Summary
Mammary stem and progenitor cells are essential for mammary gland homeostasis and are also candidates for cells of origin of mammary tumors. Here, we have investigated the function of the protein kinase p38α in the mammary gland using mice that delete this protein in the luminal epithelial cells. We show that p38α regulates the fate of luminal progenitor cells through modulation of the transcription factor RUNX1, an important controller of the estrogen receptor-positive cell lineage. We also provide evidence that the regulation of RUNX1 by p38α probably involves the kinase MSK1, which phosphorylates histone H3 at the RUNX1 promoter. Moreover, using a mouse model for breast cancer initiated by luminal cells, we show that p38α downregulation in mammary epithelial cells reduces tumor burden, which correlates with decreased numbers of tumor-initiating cells. Collectively, our results define a key role for p38α in luminal progenitor cell fate that affects mammary tumor formation.
Keywords: mammary gland, cell fate, progenitor cell, p38α, RUNX1, breast cancer, luminal cell, MSK1
Highlights
-
•
Luminal progenitor cell fate in the mammary gland is regulated by p38α
-
•
p38α controls the ER transcriptional program by modulating RUNX1
-
•
p38α regulates H3 phosphorylation at the RUNX1 promoter through the kinase MSK1
-
•
p38α promotes mammary tumorigenesis by maintaining luminal tumor-initiating cells
del Barco Barrantes and colleagues use genetically modified mice to identify p38α as a key regulator of the fate of luminal progenitor cells of the mammary gland, which impinges on the ability of these cells to initiate tumors.
Introduction
The mammary gland is a complex organ that undergoes constant remodeling during the different developmental stages (Hennighausen and Robinson, 2005). During the last decade, we have learned about the composition and cellular hierarchy of the mammary epithelium. Mammary stem cells give rise to three major cellular lineages: estrogen receptor (ER)+ cells and ER− or milk-secreting cells in the luminal compartment, and myoepithelial cells in the basal compartment (Giraddi et al., 2015, Rodilla et al., 2015, Van Keymeulen et al., 2011). The regulatory network that orchestrates the lineage specification and differentiation of the mammary epithelial cells is not fully understood. Given that stem and progenitor cells are strong candidates for cells of origin of cancer, the identification of regulatory components is crucial to improving our understanding of the mechanism that underlies breast cancer (Visvader, 2011).
The protein kinase p38α has been implicated in the control of tissue homeostasis in various organs. There is evidence that p38α signaling in mammalian epithelial cells can inhibit proliferation and induce differentiation (Cuadrado and Nebreda, 2010). A possible role for p38α in the regulation of mammary morphogenesis has been proposed, based on the treatment of established cell lines with SB203580, a compound that inhibits p38α and the related kinase p38β, as well as in the analysis of genetically modified mice that are deficient for the p38α activators MKK3 and MMK6 (Wen et al., 2011). Furthermore, p38α has been implicated in the regulation of stem cell proliferation and differentiation in lung and the hematopoietic system (Karigane et al., 2016, Ventura et al., 2007). Several reports using p38α-deficient mice have also revealed that p38α can function as a tumor suppressor in lung, liver, and colon (Gupta et al., 2014, Hui et al., 2007, Ventura et al., 2007). Moreover, the use of SB203580 or the genetic modification of p38α regulators, such as MKK3, MKK6, or the Wip1/PPM1D phosphatase, have provided evidence that p38α in mammary epithelial cells can suppress tumor initiation (Bulavin et al., 2004, Demidov et al., 2007, Wen et al., 2011). On the other hand, tumors induced by the expression of polyoma middle T antigen (PyMT) in the mouse mammary epithelia, which are initiated by luminal cells, seem to rely on p38α for normal growth based on results using chemical inhibitors (Pereira et al., 2013).
We have used mice that delete p38α in the luminal cell compartment to investigate the function of p38α in the mammary gland. We found that p38α deficiency reduces the number of mammary progenitor cells and the ER+ cell lineage. Moreover, these mice show reduced mammary tumorigenesis induced by PyMT expression, which correlates with a decreased pool of tumor-initiating cells (TICs).
Results
p38α Regulates Mammary Luminal Progenitor Cells in Homeostasis
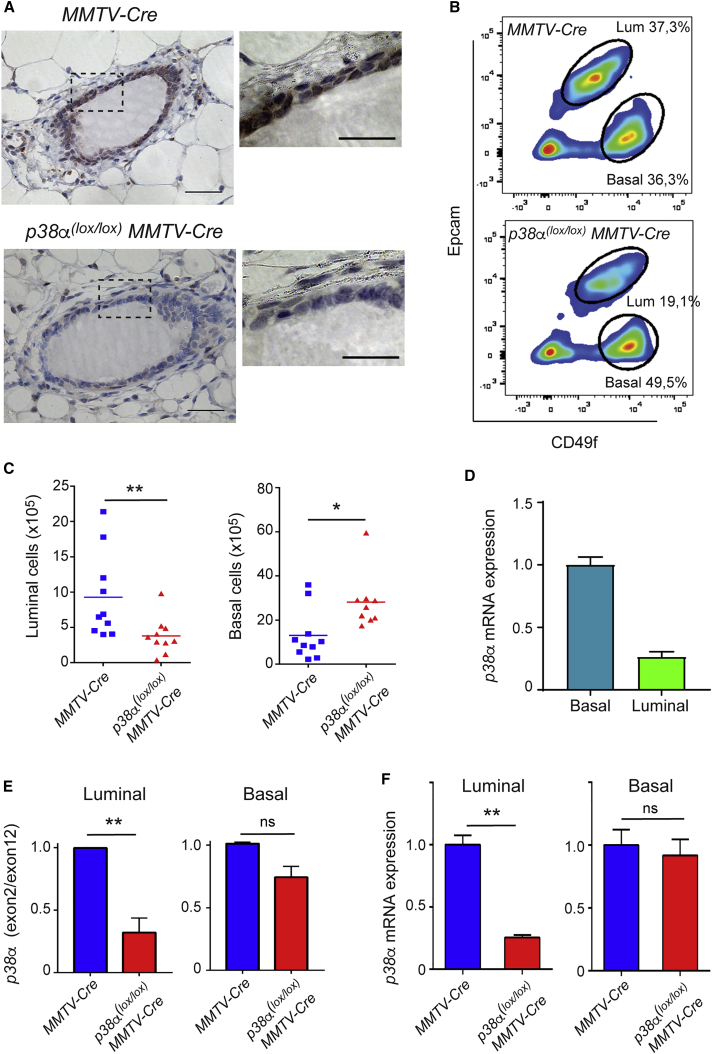
To study the role of p38α in mammary gland homeostasis, we crossed animals carrying a conditional allele of p38α (p38α(lox/lox)) (Heinrichsdorff et al., 2008, Ventura et al., 2007) with transgenic mice expressing Cre under control of the mouse mammary tumor virus (MMTV) promoter (Wagner et al., 2001). The p38α(lox/lox);MMTV-Cre mice showed downregulation of p38α in the epithelium of the mammary gland (Figure 1A). To investigate the effect of p38α depletion on the cellular composition of the mammary gland (Visvader and Stingl, 2014), we sorted EpCamhighCD49fmed (luminal) and EpCammedCD49fhigh (basal) cell populations (Prater et al., 2013) from p38α(lox/lox);MMTV-Cre and control MMTV-Cre virgin female mice (Figure 1B). We observed a reduction in the absolute number of luminal cells in p38α-deficient mammary glands (Figure 1C). In contrast, the absolute number of basal cells was increased in p38α-deficient mammary glands (Figure 1C). We detected p38α mRNA expression in both cell populations, with higher levels in basal cells (Figure 1D). However, whereas MMTV-Cre expression resulted in efficient downregulation of p38α in luminal cells, as determined by both genomic analysis of the floxed p38α exon2 and the levels of p38α mRNA, downregulation of p38α in basal cells appeared to be rather mild (Figures 1E and 1F). These observations suggest that the increased number of basal cells in p38α-deficient mammary glands is probably a consequence of the p38α depletion in luminal cells.
Figure 1.
p38α Regulates Mammary Luminal Cell Homeostasis
(A) Immunohistochemistry analysis of p38α expression in mammary ducts from animals of the indicated genotypes. Boxed areas are magnified on the right. Scale bars, 100 μm.
