporting cells used to obtain research data and for linking literature resources, standardized names are analogous to providing accurate descriptions of reagents, or genes used in experiments. Thus, use of standard names in the materials and methods section of any paper should always be enforced where hPSC lines have been used to generate published data. Any subsequent abbreviations should be clearly linked to the full cell line name. Truncating cell line names for ongoing use causes confusion in the scientific literature and should be avoided. Moreover, as pre-existing old and familiar names will persist in the literature, there may be confusion when the standard name is introduced. Although this can be minimized by generating names that maintain a visible association to a well-known cell line in widespread use (e.g., WA09 will be named WAe009-A), this is not always possible. In addition, the standard name is linked to the pre-existing name(s), which are stored as synonyms in the registry (e.g., WA09, H9 etc. become synonyms of WAe009-A). We would like to emphasize that the cell lines can be searched in hPSCreg by their synonyms, and the search results would then output all lines with their UIs. Any data/information associated with a name and all its synonyms are linked, including Biosamples ID- and Cellosaurus entry-associated data. Thus even if the pre-existing non-standard name will continue to exist as a synonym, the associated information, references, and data will be linked to the standard name in the registry. This manual and semi-automated mining process will help users to safely associate data with lines. However, because of past inconsistency, naming linking all true synonymous lines and their pre-existing publications and data will require an active process by users and journals (e.g., in the form of an erratum or by adding missing synonymous names to the registry). The cell line generators bear the primary responsibility of selecting an UI for their cell lines; however, research institutions, funding agencies, and journal editors should insist on rigorous cell line referencing, authentication, and traceable annotation to ensure that research data are reliable and reproducible. Journals such as Stem Cell Research already require hPSCreg standard nomenclature as reference.
Ambiguities of cell line names are not restricted to hPSC lines and are a widespread problem in cancer cell lines. However, a nomenclature for these cell types may require different name elements. Notably, the ICLAC group puts emphasis on tissue of origin as being part of a cell line's name, and this is highly relevant for many cell lines such as those from tumors. Our proposed nomenclature scheme and nomenclature tool can easily be adjusted to fit any specific cell type and accommodate other string elements, for example tissue of origin or species. Some ambiguities such as donor origin, however, cannot be completely eliminated by using unique names, especially when these are not systematically applied.
Resolving these remaining ambiguities requires the use of genetic identifiers such as STR or SNP profiles for authentication (Dirks and Drexler, 2011, Dirks and Drexler, 2013). Provision of such data for each line, and development of an STR/SNP resource for PSC-line authentication, should thus become a key future task, for example through using the associated BioSamples IDs as links to STR resources (Ruitberg et al., 2001). Referencing across these different data sources is not yet possible but would provide a powerful instrument for cell line authentication (Yu et al., 2015, Barrett et al., 2012). Principally, a unique name or identifier and a genetic profile are both essential elements to ensure that misauthentication of hPSC lines is controlled. A clear nomenclature and tool for generating the standardized name is a first step and is now available. Use of these names should be strictly enforced.
Author Contributions
A.K., devising and drafting of nomenclature and manuscript; A.B., providing data and revising manuscript; M.-S.B., implementing nomenclature; K.B., implementing nomenclature; A.C.-D., revising manuscript; L.C., devising nomenclature; J.M.C., revising manuscript; L.D., revising manuscript; J.D., implementing nomenclature and revising manuscript; A.F., implementing BioSample workflow; W.F., revising manuscript; A.G., design of nomenclature; D.J.H., revising manuscript; Y.-O.K., revising manuscript; J-H.K., revising manuscript; A.K.-K., revising manuscript; F.L., implementing nomenclature; G.L., revising manuscript; J.F.L., revising manuscript; T.L., designing nomenclature and revising manuscript; N.M., revising manuscript; T.M., revising manuscript; R.M., implementing nomenclature tool; H.P., devising nomenclature and revising manuscript; S.S., implementing nomenclature; M.S., revising manuscript; K.S., revising manuscript; H.S., designing nomenclature and revising manuscript; G.S., designing nomenclature and revising manuscript; I.S., implementing BioSamples tool; A.V., devising nomenclature and revising manuscript; R.-H.X., revising manuscript.