(B) Representative FACS plots showing luminal (EpCAMhighCD49fmed) and basal (EpCAMmedCD49fhigh) cell populations in mammary glands from animals of the indicated genotypes.
(C) Quantification of the absolute number of luminal and basal cell populations separated as in (B) (n = 10 animals). ∗p ≤ 0.05; ∗∗p ≤ 0.005.
(D) Relative expression of the p38α mRNA in luminal and basal cell populations separated as in (B) from MMTV-Cre mice was determined by qRT-PCR. The expression level in basal cells was given the value of 1.
(E) Genomic DNA was purified from luminal and basal cell populations separated as in (B) from the indicated mice and analyzed by qPCR with primers specific for exon 2 and exon 12 (as a control) of the p38α gene. The relative amount of exon 2 versus exon 12 in cells from MMTV-Cre mice was given the value of 1 (n = 3 animals). ∗∗p ≤ 0.005; ns, non-significant.
(F) Relative expression of the p38α mRNA in luminal and basal cell populations separated as in (B) from the indicated mice was determined by qRT-PCR (n = 3 animals). The expression levels in cells from MMTV-Cre were given the value of 1. ∗∗p ≤ 0.005; ns, non-significant.
See also Figure S1.
Next, we explored the role of p38α during mammary gland development. Whole-mount analysis of mammary glands from p38α(lox/lox);MMTV-Cre pubertal females showed a slight delay in ductal tree expansion compared with MMTV-Cre controls, although no obvious gross morphological abnormalities were observed in virgin females (Figure S1A). However, lactation glands from p38α(lox/lox);MMTV-Cre dams were histologically different from the MMTV-Cre controls, showing a flattened appearance with reduced numbers of alveolar cells and of milk globules in the alveoli (Figure S1B). The reduced number of alveolar cells correlated with decreased staining for both the luminal marker Keratin8 and phosphorylated (active) STAT5, a marker of early lactation (Liu et al., 1995) (Figures S1C and S1D), suggesting that p38α downregulation delays expansion of alveolar cells. However, despite these changes, pups from p38α(lox/lox);MMTV-Cre females survived (Figure S1E), indicating that p38α-deficient mammary glands were able to produce enough milk to support the progeny.
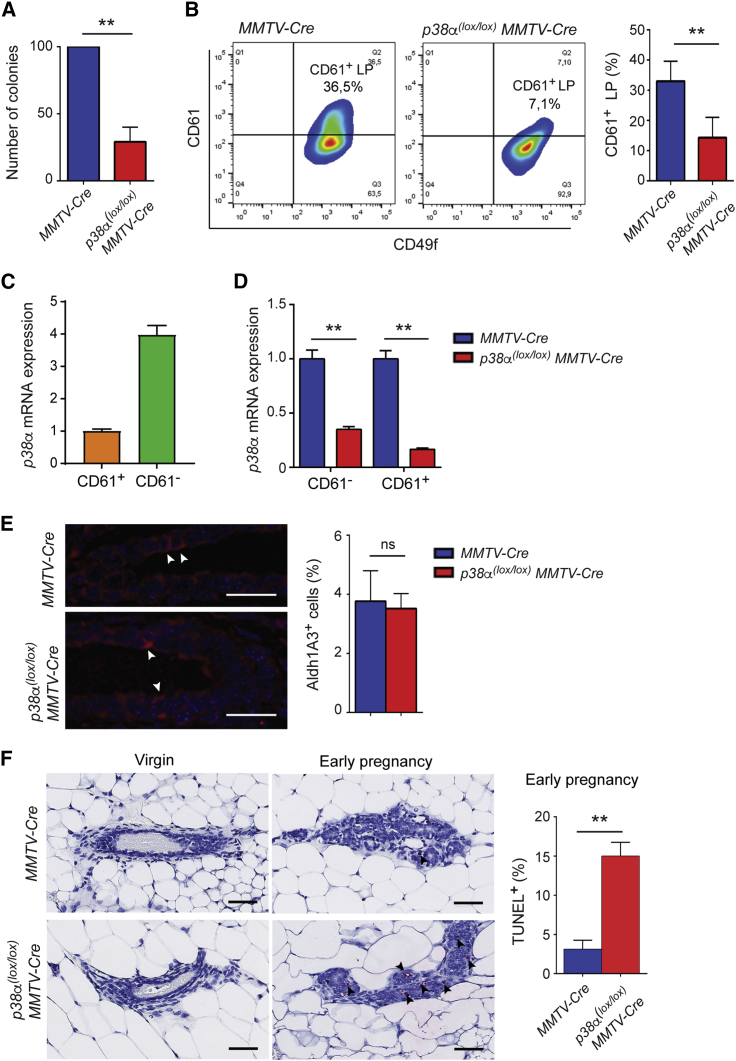
The observation that p38α downregulation reduced the luminal cell population of the mammary epithelium (Figures 1B and 1C) prompted us to explore in more detail the role of p38α in these cells. Colony-formation assays using Matrigel cultures revealed a dramatic reduction in the number and size of colonies formed by sorted luminal cells from p38α(lox/lox);MMTV-Cre mice compared with MMTV-Cre controls (Figures 2A and S2). The ability to form colonies has been associated with the numbers of CD61+ luminal progenitor cells (Asselin-Labat et al., 2007). Indeed, using a combination of CD61 and CD49f antibodies, a significant reduction in the percentage of CD61+ luminal progenitor cells was detected in p38α-deficient luminal epithelial cells (Figure 2B). We detected p38α mRNA expression in both luminal progenitor and non-progenitor cells, with higher levels in non-progenitor cells, and both cell populations showed comparable levels of p38α mRNA depletion in p38α(lox/lox);MMTV-Cre mice (Figures 2C and 2D). CD61 marks both alveolar and ductal progenitor cells, and to discriminate the progenitor cell population that was affected by p38α deletion, we examined the alveolar progenitor marker ALDH1A3 (Giraddi et al., 2015). Quantification of the stainings indicated no differences in the number of alveolar progenitor cells between p38α(lox/lox);MMTV-Cre and the MMTV-Cre control females at estrus stage (Figure 2E). However, we detected increased apoptosis by TUNEL staining in the p38α-deficient mammary epithelium during early pregnancy (Figure 2F).
Figure 2.
p38α Regulates Luminal Progenitor Cell Maintenance
(A) Colonies formed in Matrigel by EpCAMhighCD49fmed luminal cells isolated from virgin females of the indicated genotypes were quantified and values are expressed as percentage to the numbers in control MMTV-Cre mice (n = 3 independent experiments). ∗∗p ≤ 0.005.
(B) Luminal cells (EpCAMhighCD49fmed) separated as indicated in Figure 1B were further analyzed by FACS according to CD61 and CD49f expression. The histogram shows the quantification of the CD61+CD49f+ luminal progenitor cells (n = 5 animals). ∗∗p ≤ 0.005.
(C) Relative expression of the p38α mRNA in CD61+ and CD61− cell populations separated as in (B) from MMTV-Cre mice was determined by qRT-PCR. The expression level in CD61+ cells was given the value of 1.
(D) Relative expression of the p38α mRNA in CD61+ and CD61− cell populations separated as in (B) from the indicated mice was determined by qRT-PCR (n = 3 animals). The expression levels in cells from MMTV-Cre mice were given the value of 1. ∗∗p ≤ 0.005.
(E) ALDH1A3 staining for alveolar progenitor cells in virgin females of the indicated genotypes (n = 3 animals). Arrowheads indicate ALDH1A3+ cells. Scale bars, 20 μm. Quantifications are shown in the histogram. ns, not significant.
(F) Analysis of cell death by TUNEL staining in mice of the indicated genotypes, either virgin or during early pregnancy (n = 3 animals). Arrowheads indicate apoptotic cells. Scale bars, 50 μm. Quantifications during early pregnancy are shown in the histogram. ∗∗p ≤ 0.005.
See also Figure S2.
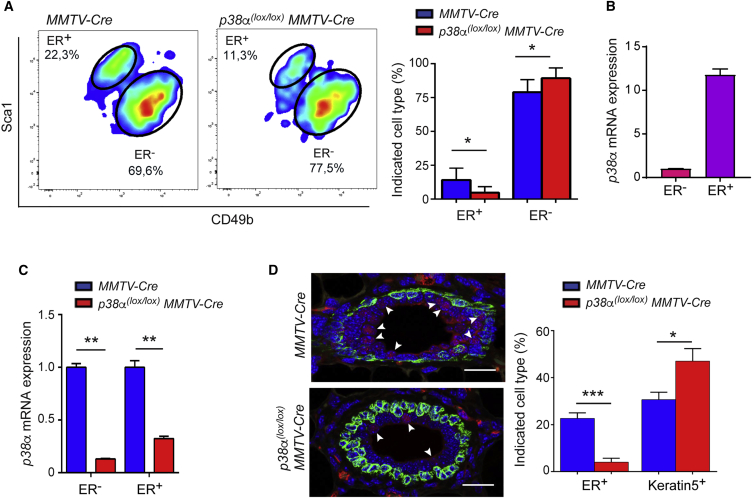
Further analysis of the mammary luminal cell compartment using previously described Sca1 and CD49b markers (Shehata et al., 2012) revealed a reduction in the ER+ cell lineage together with an increase in the ER− or milk cell lineage in p38α-deficient luminal epithelial cells compared with wild-type (WT) cells (Figure 3A). In addition, we detected substantially higher p38α mRNA expression levels in ER+ cells than in ER− cells (Figure 3B), whereas p38α depletion was similar in both ER+ and ER− cell lineages from p38α(lox/lox);MMTV-Cre mice (Figure 3C). Moreover, ER and Keratin5 immunostaining confirmed a reduced number of ER+ cells and increased number of basal cells (Keratin5+) in the p38α-deficient mammary epithelium (Figure 3D).
Figure 3.
p38α Regulates the ER+ Cell Lineage
(A) FACS plots for luminal cells isolated from the indicated mice and then separated into the ER+ and ER− cell lineages based on Sca1 and CD49b expression. Quantifications are shown in the histogram (n = 7 animals). ∗p ≤ 0.05.
(B) Relative expression of the p38α mRNA in ER+ and ER− cell populations separated as in (A) from MMTV-Cre mice was determined by qRT-PCR. The expression level in ER− cells was given the value of 1.
(C) Relative expression of the p38α mRNA in ER+ and ER− cell populations separated as in (A) from the indicated mice was determined by qRT-PCR (n = 3 animals). The expression levels in cells from MMTV-Cre were given the value of 1. ∗∗p ≤ 0.005.
(D) ER (red) and Keratin5 (green) stainings in virgin females of the indicated genotypes (n = 3 animals). Arrowheads indicate ER+ cells. Scale bars, 20 μm. Quantifications are shown in the histogram. ∗p ≤ 0.05; ∗∗∗p ≤ 0.0005.
Taken together, these results indicate that p38α downregulation in luminal epithelial cells reduces the number of ductal progenitor cells, which probably undergo apoptosis, impairing the mature ductal lineage and interfering with the expansion of alveolar progenitor cells during early pregnancy stages.
p38α Regulates the Transcriptional Program of Mammary Luminal Cells
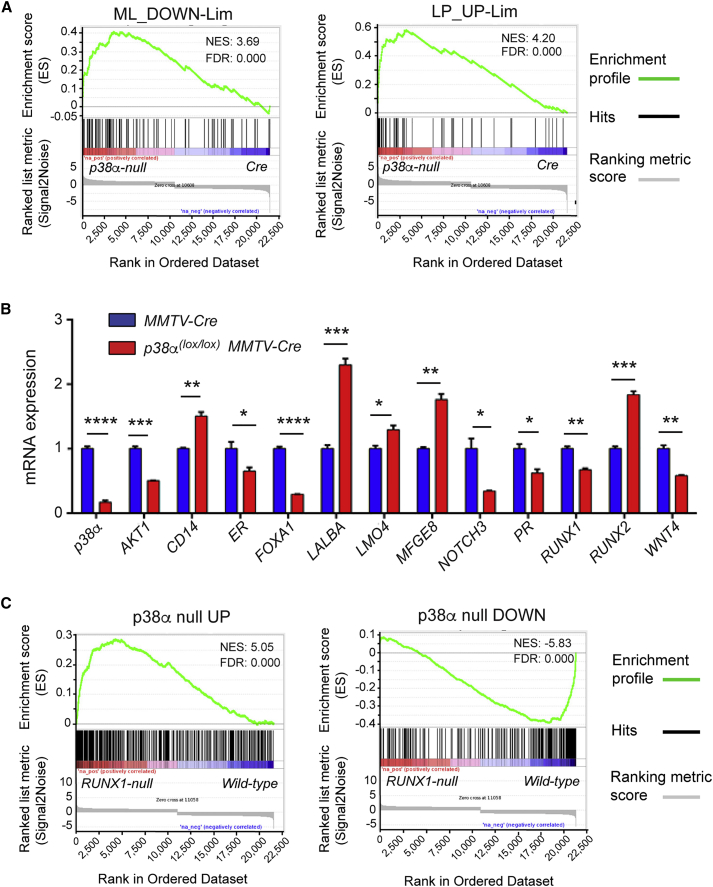
To investigate how p38α regulates luminal progenitor cell homeostasis, we analyzed global mRNA expression patterns in luminal cells isolated from mammary glands of p38α(lox/lox);MMTV-Cre and MMTV-Cre control mice. Differences in gene expression between groups were compared with published gene signatures of specific mammary gland cell populations using gene set enrichment analysis (GSEA) (Kendrick et al., 2008, Lim et al., 2010). We observed that genes overexpressed in p38α-deficient cells showed an enrichment in genes downregulated in mature luminal cells and an enrichment of the luminal progenitor cell gene signature (Figures 4A and S3), indicating that p38α is required for the maintenance of mature luminal cells. We also found that the transcriptional program of p38α-deficient luminal cells was most similar to ER− characteristics, suggesting the loss of ER+ features (Figure S3), which is consistent with our fluorescence-activated cell sorting (FACS) and immunostaining analysis (Figures 3A and 3D).
Figure 4.
p38α Regulates the Expression of Genes Related to Luminal Cell Specification
(A) Gene set enrichment analysis (GSEA) plots showing the correlation between the genetic signature of p38α null luminal cells and previously reported signatures of genes downregulated in mature luminal cells (ML_DOWN) or upregulated in luminal progenitor cells (LP_UP) (Lim et al., 2010). Hits mark the position of genes in the published signatures of ML or LP cells according to the fold changes in the p38α null cells from UP (left) to DOWN (right).
(B) Genes known to play a role in luminal cell linage specification and maintenance were analyzed by qRT-PCR in luminal cells isolated from mammary glands of mice with the indicated genotypes (n = 3 independent experiments). Values refer to the expression level of each gene in cells from MMTV-Cre mice, which was given the value of 1. ∗p ≤ 0.05; ∗∗p ≤ 0.005; ∗∗∗p ≤ 0.0005; ∗∗∗∗p ≤ 0.00005.
(C) GSEA plots showing the correlation between genes upregulated (p38α null UP) or downregulated (p38α null DOWN) in p38α null luminal cells and expression fold changes of the genes in RUNX1 null cells from published data. Hits refer to genes differentially expressed in p38α null cells, which are sorted according to the fold changes in the RUNX1 null cells from UP (left) to DOWN (right).
See also Figure S3.
We analyzed by qRT-PCR the expression of several genes implicated in the maintenance and specification of luminal cells of the mammary gland (Figure 4B). We observed that p38α-deficient luminal cells expressed reduced levels of AKT1, which has been implicated in lactogenic differentiation (Maroulakou et al., 2008) and could contribute to the alveolar compartment phenotype observed during late pregnancy in p38α(lox/lox);MMTV-Cre mice (Figures S1B–S1D). We also observed enhanced expression of markers such as CD14, LALBA, LMOA, and MFGE8 (Shehata et al., 2012), which correlated with the increased numbers of ER− cells detected. On the contrary, several luminal-specific genes, such as FOXA1, NOTCH3, and WNT4, were downregulated in p38α-deficient luminal cells. The transcription factor FOXA1 is essential for ER expression and function (Bernardo et al., 2010), whereas NOTCH3 has been implicated in maintenance of the ER+ luminal progenitor cells that give rise to the ductal lineage (Bouras et al., 2008, Lafkas et al., 2013), and WNT4 co-localizes with progesterone receptor in the ER+ cell lineage present in the ductal epithelium, as it is induced by progesterone during pregnancy (Brisken et al., 2000).