Acknowledgments
The work is supported by the European Commission grant 334502 (hPSCreg), the Innovative Medicines Initiative grant for the European bank for induced pluripotent stem cells (EBiSC), and HipSci grant WT098503. HipSci is co-funded by the Wellcome Trust and the Medical Research Council. The UK Stem Cell Bank is funded by the Medical Research Council and the Biotechnology and Biological Sciences Research Council in the UK. Contribution and support was provided by members of ICLAC, an independent scientific committee (http://iclac.org), especially Richard Neve and Ian Freschney.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.002.
Contributor Information
Andreas Kurtz, Email: andreas.kurtz@charite.de.
Stefanie Seltmann, Email: stefanie.seltmann@charite.de.
Supplemental Information
References
- Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40(DI):D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa P.A., Steeg R., Wachter E., Bruce K., King J., Hoeve M., Khadun S., McConnachie G., Holder J., Kurtz A. Rapid establishment of the European bank for induced pluripotent stem cells (EBiSC)—the hot start experience. Stem Cell Res. 2017;20:105–114. doi: 10.1016/j.scr.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Dirks W.G., Drexler H.G. Online verification of human cell line identity by STR DNA typing. Methods Mol. Biol. 2011;731:45–55. doi: 10.1007/978-1-61779-080-5_5. [DOI] [PubMed] [Google Scholar]
- Dirks W.G., Drexler H.G. STR DNA typing of human cell lines: detection of intra- and interspecies cross-contamination. Methods Mol. Biol. 2013;946:27–38. doi: 10.1007/978-1-62703-128-8_3. [DOI] [PubMed] [Google Scholar]
- Faulconbridge A., Burdett T., Brandizi M., Gostev M., Pereira R., Vasant D., Sarkans U., Brazma A., Parkinson H. Updates to BioSamples database at European Bioinformatics Institute. Nucleic Acids Res. 2014;42(DI):D50–D52. doi: 10.1093/nar/gkt1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A., Bravery C., Smith J., Chandra A., Archibald P., Gold J.D., Artzi N., Kim H.W., Barker R.W., Meissner A. Enabling consistency in pluripotent stem cell- derived products for research and development and clinical applications through material standards. Stem Cells Transl. Med. 2015;4:217–223. doi: 10.5966/sctm.2014-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasi R., Andrews P.W., Baltz J.M., Bredenoord A.L., Burton P., Chiu I.M., Hull S.C., Jung J.W., Kurtz A., Lomax G. Identifiability and privacy in pluripotent stem cell research. Cell Stem Cell. 2014;14:427–430. doi: 10.1016/j.stem.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Kurtz A., Yuan B.Z., Zeng F., Lomax G., Loring J.F., Crook J., Ju J.H., Clarke L., Inamdar M.S. Report of the International Stem Cell Banking Initiative workshop activity: current hurdles and progress in seed-stock banking of human pluripotent stem cells. Stem Cells Transl. Med. 2017;6:1956–1962. doi: 10.1002/sctm.17-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobold S., Guhr A., Löser P., Kurtz A. Human embryonic and induced pluripotent stem cell research trends: complementation and diversification of the field. Stem Cell Reports. 2015;4:914–925. doi: 10.1016/j.stemcr.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong M.X., Auerbach J., Crook J.M., Daheron L., Hei D., Lomax G., Loring J.F., Ludwig T., Schlaeger T.M., Smith K.P. A call for standardized naming and reporting of human ESC and iPSC lines. Cell Stem Cell. 2011;8:357–359. doi: 10.1016/j.stem.2011.03.002. [DOI] [PubMed] [Google Scholar]
- McKernan R., Watt F.M. What is the point of large-scale collections of human induced pluripotent stem cells? Nat. Biotechnol. 2013;31:875–877. doi: 10.1038/nbt.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Bell J., George C., Harmon S., Munsie M., Kaye J. The European General Data Protection Regulation: challenges and considerations for iPSC researchers and biobanks. Regen. Med. 2017;12:693–703. doi: 10.2217/rme-2017-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruitberg C.M., Reeder D.J., Butler J.M. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acids Res. 2001;29:320–322. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann S., Lekschas F., Müller R., Stachelscheid H., Bittner M.S., Zhang W., Kidane L., Seriola A., Veiga A., Stacey G. hPSCreg—the human pluripotent stem cell registry. Nucleic Acids Res. 2016;44(DI):D757–D763. doi: 10.1093/nar/gkv963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;15:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Selvaraj S.K., Liang-Chu M.M., Aghajani S., Busse M., Yuan J., Lee G., Peale F., Klijn C., Bourgon R. A resource for cell line authentication, annotation and quality control. Nature. 2015;520:307–311. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.