Runt-related transcription factor 1 (RUNX1) is a key regulator of the ER+ cell lineage (van Bragt et al., 2014), and was also downregulated in p38α-deficient luminal cells. On the other hand, RUNX2, which is implicated in alveolar progenitor specification and maturation (Owens et al., 2014), was upregulated. Of note, the pubertal elongation delay observed in the p38α-depleted mammary glands (Figure S1A) is very similar to the phenotype reported for mammary glands with RUNX1 loss or RUNX2 overexpression in luminal cells (Owens et al., 2014, van Bragt et al., 2014). Further support for a link between RUNX1 and p38α functions was obtained by the significant correlation in the genetic signatures reported for RUNX1-deficient luminal cells (van Bragt et al., 2014) with those of p38α-deficient luminal cells, at the level of both upregulated and downregulated genes (Figure 4C). Altogether, it seems likely that p38α regulates the ER+ cell lineage, at least in part, through RUNX1 modulation.
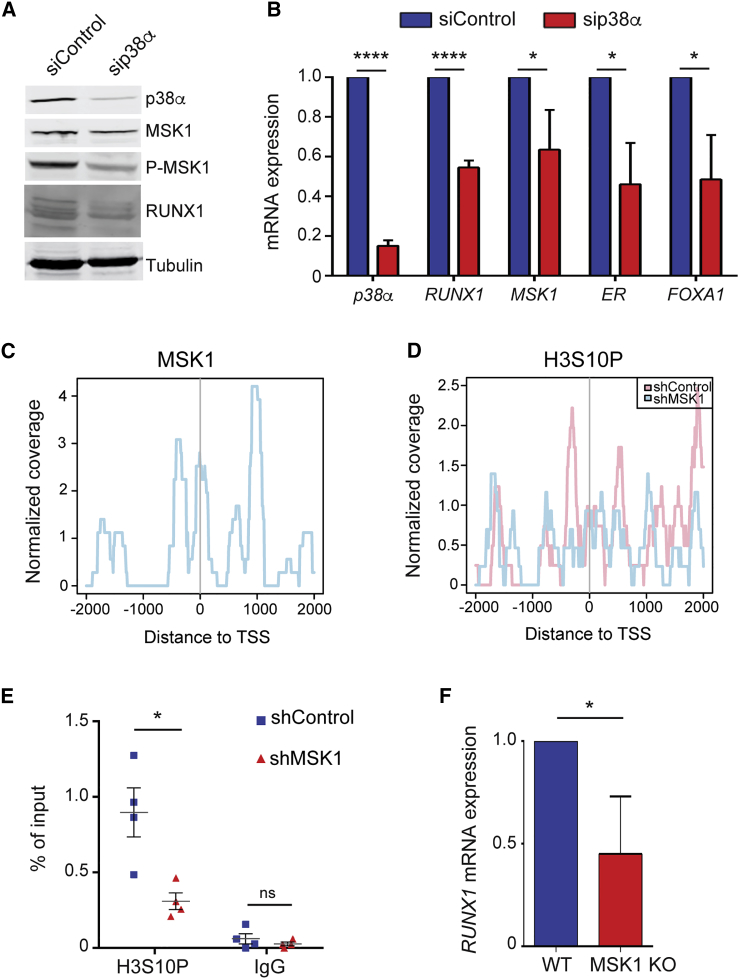
p38α Regulates RUNX1 Expression through MSK1-Dependent Chromatin Remodeling
Mitogen- and stress-activated protein kinase 1 (MSK1) is a protein kinase regulated by p38α, which has been implicated in the modulation of chromatin marks and can control the expression of genes involved in luminal cell fate (Arthur, 2008, Josefowicz et al., 2016, Reyes et al., 2014, Soloaga et al., 2003, Vermeulen et al., 2009). This makes MSK1 a possible candidate to mediate the regulation of RUNX1 by p38α. Previous studies using luminal T47D cells provided evidence for the implication of RUNX1 in the regulation of the ER+ program (van Bragt et al., 2014). We found that T47D cells treated with small interfering RNA (siRNA) against p38α showed impaired MSK1 phosphorylation on sites that are required for its kinase activity (Figure 5A), which correlated with reduced RUNX1 protein and mRNA levels (Figures 5A and 5B). Furthermore, the reduced expression of RUNX1 correlated with decreased ER and FOXA1 mRNA levels (Figure 5B), in agreement with previous reports (van Bragt et al., 2014). Interestingly, analysis of published data obtained using chromatin immunoprecipitation sequencing (ChIP-seq) (Reyes et al., 2014) indicated that MSK1 was recruited to the RUNX1 promoter (Figure 5C). MSK1 can phosphorylate histone H3 on Ser10, which in turn may affect chromatin remodeling at large (Josefowicz et al., 2016, Reyes et al., 2014, Soloaga et al., 2003). Consistent with the detection of MSK1 at the RUNX1 promoter, we observed that T47D cells that are deficient in MSK1 showed reduced levels of Ser10 phosphorylation on histone H3 at the RUNX1 promoter, as determined by ChIP-seq experiments (Figure 5D) and further confirmed by ChIP-PCR analysis (Figure 5E). The reduced phosphorylation of histone H3-Ser10 at the RUNX1 promoter correlated with decreased RUNX1 mRNA expression levels upon MSK1 depletion (Figure 5F). Taken together, these results suggest that p38α depletion results in reduced MSK1 kinase activity, which in turn probably changes the chromatin landscape at the RUNX1 promoter and impairs its transcription. Our results therefore identify p38α as an important modulator of the expression of RUNX1, a gene essential for luminal cell commitment.
Figure 5.
p38α Regulates RUNX1 Expression through MSK1
(A and B) T47D cells were treated with siRNA control or against p38α, and 48 hr later cell lysates were analyzed by immunoblotting using the indicated antibodies (A) or by qRT-PCR with primers to detect the expression levels of the indicated mRNAs (B). The expression level of each gene in siRNA control cells was given the value of 1 (n = 3 independent experiments). ∗p ≤ 0.05; ∗∗∗∗p ≤ 0.00005.
(C) Localization of MSK1 at the RUNX1 promoter of T47D cells based on ChIP-seq data analysis.
(D) Identification of regions containing histone 3 Ser10 (H3S10P) in the RUNX1 promoter of T47D cells treated with short hairpin RNA (shRNA) control or against MSK1, as determined by ChIP-seq data analysis.
(E) Analysis by ChIP-qPCR of H3S10P at the RUNX1 promoter of T47D cells treated with shRNA control or against MSK1 (n = 4 independent experiments). ∗p ≤ 0.05; ns, not significant.
(F) RUNX1 mRNA expression levels were analyzed in WT or MSK1 null T47D cells by qRT-PCR. The expression level in WT cells was given the value of 1 (n = 3 independent experiments). ∗p ≤ 0.05.
p38α Controls the Number of TICs in Mammary Tumors
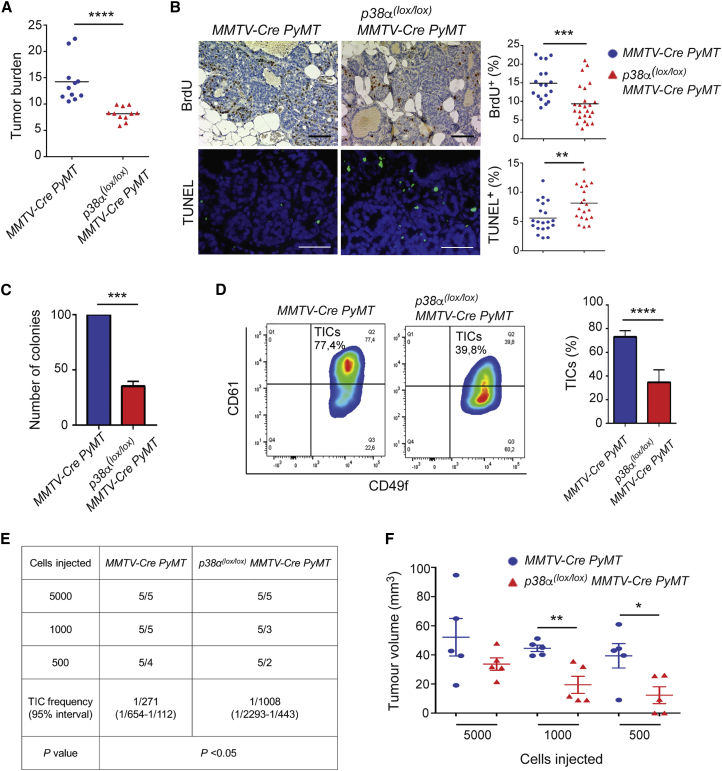
A luminal cell precursor origin has been proposed for the basal-like and luminal subtypes of breast cancer (Visvader and Stingl, 2014). Therefore, we speculated that the ability of p38α to regulate luminal progenitor cell homeostasis described above might impinge on mammary tumorigenesis. To test this hypothesis, we crossed MMTV-PyMT-expressing mice (Guy et al., 1992) with our p38α(lox/lox);MMTV-Cre mice to generate p38α(lox/lox);MMTV-Cre;PyMT animals. This is considered a valid model for the study of the multistep progression of mammary tumorigenesis, resembling differentiated human breast cancer of luminal origin (Herschkowitz et al., 2007, Maglione et al., 2001). We found that genetic deletion of p38α in the PyMT-expressing mammary epithelium reduced mammary tumor burden (Figure 6A), which correlated with reduced lung metastasis (Figure S4A). Next, we compared cell proliferation and cell death rates in pre-neoplastic lesions of p38α(lox/lox);MMTV-Cre;PyMT and MMTV-Cre;PyMT control animals, and found that p38α downregulation resulted in a decreased numbers of bromodeoxyuridine (BrdU)+ cells and increased numbers of TUNEL+ cells (Figure 6B).
Figure 6.
Depletion of p38α in Luminal Cells Impairs Mammary Tumor Growth and Reduces the Number of Tumor-Initiating Cells
(A) Tumor burden in mice of the indicated genotypes was determined at 14 weeks of age (n = 11 animals). ∗∗∗∗p ≤ 0.00005.
(B) Bromodeoxyuridine (BrdU) and TUNEL staining in mammary pre-neoplastic lesions from mice of the indicated genotypes (n = 20 animals). Quantifications are shown in the histograms. Scale bars, 10 μm. ∗∗p ≤ 0.005; ∗∗∗p ≤ 0.0005.
(C) Colonies formed in Matrigel by EpCAMhighCD49fmed luminal cells isolated from mammary pre-neoplastic lesions of mice with the indicated genotypes were quantified and values are expressed as percentage to the numbers in MMTV-Cre;PyMT mice (n = 3 independent experiments). ∗∗∗p ≤ 0.0005.
(D) FACS plots for luminal cells isolated on the basis of CD61 and CD49f expression from mammary pre-neoplastic lesions. Quantifications shown in the histogram indicate the percentage of CD61+CD49f+ tumor-initiating cells (TICs) in mice of the indicated genotypes (n = 7 animals). ∗∗∗∗p ≤ 0.00005.
(E and F) Luminal cells were isolated from mammary pre-neoplastic lesions of mice with the indicated genotypes. The indicated numbers of cells were orthotopically injected into nude mice (n = 5 animals). Tumor incidence (E) and tumor volume (F) at 7 weeks after injection are shown. ∗p ≤ 0.05; ∗∗p ≤ 0.005.
See also Figure S4.
Given the role of p38α in luminal cell maintenance, we investigated whether depletion of p38α might affect the pool of TICs that give rise to mammary tumors. We observed that p38α-deficient luminal cells isolated from pre-neoplastic lesions of p38α(lox/lox);MMTV-Cre;PyMT mice showed significantly reduced potential to form tumorspheres compared with the WT luminal cells from MMTV-Cre;PyMT mice (Figures 6C and S4B). It has been suggested that CD49f and CD61 expression can be used to identify the TICs in different mouse models of mammary tumorigenesis (Lo et al., 2012, Vaillant et al., 2008), and a cell population enriched in CD61+ cells has been reported to progressively increase in MMTV-PyMT tumors (Kouros-Mehr et al., 2008). Consistent with the tumorsphere assays, we observed a significant reduction in CD49f+ CD61+ cells in the luminal cell population isolated from pre-neoplastic lesions of p38α (lox/lox);MMTV-Cre;PyMT mice compared with MMTV-Cre; PyMT controls (Figure 6D). Furthermore, experiments based on the orthotopic injection of luminal mammary cells isolated from pre-neoplastic lesions showed a significant reduction in tumor-initiating potential of the p38α-deficient cells compared with WT cells (Figures 6E and 6F). These results indicate that p38α downregulation in luminal cells reduces the number of mammary TICs.
p38α Regulates the Transcriptional Program of Mammary TICs
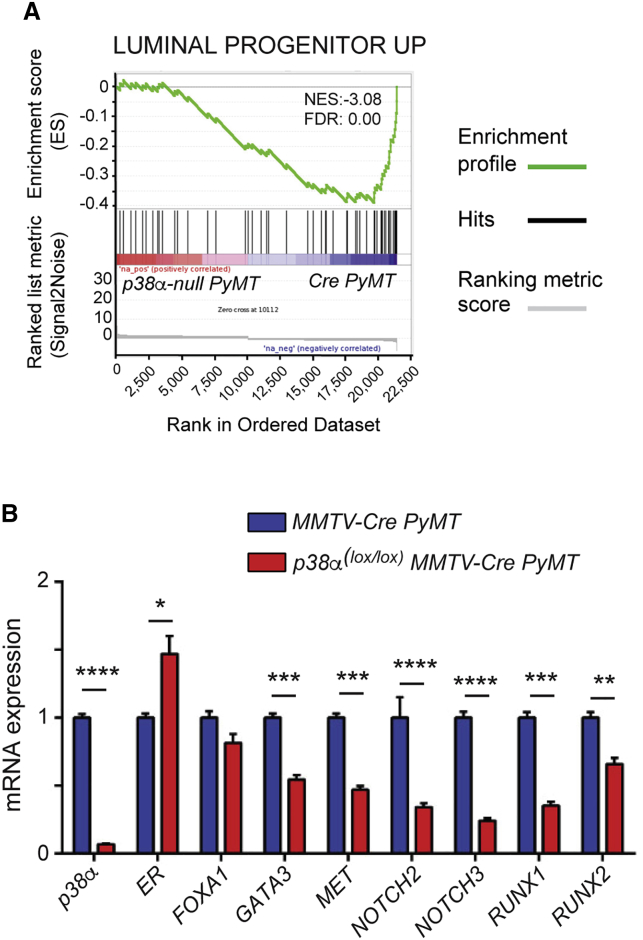
To investigate the contribution of p38α signaling in luminal cells to mammary TIC maintenance, we analyzed global gene expression changes in WT and p38α-deficient luminal cells isolated from pre-neoplastic lesions. In line with the reduced number of putative TICs observed in mice with p38α-deficient luminal cells, we found that gene sets upregulated in luminal progenitor cells isolated from tumors, which correspond to the cell of origin of PyMT-induced tumors (Lim et al., 2010), correlated better with the genetic signature of WT luminal cells than that of p38α-deficient luminal cells isolated from pre-neoplastic lesions (Figure 7A).
Figure 7.
p38α Regulates the Expression of Genes Implicated in Tumor-Initiating Cell Maintenance and Tumorigenesis
(A) GSEA plots showing a negative correlation between the genetic signature of p38α null luminal cells isolated from pre-neoplastic lesions and published signatures of luminal progenitor cells from PyMT-induced tumors (Lim et al., 2010). Ranked list ordered according to fold changes in the p38α null luminal tumor cells. Hits correspond to the position of genes from the published signatures.
(B) Genes known to play a role in TICs and tumor progression were analyzed by qRT-PCR in luminal cells isolated from mammary pre-neoplastic lesions of mice with the indicated genotypes (n = 3 independent experiments). Values refer the expression level of each gene in cells from MMTV-Cre;PyMT mice, which was given the value of 1. ∗p ≤ 0.05; ∗∗p ≤ 0.005; ∗∗∗p ≤ 0.0005; ∗∗∗∗p ≤ 0.00005.
The expression of several genes potentially important for TICs was confirmed by qRT-PCR analysis (Figure 7B). In agreement with the delayed tumor progression in p38α(lox/lox);MMTV-Cre;PyMT mice, we observed increased expression of ER, which is normally lost during PyMT-induced tumor progression (Lin et al., 2003). Additionally we observed reduced expression of the transcription factor GATA3, a well-defined marker of luminal breast cancer whose deletion leads to apoptosis and reduced tumor burden in the MMTV-PyMT model (Kouros-Mehr et al., 2008). The malignant p38α-deficient luminal cells also showed reduced expression of the MET tyrosine kinase receptor, which is highly expressed in aggressive human breast cancer and suffices to induce mammary tumors upon transgenic expression in mice (Gastaldi et al., 2010). Moreover, MET signaling regulates the growth and fate of luminal progenitor cells (Gastaldi et al., 2013), and reduced levels of MET correlate with impaired tumorigenesis and decreased numbers of TICs in the MMTV-PyMT model (Gong et al., 2015). The NOTCH2 and NOTCH3 receptors were also expressed at lower levels in p38α(lox/lox);MMTV-Cre;PyMT luminal cells. There is good evidence that NOTCH signaling is important for breast TICs (Harrison et al., 2010). In particular, NOTCH2 and NOTCH3 are expressed in the CD61+ luminal progenitor cells that are considered the cell of origin for tumors with constitutive NOTCH activation (Bouras et al., 2008), and NOTCH3 has been implicated in the survival and self-renewal of human TICs (Sansone et al., 2007). Finally, we observed reduced expression levels of RUNX1 and RUNX2, which can control mammary tumorigenesis in the MMTV-PyMT model (Browne et al., 2015, Owens et al., 2014), and have been implicated in human breast cancer (Ito et al., 2015).
Discussion
Our results identify an important function of p38α in mammary luminal cell homeostasis. We have found p38α expression in both mammary basal and luminal cells, but have not been able to address the role of this signaling pathway in the myoepithelial lineage due to the poor efficiency of deletion observed in our MMTV-Cre model. The increased number of basal cells observed in p38α deficient mammary glands could be due to paracrine signaling or reflect the conversion of some luminal cells to a basal-like phenotype upon p38α loss; lineage-tracing experiments should address these possibilities in the future. We show that p38α downregulation in luminal cells results in reduced ER+ cell fate and mature luminal cell specification with enrichment in progenitor cell gene signatures. This conclusion is supported by our gene expression analysis using the entire luminal cell population, which reflects the reduction of the ER+ luminal cell lineage observed in p38α null mammary glands, with downregulation of genes that are highly expressed in the ductal lineage but increased expression of genes mainly expressed in progenitor cells. The requirement of p38α to support the ER+ cell lineage is consistent with the higher expression of p38α detected in the ER+ cell compartment and the reduction in ER+ progenitor cells observed upon p38α downregulation.
This p38α function is probably mediated by the transcription factors RUNX1, RUNX2, and FOXA1, which drive the ER program (Bernardo et al., 2010, Owens et al., 2014, van Bragt et al., 2014). Importantly, the expression profiles of genes associated with the ER+ cell lineage are similar in p38α null and RUNX1 null luminal cells, supporting that the reduced Runx1 expression observed in p38α-deficient luminal cells is functionally significant. We also provide evidence indicating that the regulation of RUNX1 by p38α is probably mediated by MSK1, a kinase that can modulate the chromatin landscape though phosphorylation of histone H3 on Ser10 (Josefowicz et al., 2016, Reyes et al., 2014, Soloaga et al., 2003). Moreover, since FOXA1 and RUNX2 can be both regulated by RUNX1 (van Bragt et al., 2014), p38α might control the expression of both genes in luminal cells through RUNX1. At the physiological level, misregulation of RUNX1 and RUNX2 probably accounts for the mild defects in pubertal elongation observed in p38α-deficient mammary glands (van Bragt et al., 2014). Even though the initial number of alveolar progenitor cells does not seem to be affected by p38α deletion in CD61+ luminal progenitor cells, the lactation-induced expansion of the alveolar lineage is affected by a reduction in the lobulo-alveolar epithelium compatible with pup nursing. This might be due to the observed increase in apoptosis during the early events of pregnancy. The increased cell death might be the result of reduced levels of AKT1 observed in p38α-deficient luminal cells (Maroulakou et al., 2008), contributing to the late pregnancy phenotype present in p38α(lox/lox);MMTV-Cre females.
It has been reported that mice treated with SB203580, an inhibitor of p38α and p38β, and MKK3−/−;MKK6+/− mice, which are likely impaired in the activation of all p38 family members, both show defects in mammary lumen anoikis, and increased ductal tree elongation and branching (Wen et al., 2011). We did not observe these phenotypes upon p38α downregulation in luminal cells, suggesting the requirement for p38α inhibition in other cell populations of the mammary gland, or for combined inhibition of p38α and p38β, which has been shown to result in additional phenotypes in several tissues (del Barco Barrantes et al., 2011, Furlan et al., 2012, Warr et al., 2012).
The analysis of gene expression signatures obtained from different breast cancer subtypes and from several mammary epithelial cell populations has revealed that most breast tumors originate from luminal progenitor cells (Visvader and Stingl, 2014), which we show are most dependent on p38α signaling. In addition, an alveolar progenitor origin of the PyMT mammary tumors has been described (Tao et al., 2015). Comparison with previously reported genetic signatures for the cell of origin of PyMT-induced tumors supports a loss of TICs in mammary tumors from mice with p38α-deficient luminal cells. Moreover, p38α-deficient luminal cells exhibit lower tumor-initiating frequency upon orthotopic injection in mice. Our analysis reveals that p38α controls the expression of several genes that support the maintenance or expansion of TICs, such as the receptors MET and NOTCH, and the transcription factors GATA3 and RUNX (Browne et al., 2015, Gastaldi et al., 2013, Gong et al., 2015, Kouros-Mehr et al., 2008, Owens et al., 2014, Sansone et al., 2007).
Taken together, our data show a fundamental role for p38α in the maintenance of ER+ luminal progenitor cells of the mammary gland, most likely through the regulation of RUNX1.
The reduced number of ER+ luminal progenitor cells might contribute to the decreased mammary tumorigenesis observed in mice with p38α-deficient luminal cells. Of note, the decreased mammary tumorigenesis also correlates with a reduced number of TICs, which may represent an alveolar origin, due to their reduced proliferation and increased apoptosis caused by p38α loss in these cells. Our data reveal that p38α facilitates mammary tumorigenesis by maintaining or expanding the luminal progenitor cell pool.
Experimental Procedures
Mice
Mice p38α(lox/lox) carrying the floxed p38α allele (Heinrichsdorff et al., 2008, Ventura et al., 2007) were bred with MMTV-Cre mice (JAX:003553) that drive Cre expression under the control of the MMTV promoter, and with transgenic mice expressing MMTV-PyMT (Guy et al., 1992). Each line was backcrossed in an FVB/n background for at least ten generations. Mouse genotyping was performed by PCR on genomic tail DNA. Primers and conditions are available upon request. For orthotopic injections, mammary tumor cells were sorted from WT and p38α KO PyMT mice, mixed 1:1 with Matrigel (BD Biosciences #354234), and injected into the mammary fat pad of 6-week-old female nude mice as described in Supplemental Experimental Procedures. Mice were housed following national and European Union regulations, and protocols were approved by the Animal Care and Use Committee of the Barcelona Science Park (CEEA-PCB).
Whole-Mount, Immunohistochemistry, Immunofluorescence, and Image Analysis
Whole mounts of mammary glands were fixed and processed as previously described (Jones et al., 1996). For histology, immunohistochemistry, and immunofluorescence staining, mammary glands were fixed in 10% neutral buffered formalin solution and embedded in paraffin. Mammary gland sections from virgin females were stained with p38α antibody (Cell Signaling Technology, #9218) 1:50 for 2 hr at room temperature. Mammary gland sections from females at lactation day 0 were stained with H&E or were analyzed by immunofluorescence using antibodies against Keratin8 (DSHB #531826; 1:200, overnight 4°C), α-smooth muscle actin (Sigma, #A5228; 1:5,000, overnight 4°C), and phospho-STAT5 (Cell Signaling, #9314; 1:500, 2 hr at room temperature). Mammary gland sections from females at estrus stage were analyzed by immunofluorescence using antibodies against ER (Leica Biosystems, #6F11; 1:100, 1 hr at room temperature), ALDH1A3 (Sigma, #HPA04627; 1:100, 1 hr at room temperature), and Keratin5 (Abcam, #ab53121; 1:1,000, 1 hr at room temperature). The following secondary antibodies were used: anti-rabbit Alexa Fluor 594 (Life Technologies, #A21442; 1:200, 1 hr at room temperature); Alexa Fluor 488 (Molecular Probes, #A21470; 1:200, 1 hr at room temperature); horseradish peroxidase (HRP)-conjugated anti-rabbit (ImmunoLogic, #DPVR110HRP; 45 min at room temperature); goat anti-rabbit Alexa Fluor 568 (Thermo Fisher, #A21069; 1:200, 1 at hr room temperature); and VectaFluor Duet Immunofluorescence Double Labeling Kit, DyLight 488 Anti-Rabbit/DyLight 594 Anti-Mouse (VectorDK, #8818). For cell proliferation analysis, BrdU (Roche, #10280879001) was intraperitoneally injected (1 mg/10 g body weight) in 6-week-old female mice, and 1 hr later the mice were euthanized. Samples were stained with anti-BrdU antibody (BD Biosciences, #347580; 1:100, 1 hr at room temperature). The secondary antibodies used were anti-mouse (Dako, #P0447; 1:100, 30 min at room temperature) and HRP-conjugated anti-rabbit (ImmunoLogic, #DPVR110HRP, 45 min at room temperature). Apoptosis was quantified by TUNEL using the In Situ Cell Death Detection Kit, AP (Roche, #11684795910) as specified by the manufacturer. Paraffin-embedded lung sections from 14-week-old PyMT-expressing female mice were stained with H&E. Bright-field images were acquired with a NanoZoomer-2.0 HT C9600 scanner (Hamamatsu) equipped with a 20× objective. Immunofluorescent images were acquired as before but using an L11724-01 mercury lamp unit. All images were visualized with a gamma correction set at 1.8 in the image control panel of the NDP.view 2 U123888-01 software (Hamamatsu, Photonics, France). Otherwise tissue sections were imaged using the Zeiss confocal microscope LSM780. Image analysis was performed using QuPath, an open-source software for quantitative pathology.
Mammary Gland Cell Preparation, Flow-Cytometry Analysis, and Cell Sorting
Thoracic and inguinal mammary glands were dissected from 9-week-old p38α(lox/lox);MMTV-Cre and MMTV-Cre virgin females or 6-week-old p38α(lox/lox);MMTV-Cre;PyMT and MMTV-Cre;PyMT female mice. Cell suspensions were prepared as previously described (Prater et al., 2013). FACS sorting was performed with a FACSAria sorter (BD Bioscience). Data were analyzed with the FlowJo software package. The antibodies used for FACS were from Miltenyi Biotec: Epcam-APC (#130102234), Epcam-FITC (#130102214), CD49f-APC (#130097250), CD49f-PE (#130097246), BP1-biotin (#130101844), CD31-biotin (#130101955), CD45-biotin (#130101952), and Ter119-biotin (#130101882); and from Biolegend: CD61-AF480 (#104311), Sca1-PECy7 (#108114), CD49b-PE (#103506), and Streptavidin-APCCy7 (#405208). FACS gating was based on single color staining and fluorescence-minus-one controls, and was checked with isotype controls (Prater et al., 2013). Gating strategies were as described previously (Prater et al., 2013, Shehata et al., 2012). For colony-formation assays, freshly sorted cells were embedded in Matrigel (BD Pharmingen) as described previously (Shackleton et al., 2006).
Cell Culture, siRNA Transfection, and Immunoblotting
The human breast cancer cell line T47D was cultured and analyzed as described in Supplemental Experimental Procedures.
Microarray, qPCR, and ChIP-Seq Data Analysis
For microarray analysis, EpCamhighCD49fmed luminal cell populations were sorted from either normal mammary glands of 9-week-old mice or pre-neoplastic mammary lesions of 6-week-old mice expressing PyMT. RNA purification, microarray processing, qPCR, and ChIP-seq data analysis were performed as described in Supplemental Experimental Procedures.
Statistical Analyses
Data are presented as mean ± SEM. Statistical significance was determined by Student's t test using GraphPad Prism version 6.
Author Contributions
I.d.B.B. designed the study and performed most of the experiments, analyzed data, and wrote the manuscript. C.S.O.A. statistically analyzed microarray and ChIP-seq data. K.S., A.I., S. Gregorio, and S. Gawrzak performed some experiments. R.R.G. provided essential reagents and advice. A.R.N. provided funding, designed experiments, analyzed data, and wrote the paper.
Acknowledgments
We thank Salvador Aznar-Benitah, Lorenzo Rinaldi, and Aikaterini Symeonidi (IRB) for helping to generate the ChIP-seq data, John Stingl (STEMCELL Technologies, Canada) for generous discussions and useful suggestions, William Muller (McGill University, Canada) for MMTV-PyMT mice, Elisabet Llonch for performing immunostainings and help with quantifications, Lorena Ramirez for help with mouse work, and the Histology, Functional Genomics, and Advanced Digital Microscopy core facilities of IRB for technical support. This work was supported by grants from the European Commission (Advanced ERC 294665), the Spanish MINECO and FEDER funds (BFU2010-17850, SAF2016-81043-R, and SAF2016-76008-R), AGAUR (2014 SRG-535), and the Fundación BBVA. K.S., S. Gawrzak, and S. Gregorio acknowledge “La Caixa” and FPI-Severo Ochoa predoctoral fellowships. IRB Barcelona is the recipient of institutional funding from MINECO through the Centers of Excellence Severo Ochoa award and from the CERCA Program of the Catalan Government.
Published: December 28, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.021.
Contributor Information
Ivan del Barco Barrantes, Email: ivan.delbarco@irbbarcelona.org.
Angel R. Nebreda, Email: angel.nebreda@irbbarcelona.org.
Accession Numbers
The accession number for the microarray results reported in this paper is GEO: GSE92877.
Supplemental Information
References
- Arthur J.S. MSK activation and physiological roles. Front. Biosci. 2008;13:5866–5879. doi: 10.2741/3122. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M.L., Sutherland K.D., Barker H., Thomas R., Shackleton M., Forrest N.C., Hartley L., Robb L., Grosveld F.G., van der Wees J. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bernardo G.M., Lozada K.L., Miedler J.D., Harburg G., Hewitt S.C., Mosley J.D., Godwin A.K., Korach K.S., Visvader J.E., Kaestner K.H. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M.L., Oakes S.R., Lindeman G.J., Visvader J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Brisken C., Heineman A., Chavarria T., Elenbaas B., Tan J., Dey S.K., McMahon J.A., McMahon A.P., Weinberg R.A. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14:650–654. [PMC free article] [PubMed] [Google Scholar]
- Browne G., Taipaleenmaki H., Bishop N.M., Madasu S.C., Shaw L.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J. Cell. Physiol. 2015;230:2522–2532. doi: 10.1002/jcp.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin D.V., Phillips C., Nannenga B., Timofeev O., Donehower L.A., Anderson C.W., Appella E., Fornace A.J., Jr. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat. Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Nebreda A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I., Coya J.M., Maina F., Arthur J.S., Nebreda A.R. Genetic analysis of specific and redundant roles for p38alpha and p38beta MAPKs during mouse development. Proc. Natl. Acad. Sci. USA. 2011;108:12764–12769. doi: 10.1073/pnas.1015013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov O.N., Kek C., Shreeram S., Timofeev O., Fornace A.J., Appella E., Bulavin D.V. The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene. 2007;26:2502–2506. doi: 10.1038/sj.onc.1210032. [DOI] [PubMed] [Google Scholar]
- Furlan A., Lamballe F., Stagni V., Hussain A., Richelme S., Prodosmo A., Moumen A., Brun C., Del Barco Barrantes I., Arthur J.S. Met acts through Abl to regulate p53 transcriptional outcomes and cell survival in the developing liver. J. Hepatol. 2012;57:1292–1298. doi: 10.1016/j.jhep.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldi S., Comoglio P.M., Trusolino L. The Met oncogene and basal-like breast cancer: another culprit to watch out for? Breast Cancer Res. 2010;12:208. doi: 10.1186/bcr2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaldi S., Sassi F., Accornero P., Torti D., Galimi F., Migliardi G., Molyneux G., Perera T., Comoglio P.M., Boccaccio C. Met signaling regulates growth, repopulating potential and basal cell-fate commitment of mammary luminal progenitors: implications for basal-like breast cancer. Oncogene. 2013;32:1428–1440. doi: 10.1038/onc.2012.154. [DOI] [PubMed] [Google Scholar]
- Giraddi R.R., Shehata M., Gallardo M., Blasco M.A., Simons B.D., Stingl J. Stem and progenitor cell division kinetics during postnatal mouse mammary gland development. Nat. Commun. 2015;6:8487. doi: 10.1038/ncomms9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Weng D., Eguchi T., Murshid A., Sherman M.Y., Song B., Calderwood S.K. Targeting the hsp70 gene delays mammary tumor initiation and inhibits tumor cell metastasis. Oncogene. 2015;34:5460–5471. doi: 10.1038/onc.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta J., del Barco Barrantes I., Igea A., Sakellariou S., Pateras I.S., Gorgoulis V.G., Nebreda A.R. Dual function of p38alpha MAPK in colon cancer: suppression of colitis-associated tumor initiation but requirement for cancer cell survival. Cancer Cell. 2014;25:484–500. doi: 10.1016/j.ccr.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., Muller W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H., Farnie G., Brennan K.R., Clarke R.B. Breast cancer stem cells: something out of notching? Cancer Res. 2010;70:8973–8976. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- Heinrichsdorff J., Luedde T., Perdiguero E., Nebreda A.R., Pasparakis M. p38 alpha MAPK inhibits JNK activation and collaborates with IkappaB kinase 2 to prevent endotoxin-induced liver failure. EMBO Rep. 2008;9:1048–1054. doi: 10.1038/embor.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G.W. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Herschkowitz J.I., Simin K., Weigman V.J., Mikaelian I., Usary J., Hu Z., Rasmussen K.E., Jones L.P., Assefnia S., Chandrasekharan S. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L., Bakiri L., Mairhorfer A., Schweifer N., Haslinger C., Kenner L., Komnenovic V., Scheuch H., Beug H., Wagner E.F. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat. Genet. 2007;39:741–749. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Jones F.E., Jerry D.J., Guarino B.C., Andrews G.C., Stern D.F. Heregulin induces in vivo proliferation and differentiation of mammary epithelium into secretory lobuloalveoli. Cell Growth Differ. 1996;7:1031–1038. [PubMed] [Google Scholar]
- Josefowicz S.Z., Shimada M., Armache A., Li C.H., Miller R.M., Lin S., Yang A., Dill B.D., Molina H., Park H.S. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol. Cell. 2016;64:347–361. doi: 10.1016/j.molcel.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karigane D., Kobayashi H., Morikawa T., Ootomo Y., Sakai M., Nagamatsu G., Kubota Y., Goda N., Matsumoto M., Nishimura E.K. p38alpha activates purine metabolism to initiate hematopoietic stem/progenitor cell cycling in response to stress. Cell Stem Cell. 2016;19:192–204. doi: 10.1016/j.stem.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Kendrick H., Regan J.L., Magnay F.A., Grigoriadis A., Mitsopoulos C., Zvelebil M., Smalley M.J. Transcriptome analysis of mammary epithelial subpopulations identifies novel determinants of lineage commitment and cell fate. BMC Genomics. 2008;9:591. doi: 10.1186/1471-2164-9-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H., Bechis S.K., Slorach E.M., Littlepage L.E., Egeblad M., Ewald A.J., Pai S.Y., Ho I.C., Werb Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafkas D., Rodilla V., Huyghe M., Mourao L., Kiaris H., Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E., Wu D., Pal B., Bouras T., Asselin-Labat M.L., Vaillant F., Yagita H., Lindeman G.J., Smyth G.K., Visvader J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., Pollard J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Robinson G.W., Gouilleux F., Groner B., Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P.K., Kanojia D., Liu X., Singh U.P., Berger F.G., Wang Q., Chen H. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene. 2012;31:2614–2626. doi: 10.1038/onc.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione J.E., Moghanaki D., Young L.J., Manner C.K., Ellies L.G., Joseph S.O., Nicholson B., Cardiff R.D., MacLeod C.L. Transgenic Polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61:8298–8305. [PubMed] [Google Scholar]
- Maroulakou I.G., Oemler W., Naber S.P., Klebba I., Kuperwasser C., Tsichlis P.N. Distinct roles of the three Akt isoforms in lactogenic differentiation and involution. J. Cell. Physiol. 2008;217:468–477. doi: 10.1002/jcp.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens T.W., Rogers R.L., Best S.A., Ledger A., Mooney A.M., Ferguson A., Shore P., Swarbrick A., Ormandy C.J., Simpson P.T. Runx2 is a novel regulator of mammary epithelial cell fate in development and breast cancer. Cancer Res. 2014;74:5277–5286. doi: 10.1158/0008-5472.CAN-14-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Igea A., Canovas B., Dolado I., Nebreda A.R. Inhibition of p38 MAPK sensitizes tumour cells to cisplatin-induced apoptosis mediated by reactive oxygen species and JNK. EMBO Mol. Med. 2013;5:1759–1774. doi: 10.1002/emmm.201302732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prater M., Shehata M., Watson C.J., Stingl J. Enzymatic dissociation, flow cytometric analysis, and culture of normal mouse mammary tissue. Methods Mol. Biol. 2013;946:395–409. doi: 10.1007/978-1-62703-128-8_25. [DOI] [PubMed] [Google Scholar]
- Reyes D., Ballare C., Castellano G., Soronellas D., Bago J.R., Blanco J., Beato M. Activation of mitogen- and stress-activated kinase 1 is required for proliferation of breast cancer cells in response to estrogens or progestins. Oncogene. 2014;33:1570–1580. doi: 10.1038/onc.2013.95. [DOI] [PubMed] [Google Scholar]
- Rodilla V., Dasti A., Huyghe M., Lafkas D., Laurent C., Reyal F., Fre S. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P., Storci G., Giovannini C., Pandolfi S., Pianetti S., Taffurelli M., Santini D., Ceccarelli C., Chieco P., Bonafe M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloaga A., Thomson S., Wiggin G.R., Rampersaud N., Dyson M.H., Hazzalin C.A., Mahadevan L.C., Arthur J.S. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L., van Bragt M.P., Li Z. A long-lived luminal subpopulation enriched with alveolar progenitors serves as cellular origin of heterogeneous mammary tumors. Stem Cell Reports. 2015;5:60–74. doi: 10.1016/j.stemcr.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F., Asselin-Labat M.L., Shackleton M., Forrest N.C., Lindeman G.J., Visvader J.E. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68:7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- van Bragt M.P., Hu X., Xie Y., Li Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife. 2014;3:e03881. doi: 10.7554/eLife.03881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Ventura J.J., Tenbaum S., Perdiguero E., Huth M., Guerra C., Barbacid M., Pasparakis M., Nebreda A.R. p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat. Genet. 2007;39:750–758. doi: 10.1038/ng2037. [DOI] [PubMed] [Google Scholar]
- Vermeulen L., Vanden Berghe W., Beck I.M., De Bosscher K., Haegeman G. The versatile role of MSKs in transcriptional regulation. Trends Biochem. Sci. 2009;34:311–318. doi: 10.1016/j.tibs.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Visvader J.E. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K.U., McAllister K., Ward T., Davis B., Wiseman R., Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- Warr N., Carre G.A., Siggers P., Faleato J.V., Brixey R., Pope M., Bogani D., Childers M., Wells S., Scudamore C.L. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell. 2012;23:1020–1031. doi: 10.1016/j.devcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H.C., Avivar-Valderas A., Sosa M.S., Girnius N., Farias E.F., Davis R.J., Aguirre-Ghiso J.A. p38alpha signaling induces anoikis and lumen formation during mammary morphogenesis. Sci. Signal. 2011;4:ra34. doi: 10.1126/scisignal.2001684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.